Abstract
We use analyses of phylogeographic population structure across a suite of 12 mammalian, avian, amphibian, and reptilian species and species-groups to assess the role of Late Miocene to Pleistocene geological history in the evolution of a distinct Baja California Peninsular Desert biota. Comparative examination of phylogroup distributions provides support for previously hypothesized vicariant events produced by: a middle Pleistocene midpeninsular seaway, a late Pliocene northward transgression of the Sea of Cortéz, and a Pliocene seaway across the southern peninsular Isthmus of La Paz. Most of this phylogeographic architecture is cryptically embedded within widespread taxonomic species and species-groups, such that the unique evolutionary history of the Peninsular Desert has been obscured and ignored. The Peninsular Desert can no longer be considered a subset of the Sonoran Desert—it is a separate regional desert with its own unique evolutionary history, ecological arena, and conservation value.
Current depictions of species-level taxonomy and phylogeny in well-studied groups such as terrestrial mammals have been assumed, without rigorous examination, to accurately capture major patterns of biological diversity in continental biotas (previously termed an “inertial species concept”) (1), and thus provide appropriate units of analysis for biogeography, ecology, and conservation biology (2). This assumption appears to be generally incorrect in the rich, arid-adapted vertebrate biota of the western North American deserts (Fig. 1M) (3). When analyzed by using a mitochondrial DNA (mtDNA) phylogeography approach (13), widespread species in this biota often include two or more highly divergent and geographically separate mtDNA gene lineages or phylogroups (3, 10, 11). To the extent that divergent phylogroups are cryptically embedded within a single widespread species, the role of vicariance in structuring biotas will necessarily be underestimated, and the frequency and magnitude of dispersal consistently overestimated. Failure to account for vicariance-derived architecture in the assembly of biotas has wide-ranging implications in evolution, where causal connections between earth history and the geography of species formation will go unrecognized (14, 15); in ecology, where connections between climatic history and the magnitude of changes in species distributions and associations are of great interest (16, 17); and in conservation biology, where a critical problem continues to be identification and ranking of biodiversity hotspots.
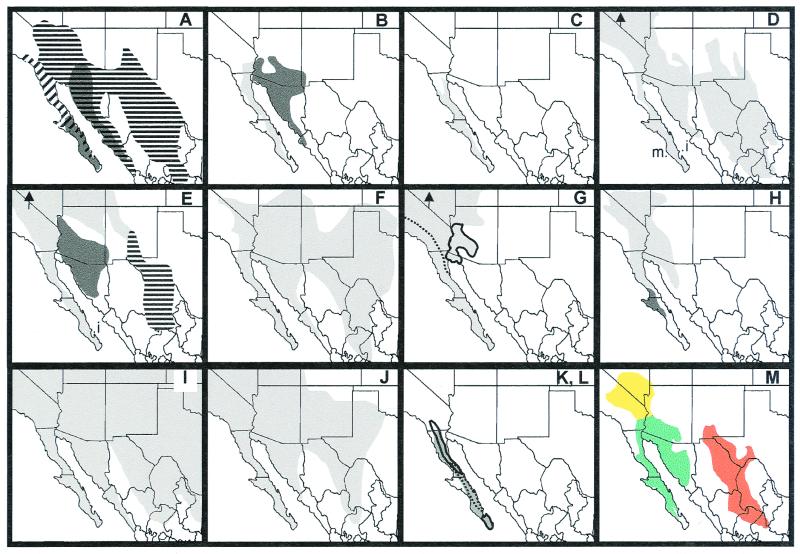
Figure 1.
Summary geographic distributions of 12 vertebrate species or species-groups evaluated in this study; data for G–L are summarized from the literature (4–9). (A) Peromyscus eremicus group, including P. eremicus (horizontal bars), P. eva (dark shading southern peninsula), P. fraterculus (diagonal bars), and P. merriami (dark shading western continental). P. eremicus was considered a single widespread species before recent phylogeographic analysis (10) that proposed western populations be designated as separate species, P. fraterculus. (B) Bailey's pocket mouse group, including Chaetodipus baileyi (dark shading) and C. rudinoris (light shading). C. baileyi was considered a single species before recent phylogeographic analysis (11) that proposed populations west of Colorado River be designated as a separate species, C. rudinoris. (C) Little desert pocket mouse, C. arenarius. (D) Merriam's, Dipodomys merriami, San José Island, D. insularis (i), and Margarita Island, D. margaritae (m) kangaroo rats. (E) Antelope ground squirrels, including Ammospermophilus leucurus (light shading), A. harrisii (dark shading), A. interpres (horizontal bars), and A. insularis (i). (F) Red-spotted toads, Bufo punctatus. (G) Desert woodrats, Neotoma lepida species-group, including N. devia (outlined) and N. lepida, with dashed line separating probable ranges of eastern and western phylogroups (4). (H) Le Conte's thrasher, Toxostoma lecontei group, considered a single species before phylogeographic analysis (5) that proposed southern peninsular populations be designated as a separate species, T. arenicola (dark shading). (I) Gopher snakes, Pituophis melanoleucus complex. (J) Side-blotched lizards, Uta stansburiana group. (K) Tree lizards, Urosaurus nigricaudus group (shaded). (L) Three species of rock lizards (9): Petrosaurus mearnsi (northernmost solid outline), P. repens (dashed outline), and P. thalassinus (southernmost solid outline). (M) Warm deserts of North America as depicted by Shreve (12): Sonoran, including peninsular (green), Mojave (yellow), Chihuahuan (red).
The Baja California peninsula includes a heterogeneous array of landscapes and habitats, from tropical deciduous forests to xeric desertscrub mountains and low arid plains. The Peninsular Desert traditionally is treated as a subset of the neighboring Sonoran Desert, on the basis of similarities in plant forms and assemblages (Fig. 1M) (12, 18). Many vertebrates in the Peninsular Desert (e.g., Fig. 1 A–L) also have been considered either conspecifics of species in the western continental warm deserts (Sonoran and Mojave) or Pleistocene-age derivatives of continental species. The alleged similarities between continental and peninsular desert taxa originally were explained, before development of plate tectonic models of earth history, as resulting from waves of dispersal from continental deserts into the Peninsular Desert coincident with late Pleistocene through Holocene climatic oscillations (19, 20). This dispersal model would be consistent with several forms of allopatric (divergence after peripheral isolation or colonization of new habitat) or parapatric (divergence with gene flow) speciation (21).
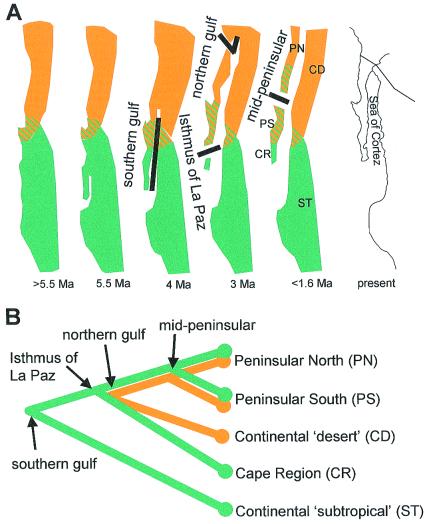
A distinctly different picture emphasizes the dynamic nature of peninsular geology over the past six million years to postulate a set of testable vicariance events in peninsular biogeographic history (22, 23). Notably, the Baja California peninsula began to separate from the Mexican mainland about 5.5 million years ago (mya) as a result of plate-boundary expansion between the North American and Pacific plates, leading to inception of the Sea of Cortéz. Continental and peninsular deserts were subsequently isolated from each other by northward transgressions of the Sea of Cortéz into low-lying deserts in southern California and Arizona. The peninsula likely was fragmented by trans-peninsular seaways (into one or more islands, and on several occasions) connecting the Pacific Ocean and Sea of Cortéz. These marine transgressions and seaways provide a basis for postulating a series of late Neogene (5.5–1 mya) vicariance events (Fig. 2A) and subsequent development of separate areas of endemism (Fig. 2B).
Figure 2.
(A) Simplified Late Miocene to middle Pleistocene geological history of the Baja California peninsula and Sea of Cortéz (8, 23) depicting four postulated vicariance events between subtropical thornscrub (green) or desert (orange) biotas: southern gulf = Southern Miocene Vicariance between subtropical thornscrub biotas across the newly developing Gulf of California; Isthmus of La Paz = Isthmus of La Paz Pliocene Vicariance isolating the Cape Region from the rest of the peninsula by a trans-peninsular seaway; northern gulf = Northern Pliocene Vicariance isolating peninsular and continental warm desert biotas through northward transgression of the Sea of Cortéz into lowlands of southern California and Arizona; mid-peninsular = Middle Pleistocene Vicariance isolating northern and southern peninsular biotas across a trans-peninsular seaway in the central Vizcaino region. Areas of endemism resulting from vicariance events are as follows: PN, Peninsular North; PS, Peninsular South; CR, Peninsular Cape Region; CD, Continental Deserts; and ST, Continental Subtropical. Ma, million years ago. (B) Postulated areas and historical relationships among areas. The south-to-north opening of the Sea of Cortéz was likely a continuous event, having progressively bisected more southerly subtropical to more northerly desert biotas (23). Nevertheless, given different time frames (late Miocene vs. late Pliocene) and predicted affinities with different continental biotas (southern subtropical vs. northern desert), it is reasonable to postulate separate southern and northern peninsular–continental vicariance events.
The models, late Pleistocene through Holocene dispersal (with possibility of parapatric peripheral isolates, or colonization of new habitat speciation) vs. late Neogene vicariance, produce alternative predictions that are testable by molecular phylogeographic examination of distinct lineages or phylogroups embedded within widespread species and species-groups. Under the former model, peninsular and western continental deserts should share a high number of taxa exhibiting little or no genetic differentiation among populations of the respective regions (other than that associated with isolation by distance or recently derived reproductive isolating mechanisms). Alternatively, under the vicariance model, populations occurring on opposite sides of a presumptive barrier (Fig. 2) will represent separate phylogroups to the extent that: (i) a taxon was widely distributed before, and thus fragmented by, the newly formed barrier; (ii) sufficient time has occurred for populations separated by the barrier to pass through predicted phases of polyphyly and paraphyly to eventual reciprocal monophyly of gene lineages (13); and (iii) subsequent dispersal across a former barrier has not eroded a signal of earlier vicariance within the gene tree. The rate at which gene lineage sorting between populations isolated by a barrier to gene flow proceeds through stages of polyphyly and paraphyly to eventual reciprocal monophyly is a function of effective population sizes of separated populations. For the haploid asexual mitochondrial genome, effective population size is one-fourth that of the nuclear genome (24). Thus, while eventual assessment of phylogeographic structure across several nonlinked gene genealogies is desirable, our expectation is that mtDNA genealogies should provide the strongest “single-linkage group” signal for biogeographic analysis. An extended and more robust prediction of the vicariance model is that multiple codistributed taxa that satisfy these three conditions should exhibit congruent patterns in the distribution of phylogroups.
We examined mtDNA sequence data of four rodent species-groups (Fig. 1 A, B, D, and E) and one anuran species (a toad; Fig. 1F) with distributions that extend from Baja California across northern continental warm deserts and in some cases into northern subtropical regions in western Mexico. In addition, we examined one rodent species endemic to the peninsula (Fig. 1C). For all six taxa, scarce fossil records preclude their use to estimate a detailed history of peninsular occupancy, although for the rodents, first appearances of fossils identifiable to genus generally occur well before the time frames of postulated vicariance: Ammospermophilus (8–12 mya) (25), Chaetodipus (16–20 mya) and Dipodomys (12–16 mya) (26), and Haplomylomys (5–8 mya) (27). We also included an additional set of six taxa (Fig. 1 G–L) in which findings from previous studies were considered relevant to the comparative phylogeographic analysis of this study.
Materials and Methods
Sampling Strategy.
Localities for each of six taxa are plotted in Fig. 3. Detailed locality data for the Peromyscus eremicus and Chaetodipus baileyi groups are given elsewhere (10, 11); all others are available from the authors on request. Sample sizes, numbers of localities, and average number of individuals per locality are as follows: Peromyscus eremicus group (73, 26, 3); Chaetodipus baileyi group (51, 12, 4); Chaetodipus arenarius (36, 7, 5); Dipodomys merriami group (60, 33, 2); Ammospermophilus (30, 17, 2); and Bufo punctatus (44, 23, 2). Our general approach was to maximize number of localities sampled across the geographic range of each taxon.
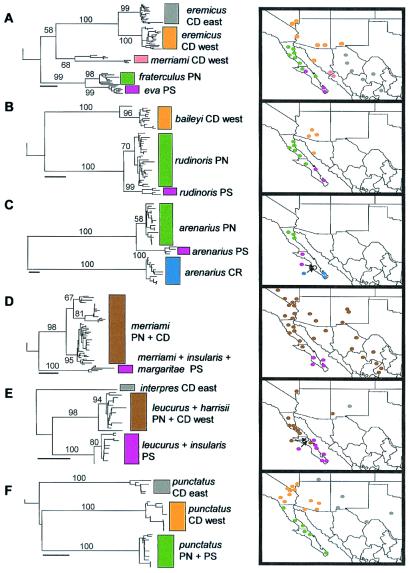
Figure 3.
Phylogeographic summaries: Left, cladograms; Right, maps. (A) Peromyscus eremicus group. (B) Chaetodipus baileyi group. (C) Chaetodipus arenarius. (D) Dipodomys merriami group. (E) Ammospermophilus. (F) Bufo punctatus. Capital letters after taxon names refer to areas as in Fig. 2B. Open circles in C and E signify localities at which both phylogroups occur. Trees shown here were produced with the neighbor-joining algorithm. Numbers along major branches refer to relative support for a clade out of 1,000 bootstrap pseudoreplications. Scale bars indicate 0.01 substitution per site.
Data and Analyses.
Data type for each of the five mammal taxa consisted of 699 base pairs (bp) from the mtDNA cytochrome oxidase subunit 3 (COIII) gene. Methods for DNA preparation, PCR amplification, and sequence generation are given elsewhere (10). The COIII gene evolves slightly more rapidly than the better-known cytochrome b gene (CYTB) in several of the rodents included in this study, and it has a similar base composition (B.R.R., unpublished data). Data type for B. punctatus consisted of 654 bp from the cytochrome b gene, and PCR was performed with primers MVZ43 (28) and a slightly modified version of MVZ16 (29). These primers and two primers designed specifically for B. punctatus cytochrome b (L16638, AGGYT ATGTT CTTCC CTGAG GACA; and H16748, CCTCA GATYC ATTGG ACAAG CTC) were used for sequencing. Phylogenetic trees were produced by using paup* 4.0b3 (30) under both neighbor-joining (31) and maximum-parsimony approaches. The neighbor-joining trees were constructed from matrices of pairwise distances by using the Tamura–Nei (32) model of substitution, which incorporates parameters for unequal nucleotide frequencies, different rates of transition and transversion substitution, and different rates of transition substitution between purines and pyrimidines. Summarized estimates of net sequence divergence among phylogroups are calculated by removing the component of overall divergence that represents divergence among haplotypes within a phylogroup (33).
Results and Discussion
Phylogeographic structure in six taxa is summarized in Fig. 3. Although only the neighbor-joining trees are shown, previously reported maximum-parsimony and maximum-likelihood results for the Peromyscus eremicus group (10) and maximum-parsimony results for the Chaetodipus baileyi group (11), either with all characters weighted equally, or after removing third codon position transition substitutions, produced the same relationships among major lineages depicted here. For all other taxa (Fig. 3 C–F), maximum-parsimony searches with all characters weighted equally produced the same relationships among major lineages as depicted here.
All five rodent groups and the toad exhibit multiple geographically separated mtDNA phylogroups (Fig. 3), demonstrating that taxonomic species frequently fail to capture the inherent geographic diversity in two ways. First, multiple phylogroups are embedded within a single widespread taxonomic species in each of the six groups. Second, the mtDNA gene genealogies suggest that taxonomic species in these groups frequently do not represent monophyletic lineages. The most serious error of this second kind for biogeographic analysis occurs in the Peromyscus eremicus group, in which the peninsular phylogroup within P. eremicus (shown as P. fraterculus in Figs. 1A and 3A; see ref. 10) is highly divergent from two continental phylogroups but is closely related to the peninsular endemic species, P. eva. Moreover, long-recognized species of Ammospermophilus on opposite sides of the Colorado River (A. leucurus and A. harrisii) possess virtually identical mtDNA gene lineages, whereas southern peninsular populations of A. leucurus form a separate phylogroup (Fig. 3E). Finally, in several cases, island species are not genetically differentiated from the most proximal phylogroup of a widespread species [Dipodomys insularis and D. margaritae within southern peninsular D. merriami phylogroup (Fig. 3D), A. insularis within the southern peninsular A. leucurus phylogroup (Fig. 3E)].
Four geographic patterns are evident in the six taxa. A pattern replicated across all five mammal groups (Fig. 3 A–E) is the occurrence of separate phylogroups in northern and southern regions of the Baja California peninsula (net average sequence divergence ranges from 1.8% to 4.1%; Table 1). At a deeper level of divergence, the P. eremicus group, the C. baileyi group, and B. punctatus exhibit separate and highly divergent continental and peninsular phylogroups (divergence ranges from 7.5% to 8.7%; Table 1). Neither the D. merriami group nor the A. leucurus group appears to be similarly differentiated between continental and northern peninsular deserts. The peninsular endemic, C. arenarius, exhibits a separate and highly divergent (12.8%) lineage in the southernmost part of the peninsula (Cape Region).
Table 1.
Summary of evidence supporting postulated vicariance events
| Taxon | Percent sequence divergence since
event
|
Data type | |||
|---|---|---|---|---|---|
| Southern gulf | Isthmus of La Paz | Northern gulf | Midpeninsular | ||
| Peromyscus eremicus group | — | — | 8.7 | 3.1 | mtDNA COIII |
| Chaetodipus baileyi group | — | — | 8.7 | 1.8 | mtDNA COIII |
| Chaetodipus arenarius | ? | 12.8 | — | 2.2 | mtDNA COIII |
| Dipodomys merriami group | — | — | — | 4.1 | mtDNA COIII |
| Ammospermophilus leucurus group | — | — | — | 3.3 | mtDNA COIII |
| Bufo punctatus | — | — | 7.5 | — | mtDNA CYTB |
| Neotoma lepida group | — | — | Yes | ? | mtDNA RFLP |
| Toxostoma lecontei group | — | — | — | 2.5 | mtDNA CYTB |
| Pituophis melanoleucus complex | — | — | — | 5.0 | mtDNA ND4 |
| Uta stansburiana group | — | — | — | 5.5 | mtDNA CYTB |
| Urosaurus nigricaudus group | Yes | Yes | — | Yes | Nuclear DNA allozymes |
| Petrosaurus | — | ? | — | Yes | Nuclear DNA allozymes |
Estimates of percent sequence divergence (net average values) (33) among sister groups separated by postulated vicariant event were calculated by using the Tamura–Nei (32) model of sequence evolution (first six this study, remaining three recalculated from studies referenced in Fig. 1 or ref. 43). Yes, evidence available, but calculations of sequence divergence values not attainable given data; ?, postulated but evidence not yet available in sufficient detail. RFLP, restriction fragment length polymorphism; ND4, NADH dehydroxygenase subunit 4 gene.
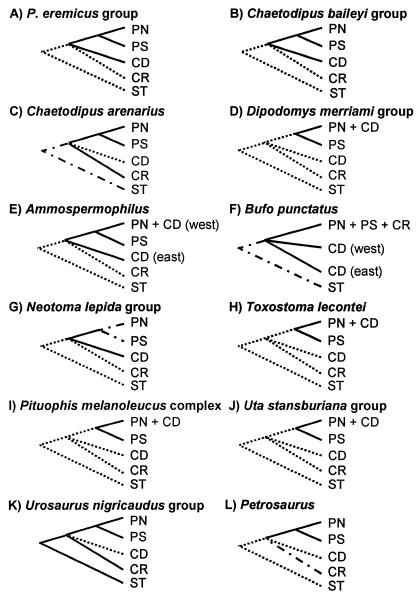
Phylogroup distributions of additional taxa previously reported in the literature (Fig. 1 G–L) provide further evidence of historical vicariance in one bird, one snake, three lizards, and one additional rodent (Table 1). A summary of area relationships derived from all 12 taxa (Fig. 4) strengthens support for the vicariance events depicted in Fig. 2B as follows: (i) a separation of populations on the Baja California peninsula into northern and southern phylogroups consistent with the proposed midpeninsular vicariance event is present in at least 10 of 12 taxa, including all mammals thus far analyzed (data not yet available for southern peninsular populations of the Neotoma lepida group), all reptiles, and the bird; (ii) a subdivision of peninsular and continental desert populations into separate phylogroups consistent with the northern gulf vicariance event is present in four taxa, including three mammals and the anuran; and (iii) separation of Cape Region from remaining peninsular populations consistent with the Isthmus of La Paz vicariant event is present in at least two taxa, including one mammal and one reptile (data not yet available for the Cape Region species Petrosaurus thalassinus). Five taxa that exhibit no signature of separate phylogroups for northern peninsular and at least the western regions of the continental deserts each exhibit a separate southern peninsular phylogroup. Evidence for a peninsular–continental separation consistent with a southern gulf vicariance event was postulated by other researchers for the U. nigricaudus group of tree lizards (8, 22) and is supported by the high degree of reptilian endemism in the Cape Region (23). Further evidence for the southern gulf event is provisionally indicated in the peninsular endemic rodent, C. arenarius. The separate Cape Region phylogroup in C. arenarius (and presumably in the U. nigricaudus group) indicates occupancy of the peninsula before the Isthmus of La Paz event. As additional support for this interpretation, previously reported (11) phylogenetic analysis using 14 extant species of Chaetodipus indicated that C. arenarius has no closely related sister group, and it is included in an unresolved polychotomy between seven separate lineages distributed across desert, subtropical thornscrub, and chaparral habitats in western North America. The essentially simultaneous origination of these lineages is consistent with the late Miocene to Pliocene aridification and increased provinciality of western North American biotas (34, 35), and thus supports the hypothesis that C. arenarius represents divergence resulting from the southern gulf vicariance event (Fig. 2).
Figure 4.
Summary of postulated area relationships (see Fig. 2B) within 12 vertebrate species or species-groups inferred from distributions of mtDNA phylogroups (see Fig. 3): A–F from this study; G–L from the literature. Distinct phylogroup or taxon distributions are depicted as present (solid lines), missing (dotted lines), or unknown but postulated (dot–dash lines) in each area.
Results of this study suggest a pervasive history of biogeographic and evolutionary independence of arid lands vertebrate biotas in northern and southern regions of the Baja California Peninsular Desert, probably dating to a middle Pleistocene time frame as earlier suggested through analysis of side-blotched lizards (8). Proposed timing for the midpeninsular vicariance event (about 1 mya) (8) is further supported indirectly by the fact that in at least three taxa (Fig. 4 A–C) divergence estimates consistent with Pliocene vicariance events (northern gulf and Isthmus of La Paz) are 3 to 6 times higher than those associated with the midpeninsular event (Table 1; Fig. 3). Clearly, a large number of desert taxa, ranging across mammalian, avian, and reptilian members of the biota, persisted on the southern Peninsula throughout the later Pleistocene glacial periods. The absence of subdivision within B. punctatus in accord with the midpeninsular event is an anomaly given the pervasiveness of the general pattern displayed across other taxa.
Although not as general as the midpeninsular event, the northern gulf event appears to have played a significant role in separating peninsular from continental desert populations in four taxa (Fig. 4 A, B, F, and G), probably within a late Pliocene time frame (Fig. 2A). Species-level taxonomy, for the most part, failed to recognize the long history of isolation of peninsular from continental populations in these four taxa, each having been represented traditionally as a single widespread species (P. eremicus, C. baileyi, B. punctatus, N. lepida). Subspecific taxonomic boundaries within C. baileyi and N. lepida are coincident with boundaries of peninsular and continental phylogroups, indicating that to some extent previously recognized morphological patterns of divergence are interpretable within a phylogeographic framework. The five taxa without obvious northern peninsular and continental separations (Fig. 4 D, E, H, I, and J) are postulated either to have not been widely distributed between northern peninsular and continental deserts before the northern gulf event, or subsequent dispersal and lineage extinction have eroded a prior history of isolation and divergence. The latter hypothesis seems less likely, given that the signature of southern peninsular isolation and divergence remains intact in each of these taxa, although we cannot speculate further on the original area of endemism in either northern peninsular or continental deserts. Finally, C. arenarius, with an extremely deep separation between Cape Region and remaining peninsular phylogroups, presents a striking case of cryptic divergence consistent with Pliocene vicariance across the Isthmus of La Paz.
The Baja California Peninsular Desert embraces a rich assemblage of biological diversity that has evolved in step with the origination and subsequent geological history of the peninsula itself. Unfortunately, sole reliance on species-level taxonomy and unquestioned adherence to dated definitions of regional deserts has obscured this biodiversity, as the fundamental record of peninsular evolutionary history is cryptically embedded within widespread species and species-groups. This failure to appreciate the distinctiveness of the Peninsular Desert has in turn led to false notions of interregional similarity (36) and temporally shallow models that purport to explain the regional biogeographic history in terms of late Pleistocene–Holocene dispersal (19, 20), the peninsular effect (37–39), and recent isolation on islands in the Sea of Cortéz (40, 41). Samples representing the Peninsular Desert are wholly absent from the data set that has served as the basis for a recent series of papers concerning desert mammals (36, 42), rendering conclusions made in those studies regarding community composition, comparisons among North American regional deserts, and comparisons with deserts of other continents incomplete.
Although it is customary to use species distribution lists to characterize and prioritize biodiversity hotspots across the earth, we caution that an over-reliance on species for which phylogeographic data are currently unavailable could introduce a serious bias in the interpretation of biogeographic pattern and processes. For example, while the Baja California Peninsular Desert (and particularly the southern portion of that region) appears to have been an area of high significance in the historical diversification of North American desert biotas, that contribution will go undetected to the extent that currently recognized widespread species mask the underlying general patterns in the distribution of biological diversity.
Acknowledgments
We thank R. Snook for comments on an earlier draft; L. L. Grismer for helpful discussion; S. T. Alvarez-Castañeda for arranging collecting permits; and D. Bradford, Q. Bradwisch, G. Dayton, R. Fisher, R. Jennings, M. Jorgenson, Z. Marshall, J. Mendelson, D. Mulcahy, T. LaDuc, and the Museum of Vertebrate Zoology at the University of California, Berkeley, for Bufo punctatus tissue. Financial support was provided through grants from the National Science Foundation to B.R.R. (DEB-9629787) and D.J.H. (DEB-9629840).
Abbreviation
- mya
million years ago
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY009173–AY009237, AY009253–AY009316, and AY010121–AY010230).
See commentary on page 14017.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250413397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250413397
References
- 1.Good D A. Herp Monogr. 1994;8:180–202. [Google Scholar]
- 2.Brown J H. Macroecology. Chicago: Univ. of Chicago Press; 1995. [Google Scholar]
- 3.Riddle B R, Hafner D J. Global Ecology and Biogeography. 1999;8:433–441. [Google Scholar]
- 4.Planz J V. Ph.D. dissertation. Denton: Univ. of North Texas; 1992. [Google Scholar]
- 5.Zink R M, Blackwell R C, Rojas-Soto O. Condor. 1997;99:132–138. [Google Scholar]
- 6.Rodriquez-Robles J A, De Jesús-Escobar J M. Mol Phylogenet Evol. 2000;14:35–50. doi: 10.1006/mpev.1999.0698. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre G, Morafka D J, Murphy R W. Herpetologica. 1999;55:369–381. [Google Scholar]
- 8.Upton D E, Murphy R W. Mol Phylogenet Evol. 1997;8:104–113. doi: 10.1006/mpev.1996.0392. [DOI] [PubMed] [Google Scholar]
- 9.Grismer L L. Herpetologica. 1999;55:446–469. [Google Scholar]
- 10.Riddle, B. R., Hafner, D. J. & Alexander, L. F. (2000) Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 11.Riddle, B. R., Hafner, D. J. & Alexander, L. F. (2000) Mol. Phylogenet. Evol., in press. [DOI] [PubMed]
- 12.Shreve F. Bot Rev. 1942;8:195–246. [Google Scholar]
- 13.Avise J C. Phylogeography: The History and Formation of Species. Cambridge, MA: Harvard Univ. Press; 2000. [Google Scholar]
- 14.Cracraft J. Am Zool. 1994;34:33–47. [Google Scholar]
- 15.Vrba E S. In: Paleoclimate and Evolution, with Emphasis on Human Origins. Vrba E S, Denton G H, Partridge T C, Burckle L H, editors. New Haven, CT: Yale Univ. Press; 1995. pp. 24–45. [Google Scholar]
- 16.FAUNMAP Working Group. Science. 1996;272:1601–1606. [Google Scholar]
- 17.Riddle B R. Ecography. 1998;21:437–446. doi: 10.1111/ecog.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacMahon J A. In: North American Terrestrial Vegetation. Barbour M G, Billings W D, editors. Cambridge, U.K.: Cambridge Univ. Press; 1988. pp. 212–264. [Google Scholar]
- 19.Savage J M. Syst Zool. 1960;9:184–212. [Google Scholar]
- 20.Orr R T. Syst Zool. 1960;9:171–179. [Google Scholar]
- 21.Patton J L, da Silva M N F. In: Endless Forms. Howard D J, Berlocher S H, editors. New York: Oxford University Press; 1997. pp. 202–213. [Google Scholar]
- 22.Murphy R W. Occas Pap Calif Acad Sci. 1983;137:1–48. [Google Scholar]
- 23.Grismer L L. Herp Nat Hist. 1994;2:51–106. [Google Scholar]
- 24.Moore W S. Evolution. 1995;49:718–726. doi: 10.1111/j.1558-5646.1995.tb02308.x. [DOI] [PubMed] [Google Scholar]
- 25.James G T. Univ Cal Publ Geol Sci. 1963;45:1–170. [Google Scholar]
- 26.Wahlert J H. Spec Publ Am Soc Mammal. 1993;10:1–37. [Google Scholar]
- 27.Hibbard C W. Spec Publ Am Soc Mammal. 1968;2:1–593. [Google Scholar]
- 28.Graybeal A. Mol Phylogenet Evol. 1993;2:256–269. doi: 10.1006/mpev.1993.1024. [DOI] [PubMed] [Google Scholar]
- 29.Moritz C, Schneider C J, Wake D B. Syst Biol. 1992;41:273–291. [Google Scholar]
- 30.Swofford D L. paup*, Phylogenetic Analysis Using Parsimony (and Other Methods) Sunderland, MA: Sinauer; 2000. [Google Scholar]
- 31.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Nei M. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 33.Edwards S V. In: Avian Molecular Systematics and Evolution. Mindell D P, editor. New York: Academic; 1997. pp. 251–278. [Google Scholar]
- 34.Axelrod D I. Occas Pap Calif Acad Sci. 1979;132:1–74. [Google Scholar]
- 35.Webb S D. In: Coevolution. Nitecki M H, editor. Chicago: Univ. of Chicago Press; 1983. pp. 267–306. [Google Scholar]
- 36.Kelt D A, Brown J H, Heske E J, Marquet P A, Morton S R, Reid J R W, Rogouin K A, Shenbrot G. Ecology. 1996;77:746–761. [Google Scholar]
- 37.Taylor R J, Regal P J. Am Nat. 1978;112:583–593. [Google Scholar]
- 38.Lawlor T E. Am Nat. 1983;121:432–439. [Google Scholar]
- 39.Seib R L. Am Nat. 1980;115:613–620. [Google Scholar]
- 40.Case T J. In: Island Biogeography in the Sea of Cortéz. Case T J, Cody M L, editors. Berkeley: Univ. of California Press; 1983. pp. 159–209. [Google Scholar]
- 41.Lawlor T E. In: Island Biogeography in the Sea of Cortéz. Case T J, Cody M L, editors. Berkeley: Univ. of California Press; 1983. pp. 265–289. [Google Scholar]
- 42.Brown J H, Kurzius M A. Ann Zool Fenn. 1987;24:227–237. [Google Scholar]
- 43.Angeles-Aguilars M, Sites J W, Jr, Murphy R W. J Herpetol. 1988;22:135–145. [Google Scholar]