Abstract
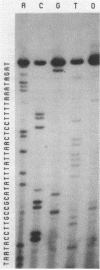
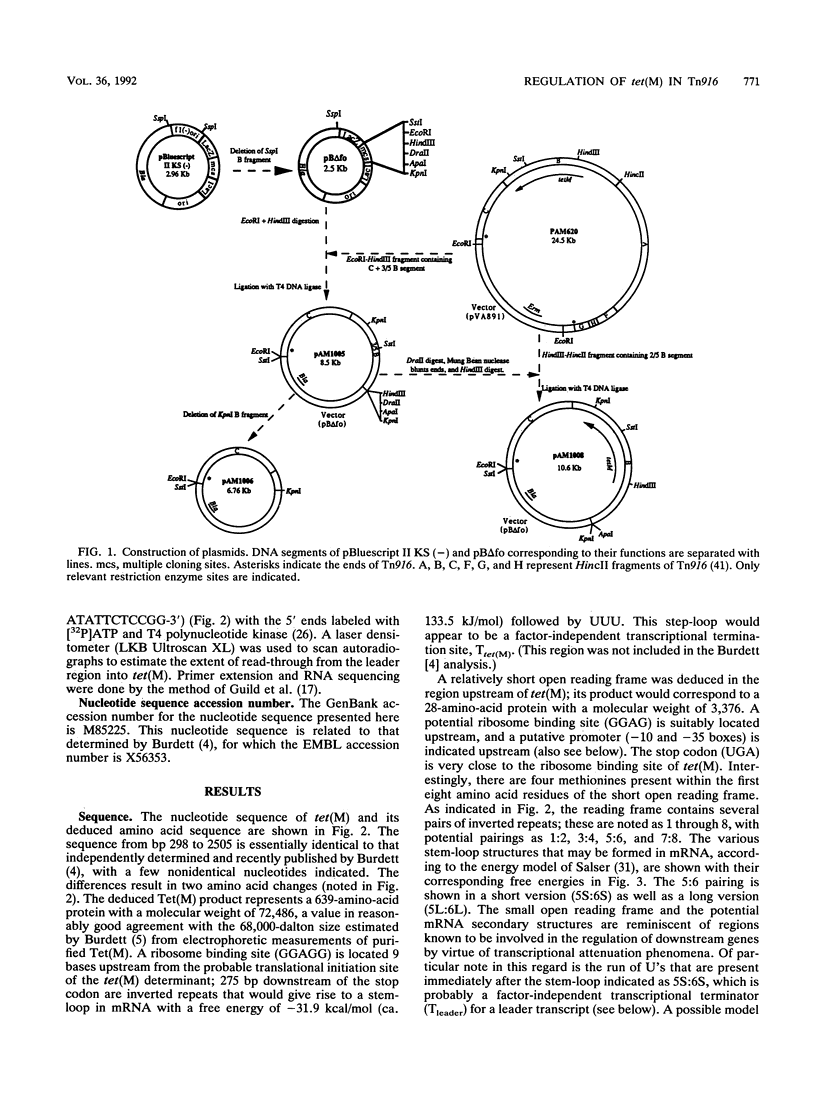
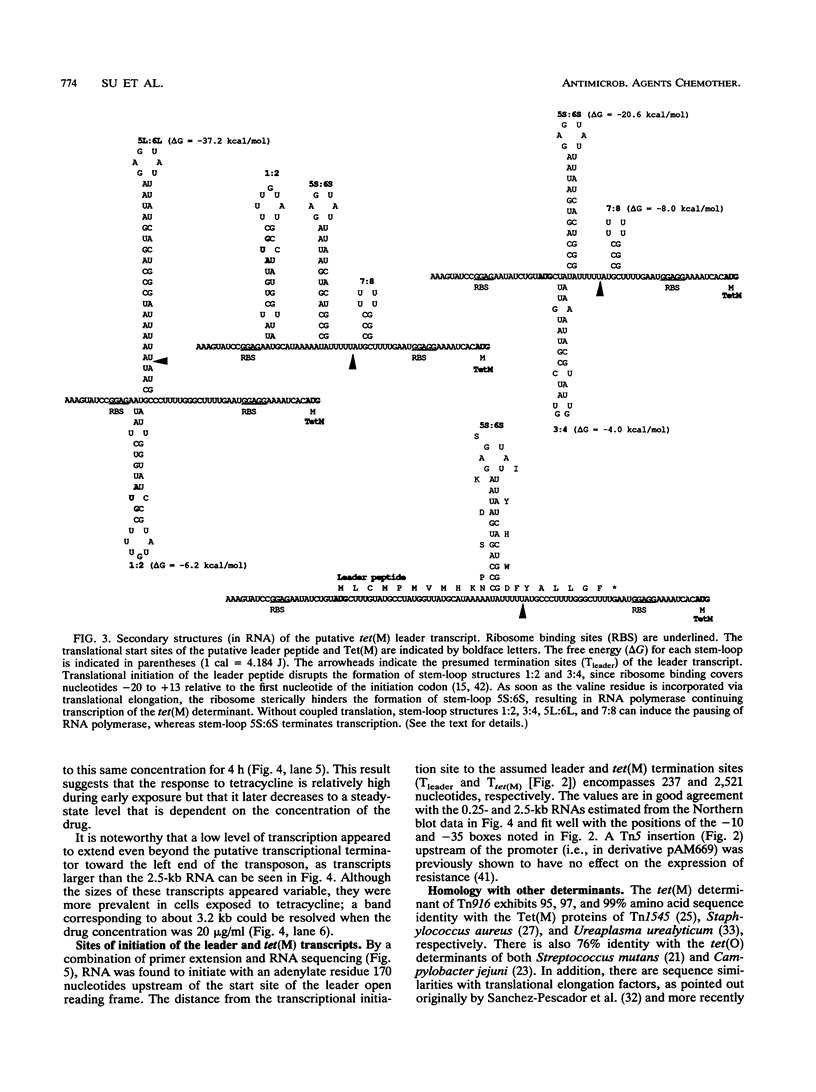
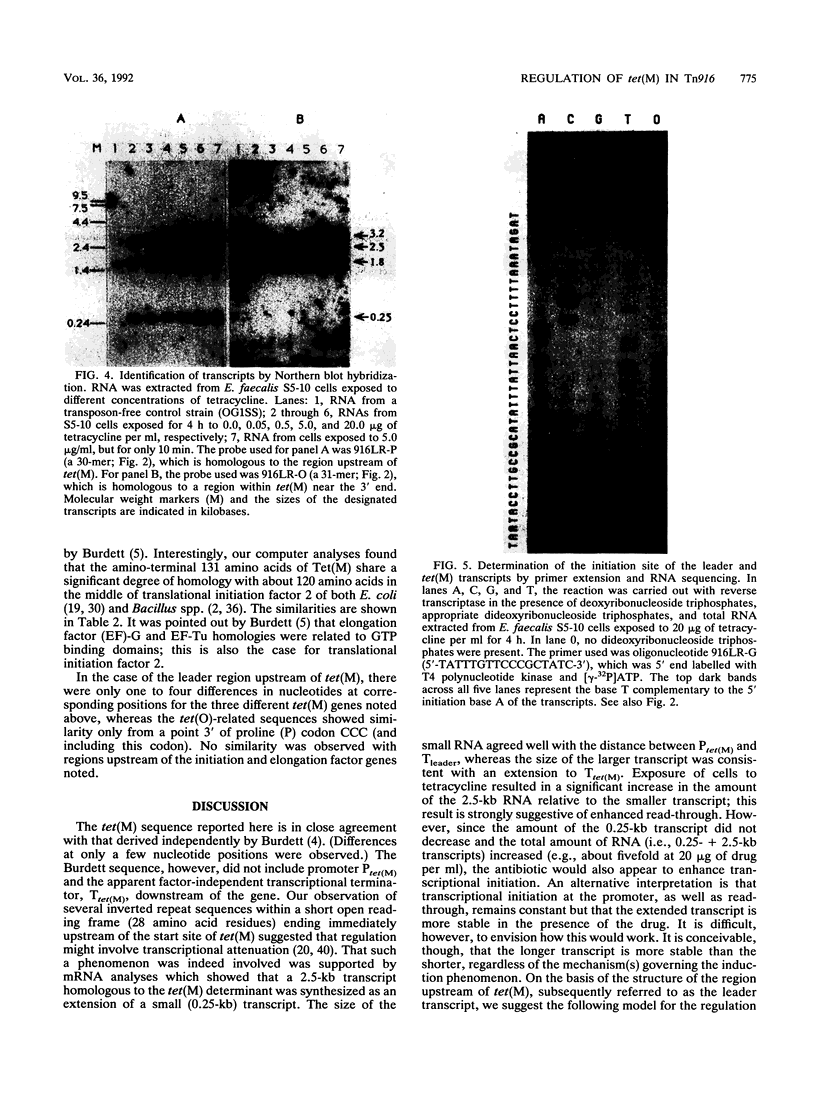
The nucleotide sequence of the tetracycline resistance determinant tet(M), located on conjugative transposon Tn916 of Enterococcus faecalis, was determined and found to encode a 72,486-dalton protein exhibiting a high degree of homology with other tet(M) determinants. A short open reading frame corresponding to a 28-amino-acid peptide and containing a number of inverted repeat sequences was noted immediately upstream of tet(M), suggesting that regulation might occur by a mechanism involving transcriptional attenuation. Transcription analyses found this to indeed be the case, showing that the expression of tet(M) resulted from an extension of a small transcript representing the upstream leader region into the resistance determinant. Exposure of cells to tetracycline resulted in a significant increase in the amount of tet(M) transcription; this increase could be explained on the basis of increased transcriptional read-through from the upstream transcript. A model suggesting how transcriptional attenuation might operate in this system is presented.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brombach M., Gualerzi C. O., Nakamura Y., Pon C. L. Molecular cloning and sequence of the Bacillus stearothermophilus translational initiation factor IF2 gene. Mol Gen Genet. 1986 Oct;205(1):97–102. doi: 10.1007/BF02428037. [DOI] [PubMed] [Google Scholar]
- Burdett V., Inamine J., Rajagopalan S. Heterogeneity of tetracycline resistance determinants in Streptococcus. J Bacteriol. 1982 Mar;149(3):995–1004. doi: 10.1128/jb.149.3.995-1004.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett V. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J Biol Chem. 1991 Feb 15;266(5):2872–2877. [PubMed] [Google Scholar]
- Burdett V. Streptococcal tetracycline resistance mediated at the level of protein synthesis. J Bacteriol. 1986 Feb;165(2):564–569. doi: 10.1128/jb.165.2.564-569.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Movable genetic elements and antibiotic resistance in enterococci. Eur J Clin Microbiol Infect Dis. 1990 Feb;9(2):90–102. doi: 10.1007/BF01963632. [DOI] [PubMed] [Google Scholar]
- Cruz-Rodz A. L., Gilmore M. S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990 Oct;224(1):152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- Dever T. E., Glynias M. J., Merrick W. C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman I., Zamir A. The nucleotide sequence of the ferrochelatase and tRNA(val) gene region from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 Oct 25;18(20):6130–6130. doi: 10.1093/nar/18.20.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L., Pribnow D., Schneider T., Shinedling S., Singer B. S., Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- Guild N., Gayle M., Sweeney R., Hollingsworth T., Modeer T., Gold L. Transcriptional activation of bacteriophage T4 middle promoters by the motA protein. J Mol Biol. 1988 Jan 20;199(2):241–258. doi: 10.1016/0022-2836(88)90311-7. [DOI] [PubMed] [Google Scholar]
- Ishii S., Ihara M., Maekawa T., Nakamura Y., Uchida H., Imamoto F. The nucleotide sequence of the cloned nusA gene and its flanking region of Escherichia coli. Nucleic Acids Res. 1984 Apr 11;12(7):3333–3342. doi: 10.1093/nar/12.7.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D. J., Lee L. N., Titmas B. M., Smith C. J., Tenover F. C. Nucleotide sequence analysis of tetracycline resistance gene tetO from Streptococcus mutans DL5. J Bacteriol. 1988 Aug;170(8):3618–3626. doi: 10.1128/jb.170.8.3618-3626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavathu E. K., Hiratsuka K., Taylor D. E. Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene. 1988;62(1):17–26. doi: 10.1016/0378-1119(88)90576-8. [DOI] [PubMed] [Google Scholar]
- Martin P., Trieu-Cuot P., Courvalin P. Nucleotide sequence of the tetM tetracycline resistance determinant of the streptococcal conjugative shuttle transposon Tn1545. Nucleic Acids Res. 1986 Sep 11;14(17):7047–7058. doi: 10.1093/nar/14.17.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubens C. E., Heggen L. M. Tn916 delta E: a Tn916 transposon derivative expressing erythromycin resistance. Plasmid. 1988 Sep;20(2):137–142. doi: 10.1016/0147-619x(88)90016-9. [DOI] [PubMed] [Google Scholar]
- Sacerdot C., Dessen P., Hershey J. W., Plumbridge J. A., Grunberg-Manago M. Sequence of the initiation factor IF2 gene: unusual protein features and homologies with elongation factors. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7787–7791. doi: 10.1073/pnas.81.24.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Brown J. T., Roberts M., Urdea M. S. Homology of the TetM with translational elongation factors: implications for potential modes of tetM-conferred tetracycline resistance. Nucleic Acids Res. 1988 Feb 11;16(3):1218–1218. doi: 10.1093/nar/16.3.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Brown J. T., Roberts M., Urdea M. S. The nucleotide sequence of the tetracycline resistance determinant tetM from Ureaplasma urealyticum. Nucleic Acids Res. 1988 Feb 11;16(3):1216–1217. doi: 10.1093/nar/16.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senghas E., Jones J. M., Yamamoto M., Gawron-Burke C., Clewell D. B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988 Jan;170(1):245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shazand K., Tucker J., Chiang R., Stansmore K., Sperling-Petersen H. U., Grunberg-Manago M., Rabinowitz J. C., Leighton T. Isolation and molecular genetic characterization of the Bacillus subtilis gene (infB) encoding protein synthesis initiation factor 2. J Bacteriol. 1990 May;172(5):2675–2687. doi: 10.1128/jb.172.5.2675-2687.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Su Y. A., Sulavik M. C., He P., Makinen K. K., Makinen P. L., Fiedler S., Wirth R., Clewell D. B. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun. 1991 Jan;59(1):415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Jones J. M., Senghas E., Gawron-Burke C., Clewell D. B. Generation of Tn5 insertions in streptococcal conjugative transposon Tn916. Appl Environ Microbiol. 1987 May;53(5):1069–1072. doi: 10.1128/aem.53.5.1069-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Yager T. D. Transcript elongation and termination are competitive kinetic processes. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2307–2311. doi: 10.1073/pnas.88.6.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]