Abstract
Sexual reproduction allows deleterious transposable elements to proliferate in populations, whereas the loss of sex, by preventing their spread, has been predicted eventually to result in a population free of such elements [Hickey, D. A. (1982) Genetics 101, 519–531]. We tested this expectation by screening representatives of a majority of animal phyla for LINE-like and gypsy-like reverse transcriptases and mariner/Tc1-like transposases. All species tested positive for reverse transcriptases except rotifers of the class Bdelloidea, the largest eukaryotic taxon in which males, hermaphrodites, and meiosis are unknown and for which ancient asexuality is supported by molecular genetic evidence. Mariner-like transposases are distributed sporadically among species and are present in bdelloid rotifers. The remarkable lack of LINE-like and gypsy-like retrotransposons in bdelloids and their ubiquitous presence in other taxa support the view that eukaryotic retrotransposons are sexually transmitted nuclear parasites and that bdelloid rotifers evolved asexually.
Transposons are commonly thought to be of universal occurrence in eukaryotes, even though only a few major phyla have actually been examined. We tested representatives of the majority of animal phyla for the most prominent superfamilies of the two classes of eukaryotic transposons: (i) retrotransposons, which transpose via an RNA intermediate copied into DNA by an element-encoded reverse transcriptase (RTase), and (ii) DNA transposons, which transpose as DNA by a cut-and-paste mechanism using an element-encoded transposase (1, 2). Amino acid sequences of RTases or transposases exhibit more similarity within the same superfamily, even in distantly related host species, than between superfamilies, even within the same host. This similarity allows the design of PCR screens capable of detecting transposons belonging to a given superfamily in diverse taxa. Even within relatively conserved domains, however, sequence diversity necessitates the use of highly degenerate primer pools, in most cases giving no transposon-specific bands when only a single pair of domains is targeted. To achieve the necessary specificity, we developed a two-step PCR procedure that takes advantage of the presence of more than two conserved domains in each enzyme. This procedure was used to screen for RTases of LINE-like and gypsy-like superfamilies and DNA transposases of the mariner/Tc1-like superfamily.
Materials and Methods
DNA Isolation.
Live specimens were purchased from (a) Gulf Specimen Marine Laboratories (Panacea, FL), (b) Carolina Biological Supply (Burlington, NC), (c) Marine Biological Laboratory (Woods Hole, MA), and (d) Florida Aqua Farms (Dade City, FL). Specimens also were provided by (e) D. McHugh (Colgate University, Hamilton, NY), (f) D. Rowell (Australian National University, Canberra) or (h) were from our laboratory cultures. DNA was prepared by SDS-proteinase K digestion and phenol-chloroform extraction, using thoroughly washed whole animals or dissected tissues. DNA samples were obtained from: (i) J. Samuelson (Harvard School of Public Health), (k) D. Mark Welch (Harvard University), (l) R. Horvitz (Massachusetts Institute of Technology, Cambridge), (m) W. Gilbert (Harvard University), (n) A. McMahon (Harvard University), (o) D. Melton (Harvard University), (r) M. Evgen'ev (Institute of Molecular Biology, Moscow), (s) S. Palumbi (Harvard University), and (t) Sigma. Letter designations correspond to superscripts in Table 1.
Table 1.
Tests for LINE-like and gypsy-like RTase sequences and mariner/Tc1-like transposase sequences in diverse species
| Phylum | Species | No.* | Mariner | Tc1 | LINE‡ | Gypsy‡ |
|---|---|---|---|---|---|---|
| Sarcomastigophora | Giardia lamblial | 1 | +S | |||
| Porifera | Halichondria bowerbankia | 2 | + | + | +S | |
| Spongilla sp.b | 3 | + | + | +S | ||
| Cnidaria (L,M)† | Hydra littoralisb | 4 | +S | +S | + | − |
| Aurelia auritac | 5 | + | + | |||
| Ctenophora | Condylactus sp.a | 6 | − | + | + | |
| Platyhelminthes (M) | Dugesia tigrinab | 7 | +S | +S | +S | |
| Rotifera (Acanthocephala) | Moniliformis moniliformisk | 8 | − | − | +S | + |
| Rotifera (Monogononta) | Brachionus plicatilisd | 9 | + | − | +S | +S |
| Brachionus calyciflorusd | 10 | − | − | + | ||
| Sinantherina socialisk | 11 | − | + | +S | + | |
| Monostyla sp.b | 12 | − | − | +S | ||
| Rotifera (Bdelloidea) | Philodina roseolab | 13 | + | − | − | − |
| Philodina rapidah | 14 | + | − | − | − | |
| Habrotrocha constrictah | 15 | +S | − | − | − | |
| Adineta vagak | 16 | +S | − | − | − | |
| Macrotrachela quadricorniferah | 17 | +S | − | − | ||
| Gastrotricha | Lepidodermella sp.b | 18 | + | + | +S | − |
| Nemertea | Lineus sp.c | 19 | + | + | +S | + |
| Priapulida | Priapulus caudatuse | 20 | − | +S | + | |
| Sipuncula | Themiste alutaceaa | 21 | − | +S | + | |
| Annelida | Glycera sp.e | 22 | − | +S | + | |
| Echiura | Lissomyema mellitaa | 23 | + | − | +S | +S |
| Mollusca (L) | Chione cancellataa | 24 | +S | +S | + | |
| Brachiopoda | Glottidea pyramidataa | 25 | − | +S | + | |
| Bryozoa | Amathia convolutaa | 26 | + | + | +S | +S |
| Phoronida | Phoronis architectaa | 27 | + | + | + | |
| Nematoda (L,G,M,T) | Caenorhabditis elegansl | 28 | + | + | +S | +S |
| Onychophora | Euperipatoides rowellif | 29 | + | +S | +S | +S |
| Arthropoda (L,G,M,T) | Drosophila melanogasterh | 30 | − | + | +S | +S |
| Drosophila pseudoobscurah | 31 | + | + | + | +S | |
| Drosophila virilisr | 32 | + | + | + | +S | |
| Lasius nigerr | 33 | +S | + | |||
| Formica polyctenumr | 34 | + | +S | |||
| Aphis sp.r | 35 | +S | + | |||
| Tardigrada | Milnesium sp.b | 36 | + | + | + | |
| Chaetognatha | Sagitta sp.c | 37 | + | +S | + | |
| Echinodermata (G) | Echinometra mathaeis | 38 | + | +S | ||
| Strongylocentrotus purpuratuss | 39 | + | + | |||
| Hemichordata | Saccoglossus kowalevskiic | 40 | + | +S | + | |
| Chordata (L,G,M,T) | Branchiostoma floridaea | 41 | +S | + | ||
| Danio reriom | 42 | + | +S | |||
| Onchorhynchus ketat | 43 | + | + | |||
| Xenopus laeviso | 44 | + | + | |||
| Mus musculusn | 45 | + | +S | |||
| Bos taurust | 46 | + | + |
Superscript letters under species column refer to suppliers of specimens as indicated in Materials and Methods.
For reference to Fig. 2, each species is assigned a number.
Superfamilies previously reported to be present in representatives of a phylum are indicated in parentheses: L, LINE; G, gypsy; M, mariner; T, Tc1.
Presence or absence of diagnostic PCR bands is indicated by + or −, respectively; S, verified by sequencing; blank, not done.
PCR, Cloning, and Sequencing.
RTase sequences were first amplified with primers GAYITIINNNVNGSNTWY (A) and ANININAINCCNARRWM (E) for 5 min at 95°C, 10 × (1 min at 94°C, 1 min at 47°C, 1 min at 72°C), 50 × (20 sec at 94°C, 45 sec at 52°C, 45 sec at 72°C), 10 min at 72°C. Second-step primers were INGGNIBNCSNCARGG (B-LINE) and RNNRNRTCRTCNGCRWA (C-LINE) or HIIDBNNTNCCNTTYGG (B-gyp) and ANNANRTCRTCNANRTA (C-gyp). For LINE-like RTases, the second step was performed for 2 min at 95°C, 10 × (1 min at 94°C, 45 sec at 50°C, 45 sec at 72°C), 35 × (20 sec at 94°C, 30 sec at 55°C, 20 sec at 72°C) and 20 min at 72°C. For gypsy-like RTases, generally present in lower copy numbers, the second step included 20 additional cycles and annealing was at 48°C and 53°C. Mariner-like transposases were first amplified with primers TNNTNWBNDBNGAYGARA (DE) and RWARTYNVWNGGNGC (mar-I) and then with primers TNNTNYWNGAYAAYGM (D) and GCNARRTCNGGNSWRTA (mar-II); Tc1-like transposases with DE and WNYTCDATNGKRTT (Tc-I) followed by D and RTTNARRTCNGGNSWYTG (Tc-II). First- and second-step amplification conditions were essentially the same as for LINE-like RTases. Amplification reactions contained primers at 35 nM/ml (first step) or 12.5 nM/ml (second step), 0.5 mM dNTPs, Taq DNA polymerase (Qiagen, Chatsworth, CA), and the buffer provided by the manufacturer. All primers contained XbaI (A, B-LINE, B-gyp, DE, D) or EcoRI (E, C-LINE, C-gyp, mar-I, mar-II, Tc-I, Tc-II) recognition sites at their 5′ ends. PCR products were cloned with the TA or TOPO-TA cloning kits (Invitrogen) from total amplification reactions or by standard cloning procedures from excised bands, sequenced with the Big Dye Terminator Cycle Sequencing Kit (Perkin–Elmer Applied Biosystems), and analyzed on an Applied Biosystems Prism 310 Genetic Analyzer.
Sequence Analysis.
Sequence comparisons were done with Wisconsin Package Version 10.0, GCG. Uncorrected pairwise distances were determined for groups of aligned amino acid sequences with the distances program. The degree of sequence similarity is graphically represented by dendrograms in Fig. 3, obtained by neighbor-joining with the growtree program. GenBank accession numbers for sequences in Fig. 3 are as follows: SART1, D85594; RT1, M93690; TRAS, D38414; Jockey, P21328; CR1, AF086712; Frodo2, Z48009; T1, M93689; Amy, U07847; R2Bm, M16558; R2Dm, P16423; L1, P08547; Swimmer, AF055640; DRE, X57034; R4, U29445; Dong, L08889; RTE2, U58755; JAM1, Z86117; sushi, AF030881; GRT(Xl), AJ243723; Osvaldo, S75260; pol(Gm), AF104899; gypsy, M12927; CerR03D7, Z46828; micropia, X13304; GRT(Lf), AJ243733; ZAM, AJ000387; Mar8.2, L10493; MarR4, U51175; MarR11, U51174; Hsmar2, U49974; Himar3.4, L10463; Hsmar1, U52077; Hcmar1.2, L10444; and Cemar2, AL132949. Sequences MarR5(Sz), MarR6(Sz), and MarR15(Dt) are from ref. 3.
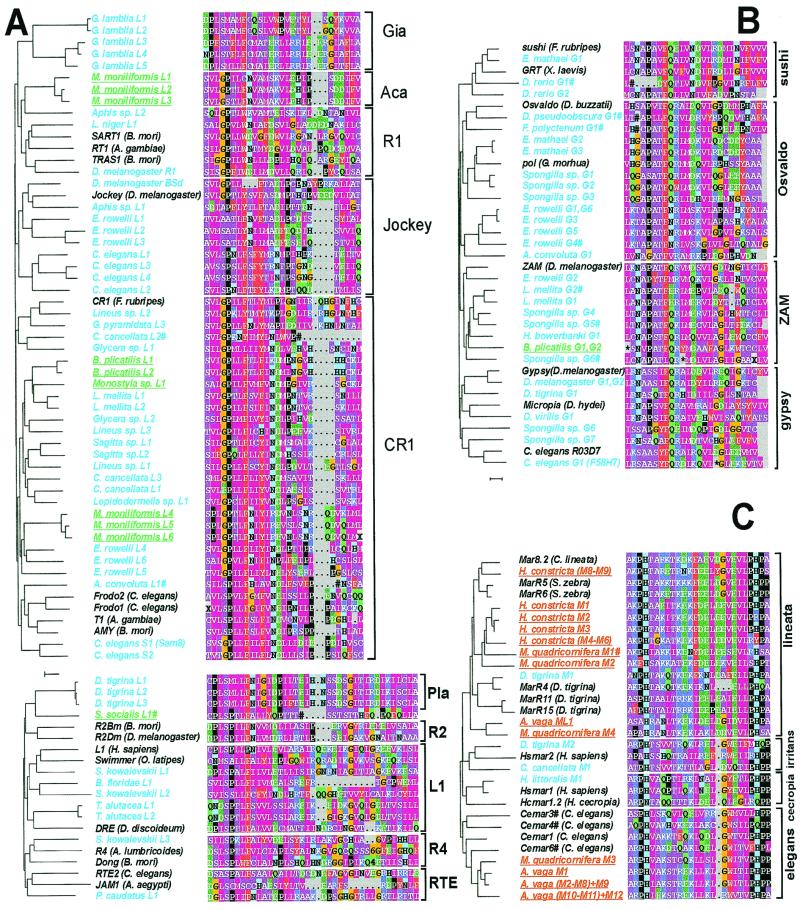
Figure 3.
Alignments of deduced (A) LINE-like RTase, (B) gypsy-like RTase, and (C) mariner-like transposase amino acid sequences with similarity to previously identified clades (6–8); newly designated LINE-like clades Gia, Aca and Pla; or known mariner subfamilies (3). Except for primer sequences, which are not included, the regions of RTases and transposases depicted correspond to the filled bars in Fig. 1. Rotifer species are underlined, with bdelloids in red and other rotifers in green. All other RTase or transposase sequences we determined are in blue. Three LINE-like sequences and nine gypsy-like sequences contained stop codons and/or frame shifts (*, stop codon; #, frameshift). LINE-like sequences are shown in two separate groups, corresponding to their difference in the B-C distance. Clones within parentheses have identical nucleotide sequences. Dendrograms obtained by neighbor-joining illustrate sequence similarity. For comparison, sequences of two or three known members of each clade are included in black. (Scale bar: 10% amino acid difference.)
Results
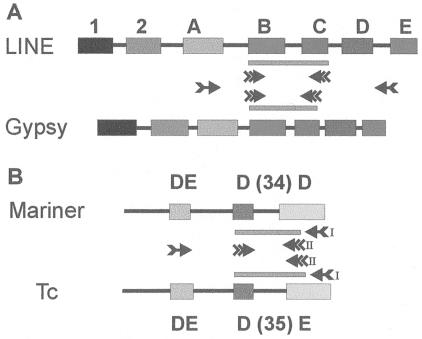
For enrichment of both RTase superfamilies, the first amplification used primer pools targeted to conserved residues in domains A and E (4, 5) (Fig. 1A). An aliquot of the reaction product was subjected to a second amplification with either LINE-specific or gypsy-specific primer pools targeted to conserved residues in domains B and C. This procedure typically yielded bands of sizes diagnostic for the corresponding superfamily. The identity of such bands then could be confirmed by cloning, sequencing, and comparison with known RTase clades (6–8), for which the region between the second-step primers exhibits clade-specific conservation of amino acid sequences. A similar strategy was used for amplification of DNA transposases, using primer pools directed against conserved residues in domains depicted in Fig. 1B.
Figure 1.
Domain structure of RTases (A) and transposases (B) indicating regions targeted for nested PCR. Conserved domains are denoted by filled boxes, interdomain regions by lines. Domains A-E (4) correspond to RTase domains 3–7, respectively (5). For DNA transposases, two primers (I and II) cover conserved residues in the C-terminal domain. Single-feathered and double-feathered arrows indicate first- and second-step amplification primers, respectively. Filled bars represent second-step amplicons. Not to scale.
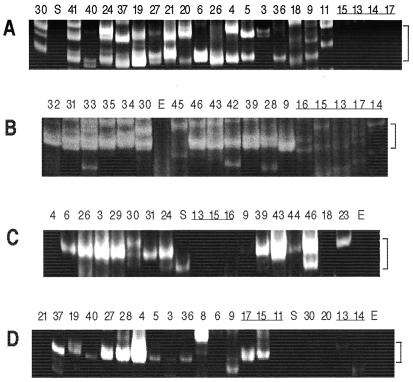
Representatives of all 24 phyla tested by PCR for LINE-like RTase sequences yielded one or more prominent bands of 120–170 bp, the size range of the B-C interval in known LINE-like RTase clades (Fig. 2A; Table 1). Of 70 amplicons in this size range cloned from 23 species, in at least one amplicon from each species except Giardia lamblia the 60-bp region of the B domain adjacent to the primer coded for an amino acid sequence whose closest matches in GenBank always included members of known RTase clades.
Figure 2.
Nested PCR assays for (A and B) LINE-like or (C) gypsy-like RTases, and (D) mariner-like transposases. Twenty additional cycles of second-step amplification were done for B. Brackets indicate expected size ranges. Lane numbers correspond to species numbers in Table 1. Bdelloid rotifers are underlined. E, Escherichia coli; S, Saccharomyces cerevisiae.
Dendrograms based on distance matrix comparisons of amino acid sequences in the B-C interval, excluding the primers, place most sequences in the known LINE-like clades R4, L1, RTE, R1, Jockey, or CR1 (6), of which CR1 is the most widely represented (Fig. 3A). Groups of LINE-like RTase sequences apparently representing new clades, found in an acanthocephalan (Rotifera), a flatworm (Platyhelminthes), and a diplomonad protozoan (Sarcomastigophora), are designated Aca, Pla, and Gia, respectively.
Similarly, bands of 120–130 bp, diagnostic for gypsy-like RTases, were found in two-step amplifications of DNA from 35 species, representing 21 of the 23 phyla tested (Fig. 2C; Table 1). Sequenced amplicons from 14 of these species could be assigned to known clades of gypsy-like RTases (7, 8), designated gypsy, Osvaldo, sushi, and ZAM, according to the name of a known representative (Fig. 3B; Table 1).
The validity of our screen is supported by the finding that all sequences identified as RTase-like amplified from species whose genomic sequence is almost completely known (Caenorhabditis elegans, Drosophila melanogaster, and G. lamblia) exactly or nearly exactly matched the corresponding region of an authentic RTase in the respective database, even in the case of all five of the highly atypical LINE-like RTase nucleotide sequences identified in the protozoan G. lamblia.
Evidence that LINE-like and gypsy-like elements are present in multiple copies in each genome is seen in the observations that several similar but not identical LINE-like or gypsy-like RTase sequences typically were found in individual species (Fig. 3 A and B) and that the yields of the corresponding RTase amplicons were generally comparable to those obtained for C. elegans and D. melanogaster, species whose genomes contain many copies of these retrotransposons (9, 10).
In striking contrast to the occurrence of RTase sequences in all other species tested, amplifications conducted under identical experimental conditions revealed no bands diagnostic for either LINE-like or gypsy-like RTases in any of the five species representing three bdelloid families that we tested (Fig. 2; Table 1). For comparison, we also tested five species of rotifers belonging to classes in which sexual reproduction is either constitutive (Acanthocephala) or facultative (Monogononta) (11, 12). Nested PCR revealed bands of size diagnostic for LINE-like RTases in all five species of nonbdelloid rotifers, an identification confirmed by sequencing for the monogononts Brachionus plicatilis, Monostyla sp., and Sinantherina socialis and the acanthocephalan Moniliformis moniliformis (Figs. 2A and 3A; Table 1). Similarly, amplifications for gypsy-like RTases gave bands of the expected size for each nonbdelloid rotifer tested—B. plicatilis, S. socialis, and M. moniliformis—confirmed by sequencing for B. plicatilis (Figs. 2C and 3B; Table 1).
As a further test for retrotransposons in bdelloids, 20 additional cycles of second-step amplification were performed for LINE-like RTases, giving a broad distribution of amplicons, including some in the size range expected for RTases (Fig. 2B). Of the 95 amplicons cloned from this region or from the total second-step amplification product of Philodina roseola, Philodina rapida, and Habrotrocha constricta, none had a sequence resembling RTase genes except for one clone from P. rapida with similarity to an RTase of Escherichia coli, the food on which we maintain rotifer cultures. The same procedure applied to nonbdelloid species consistently gave clones the majority of which had sequences characteristic of LINE-like RTases. It is evident that few, if any, copies of LINE-like or gypsy-like RTase sequences are present in the bdelloid genomes we examined.
Although RTase sequences were not detected in bdelloids, amplicons of size diagnostic for the mariner family of DNA transposons were clearly evident in two-step amplifications of DNA from the bdelloids Adineta vaga, H. constricta, and Macrotrachela quadricornifera, and weaker bands of the expected size were seen for P. roseola and P. rapida (Fig. 2D). Mariner-like transposase sequences of the lineata subfamily (5) were identified in amplicons cloned from each of the three species that gave the strongest bands, and sequences of the elegans subfamily were found in amplicons from A. vaga and M. quadricornifera (Fig. 3C). Altogether, amplicons diagnostic for mariner-like transposases were clearly evident in 24 of the 34 species tested (Table 1), exhibiting a patchy taxonomic distribution as seen in other investigations (3, 13).
An unusual feature of the mariner-like transposase sequences in bdelloids, not known in other metazoans, is a pronounced bias toward synonymous nucleotide substitution in cloned amplicons of both subfamilies of mariner-like transposases, as indicated by excess polymorphism at codon third positions. Of 88 polymorphic sites in the five nonidentical lineata sequences from H. constricta, 18, 18, and 52 were at first, second, and third codon positions, respectively. The corresponding values for the 39 polymorphic sites in the five nonidentical elegans sequences from A. vaga were 12, 3, and 24. The same bias was observed in comparisons of the rates of synonymous and nonsynonymous substitution (14). Such bias indicates that these sequences are or recently were under prolonged selection to preserve amino acid sequence within either the present or a previous host. The only other case of which we are aware in which mariner/Tc1-like transposase appears to be under such selection is that of the ciliated protozoans Oxytricha fallax and O. trifallax, where it has been suggested that these transposases are involved in programmed genome rearrangement (15), although other possible functions include inhibition of transposition by dominant-negative complementation or by overproduction inhibition, phenomena known in Drosophila (16).
Discussion
Deleterous transposable elements, if not too harmful, can spread through a sexually reproducing population if a copy present in one parent is transmitted, on average, to more than half the progeny (17, 18). If sexual reproduction ceases, however, spread within a population is restricted to rare horizontal transmission to individuals and their clonal descendants. Eventually, if the population has not become extinct and if horizontal transmission is negligible, mutation, including deletion, and selection against clones in which active elements have increased the deleterious load will result in a population without active elements. As the bdelloid lineage appears to have abandoned sexual reproduction many millions of years ago (19), any such elements it once harbored should have been lost or have become so greatly depleted or diverged as to be undetectable by our PCR screen.
Retrotransposons rarely move horizontally, can disrupt the expression of genes into which they transpose, and contain signals that can interfere with proper expression of genes nearby (1, 2). In addition, retrotransposon-encoded RTase can mediate deleterious germ-line insertions of cDNA copies of diverse transcripts. In contrast, mariner-like transposons often are transmitted horizontally (16), lack potentially interfering signals, do not code for RTase, and, as perhaps indicated by the observed conservation of mariner amino acid sequence in bdelloids, may not be significantly detrimental to bdelloid hosts. These differences may explain why mariner-like elements were found in bdelloids while retrotransposons were not.
The finding of retrotransposon RTase sequences in all species tested, except bdelloid rotifers, but including other members of the phylum as well as representatives of more basal phyla, indicates that active retrotransposons were lost after the separation of bdelloids from other rotifers and, if such loss was monophyletic, before the radiation of modern bdelloids, during approximately the same interval in which sexual reproduction appears to have ceased (19).
Although the lack of retrotransposons in bdelloid rotifers may be understood as a consequence of long-term asexuality, it is also possible that their loss has allowed bdelloid rotifers to avoid the early extinction that typically follows the abandonment of sex in other taxa. This could be the case if sexual reproduction plays a significant role in limiting the load of deleterious insertions, by mechanisms involving recombination (20–22), or by other mechanisms dependent on sexual reproduction or meiosis. Indeed, a major advantage of sex may be in limiting the deleterious load imposed by retrotransposon insertions and by the action of the RTase they encode (ref. 3, pp. 122–123). If so, the abandonment of sex ordinarily could allow retrotransposons and other retro-insertions to increase, driving the population to extinction. The bdelloid lineage may be unusual in having escaped this fate by somehow becoming free of active retrotransposons and RTases, either before or not long after abandoning sex. [That active retrotransposons may in rare instances be lost even from sexually reproducing species is suggested by the apparent lack of active LINE-1 elements from certain species of rice rats (Oryzomys) (23).]
Speculation aside, our evidence for the presence of retrotransposons in representatives of all 24 animal phyla tested, including 16 phyla not previously investigated, demonstrates the virtually universal occurrence of retrotransposons throughout the animal kingdom, whereas their apparent absence only in bdelloid rotifers strongly supports the view that retrotransposons are sexually transmitted nuclear parasites and that bdelloid rotifers are ancient asexuals.
Acknowledgments
We thank the individuals listed in Materials and Methods for providing animal specimens or DNA, T. Bestor, M. Cummings, D. Mark Welch, J. Mark Welch, B. Normark, and two anonymous reviewers for critically reading the manuscript, M. Sogin and A. McArthur for help with searches of the G. lamblia genome project database, and the Eukaryotic Genetics Program of the U.S. National Science Foundation for support.
Abbreviation
- RTase
reverse transcriptase
Footnotes
References
- 1.Arkhipova I, Lyubomirskaya N, Ilyin Y. Drosophila Retrotransposons. Austin, TX: Landes; 1995. [Google Scholar]
- 2.Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Austin, TX: Landes; 1998. [Google Scholar]
- 3.Robertson H M. J Hered. 1997;88:195–201. doi: 10.1093/oxfordjournals.jhered.a023088. [DOI] [PubMed] [Google Scholar]
- 4.Poch O, Sauvaget I, Delarue M, Tordo N. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong Y, Eickbush T H. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik H S, Burke W D, Eickbush T H. Mol Biol Evol. 1999;16:793–805. doi: 10.1093/oxfordjournals.molbev.a026164. [DOI] [PubMed] [Google Scholar]
- 7.Miller K, Lynch C, Martin J, Herniou E, Tristem M. J Mol Evol. 1999;49:358–366. doi: 10.1007/pl00006559. [DOI] [PubMed] [Google Scholar]
- 8.Malik H S, Eickbush T H. J Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 10.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 11.Mark Welch D. Invert Biol. 2000;119:17–26. [Google Scholar]
- 12.Gilbert J. In: Reproductive Biology of the Invertebrates. Adiyodi K, Adiyodi R, editors. 4, Part A. New York: Wiley; 1989. pp. 179–199. [Google Scholar]
- 13.Avancini R M, Walden K K, Robertson H M. Genetica. 1996;98:131–140. doi: 10.1007/BF00121361. [DOI] [PubMed] [Google Scholar]
- 14.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 15.Witherspoon D J, Doak T G, Williams K R, Seegmiller A, Seger J, Herrick G. Mol Biol Evol. 1997;14:696–706. doi: 10.1093/oxfordjournals.molbev.a025809. [DOI] [PubMed] [Google Scholar]
- 16.Hartl D, Lohe A R, Lozovskaya E R. Annu Rev Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 17.Hickey D A. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeyl C, Bell G. Trends Ecol Evol. 1996;11:10–15. doi: 10.1016/0169-5347(96)81058-5. [DOI] [PubMed] [Google Scholar]
- 19.Mark Welch D, Meselson M. Science. 2000;288:1211–1215. doi: 10.1126/science.288.5469.1211. [DOI] [PubMed] [Google Scholar]
- 20.Kondrashov A S. J Hered. 1993;84:372–387. doi: 10.1093/oxfordjournals.jhered.a111358. [DOI] [PubMed] [Google Scholar]
- 21.Crow J F. Dev Genet. 1994;15:205–213. doi: 10.1002/dvg.1020150303. [DOI] [PubMed] [Google Scholar]
- 22.Muller H J. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 23.Casavant N C, Scott L, Cantrell M A, Wiggins L E, Baker R J, Wichman H A. Genetics. 2000;154:1809–1817. doi: 10.1093/genetics/154.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]