Abstract
Recent research has emphasized the importance of the metabolic cluster, which includes glucose intolerance, dyslipidemia, and high blood pressure, as a strong predictor of the obesity-related morbidities and premature mortality. Fundamental to this association, commonly referred to as the metabolic syndrome, is the close interaction between abdominal fat patterning, total body adiposity, and insulin resistance. As the initial step in identifying major genetic loci influencing these phenotypes, we performed a genomewide scan by using a 10-centiMorgan map in 2,209 individuals distributed over 507 nuclear Caucasian families. Pedigree-based analysis using a variance components linkage model demonstrated a quantitative trait locus (QTL) on chromosome 3 (3q27) strongly linked to six traits representing these fundamental phenotypes [logarithm of odds (lod) scores ranged from 2.4 to 3.5]. This QTL exhibited possible epistatic interaction with a second QTL on chromosome 17 (17p12) strongly linked to plasma leptin levels (lod = 5.0). Situated at these epistatic QTLs are candidate genes likely to influence two biologic precursor pathways of the metabolic syndrome.
Obesity is a common and chronic disorder associated with decreased longevity and increased morbidity from a variety of diseases, including type 2 diabetes mellitus, hypertension, stroke, and coronary heart disease (1). Fat distribution, specifically the pattern known as upper-body, abdominal, or visceral obesity, is a major predictor of the adverse metabolic profile predisposing to these health risks (2). Thus, abdominal-visceral fat size has emerged as a significant precursor of glucose intolerance, hyperinsulinemia, elevated plasma triglycerides, decreased high density lipoprotein-cholesterol, and increased blood pressure (3). Fundamental to this metabolic milieu are close interactions between total body adiposity, abdominal-visceral fat size, and insulin resistance. Reaven (4) provided evidence to suggest that resistance to insulin-stimulated glucose uptake is associated with a series of related metabolic variables, termed “syndrome X,” which cluster in the same individual and include glucose intolerance, disturbed plasma lipids, and high blood pressure (4). Because of close similarities of these features with those associated with abdominal obesity, the more collective term metabolic syndrome was introduced (5).
The etiology of the abdominal obesity-metabolic syndrome is complex and is thought to involve metabolic, neuroendocrine, and genetic interactions. A metabolic-neuroendocrine cascade has been proposed in which increased free fatty acid flux from the highly lipolytic visceral adipocytes, together with imbalances in sex hormones, could cause the insulin resistance and hyperinsulinemia, with their metabolic consequences (6). Weight gain with preferential deposition of adipocytes in the abdominal-visceral region is considered secondary to adoption of Westernized diet, activity lifestyle, and reactivity to emotional, intellectual, and physical stresses (5). Meanwhile, prospective twin studies, familial segregation, and intercorrelation analyses have supported the existence of genetic influences (7–10). It is not known, however, whether this occurs through a major locus or multiple, distinct loci acting in concert, perhaps in response to common metabolic and/or neuroendocrine factors. The purpose of this study was to identify genes with measurable influence on the quantitative expression of phenotypes fundamental to the abdominal obesity-metabolic syndrome. We report results of a genomewide linkage scan on a large cohort of Caucasian families. Our findings reveal two quantitative trait loci (QTLs) in chromosomal regions containing candidate genes that could affect expression of the metabolic syndrome, with possible epistatic interaction between them.
Methods
Subjects.
The Metabolic Risk Complications of Obesity Genes project was initiated in 1994 with the formulation of a nine-page questionnaire that collected information on family structure, health and behavior status, and detailed family and personal history of obesity and its health complications. These families were recruited from the TOPS (Take Off Pounds Sensibly, Inc.) membership. TOPS provided mailing material on membership attending its chapters in 10 states (Wisconsin, Illinois, Michigan, Iowa, Minnesota, Ohio, West Virginia, Missouri, Kentucky, and Indiana). Questionnaire data received from 60,000 respondents were verified and entered into the TOPS Obesity and Metabolic Research Center databases at the Medical College of Wisconsin. Families with at least two obese sibs [body mass index (BMI) ≥ 30 mg/k (2)], availability of one (preferably both) parents, as well as at least one never-obese sib and/or parent [BMI ≤ 27 mg/k (2)] were identified and contacted for ascertainment. Families were scheduled to visit satellites (4–6 per state), where an experienced team undertook the phenotypic procedures. A more detailed questionnaire garnered personal data (date of birth, race, marital status), health history (asthma, kidney or liver disease, hypertension, heart disease, stroke, hyperlipidemia, thyroid disorders, diabetes, medications, menopausal status, and hormonal replacement therapy), weight history (age of onset, maximum and minimum adult weight, dietary and exercise profiles, smoking history), as well as availability and information about family relatives (twins, sibs, adopted, fostered, stepchildren). Exclusion criteria included: pregnancy, type 1 diabetes mellitus, history of cancer, renal or hepatic disease, severe coronary artery disease, substance abuse, corticosteroids or thyroid medications above replacement dose, and history of weight loss of more than 10% of body weight in the preceding 12 months. Research protocols were approved by the Institutional Review Board of the Medical College of Wisconsin.
Data for the genetic analyses presented here included 2,209 individuals distributed across 507 Caucasian families of predominantly northern European ancestry and residing in the United States. Recruitment was initiated via an obese proband, with one sib who is also obese, availability of one parent (preferably both), and one never-obese sib and/or parent. Table 1 shows the pair-wise relationships represented by this data set.
Table 1.
Distribution of relative pairs
| Relationship | Number of pairs |
|---|---|
| Parent–offspring | 2,177 |
| Siblings | 2,198 |
| Grandparent–grandchild | 96 |
| Avuncular | 498 |
| Half siblings | 61 |
| Grand avuncular | 4 |
| Half avuncular | 7 |
| First cousins | 331 |
| First cousins, once removed | 16 |
| Monozygotic twins | 11 |
| Total | 5,399 |
Phenotypes.
Waist and hip circumferences were measured according to the World Health Organization criteria: with participants wearing light clothing, waist circumference was the minimal measurement at the navel region, and hip circumference the widest measurement at the hip and buttocks. Serum glucose concentrations were measured with a Glucose Analyzer II (Beckman Instruments, Brea, CA), using a glucose oxidase procedure. Replicate readings were repeated to within 3 mg/dl in triplicate. A double-antibody, equilibrium RIA (Linco Research, St. Louis) was used for the measurement of plasma insulin, using antibody specific to human insulin. RIA of leptin also was performed by using a specific antibody to human leptin (Linco Research). Quality controls were performed to assure stability and reliability of the assays. Five pool sera of increasing peptide concentrations were used to evaluate the intra-assay and interassay coefficients of variance. For linkage analysis with insulin and insulin/glucose, subjects with diabetes were excluded.
Genotyping.
Whole blood was obtained from all consenting family members for DNA extraction. DNA was prepared by using commercial kits (Puregene, Gentra Systems, Minneapolis), which use a nonphenol-based method involving RNase A treatment. The DNA samples were stored in aliquots at 4°C, and back-up DNA samples were stored in ethanol at −70°C. Additional whole blood aliquots were stored at −70°C for further DNA extraction. More than 200 μg genomic DNA with a 260/280 ratio of 1.8–2.1 was available from each family member.
Genotyping was performed at the Marshfield (Wisconsin) Medical Research Foundation by using the Weber screening set 9 (Research Genetics, Huntsville AL). This procedure used 387 markers, representing tandem repeat polymorphisms, including 366 autosomal as well as 17 X-linked and 4 Y-linked markers (11), and yielded an average map density of 10 centiMorgans (cM). Initial analyses included validation of the reported relationships between individuals, checked by calculating likelihoods of the relationships based on the autosomal genotype data (12). Clear errors in the relations between individuals were ascertained, and as a result data on eight proband families were discarded. The genotypic data also were examined for Mendelian inconsistencies, and those genotypes proven to be inconsistent also were removed. The autosomal genotype data were 97.6% complete. The average (±SD) heterozygosity of these markers used was 0.79 ± 0.06, and the sex-averaged genetic spacing was 9.1 ± 3.8 cM. DNA was screened by using fluorescently labeled primers from Research Genetics. The PCR assay mixture contained 45 ng DNA, 0.075 μM fluorescently labeled primers, 0.12 units AmpliTaq Polymerase (Sigma), 100 μM each dNTP, 25 mM MgCl2, and buffer. PCR conditions included 27 cycles of denaturation (95°C for 30 s), annealing (55°C for 75 s), and elongation (72°C for 30 s), followed by a final 6-min elongation period. Samples were analyzed through automated high-throughput scanning fluorescence detectors, each simultaneously detecting three separate dyes.
Variance Components Linkage Analysis.
A variance component model applied to extended family data was used to test for evidence of linkage of QTLs for phenotypes related to the metabolic syndrome with short tandem repeat loci by using a 10-cM genomewide map. An extension of the strategy developed by Amos (13) was used to estimate the genetic variance attributable to a specific chromosomal location (14). This approach is based on specifying the expected genetic covariances between arbitrary relatives as a function of the identity-by-descent (IBD) relationships at a given marker locus. Additionally unbiased estimates of the effects of covariates are simultaneously obtained.
The basic method of variance component linkage analysis also includes a QTL-specific component, which is used to test for linkage. Using a variance component model (15), we tested the null hypothesis that, the additive genetic variance because of a QTL (σq2), equals zero (no linkage) by comparing the likelihood of this restricted model with that of a model in which σq2 is estimated. The difference between the two log10 likelihoods produces a logarithm of odds (lod) score that is the equivalent of the classical lod score of linkage analysis. Twice the difference in loge likelihoods of these models yields a test statistic that is asymptotically distributed as a 1/2:1/2 mixture of a χ2 variable and a point mass at zero (16). This quantitative trait linkage method has been implemented in the program package solar (14), which determines whether genetic variation at a specific chromosomal location can explain the variation in the phenotype (13, 15, 17).
Extensive simulation suggests that the likelihood ratio test used in this approach yields expected nominal P values for a wide variety of reasonable trait distributions (18); however, as an added precaution we also obtained the empirical distribution for the lod scores for this sample by simulation over 10,000 replicates (19, 20). This approach allows for the calculation of the expected lod score that is then regressed on the observed lod score to obtain the appropriate correction constant (19, 20). This approach is guaranteed to provide robust lod scores. In addition, genomewide P values were calculated by using the method of Feingold and colleagues (21) allowing for chromosome specific marker densities.
The use of the variance component approach requires an estimate of the IBD matrix. For the relatively simple TOPS pedigrees, a pairwise maximum likelihood-based procedure was used to estimate IBD probabilities (14). To permit multipoint analysis for QTL mapping, an extension (16) of the technique of Fulker and colleagues (22) was used. Estimates of the IBD probabilities were generated at any point on a chromosome by using a constrained linear function of observed IBD probabilities of markers at known locations within the region. This multipoint procedure, which yields substantially greater power to localize QTLs than two-point methods, enabled direct localization of the QTL and construction of confidence intervals. One lod unit support intervals are obtained by identifying the peak for the maximum lod score on the plot of the multipoint results, dropping down one lod unit and finding the chromosomal region defined by the shoulders of the curve. For the current data set, a lod-score evaluation was performed every cM along the chromosome, the distances between markers having been determined by using cri-map (23).
As a hypothesis generating exercise, we also looked for evidence of epistasis in the expression of these highly intercorrelated phenotypes. For computational simplicity and to minimize the number of tests performed, we only considered the two chromosomal regions with the strongest marginal lod scores as revealed in the original genome scan. An additive two-locus model and an epistatic model that allowed an additional variance component for (additive × additive) genetic interactions (14, 24) were examined. Each additional variance component was examined by using a likelihood ratio test.
Results
Table 2 shows the means, standard deviations, and total additive genetic heritabilities of the phenotypes included in this study. Values for kurtosis, the distributional attribute most likely to contribute to inflation of type I error, were found to range from 0.2 to 1.8. The total additive genetic heritabilities ranged from 16% to 32% and were estimated after allowing for the covariate effects listed in Table 3.
Table 2.
Means and additive genetic heritabilities for abdominal obesity-metabolic syndrome-related phenotypes
| Trait | Mean | Standard deviation | Sample size | Additive genetic heritability |
|---|---|---|---|---|
| BMI, kg/m2 | 32.0 | 7.6 | 2,154 | 0.24 ± 0.06 |
| Waist, cm | 103 | 19.0 | 2,205 | 0.28 ± 0.06 |
| Hip, cm | 117 | 17.0 | 2,160 | 0.32 ± 0.06 |
| Weight, kg | 90.0 | 22.7 | 2,163 | 0.29 ± 0.06 |
| Insulin, pmol/liter | 84.7 | 40.4 | 1,620 | 0.16 ± 0.07 |
| Insulin/glucose | 9.97 | 4.4 | 1,604 | 0.19 ± 0.06 |
| Leptin, ng/ml | 26.2 | 19.9 | 2,044 | 0.32 ± 0.06 |
Table 3.
Covariate analysis
| Covariate | BMI | Hip | Waist | Weight | Insulin | Insulin/glucose | Leptin |
|---|---|---|---|---|---|---|---|
| Sex | 3.8 (0.8) | 10.2 (0.1) | −1.1 (2.1) | −5.4 (2.5) | 2.5 (5.7) | 0.4 (0.6) | 24.9 (2.1) |
| Male age | 0.0 (0.04) | 0.1 (0.1) | 0.3 (0.1) | −0.1 (0.1) | 0.3 (0.2) | 0.0 (0.02) | 0.0 (0.1) |
| Female age | −0.1 (0.03) | −0.1 (0.1) | 0.0 (0.1) | −0.4 (0.1) | 0.1 (0.2) | 0.0 (0.02) | −0.1 (0.1) |
| Menstrual status | −1.3 (0.7) | −1.3 (1.4) | −2.3 (1.7) | −2.7 (2.0) | −4.5 (4.7) | −0.7 (0.5) | −4.2 (1.6) |
| Birth control medication/ estrogen replacement | 2.3 (0.7) | 3.2 (1.4) | −5.6 (1.6) | −6.2 (1.9) | −5.6 (4.5) | −0.6 (0.5) | −1.6 (1.6) |
| Smoking | −2.4 (0.6) | −5.0 (1.24) | −4.6 (1.5) | −7.1 (1.7) | −6.8 (4.0) | −1.0 (0.4) | −5.9 (1.4) |
Data represent mean effects, with standard errors in parentheses.
Table 4 gives the phenotypic correlations among this set of phenotypes that all relate to the metabolic syndrome. These values range from 0.39 for the correlation between hip circumference and the insulin/glucose ratio to 0.94 between insulin and the insulin/glucose ratio. However, as can be seen from Table 4 the majority of the correlations fall between 0.43 and 0.86, suggesting strong, but not complete correlation among this set of traits.
Table 4.
Phenotypic correlations
| Weight | Waist circumference | Leptin | Insulin | Insulin/ glucose ratio | Hip circumference | |
|---|---|---|---|---|---|---|
| Waist circumference | 0.82 | |||||
| Leptin | 0.66 | 0.59 | ||||
| Insulin | 0.47 | 0.47 | 0.43 | |||
| Insulin/glucose | ||||||
| Ratio | 0.43 | 0.44 | 0.40 | 0.94 | ||
| Hip circumference | 0.87 | 0.82 | 0.65 | 0.43 | 0.39 | |
| BMI | 0.93 | 0.81 | 0.69 | 0.49 | 0.45 | 0.87 |
Robust lod scores were calculated by using the method detailed by Blangero and colleagues (19, 20). Basically, traits exhibiting markedly leptokurtic distributions can yield linkage results showing excessive type I error if analyzed naively. For the traits considered here only insulin and the insulin/glucose ratio required robustness correction. The estimated constants for obtaining robust lod scores were 0.9 and 0.8, respectively, for these two traits. As a result of these findings all lod scores reported here represent the appropriately corrected lod score (i.e., original lod score × lod correction constant).
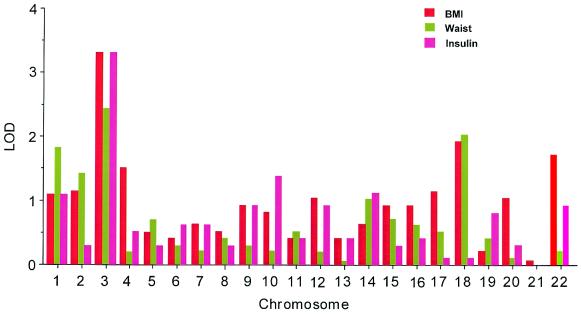
Fig. 1 displays results of the linkage analysis of the total genome scan for three phenotypes, representing total adiposity (BMI), abdominal fat patterning (waist circumference), and insulin sensitivity (fasting plasma insulin). The highest lod scores were detected at a common site on chromosome 3. Similar results were found for the remaining three of this set of phenotypic measures (body weight, hip circumference, and insulin/glucose, data not shown). Other loci meeting the criterion for suggestive linkage (lod ≥ 1.9) (25) were on chromosome 1 (total body weight, lod = 2.3) and chromosome 18 (BMI, lod = 1.9; waist circumference, lod = 2.0).
Figure 1.
Multipoint linkage lod scores by chromosome for BMI, waist circumference, and plasma insulin levels.
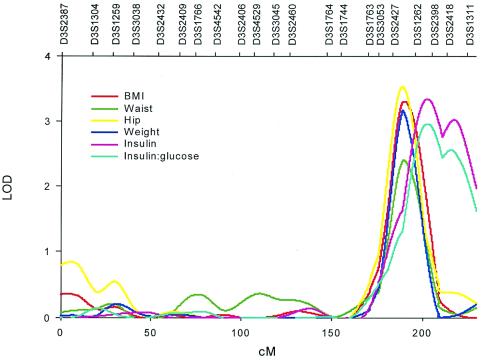
Fig. 2 shows the multipoint linkage analysis plots of the six phenotypes clustered on the chromosome 3 locus (189–203 cM from pter). Lod scores for these traits ranged from 2.4 for waist circumference to 3.5 for hip circumference (with genomewide P values from 0.15 to 0.009). The 1-lod unit confidence intervals surrounding the peak lod scores for BMI, hip and waist circumferences, and body weight spanned from 182 to 200 cM. The peak lod scores for these four phenotypes occurred near 190 cM, closest to the marker D3S2427. Insulin and insulin/glucose demonstrated their highest lod scores between 192 and 227 cM, with maxima at 202 cM, nearest to markers D3S2398 and D3S2418. The confidence intervals surrounding the peak lod scores for the two sets of phenotypes overlapped completely, suggesting that all six phenotypes are linked to the same QTL (Table 5).
Figure 2.
Multipoint linkage map and lod scores on chromosome 3 for phenotypic cluster fundamental to abdominal obesity-metabolic syndrome.
Table 5.
Multipoint linkage results on chromosomes 3 and 17
| Trait | Location | lod score | Nominal P value | Genomewide P value | One lod unit support interval |
|---|---|---|---|---|---|
| BMI | 3 @ 190 cM | 3.30 | 0.00005 | 0.016 | 183–200 cM |
| Waist circumference | 3 @ 190 cM | 2.40 | 0.0004 | 0.145 | 182–200 cM |
| Hip circumference | 3 @ 189 cM | 3.54 | 0.00003 | 0.009 | 182–197 cM |
| Weight | 3 @ 189 cM | 3.17 | 0.00007 | 0.022 | 183–200 cM |
| Insulin | 3 @ 202 cM | 3.01 | 0.0001 | 0.032 | 193–227 cM |
| Insulin/glucose | 3 @ 203 cM | 2.37 | 0.0005 | 0.156 | 192–227 cM |
| Ratio | |||||
| Leptin | 17 @ 38 cM | 4.97 | 0.0000009 | 0.0003 | 31–45 cM |
Plasma leptin levels showed its strongest linkage signal on chromosome 17 (lod = 5.0 at 38 cM; genomewide P value = 0.0003) near marker D17S947. The 1-lod unit confidence interval surrounding the peak spanned from 31 to 45 cM (Table 5). In addition, both waist and hip circumference showed marginal evidence of linkage in this same region (lod = 1.1; nominal P value = 0.01).
Based on the strength of the linkage signals for the QTLs on both chromosomes 3 and 17, and the extent of correlation among these phenotypes, we performed oligogenic analysis using both additive and epistatic two-locus models to look for evidence of joint effects. Table 6 shows the results of this analysis. The P values obtained from the likelihood ratio test of an additive two-locus model compared with the one-locus model was only significant for hip circumference. On the other hand, the P value from the likelihood ratio of an epistatic (i.e., additive × additive) model compared with a one-locus model was significant (P < 0.05) for BMI and hip circumference, and marginally significant (P < 0.10) for body weight and insulin. This analysis suggests possible gene–gene interactions occurring between the QTLs on chromosomes 3 and 17 with respect to phenotypes related to the metabolic syndrome.
Table 6.
Tests of two-locus models: Chromosomes 3 and 17 QTLs interaction effects
| Trait | Two-locus
component
|
|
|---|---|---|
| Additive model P value | Epistatic model P value | |
| BMI | 0.086 | 0.011 |
| Waist | 0.127 | 0.196 |
| Hip | 0.033 | 0.042 |
| Weight | 0.106 | 0.064 |
| Insulin | 0.500 | 0.098 |
| Insulin/glucose | 0.500 | 0.130 |
| Leptin | 0.407 | 0.248 |
Discussion
Although the use of genome scans has increased over the last several years, the number of published scans with obesity-related phenotypes as the primary focus is still rather small. Despite the relatively small number of scans thus far published there have already been several significant findings (26–32), some of which have now been replicated across several populations (27, 33). Although to date, none of the published scans for obesity-related traits have reported significant linkages on either chromosome 3 or 17 as reported here, at least two have reported marginally significant linkages in this same region of chromosome 3 (27, 32). The present study clearly demonstrates the existence of a QTL on chromosome 3 (3q27), which influences phenotypes fundamental to the abdominal obesity-metabolic syndrome. The size of the sample, the selection of families through obese and never-obese members, the consistent linkage to six independently measured surrogate phenotypes, and the magnitude of the lod scores emphasize the significance of this locus. Additionally, there is possible evidence of epistatic interaction between this QTL and a second QTL on chromosome 17 (17p12) that exhibited significant linkage with plasma leptin levels. These results provide insight into the genetic etiology of the metabolic syndrome.
Recent research supports major genetic influences mediating phenotypes of the abdominal obesity-metabolic syndrome. The San Antonio Family Heart Study proposes the existence of a common set of genes influencing plasma insulin levels, a surrogate of insulin resistance (7). The Atherosclerosis Risk in Communities Study reports significant associations between parental history of the metabolic syndrome and clustering of its phenotypes in their offspring (8). The Swedish Adoption/Twin Study of Aging shows common genetic influences on five principal phenotypes of the metabolic syndrome (9), whereas the Heritage Family Study concludes that major genetic components influence the association of abdominal-visceral fat size with insulin insensitivity (10).
The most recent update on the human obesity gene map includes 89 reports of positive associations identifying 40 candidate genes, with well over 200 genes, markers, and chromosomal regions putatively linked with BMI, body fat, or other obesity phenotypes (34). Our study demonstrates the existence of a common QTL linked to six phenotypes representing not only total adiposity, but also body fat patterning and insulin resistance. We do recognize that the phenotypes are all obesity-related and that identification of the primary phenotype(s) mediating this linkage requires further analysis. Furthermore, the phenotypes used represent distant surrogates, although highly correlated with direct measurements of total body fat, visceral fat size, and insulin sensitivity.
Leptin is a satiety hormone produced by adipocytes under the control of hormonal and metabolic stimuli (35). Besides its influences on food intake, leptin regulates energy expenditure, fuel partitioning, and cellular processing of lipids and carbohydrates (36). These events are mediated by signaling at its specific receptor. Although genetic defects causing leptin deficiency can produce massive obesity in mice, there is no convincing evidence that genetic mutations or polymorphisms in this receptor account for the common forms of human obesity (37). Nonetheless, the possible existence of an interaction between the QTL linked to the phenotypic cluster on chromosome 3 and the QTL linked to leptin on chromosome 17 suggest an association between leptin levels and some aspects of the metabolic syndrome.
The biologic precursors of the abdominal obesity-metabolic syndrome have been extensively researched. Our early studies unraveled the fundamental role of insulin resistance as a primary feature (2). Subsequent studies have shown that this insulin resistance is highly expressed in skeletal muscle and involves early rate-determining steps in insulin-mediated glucose disposal (38). Basal free fatty acid flux into the plasma is increased, and its suppressibility by insulin is impaired (39). Pancreatic insulin secretion sensorship is blunted, with delayed first-phase response and disrupted periodic pulsatilities entrainment with plasma glucose (40). Whereas overall insulin production is increased, reduced hepatic insulin clearance is the main determinant of hyperinsulinemia (41). Unsuppressed portal and systemic free fatty acid flux, together with accompanying sex hormone imbalances, appear to perpetuate these events (6). Enhanced responsiveness of a hypothalamic arousal system results in overactivity of the adrenocortical axis and chronic body exposure to glucocorticoids (5, 42). Visceral adipocytes are known to express higher glucocorticoid binding capacity than other regions (43). Increased activity of this system as a result of chronic stress may promote preferential deposition of intraabdominal fat, as well as precipitate aberrations in insulin-glucose homeostasis (44).
The intriguing question, then, is how to reconcile this biology with the QTLs on chromosomes 3 and 17. The potential epistatic relationship between the two QTLs suggests that pathways regulated by interactive genes residing in the two loci may influence variations in total adiposity, fat patterning, and insulin sensitivity. In this regard we have identified two sets of positional candidate genes. The first encodes proteins known to influence glucose-insulin homeostasis. These include the solute carrier family 2 of the facilitated glucose transporter (GLUT2, at 3q26-q27); the catalytic α polypeptide of phosphoinositide 3-kinase (at 3q26.3); and the solute carrier family 4 of the insulin-specific facilitated glucose transporter (GLUT4, at 17p13). The second encodes proteins thought to influence fat partitioning, lipid homeostasis, and energy balance. These include the adipose tissue-secreted protein adiponectin (synonyms: AdipoQ; adipose most abundant gene transcript 1, or apM1; gelatin-binding protein of 28 kDa, or GBP28, at 3q27), the receptor protein known to bind to globular “heads” of the complement C1q (gC1qR, at 17p13.3), and the peroxisome proliferative-activated receptor α (at 17p12-p11.2).
Within the first set, GLUT2 influences pancreatic sensorship and the temporal release of insulin in response to glucose (45). The phosphoinositide 3-kinase (PI3-kinase) gene product is a key component of the insulin signaling cascade, and polymorphisms in PI3-kinase genes have been associated with insulin resistance (46). GLUT4 is the main insulin-responsive glucose transporter in skeletal muscle (47), and polymorphisms affecting its function could aggravate insulin resistance.
Within the second set, adiponectin is the most abundant gene transcript specific to adipose tissue (48). It encodes a secreted protein that circulates in serum of normal individuals. Its circulating level is inversely correlated with BMI (49), and its mRNA level is suppressed in adipose tissue of obese animals and humans (50). Its genomic structure suggests close links to the complement C1q and cytokine tumor necrosis factor families (51). The promoter region of the gene encoding adiponectin contains consensus sequences for both peroxisome proliferative-activated receptor α (PPARα) and glucocorticoid receptor binding (52), and hence could be subject to regulation by such metabolic and neuroendocrine adaptations as increased free fatty acid (via PPARα) and stress (via glucocorticoids). With considerable structural similarity to C1q, including the globular heads, adiponectin could use gC1qR to exert its biologic function (53). Indeed, adiponectin has demonstrated high affinity binding to cells enriched with this receptor (52). In addition to specific clones of immune cells, gC1qR is highly expressed in endothelium, smooth muscle, and perhaps most importantly in hepatocytes (54). The precise actions of the adiponectin pathway and how it might influence energy expenditure, fat partitioning, and insulin sensitivity remain unclear. The PPARα gene, abundantly expressed in liver and muscle, is known to influence cellular partitioning of fatty acids between oxidation and storage (55, 56). Reduced expression of this gene has been associated with obesity in animals (57).
In summary, we have identified two QTLs with significant lod scores that influence several phenotypic aspects of the metabolic syndrome. We have identified positional candidate genes of two pathways that could contribute to its underlying biology. Clearly, further work is needed to clarify the functions of proteins encoded by these genes and polymorphisms that could influence the phenotypic expression of the metabolic syndrome.
Acknowledgments
This work was supported by Grants HL34989, DK54026, MH59490, and RR00058 from the National Institutes of Health. The genotyping was undertaken through the auspices of the Mammalian Genotyping Service of the National Institutes of Health, funds being allocated to the Marshfield Medical Research Foundation. TOPS, Inc. provided funds for establishment of the family database, phenotyping, and linkage analysis. Part of the phenotyping costs was also provided through a collaborative research agreement with Millennium Pharmaceuticals, Inc.
Abbreviations
- QTL
quantitative trait locus
- BMI
body mass index
- cM
centiMorgan
- IBD
identity-by-descent
- TOPS
Take Off Pounds Sensibly
- lod
logarithm of odds
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kissebah A H, Freedman D S, Peiris A N. Med Clin N Am. 1989;73:111–138. doi: 10.1016/s0025-7125(16)30695-2. [DOI] [PubMed] [Google Scholar]
- 2.Kissebah A, Vydelingum N, Murray R W, Evans D J, Hartz A J, Kalkhoff R K, Adams P W. J Clin Endocrinol Metab. 1982;54:254–260. doi: 10.1210/jcem-54-2-254. [DOI] [PubMed] [Google Scholar]
- 3.Peiris A N, Sothmann M S, Hoffmann R G, Hennes M M, Wilson C R, Gustafson A B, Kissebah A H. Ann Intern Med. 1989;110:867–882. doi: 10.7326/0003-4819-110-11-867. [DOI] [PubMed] [Google Scholar]
- 4.Reaven G. Diabetes. 1988;37:1592–1607. [Google Scholar]
- 5.Björntorp P. Diabetes Care. 1991;14:1132–1143. doi: 10.2337/diacare.14.12.1132. [DOI] [PubMed] [Google Scholar]
- 6.Kissebah A H, Evans D J, Peiris A, Wilson C R. In: Metabolic Complications of Human Obesities. Vague J, Björntorp P, Guy-Grand B, Rebuffé-Scrive M, Vague P, editors. Amsterdam: Excerpta Medica; 1985. pp. 115–130. [Google Scholar]
- 7.Mitchell B D, Kammerer C M, Mahaney M C, Blangero J, Comuzzie A G, Atwood L D, Haffner S M, Stern M P, MacCluer J W. Arterioscler Thromb Vasc Biol. 1996;16:281–288. doi: 10.1161/01.atv.16.2.281. [DOI] [PubMed] [Google Scholar]
- 8.Liese A D, Mayer-Davis E J, Tyroler H A, Davis C E, Keil U, Schmidt M I, Brancati F L, Heiss G. Diabetologia. 1997;40:963–970. doi: 10.1007/s001250050775. [DOI] [PubMed] [Google Scholar]
- 9.Hong Y, Pedersen N L, Brismar K, de Faire U. Am J Hum Genet. 1997;60:143–152. [PMC free article] [PubMed] [Google Scholar]
- 10.Hong Y, Rice T, Gagnon J, Després J P, Nadeau A, Pérusse L, Bouchard C, Leon A S, Skinner J S, Wilmore J H, et al. J Clin Endocrinol Metab. 1998;83:4239–4245. doi: 10.1210/jcem.83.12.5312. [DOI] [PubMed] [Google Scholar]
- 11.Yuan B, Vaske D, Weber J L, Beck J, Sheffield V C. Am J Hum Genet. 1997;60:459–460. [PMC free article] [PubMed] [Google Scholar]
- 12.Boehnke M, Cox N J. Am J Hum Genet. 1997;61:423–429. doi: 10.1086/514862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos C I. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 14.Almasy L, Blangero J. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blangero J, Almasy L. Genet Epidemiol. 1997;14:959–964. doi: 10.1002/(SICI)1098-2272(1997)14:6<959::AID-GEPI66>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 16.Hopper J L, Matthews J D. Ann Hum Genet. 1982;46:373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldgar D E. Am J Hum Genet. 1990;47:957–967. [PMC free article] [PubMed] [Google Scholar]
- 18.Allison D B, Heo M, Kaplan N, Martin E R. Am J Hum Genet. 1999;64:1754–1764. doi: 10.1086/302404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blangero J, Williams J T, Almasy L. Genet Epidemiol. 2000;19:S8–S14. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 20.Blangero J, Williams J T, Almasy L. Adv Genet. 2001;42:151–181. doi: 10.1016/s0065-2660(01)42021-9. [DOI] [PubMed] [Google Scholar]
- 21.Feingold E, Brown P O, Siegmund D. Am J Hum Genet. 1993;53:234–251. [PMC free article] [PubMed] [Google Scholar]
- 22.Fulker D W, Cherny S S, Cardon L R. Am J Hum Genet. 1995;56:1224–1233. [PMC free article] [PubMed] [Google Scholar]
- 23.Green P, Falls K, Crooks S. Documentation for CRI-MAP. St. Louis: Washington University; 1990. , Version 2.4. [Google Scholar]
- 24.Mitchell B D, Ghosh S, Schneider J L, Birznieks G, Blangero J. Genet Epidemiol. 1997;14:1017–1022. doi: 10.1002/(SICI)1098-2272(1997)14:6<1017::AID-GEPI76>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Lander E, Kruglyak L. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 26.Comuzzie A G, Hixson J E, Almasy L, Mitchell B D, Mahaney M C, Dyer T D, Stern M P, MacCluer J W, Blangero J. Nat Genet. 1997;15:273–276. doi: 10.1038/ng0397-273. [DOI] [PubMed] [Google Scholar]
- 27.Hager J, Dina C, Francke S, Dubois S, Houari M, Vatin V, Vaillant E, Lorentz N, Basdevant A, Clement K, et al. Nat Genet. 1998;20:304–308. doi: 10.1038/3123. [DOI] [PubMed] [Google Scholar]
- 28.Lee J H, Reed D R, Li W D, Xu W, Joo E J, Kilker R L, Nanthakumar E, North M, Sakul H, Bell C, et al. Am J Hum Genet. 1999;64:196–209. doi: 10.1086/302195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell B D, Cole S A, Comuzzie A G, Almasy L, Blangero J, MacCluer J W, Hixson J E. Diabetes. 1999;48:1863–1867. doi: 10.2337/diabetes.48.9.1863. [DOI] [PubMed] [Google Scholar]
- 30.Norman R A, Thompson D B, Foroud T, Garvey W T, Bennett P H, Bogardus C, Ravussin E. Am J Hum Genet. 1997;60:166–173. [PMC free article] [PubMed] [Google Scholar]
- 31.Norman R A, Tataranni P A, Pratley R, Thompson D B, Hanson R L, Prochazka M, Baier L, Ehm M G, Sakul H, Foroud T, et al. Am J Hum Genet. 1998;62:659–668. doi: 10.1086/301758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walder K, Hanson R L, Kobes S, Knowler W C, Ravussin E. Int J Obes Relat Metab Disord. 2000;24:559–565. doi: 10.1038/sj.ijo.0801197. [DOI] [PubMed] [Google Scholar]
- 33.Rotimi C N, Comuzzie A G, Lowe W L, Luke A, Blangero J, Cooper R S. Diabetes. 1999;48:643–644. doi: 10.2337/diabetes.48.3.643. [DOI] [PubMed] [Google Scholar]
- 34.Chagnon Y C, Pérusse L, Weisnagel S J, Rankinen T, Bouchard C. Obesity Res. 2000;8:89–117. doi: 10.1038/oby.2000.12. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman J M. Nature (London) 1994;387:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 36.Frühbeck G, Jebb S A, Prentice A M. Clin Physiol. 1998;18:399–419. doi: 10.1046/j.1365-2281.1998.00129.x. [DOI] [PubMed] [Google Scholar]
- 37.Carlsson B, Lindell K, Gabrielsson B, Karlsson C, Bjarnason R, Westphal O, Karlsson U, Sjöström L, Carlsson L M S. Obesity Res. 1996;5:30–35. doi: 10.1002/j.1550-8528.1997.tb00280.x. [DOI] [PubMed] [Google Scholar]
- 38.Peiris A N, Struve M F, Mueller R A, Lee M B, Kissebah A H. J Clin Endocrinol Metab. 1988;67:760–767. doi: 10.1210/jcem-67-4-760. [DOI] [PubMed] [Google Scholar]
- 39.Hennes M M I, O'Shaughnessy I M, Kelly T M, LaBelle P, Egan B M, Kissebah A H. Hypertension. 1996;28:120–126. doi: 10.1161/01.hyp.28.1.120. [DOI] [PubMed] [Google Scholar]
- 40.Sonnenberg G E, Hoffmann R G, Mueller R A, Kissebah A H. Diabetes. 1994;43:468–477. doi: 10.2337/diab.43.3.468. [DOI] [PubMed] [Google Scholar]
- 41.Peiris A, Mueller R A, Smith G A, Struve M F, Kissebah A H. J Clin Invest. 1986;78:1648–1657. doi: 10.1172/JCI112758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mårin P, Darin N, Amemiya T, Andersson B, Jern S, Björntorp P. Metabolism. 1992;41:882–886. doi: 10.1016/0026-0495(92)90171-6. [DOI] [PubMed] [Google Scholar]
- 43.Rebuffé-Scrive M, Lundholm K, Björntorp P. Eur J Clin Invest. 1992;15:267–271. doi: 10.1111/j.1365-2362.1985.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 44.Rebuffé-Scrive M, Walsh U A, McEwan B, Rodin J. Physiol Behav. 1992;52:583–590. doi: 10.1016/0031-9384(92)90351-2. [DOI] [PubMed] [Google Scholar]
- 45.Unger R H. Science. 1991;251:1200–1205. doi: 10.1126/science.2006409. [DOI] [PubMed] [Google Scholar]
- 46.Pederson O. Exp Clin Endocrinol Diabetes. 1999;107:113–118. doi: 10.1055/s-0029-1212085. [DOI] [PubMed] [Google Scholar]
- 47.Shepherd P R, Kahn B B. N Engl J Med. 1999;341:248–257. doi: 10.1056/NEJM199907223410406. [DOI] [PubMed] [Google Scholar]
- 48.Maeda K, Okubo K, Shimomura I, Funahashi T, Matsuzawa Y, Matsubara K. Biochem Biophys Res Commun. 1996;221:286–289. doi: 10.1006/bbrc.1996.0587. [DOI] [PubMed] [Google Scholar]
- 49.Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J-i, Hotta K, Shimomura I, Nakamura T, Miyaoka K, et al. Biochem Biophys Res Commun. 1999;257:79–83. doi: 10.1006/bbrc.1999.0255. [DOI] [PubMed] [Google Scholar]
- 50.Hu E, Liang P, Spiegelman B M. J Biol Chem. 1996;271:10697–10703. doi: 10.1074/jbc.271.18.10697. [DOI] [PubMed] [Google Scholar]
- 51.Saito K, Tobe T, Minoshima S, Asakawa S, Sumiya J, Yoda M, Nakano Y, Shimizu N, Tomita M. Gene. 1999;229:67–73. doi: 10.1016/s0378-1119(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 52.Schäffler A, Orsó E, Palitzsch K-D, Büchler C, Drobnik W, Fürst A, Schölmerich J, Schmitz G. Biochem Biophys Res Commun. 1999;260:416–425. doi: 10.1006/bbrc.1999.0865. [DOI] [PubMed] [Google Scholar]
- 53.Shapiro L, Scherer P E. Curr Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- 54.Ghebrehiwet B, Lim B-L, Peerschke E I B, Willis A C, Reid K B M. J Exp Med. 1994;179:1809–1821. doi: 10.1084/jem.179.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brun S, Carmona M C, Mampel T, Viñas O, Giralt M, Iglesias R, Villarroya F. Diabetes. 1999;48:1217–1222. doi: 10.2337/diabetes.48.6.1217. [DOI] [PubMed] [Google Scholar]
- 56.Kersten S, Seydoux J, Peters J M, Gonzalez F J, Desvergne B, Wahli W. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costet P, Legendre C, More J, Edgar A, Galtier P, Pineau T. J Biol Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]