Abstract
This study provides genetic evidences at the chromosome, DNA content, DNA fragment and sequence, and morphological levels to support the successful establishment of the polyploid hybrids of red crucian carp × blunt snout bream, which belonged to a different subfamily of fish (Cyprininae subfamily and Cultrinae subfamily) in the catalog. We successfully obtained the sterile triploid hybrids and bisexual fertile tetraploid hybrids of red crucian carp (RCC) (♀) × blunt snout bream (BSB) (♂) as well as their pentaploid hybrids. The triploid hybrids possessed 124 chromosomes with two sets from RCC and one set from BSB; the tetraploid hybrids had 148 chromosomes with two sets from RCC and two sets from BSB. The females of tetraploid hybrids produced unreduced tetraploid eggs that were fertilized with the haploid sperm of BSB to generate pentaploid hybrids with 172 chromosomes with three sets from BSB and two sets from RCC. The ploidy levels of triploid, tetraploid, and pentaploid hybrids were confirmed by counting chromosomal number, forming chromosomal karyotype, and measuring DNA content and erythrocyte nuclear volume. The similar and different DNA fragments were PCR amplified and sequenced in triploid, tetraploid hybrids, and their parents, indicating their molecular genetic relationship and genetic markers. In addition, this study also presents results about the phenotypes and feeding habits of polyploid hybrids and discusses the formation mechanism of the polyploid hybrids. It is the first report on the formation of the triploid, tetraploid, and pentaploid hybrids by crossing parents with a different chromosome number in vertebrates. The formation of the polyploid hybrids is potentially interesting in both evolution and fish genetic breeding.
DISTANT crossing is defined as the interspecific or above-specific hybridization. It is a useful strategy for hybrid offspring to change genetic composition and phenotype. Interspecific hybridization normally results in genome-level alterations, including the occurrence of triploid hybrids and tetraploids. For example, F1 hybrids of grass carp (2n = 48) × big head carp (2n = 48) were triploids (3n = 72) (Marian and Kraszai 1978; Beck et al. 1984). In reproduction, the triploid hybrids were sterile while the tetraploid hybrids were fertile. In general, the fertile tetraploid hybrids were used to generate the sterile hybrids, which had some advantages such as faster growth and higher anti-disease ability (Liu et al. 2001). Until now, reports on the formation of living polyploidy hybrids produced by distant parents with different chromosome number rarely existed. To create a new type of polyploidy fish, in this study we made the distant crosses by choosing different parents that belonged to a different subfamily and had a different chromosome number. In the catalog, the red crucian carp (RCC) with 100 chromosomes belonged to the Cyprininae subfamily, and the blunt snout bream (Megalobrama amblycephala) (BSB) with 48 chromosomes belonged to the Cultrinae subfamily (Yu 1989). In feeding habit, the red crucian carp is omnivorous whereas the blunt snout bream is herbivorous. In summary, in chromosome number, phenotype, and feeding habit, both BSB and RCC are quite different. In this study, we successfully obtained bisexual fertile allotetraploid hybrids and the sterile triploid hybrids of RCC ♀ × BSB ♂, as well as the pentaploid hybrids of the tetraploid hybrids ♀ × BSB ♂. In evolution, the fertile tetraploid hybrids are very valuable because their fertility guarantees the formation of a new type of offspring with a new chromosome number. In fish genetic breeding, the sterile triploid hybrids have potential value because they will not disturb the fish genetic resources in the natural environment.
In this study, we not only investigated the polyploid hybrids on the cytogenetic level, but also analyzed the genetic relationship between the polyploid hybrids and their parents on the molecular level. We designed the primers on the basis of the sequences of the conserved HMG box of Sox genes and conducted polymerase chain reaction (PCR) using the standard method. Sox genes are those genes that are characterized by a conserved DNA sequence encoding a domain of 80 amino acids. This domain, called the high mobility group (HMG), is responsible for specific DNA sequence binding. We amplified and sequenced the conserved HMG box of Sox genes in polyploidy hybrids and their parents and found that the tetraploid and triploid hybrids, respectively, could be distinguished from their parents by presenting different PCR products. Sequence homology among the fragments amplified from polyploid hybrids and their parents was compared to analyze their genetic relationship.
In summary, the formation of the triploid, tetraploid, and pentaploid hybrids is significant in both evolution and fish genetic breeding. This study is the first report on the formation of the polyploid hybrids by crossing parents with a different chromosome number in vertebrates.
MATERIALS AND METHODS
Animals and crosses:
BSB and RCC were obtained from the Protection Station of Polyploidy Fish in Hunan Normal University. During the reproductive seasons (from April to June) in 2004, 2005, and 2006, 15 mature females and 15 mature males of both RCC and BSB were chosen as the maternals or paternals, respectively. The crossings were performed by two groups. In the first group, the RCC was used as the maternal, and the BSB was used as the paternal. In the second group, the maternals and paternals were reversed. The mature eggs and sperm of RCC and BSB were fertilized and the embryos developed in the culture dishes at the water temperature of 19°–20°. In each cross, 2000 embryos were taken at random for the examination of the fertilization rate (number of embryos at the stage of gastrula/number of eggs × 100%) and the hatching rate (number of hatched fry/number of eggs × 100%). The hatched fry were transferred to the pond for further culture.
Because in the cross RCC ♀ × BSB ♂ there existed the offspring of triploid and tetraploid hybrids while in the reverse cross BSB ♀ × RCC ♂ there were no living progeny, in this study we abbreviate triploid hybrids of RCC ♀ × BSB ♂ as 3nRB hybrids and tetraploid hybrids of RCC ♀ × BSB ♂ as 4nRB hybrids.
At the age of 1 year, no 3nRB and 4nRB hybrids were found to be fertile. At the age of 2 years, the 3nRB hybrids were still found to be sterile. However, the 2-year-old female 4nRB hybrids produced mature eggs whereas the 2-year-old male tetraploid hybrids produced only water-like semen. The female 4nRB hybrids produced two sizes of eggs. The diameters of 200 larger eggs and 200 smaller eggs were measured. The eggs of 4nRB hybrids were fertilized with the semen of BSB to produce pentaploid hybrids. We abbreviate pentaploid hybrids of 4nRB ♀ × BSB ♂ as 5nRB hybrids. On the other hand, the eggs and semen of 4nRB hybrids were mated to each other to produce the next generation. All crossing procedures and formation of the polyploid hybrids are illustrated in Figure 1.
Figure 1.—
Crossing procedure and formation of the polyploid hybrids.
Preparation of chromosome spreads:
To determine ploidy, chromosome counts were done on kidney tissue for 10 RCC, 10 BSB, 10 3nRB, 10 4nRB, and 10 5nRB hybrids at 1 year of age. After culture for 1–3 days at the water temperature of 18°–22°, the samples were injected with concanavalin one to three times at a dose of 2–8 μg/g body weight. The interval time of injection was 12–24 hr. Six hours prior to dissecting, each sample was injected with colchicine at a dose of 2–4 μg/g body weight. The kidney tissue was ground in 0.9% NaCl, followed by hypotonic treatment with 0.075 m KCl at 37° for 40–60 min and then fixed in 3:1 methanol–acetic acid for three changes. Cells were dropped on cold, wet slides and stained for 30 min in 4% Giemsa. The shape and number of chromosomes were analyzed under microscope. For each type of fish, 200 metaphase spreads (20 metaphase spreads in each sample) of chromosomes were analyzed. Preparations were examined under an oil lens at a magnification of ×330. Good-quality metaphase spreads were photographed and used for analysis of karyotypes. Lengths of entire chromosomes, long and short arms, were measured. Chromosomes were classified on the basis of their long-arm to short-arm ratios according to the reported standards (Levan et al. 1964): values of 1.0–1.7 were classified as metacentric (m), 1.7–3.0 as submetacentric (sm), 3.1–7.0 as subtelocentric (st), and 7.1 as telocentric (t) chromosome.
Measurement of DNA content:
To measure the DNA content of erythrocytes of 3nRB, 4nRB, and 5nRB hybrids and RCC and BSB, the 1–2 ml of red blood cells were collected from the caudal vein of the above fish into syringes containing ∼200–400 units of sodium heparin. The blood samples were treated with the nuclei extraction and DAPI DNA staining solution, cystain DNA 1 step (Partec). Then all the samples were filtered. A flow cytometer (cell counter analyzer, Partec) was used to measure the DNA content. Under the same conditions, the DNA content of each sample was measured. To calculate the probabilities of the ratios of the DNA content of the polyploid hybrids to the sum of that of RCC and BSB, the χ2 test with Yate's correction was used for testing deviation from expected ratio values.
Measurement of nuclear volume and appearance of erythrocytes:
Nuclear micromeasurements of erythrocytes were also used to further ascertain ploidy. A 1- to 2-ml blood sample was drawn from 3nRB, 4nRB, and 5nRB hybrids and from RCC and BSB. Blood smears were fixed in ethanol and stained with Giemsa solution. To measure nuclear diameters, 20 erythrocytes from each fish were observed under oil at ×330 using an ocular micrometer. Nuclear volume was calculated by 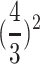 , where a is the major semi-axis and b is the minor semi-axis of a perfect ellipsoid. Volume may have been slightly overstimulated due to cell flattening. To calculate the probabilities of the ratios of nuclear volume of the polyploid hybrids to the sum of that of RCC and BSB, the χ2 test with Yate's correction was used for testing deviation from expected ratio values. The appearance of the erythrocytes' nuclei was also observed under microscope.
, where a is the major semi-axis and b is the minor semi-axis of a perfect ellipsoid. Volume may have been slightly overstimulated due to cell flattening. To calculate the probabilities of the ratios of nuclear volume of the polyploid hybrids to the sum of that of RCC and BSB, the χ2 test with Yate's correction was used for testing deviation from expected ratio values. The appearance of the erythrocytes' nuclei was also observed under microscope.
Morphological traits and feeding habit:
At 1 year of age, 20 RCC, 20 BSB, 20 3nRB, 20 4nRB, and 20 5nRB hybrids, respectively, were morphologically examined. The examined measurable traits included the average values of the whole length, the body length and width, the head length and width, and the tail length and width. The average ratios of whole length to body length (WL/BL), body length to body width (BL/BW), body length to head length (BL/HL), head length to head width (HL/HW), tail length to tail width (TL/TW), and body width to head width (BW/HW) were calculated. The examined countable traits included the number of dorsal fins, abdominal fins, anal fins, lateral scales, upper and lower lateral scales.
For both measurable and countable data, we used the software of SPSS to analyze the covariance of the data between two kinds of fish in 3nRB, 4nRB, and 5nRB hybrids and in RCC and BSB. The feeding habit of 3nRB, 4nRB, and 5nRB hybrids was also investigated.
PCR:
The HMG box of Sox genes was amplified by PCR using the degenerate primers P (+) [5′-TGAAGCGACCCATGAA(C/T)G-3′] and P (−) [5′-AGGTCG(A/G)TACTT(A/G)TA(A/G)T-3′]. The genomic DNAs extracted from the blood cells of RCC, BSB, 3nRB, and 4nRB hybrids by routine approaches (Sambrook et al. 1989) were used as templates. The PCR was performed in a volume of 25 μl with ∼80 ng genomic DNA, 1.5 mm of MgCl2, 200 μm of each dNTP, 0.3 μm of each primer, and 0.9 unit of Taq polymerase (Takara). The cycling program included 35 cycles of 94° for 30 sec, 52° for 30 sec, and 72° for 1 min, with a final extension of 7 min at 72°. All PCR products, including three DNA fragments in RCC, two DNA fragments in BSB, three DNA fragments in 3nRB hybrids, and four DNA fragments in 4nRB hybrids were separated in 1.5% agarose gels.
Sequencing:
After electrophoresis, all fragments of PCR products in 3nRB and 4nRB hybrids and their parents were purified using a gel extraction kit (Sangon) and ligated into the pMD-18T vector. The plasmids were amplified in DH5α. The inserted DNA fragments in the pMD-18T vector were sequenced by an automated DNA sequencer (ABI PRISM 3730). The sequences were aligned with the corresponding sequences of Sox genes in zebrafish, rainbow trout, medaka, rice field eel, mouse, and the common carp, etc., derived from the NCBI database using Jellyfish (2.1) software and named accordingly. Sequence homology and variation among the fragments amplified from RCC, BSB, 3nRB, and 4nRB hybrids were analyzed using Clustal W software (http://www.ebi.ac.uk/clustalw/intex.html).
RESULTS
Formation of triploid, tetraploid, and pentaploid hybrids:
In the crossings of RCC ♀ × BSB ♂, we observed a higher fertilization rate (>60%) and hatching rate (>50%) and obtained ∼5000, 20,000, and 100,000 living hybrids in 2004, 2005, and 2006, respectively. In the reverse crossing, RCC ♂ × BSB ♀, there was no survival. In 2006, we obtained 25,000 living hybrids with a 67% fertilization rate and a 51% hatching rate in the cross of the mature eggs from tetraploid hybrids of RCC ♀ × BSB ♂ and the white semen from BSB, and only 8 living hybrids generated by fertilizing the mature eggs with the water-like semen of the tetraploid (4nRB) hybrids of RCC ♀ × BSB ♂. All the fish grew well, except the 8 hybrids produced by male–female mating of the 4nRB hybrids, which were dead at the age of 4 months due to accidental oxygen loss in the water.
Examination of chromosome number and formation of karyotypes:
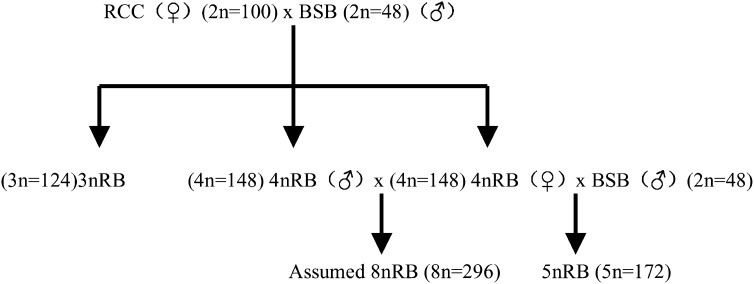
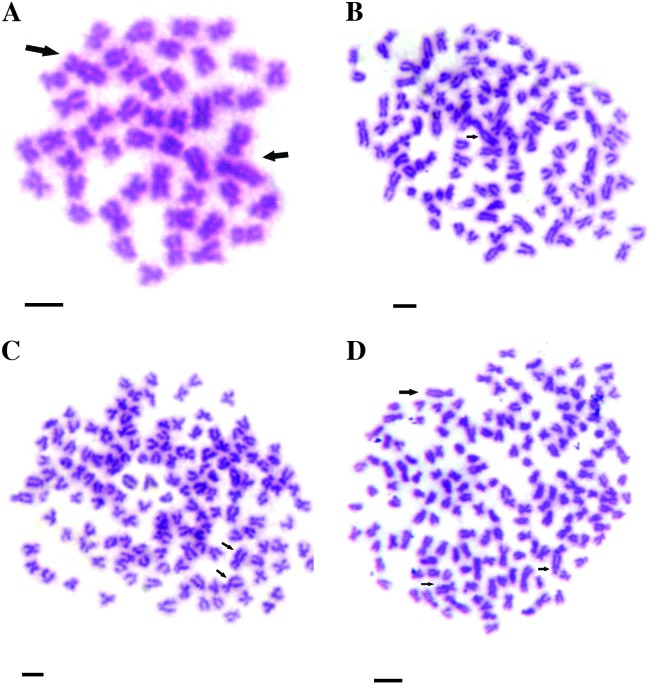
Table 1 presents the distribution of chromosome number in BSB, RCC, 3nRB, 4nRB, and 5nRB hybrids. Of all examined samples in BSB, 93% of chromosomal metaphases possessed 48 chromosomes (Table 1), indicating that they were diploids with 48 chromosomes (2n = 48) (Figure 2A) with the karyotype of 18 m + 22 sm + 8 st. A pair of the largest submetacentric chromosomes in BSB was observed, which could be used as the chromosomal marker to identify this species (Figure 2A). Of all examined examples in RCC, 96% of chromosomal metaphases had 100 chromosomes (Table 1), indicating that they were diploids with 100 chromosomes with the karyotype of 22 m +34 sm +22 st + 22 t, which was the same as described in our previous study (Liu et al. 2001). In the chromosomes of RCC, no special larger submetacentric chromosome was found. In the offspring of RCC ♀ × BSB ♂ without the barbells in the phenotype, 77% of chromosomal metaphases had 124 chromosomes (Table 1, Figure 2B), showing that they were triploids (3nRB). In these metaphases, only one large submetacentric chromosome, similar to the two found in BSB, was observed, suggesting that the 3nRB hybrids possessed 24 chromosomes from BSB and 100 chromosomes from RCC. In the offspring of RCC ♀ × BSB ♂, which bore the barbells in appearance, 73% of chromosomal metaphases had 148 chromosomes (Table 1, Figure 2C), indicating that they were tetraploids (4nRB). In these metaphases, the two largest submetacentric chromosomes, similar to the two in BSB, were found, suggesting that the 4nRB hybrids possessed 48 chromosomes from BSB and 100 chromosomes from RCC. The karyotype (Figure 3A) of 40 m + 56 sm + 30 st + 22 t in 4nRB hybrids indicated that their 40 m chromosomes contained 22 m chromosomes from RCC and 18 m chromosomes from BSB; their 56 sm chromosomes contained 34 sm chromosomes from RCC and 22 sm chromosomes from BSB; their 30 st chromosomes contained 22 st chromosomes from RCC and 8 st chromosomes from BSB; and their 22t chromosomes contained 22 t chromosomes from RCC. In the offspring of 4nRB ♀ × BSB ♂, 91% of chromosomal metaphases had 172 chromosomes (Table 1, Figure 2D), suggesting that they were pentaploids (5nRB). In these metaphases, the three largest submetacentric chromosomes, similar to those found in BSB (Figure 2A), were observed, indicating that the 5nRB hybrids had three sets of chromosomes of BSB and two sets of chromosomes of RCC. The karyotype of 5nRB hybrids (Figure 3B) showed that they possessed 49 m chromosomes (22 m from RCC and 27 m from BSB), 67 sm chromosomes (34 sm from RCC and 33 sm from BSB), 34 st chromosomes (22 st from RCC and 12 st from BSB), and 22 t chromosomes (22 t from RCC).
TABLE 1.
Examination of chromosome number in BSB, RCC, 3nRB, 4nRB, and 5nRB hybrids
| Distribution of chromosome number |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fish type | No. in metaphase | <48 | 48 | <100 | 100 | <124 | 124 | <148 | 148 | <172 | 172 |
| BSB | 200 | 14 | 186 | ||||||||
| RCC | 200 | 8 | 192 | ||||||||
| 3nRB | 200 | 46 | 154 | ||||||||
| 4nRB | 200 | 54 | 146 | ||||||||
| 5nRB | 200 | 18 | 182 | ||||||||
Figure 2.—
Chromosome spreads at metaphase in BSB, 3nRB, 4nRB, and 5nRB hybrids. (A) The 48 chromosomes of BSB, in which a pair of the largest submetacentric chromosomes (arrows) are indicated. (B) The 124 chromosomes of 3nRB hybrids of RCC × BSB in which a piece of the largest submetacentric chromosomes (arrow) is indicated. (C) The 148 chromosomes of 4nRB hybrids of RCC × BSB in which a pair of the largest submetacentric chromosomes (arrows) are indicated. (D) The 172 chromosomes of 5nRB hybrids produced by crossing the females of 4nRB hybrids with the males of BSB, in which the three largest submetacentric chromosomes (arrows) are indicated. Bar in A–D, 4 μm.
Figure 3.—
Karyotypes of 4nRB and 5nRB hybrids. (A) The karyotype of 4nRB hybrids, 40 m + 56 sm + 30 st + 22t, which consists of two sets of chromosomes from RCC and two sets of chromosomes from BSB. The arrows indicate a pair of the largest submetacentric chromosomes similar to the two in BSB. (B) The karyotype of 5nRB hybrids contained two sets of chromosomes from RCC and three sets of chromosomes from BSB. The arrows indicate the three largest submetacentric chromosomes similar to the two in BSB. Bar in A and B, 4 μm.
Measurement of DNA content:
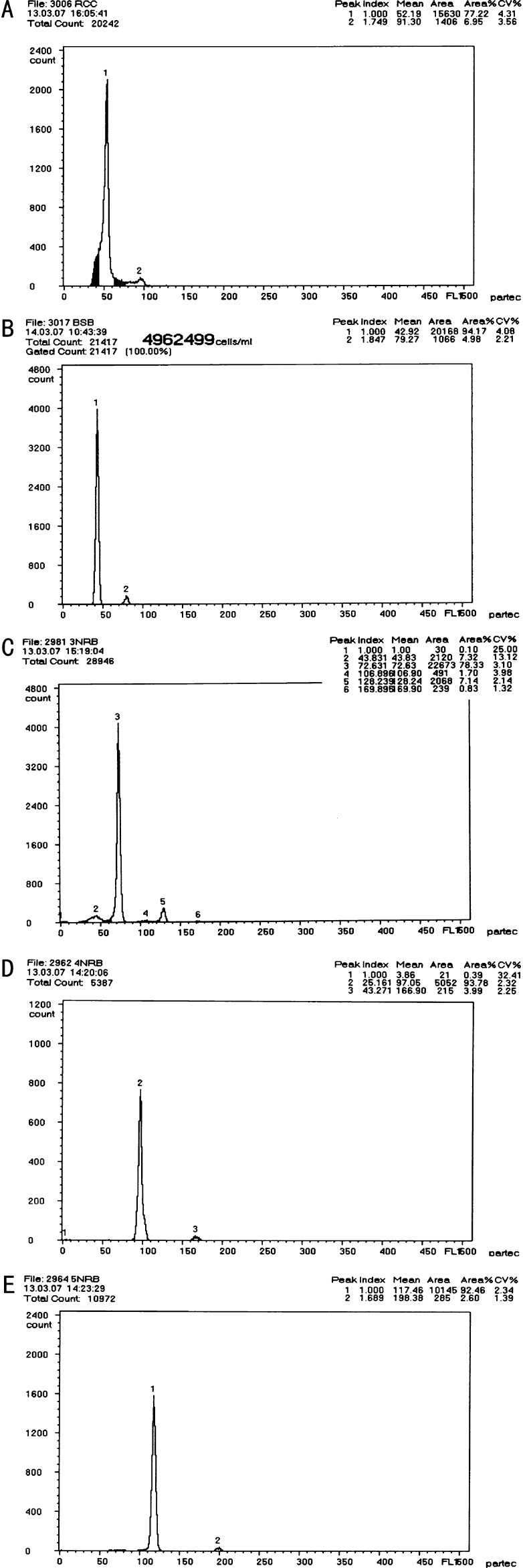
We used the sum of the DNA content of RCC and BSB as the controls. The results of the distribution of DNA content of all the samples are shown in Table 2 and Figure 4, A–E. The mean DNA content of 3nRB hybrids was equal (P > 0.01) to the sum of that of RCC and half of BSB, suggesting that 3nRB hybrids had two sets of chromosomes from RCC and one set of chromosomes from BSB. The mean DNA content of 4nRB hybrids was equal (P > 0.01) to the sum of that of RCC and BSB, indicating that 4nRB hybrids had two sets of chromosomes from RCC and two sets of chromosomes from BSB. The mean DNA content of 5nRB hybrids was equal (P > 0.01) to the sum of that of RCC and one and half of BSB, showing that 5nRB hybrids had two sets of chromosomes from RCC and three sets of chromosomes from BSB.
TABLE 2.
Mean DNA content in RCC, BSB, 3nRB, 4nRB, and 5nRB hybrids
| Fish type | Mean DNA content | Ratio |
|
|---|---|---|---|
| Observed | Expected | ||
| BSB | 42.92 | ||
| RCC | 52.19 | ||
| 3nRB | 72.63 | 3nRB/(RCC + 0.5 BSB) = 0.99a | 1 |
| 4nRB | 97.05 | 4nRB/(RCC + BSB) = 1.02a | 1 |
| 5nRB | 117.46 | 5nRB/(RCC + 1.5 BSB) = 1.01a | 1 |
The observed ratio was not significantly different (P > 0.05) from the expected ratio.
Figure 4.—
Cytometric histograms of DNA fluorescence for BSB, RCC, and polyploid hybrids. (A) The mean DNA content of RCC (peak 1: 52.19). (B) The mean DNA content of RCC (peak 1: 42.92). (C) The mean DNA content of 3nRB hybrid (peak 3: 72.63). (D) The mean DNA content of 4nRB hybrid (peak 2: 97.05). (E) The mean DNA content of 5nRB hybrid (peak 1: 117.46).
Measurement of the nuclear volume of erythrocytes:
The results of the measurements of the mean erythrocyte nuclear volume for BSB, RCC, 3nRB, 4nRB, and 5nRB hybrids are shown in Table 3 and Figure 5, A–F. With the increase of the polyploid level, the mean erythrocyte nuclear volume regularly increased among 3nRB, 4nRB, and 5nRB hybrids. The mean erythrocyte nuclear volume ratio of the 3nRB hybrid to the sum of that of RCC and half of BSB was not significantly different (P > 0.05) from the 1:1 ratio, suggesting that 3nRB hybrids were triploids. The mean erythrocyte nuclear volume ratio of the 4nRB hybrid to the sum of that of RCC and BSB was not significantly different (P > 0.05) from the 1:1 ratio, indicating that 4nRB hybrids were tetraploids. The mean erythrocyte nuclear volume ratio of the 5nRB hybrid to the sum of that of RCC and one and one-half of BSB was not significantly different (P > 0.05) from the 1:1 ratio, indicating that 5nRB hybrids were pentaploids.
TABLE 3.
Mean erythrocyte nuclear volume measurements for BSB, RCC, 3nRB, 4nRB, and 5nRB hybrids
| Volume ratio |
|||||
|---|---|---|---|---|---|
| Fish type | Major axis (μm) | Minor axis (μm) | Volume (μm3) | Observed | Expected |
| BSB | 5.07 ± 0.47 | 2.44 ± 0.80 | 15.76 ± 1.92 | ||
| RCC | 4.99 ± 0.27 | 2.95 ± 0.20 | 22.71 ± 2.42 | ||
| 3nRB | 6.96 ± 0.70 | 3.22 ± 0.33 | 37.64 ± 6.32 | 3nRB/(RCC + 0.5 BSB) = 1.23a | 1 |
| 4nRB | 7.46 ± 0.56 | 3.27 ± 0.34 | 42.01 ± 8.31 | 4nRB/(RCC + BSB) = 1.09a | 1 |
| 5nRB | 7.71 ± 0.81 | 3.35 ± 0.23 | 45.56 ± 8.18 | 5nRB/(RCC + 1.5 BSB) = 0.98a | 1 |
The observed ratio was not significantly different (P > 0.05) from the expected ratio.
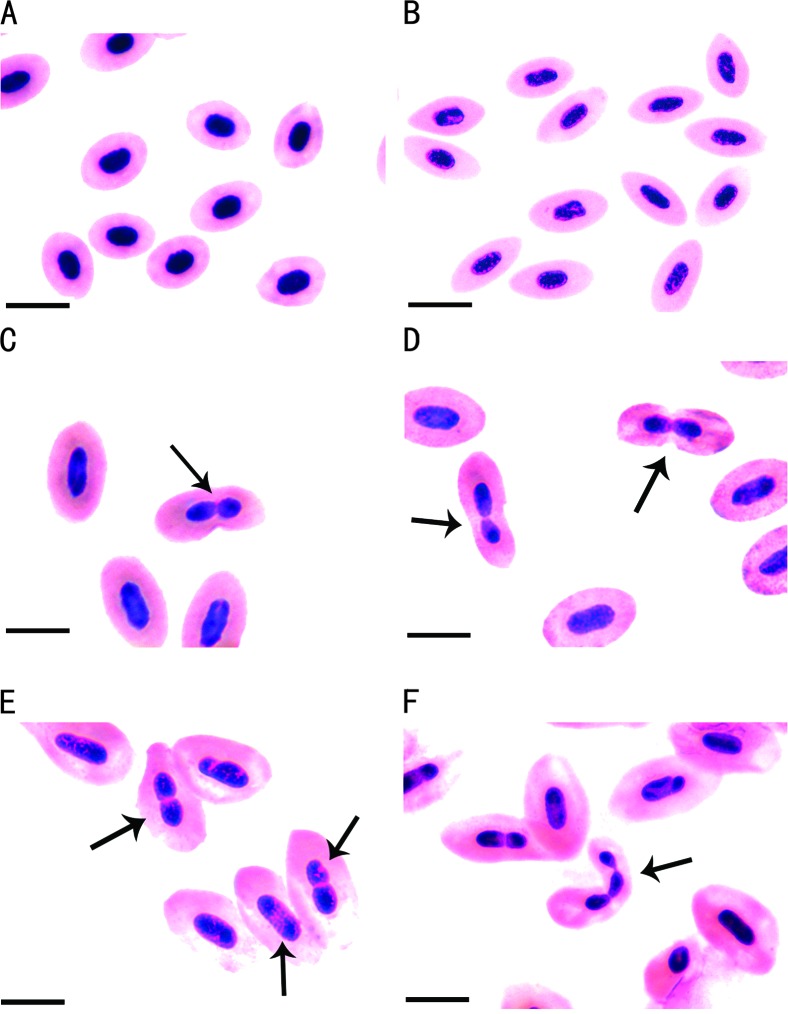
Figure 5.—
Erythrocytes of the RCC, BSB, and polyploid hybrids. (A) Normal erythrocytes with one nucleus in RCC. (B) Normal erythrocytes with one nucleus in BSB. (C) Normal erythrocytes with one nucleus and unusual erythrocytes with two nuclei (arrows) in a 3nRB hybrid. (D) Normal erythrocytes with one nucleus and unusual erythrocytes with two nuclei (arrows) in a 4nRB hybrid. (E) Normal erythrocytes with one nucleus and unusual erythrocytes with two nuclei (arrows) in a 5nRB hybrid. (F) Normal erythrocytes with one nucleus and unusual erythrocytes with two nuclei or three nuclei (arrow) in a 5nRB hybrid. Bar in A–F, 0.01 mm.
Nuclear traits of erythrocytes:
Figure 5 showed the nuclear traits of erythrocytes in BSB, RCC, 3nRB, 4nRB, and 5nRB hybrids. The size of the nuclei of erythrocytes in 3nRB, 4nRB, and 5nRB hybrids was larger than that of BSB or RCC. With the increase of the ploidy level, the size of the nuclei of erythrocytes regularly enlarged among 3nRB, 4nRB, and 5nRB hybrids. In addition to the difference in nuclear size, there also existed the difference in nuclear appearance between the polyploidy hybrids and their parents. For example, only the normal erythrocytes with one nucleus, but no unusual erythrocytes with two nuclei, were observed in both RCC and BSB (Figure 5, A and B). However, unusual erythrocytes with two nuclei were found in 3nRB, 4nRB, and 5nRB hybrids (Figure 5, C–F). The unusual erythrocytes with three nuclei were also observed in 5nRB hybrids (Figure 5F). The two-nucleus erythrocytes accounted for 3.4, 9.4, and 21.8% in the 3nRB, 4nRB, and 5nRB hybrids, respectively, indicating that, with the increase of ploidy level, the percentage of unusual erythrocytes with two nuclei increased. The unusual erythrocytes with three nuclei in 5nRB hybrids accounted for 0.7%.
Morphological traits and feeding habit:
Tables 4 and 5 present the examined measurable traits and countable traits in 3nRB, 4nRB, 5nRB hybrids and in RCC and BSB. For the measurable traits between 3nRB hybrids and BSB, all ratios were significantly different. Between 3nRB hybrids and RCC, apart from the ratio of BL/HL, which was not significantly different (P > 0.01), other ratios were significantly different. Between 4nRB hybrids and BSS, all ratios were significantly different. Between 4nRB hybrids and RCC, apart from the ratios of BL/BW and HL/HW, which were not significantly different (P > 0.01), other ratios were significantly different. Between 5nRB hybrids and BSB, apart from the ratio of BL/BW, which was not significantly different (P > 0.01), other ratios were significantly different. Between 5nRB hybrids and RCC, all ratios were significantly different. On the other hand, between 3nRB and 4nRB hybrids, apart from the ratio of HL/HW, which was not significantly different (P > 0.01), other ratios were significantly different. Between 3nRB and 5nRB hybrids, all ratios were significantly different. Between 4nRB and 5nRB hybrids, all ratios were significantly different.
TABLE 4.
Comparison of the measurable traits between the hybrid offspring and RCC and BSB
| Fish type | WL/BL | BL/BW | BL/HL | HL/HW | TL/TW | BW/HW |
|---|---|---|---|---|---|---|
| 3nRB | 1.31 ± 0.03 | 1.67 ± 0.03 | 3.70 ± 0.03 | 1.11 ± 0.04 | 0.71 ± 0.04 | 2.31 ± 0.04 |
| 4nRB | 1.18 ± 0.02 | 2.18 ± 0.02 | 3.83 ± 0.03 | 1.08 ± 0.04 | 0.75 ± 0.04 | 1.92 ± 0.02 |
| 5nBRB | 1.25 ± 0.03 | 2.40 ± 0.06 | 3.35 ± 0.03 | 1.25 ± 0.03 | 1.11 ± 0.03 | 1.66 ± 0.03 |
| RCC | 1.22 ± 0.02 | 2.18 ± 0.02 | 3.72 ± 0.03 | 1.07 ± 0.03 | 0.82 ± 0.03 | 1.84 ± 0.03 |
| BSB | 1.19 ± 0.03 | 2.37 ± 0.03 | 4.75 ± 0.04 | 1.14 ± 0.03 | 1.08 ± 0.04 | 2.09 ± 0.04 |
TABLE 5.
Comparison of the countable traits between the polyploids and their parents
| Fish type | No. of lateral scales | No. of upper lateral scales | No. of lower lateral scales | No. of dorsal fins | No. of abdominal fins | No. of anal fins |
|---|---|---|---|---|---|---|
| 3nRB | 29.45 ± 0.51 (29–30) | 6.60 ± 0.50 (6–7) | 7.70 ± 0.47 (7–8) | III + 18.10 ± 0.79 (III + 17–19) | 8.35 ± 0.49 (8–9) | III + 6.30 ± 0.47 (III + 6–7) |
| 4nRB | 31.65 ± 0.49 (31–32) | 6.55 ± 0.51 (6–7) | 6.45 ± 0.51 (6–7) | III + 18.70 ± 0.98 (III + 17–20) | 8.60 ± 0.50 (8–9) | III + 6.40 ± 0.68 (III + 5–7) |
| 5nRB | 35.20 ± 0.77 (34–36) | 7.60 ± 0.50 (7–8) | 7.70 ± 0.47 (7–8) | III + 19.15 ± 1.23 (III + 17–21) | 10.75 ± 1.07 (9–12) | III + 6.55 ± 0.51 (III + 6–7) |
| RCC | 29.20 ± 0.70 (28–30) | 5.60 ± 0.50 (5–6) | 5.70 ± 0.47 (5–6) | III + 18.65 ± 0.49 (III + 18–19) | 8.55 ± 0.51 (8–9) | III + 5.65 ± 0.49 (III + 5–6) |
| BSB | 50.90 ± 0.91 (52–49) | 9.65 ± 0.49 (9–10) | 10.05 ± 0.69 (9–11) | III + 8.65 ± 0.49 (III + 8–9) | 9.10 ± 0.55 (8–10) | III + 25.85 ± 0.59 (III + 25–27) |
For the countable traits between 3nRB hybrids and BSB, all data were significantly different. Between 3nRB hybrids and RCC, apart from the number of lateral scales, dorsal fins, and abdominal fins, which were not significantly different (P > 0.01), other data were significantly different. Between 4nRB hybrids and BSS, all data were significantly different. Between 4nRB hybrids and RCC, apart from the number of dorsal fins and abdominal fins, which were not significantly different (P > 0.01), other data were significantly different. Between 5nRB hybrids and BSB, all data were significantly different. Between 5nRB hybrids and RCC, apart from the number of dorsal fins, which was not significantly different (P > 0.01), other data were significantly different. On the other hand, between 3nRB and 4nRB hybrids, apart from the number of lateral scales and lower lateral scales, which were significantly different, other data were not significantly different (P > 0.01). Between 3nRB and 5nRB hybrids, apart from the number of lower lateral scales and buttock fins, which were not significantly different (P > 0.01), other data were significantly different. Between 4nRB and 5nRB hybrids, apart from the number of dorsal fins and anal fins, which were not significantly different (P > 0.01), other data were significantly different.
In morphological traits, the 3nRB (Figure 6A), 4nRB (Figure 6B), and 5nRB hybrids (Figure 6C) showed obvious differences from RCC and BSB. Tables 4 and 5 indicated that most of the morphological data in 3nRB, 4nRB, and 5nRB hybrids were significantly different from those in both RCC and BSB, suggesting that the variation traits occurred in the polyploid hybrid offspring. For example, the ratio of the body length to width length in 3nRB hybrids was 1.67 whereas it was 2.18 in RCC and 2.37 in BSB. The most interesting variation trait, the presence of the barbells, occurred in 4nRB hybrids while there was no barbell in their parents, RCC and BSB. On the other hand, there were some significantly different traits between 3nRB and 4nRB hybrids. For example, the 4nRB hybrids had a pair of barbells on the mouth, and their lateral scale number was 31–33 whereas the 3nRB hybrids did not present any barbell on the mouth, and their lateral scale number was 28–30. In the offspring of RCC ♀ × BSB ♂, 4nRB hybrids accounted for 77% and 3nRB hybrids accounted for 23%. Furthermore, some significantly different traits between 4nRB and 5nRB hybrids also existed. For example, the 5nRB hybrids did not possess any barbell on the mouth, and their lateral scale number was 34–36. For the feeding habit, like BSB, 3nRB, 4nRB, and 5nRB hybrids were herbivorous.
Figure 6.—
Appearance of 3nRB, 4nRB, and 5nRB hybrids. (A) The 3nRB hybrid did not present any barbell on the mouth. Its ratio of body length to body width was 1.67, and its lateral scale number was 28–30. (B) The 4nRB hybrid had a pair of barbells on the mouth. Its ratio of body length to body width was 2.19, and its lateral scale number was 31–33. (C) The 5nRB hybrid did not possess any barbell on the mouth. Its ratio of body length to body width was 2.42, and its lateral scale number was 34–36. Bar in A–C, 1 cm.
Sizes of eggs produced by 4nRB hybrids:
The females of 4nRB hybrids produced two sizes of eggs. The average diameter of the larger eggs was 0.20 cm, which accounted for 95%, and the average diameter of the smaller eggs was 0.17 cm, which accounted for 5% (Figure 7).
Figure 7.—

Eggs produced by 4nRB hybrids. There are two sizes of eggs produced by 4nRB hybrids. The larger eggs, each with a diameter of 0.2 cm, were considered 4n eggs, and the smaller eggs (arrows), each with the diameter of 0.17 cm, were considered 2n eggs because their size was the same as that of the diploid eggs produced by the tetraploid hybrids of red crucian carp ♀ × common carp ♂ as described in our previous study (Liu et al. 2001). Bar, 0.4 cm.
Sterility of 3nRB hybrids and fertility of 4nRB hybrids:
During the reproductive season, no egg or semen could be squeezed out from 3nRB hybrids at the ages of 1 and 2 years. No egg or semen could be stripped out from 4nRB hybrids at the age of 1 year. However, the mature eggs (Figure 7) or water-like semen were squeezed out from the females and males of the 4nRB hybrids, respectively, at the age of 2 years. The eggs of 4nRB hybrids were mated with the semen of the BSB to generate ∼25,000 living pentaploid hybrids. However, after the water-like semen of 4nRB hybrids was fertilized with the eggs of 4nRB hybrids, only 8 living hybrids were produced, suggesting that, at the age of 2 years, the female 4nRB hybrids had stronger fertility than that of the male 4nRB hybrids.
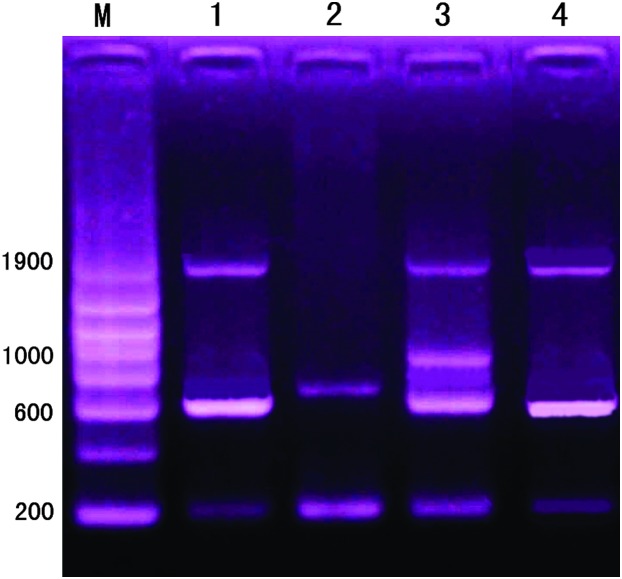
DNA fragments based on the primers of the HMG box of Sox genes:
The PCR results based on the primers of HMG of Sox genes and the sequencing results showed that there were three DNA fragments (215, 617, and 1958 bp) in RCC, two DNA fragments (215 and 712 bp) in BSB, three fragments (215, 616, and 1955 bp) in 3nRB hybrids, and four fragments (213, 616, 918, and 1959 bp) in 4nRB hybrids (Figure 8). All the sequences of the PCR products have been submitted to GenBank and their accession numbers are in Table 6. By comparing the sequences, we confirmed that a 215-bp DNA fragment that existed in RCC, BSB, and 3nBB hybrids belonged to Sox 11 whereas a 213-bp DNA fragment in 4nRB hybrids represented the Sox 1 gene. The 616- and 617-bp DNA fragments existed in RCC, 4nRB, and 3nRB hybrids and the 712-bp fragment in BSB represented Sox 9a gene. The 918-bp fragment in 4nRB hybrids was newly formed and represented Sox 9b gene. The 1955-, 1958-, and 1959-bp DNA fragments in the 3nRB hybrid, RCC, and the 4nRB hybrid were from the Sox 4 gene (Table 7).
Figure 8.—
Amplified DNA fragments resulting from PCR based on the primers of HMG of Sox genes in BSB, RCC, 3nRB, and 4nRB hybrids. M, DNA ladder markers with an increase of 200 bp. In lane1, there are three DNA fragments (215, 617, and 1958 bp) in RCC; in lane 2, two DNA fragments (215 and 712 bp) in BSB; in lane 3, four fragments (213, 616, 918, and 1959 bp) in 4nRB hybrids; in lane 4, three fragments (215, 616, and 1955 bp) in 3nRB hybrids.
TABLE 6.
GenBank accession numbers of the sequences in 3nRB and 4nRB and of their parents, RCC and BSB
TABLE 7.
Nucleotide similarities of separate regions of the DNA fragments produced by PCR with the primers of the HMG box of Sox genes in 3nRB, 4nRB, and their parents, RCC and BSB
| DNA fragments | RCC and BSB | RCC and 3nRB | BSB and 3nRB | RCC and 4nRB | BSB and 4nRB |
|---|---|---|---|---|---|
| 213 or 215 bp | 84 | 96 | 86 | 65 | 69 |
| 616, 617, or 712 bp | 72 | 99 | 73 | 99 | 72 |
| 918 bp | Absent in both | Absent in both | Absent in both | Absent in RCC | Absent in BSB |
| 1955, 1958, or 1959 bp | Absent in BSB | 99 | Absent in BSB | 100 |
Numbers are percentages.
Table 7 indicates the percentage of nucleotide similarities of separate regions of the DNA fragments produced by PCR with the primers of the HMG box of Sox genes in 3nRB and 4nRB and their parents, RCC and BSB. In the sequences of the 215-bp DNA fragments in RCC, BSB, and 3nRB hybrids and the 213-bp DNA fragment in 4nRB hybrids, 84% identity between RCC and BSB, 86% identity between BSB and 3nRB hybrids, and 96% identity between RCC and 3nRB hybrids were found, indicating that the sequence of this DNA fragment in 3nRB hybrids was highly homologous to that of RCC (Table 7). The sequence identity between 4nRB and BSB (69%) is higher than that between 4nRB and RCC (65%), indicating that the sequence of this DNA fragment in 4nRB hybrids was similar to that of BSB (Table 7).
As for the sequences of the DNA fragments of 617 bp in RCC, 712 bp in BSB, and 616 bp in 3nRB and 4nRB hybrids, a 72% similarity between RCC and BSB, 73% similarity between BSB and 3nRB hybrids, and 99% similarity between RCC and 3nRB hybrids existed, indicating that the sequence of this DNA fragment in 3nRB hybrids was similar to that of RCC (Table 7). A 72% similarity between BSB and 4nRB hybrids and a 99% similarity between RCC and 4nRB existed, suggesting that the sequence of this DNA fragment in 4nRB hybrids was similar to that of RCC (Table 7).
As for the sequences of the DNA fragment of 1958 bp in RCC, 1955 bp in 3nRB hybrids, and 1959 bp in 4nRB hybrids, 99% similarity between RCC and 3nRB hybrids and 100% similarity between RCC and 4nRB hybrids existed, suggesting that the sequence in this DNA region of both 3nRB and 4nRB hybrids was very close to that of RCC. In the 4nRB hybrids, the 918-bp DNA fragment was newly formed, and the sequence was deposited in GenBank under accession no. EF370033 (Table 6).
DISCUSSION
The distant crossing is an important and effective means of increasing the possibility of variation in the hybrid progeny. With this method, it is possible that the whole haploid genome of one species could be translated into another species. The distant genome translation not only will result in changes of the ploidy levels in chromosomes, but also will lead to changes in phenotypes. Furthermore, it will lead to changes in reproductive behavior, for example, the formation of the sterile triploid hybrids.
In this study, the ploidy levels of the crossing offspring were confirmed by counting chromosomal number (Figure 2), forming chromosomal karyotype (Figure 3), and measuring DNA content (Figure 4, Table 2) and the erythrocyte nuclear volume (Figure 5, Table 3). All of the above results were in agreement that 3nRB, 4nRB, and 5nRB hybrids were triploids, tetraploids, and pentaploids, respectively. Counting the chromosome number is a direct and accurate method of determining the ploidy of the samples. Using the instrument of the flow cytometer to examine the DNA content of samples is a rapid and simple as well as accurate method. The erythrocyte nuclear measurement is also a simple and useful method for determining the ploidy.
The 3nRB, 4nRB, and 5nRB hybrids were different from RCC and BSB in chromosome number, largest chromosome marker, DNA content, nuclear volume of erythrocytes, and morphological traits, indicating that they were from hybridization rather than from gynogenesis or androgenesis. The feeding habit in 3nRB, 4nRB, and 5nRB hybrids, like that of BSB, was herbivorous, which also suggested that the polyploid offspring had chromosomes from BSB and that they were not from gynogenesis. Their herbivorous feeding habit indicated that they had wider feeding resources, which was very beneficial in aquaculture.
At the chromosome level, the chromosomal number and the special chromosome(s) can be used as the marker(s) for analyzing the changes of the ploidy levels and the origin of the chromosomes in the hybrid progeny. In this study, the diploid BSB had 48 chromosomes with one pair of the largest submetacentric chromosomes (Figure 2A), which could be used as markers for identifying BSB from RRC possessing 100 chromosomes without an obviously large submetacentric chromosome. With 124 chromosomes and the presence of one submetacentric largest chromosome similar to the two in BSB, the 3nRB hybrids were considered to have two sets of chromosomes from RCC and one set of chromosomes from BSB (Figure 2B). On the basis of the chromosomal number (148) and one pair of the largest chromosomes similar to those in BSB, we concluded that 4nRB hybrids contained two sets of chromosomes from BSB and two sets of chromosomes from RCC (Figure 2C; Figure 3A). With 172 chromosomes and the three largest submetacentric chromosomes similar to those in BSB (Figure 2D), the 5nRB hybrids proved to have two sets of chromosomes from RCC and three sets of chromosomes from BSB (Figure 3B).
At the DNA fragment level, the RCC presented three different DNA fragments whereas the BSB presented only two different DNA fragments (Figure 8), indicating that these two different species possessed different DNA characteristics. As the hybrid offspring of RCC and BSB, the 4nRB hybrids not only shared all the DNA fragments found in both RCC and BSB, but also possessed the newly formed 918-bp DNA fragment, indicating that the 4nRB hybrids not only changed in chromosomal ploidy levels, but also changed in DNA sequences. The 4nRB hybrids shared the common and different fragments found in RCC and BSB (Figure 8), providing the genetic evidence that the 4nRB hybrids came from RCC and BSB, but were different from RCC and BSB. On the other hand, the specific 712-bp DNA fragment (Figure 8) of BSB was absent in 3nRB and 4nRB hybrids, providing further genetic evidence that the 3nRB and 4nRB hybrids were different from BSB (Figure 8). Table 7 indicated that most of the sequences in the amplified DNA fragments were similar to those of RCC, consistent with the result that 3nRB and 4nRB hybrids had more chromosomes from RCC than from BSB.
The presence of the unusual erythrocytes with two nuclei could be used as one of the cytological markers to distinguish the polyploid hybrids from their diploid parents (Figure 5). With the increase of the ploidy level, the percentage of the unusual erythrocytes with two nuclei increased, suggesting that the higher the ploidy level, the higher the possibility of the presence of the unusual erythrocytes in polyploid hybrids. In our previous study (Liu et al. 2003), 32.4% of unusual erythrocytes with two nuclei were also found in the tetraploid hybrids of red crucian carp ♀ × common carp ♂. It was concluded that the higher DNA content made it easier for the nuclei to divide.
In this study, only 3nRB and 4nRB hybrids, but no diploid hybrids, were found to survive in the cross of RCC ♀ × BSB ♂. However, we still observed diploid embryos with 74 chromosomes by examining the chromosomal number (data not shown), but no survival diploid hybrid. This means that the diploid hybrids could not survive probably due to the larger difference in chromosomal number between the maternal RCC (100) and the paternal BSB (48), which inhibits the embryos from developing into living diploid hybrids. The tetraploidization and triploidization were helpful to the hybrids in overcoming the obstacle to developing into living hybrids. The 3nRB hybrids resulted from the retention of the second polar body of the fertilized eggs and the 4nRB hybrids resulted from the inhibition of the first cleavage of the fertilized eggs.
In general, animals generate the half-reduced gametes through meiosis. For example, most of diploid fish produce haploid gametes. However, there were a few reports about the production of diploid gametes generated by diploid hybrids. For example, the diploid female hybrids of crucian carp (Carassius auratus gibelio) (♀) × common carp (Cyprinus carpio L.) (♂) (interspecific crossing) produced unreduced diploid eggs (Cherfas et al. 1994). The diploid female hybrid of medaka between Oryzias latipes and O. curvinotus also spawned diploid eggs (Shimizu et al. 2000). Some diploid male hybrids of Rutilus alburnoides (Teleostei, Cyprinidae) produced unreduced diploid sperm (Alves et al. 1999). In our previous study (Liu et al. 2001), we made the distant crossing of RCC ♀ × common carp ♂ and found that both F1 and F2 hybrids were diploids with 100 chromosomes. However, some females and males of F2 hybrids were able to generate diploid eggs and diploid sperm, respectively, which fertilized each other to form the bisexual tetraploid hybrids in F3. Until now the population of F3–F16 was proved to be tetraploid hybrids, indicating that their tetraploidy could be stably inherited from one generation to another (Liu et al. 2001, 2003, 2004; Sun et al. 2003; Guo et al. 2006). In that study, the formation of unreduced diploid gametes was due to the premeiotic endoreduplication of oogonia or spermatogonia of females or males of the F2 (Liu et al. 2001). The fertile males and females of F2 hybrids reached maturity when they were 2 years old. In this study, the 2-year-old female 4nRB hybrids reached maturity and produced many mature eggs whereas the male 4nRB hybrids produced only the water-like semen rather than the normal white semen as the male RCC or BSB did, indicating that the females of 4nRB hybrids matured early and better than the males of 4nRB hybrids. In contrast, no egg and semen were squeezed from either 1- or 2-year-old 3nRB hybrids. Interestingly, the females of 4nRB hybrids generated two sizes of eggs. The larger eggs, each with a diameter of 0.2 cm (Figure 7), were considered as tetraploid eggs because the pentaploid hybrids were found in 4nRB ♀ × BSB ♂. The smaller eggs, each with a diameter of 0.17 cm, were considered as diploid eggs because their size was similar to that of the diploid eggs of the tetraploid hybrids of RCC ♀ × common carp ♂ (Liu et al. 2001). In the progeny of 4nRB ♀ × BSB ♂, we found only the pentaploid hybrids, but no triploid hybrids. It is probably because 95% of the eggs produced by 4nRB hybrids were tetraploid eggs and only 5% were diploid eggs. We thought the formation of the unreduced tetraploid eggs was also due to the premeiotic endoreduplication of oogonia of 4nRB hybrids. The similarity between this study and our previous study was the formation of unreduced gametes produced by the distant crossing hybrids. The difference between this study and our previous study is that, in our previous study, the unreduced diploid gametes resulted from the diploid hybrids and, in this study, the unreduced tetraploid eggs resulted from the tetraploid hybrids. On the other hand, in our previous study, the tetraploids' maternal and paternal had the same chromosomal number (2n = 100) and belonged to the same subfamily and different genus in the catalog. In this study, the RCC and BSB had different chromosomal numbers (2n = 100 and 2n = 48) and belonged to different subfamilies in the catalog. The formation of 4nRB hybrids resulted from the inhibition of the first cleavage of the fertilized eggs, but not from the fertilization of the diploid eggs and sperm produced by F2 hybrids of RCC ♀ × common carp ♂. Both the fertile males and females of the 4nRB hybrids were found when they were 2 years old and many pentaploid hybrids were found in the progeny of 4nRB ♀ × BSB ♂, suggesting that it is very possible to obtain the octoploid hybrids with 296 chromosomes if we obtain more fertile male 4nRB hybrids in 2007.
The 4nRB hybrids could be identified from 3nRB hybrids by traits such as the presence of the barbells, a different chromosomal number, a different DNA content, and different molecular markers of DNA fragments of Sox genes. The 3nRB hybrids presented a similar number and size of DNA fragments to their maternal RCC, suggesting that two sets of chromosomes of RCC played a major role in inheritance material, which resulted in the 3nRB hybrid phenotype being similar to that of RCC. The appearance and the DNA markers also suggested that 4nRB hybrids showed more hybrid traits than 3nRB hybrids displayed. Furthermore, the 213-bp DNA fragment of 4nRB hybrids belonged to Sox 1, rather than to Sox 11, as happened in RCC, BSB, and 3nRB hybrids, suggesting that more DNA variation occurred in 4nRB hybrids than in 3nRB hybrids.
In this study, compared with their parents, obvious differences occurred in 3nRB, 4nRB, and 5nRB hybrids in the measurable and countable data, indicating the distant hybridizing effect. The most obvious variation trait was the presence of the barbells in 4nRB hybrids, which were absent in their parents, RCC and BSB. The changes in the phenotype traits resulted from the genomic changes of the chromosomes, DNA fragments, or sequences.
Also, in this study, we found that both females and males of the 4nRB hybrid were fertile. Therefore, the changes in chromosomes and sequences could be inherited from one generation to another. In evolution, the fertility of the 4nRB hybrid makes it available to create a new type of offspring with a changed chromosome number. For example, we have created the 5n hybrids, and it is very possible to create the octoploid hybrids by crossing the females and males of the 4nRB hybrids. In theory, the octoploid hybrids should also be fertile. In aquaculture, the 3nRB hybrids combined the biological characteristics between RCC and BSB and they will become a new fish product in the market. Their sterility ensures that they will not disturb the natural fish genetic resources. In a word, the bisexual fertile 4nRB hybrids and sterile 3nRB hybrids are significant in both evolution and aquaculture.
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (nos. 30330480 and 30571444), the Program for Changjiang Scholars and Innovative Research Team in University (no. IRT0445), the Key Item of Science and Technology of Hunan Province (no. 2006NK2008), and the Training Project of Excellent Young Researchers of the State Education Ministry of China (no. 200248).
References
- Alves, M. J., M. M. Coelho, M. I. Prospero and M. J. Collares-Pereira, 1999. Production of fertile unreduced sperm by hybrid males of the Rutilus alburoides complex (Teleost, Cyprinidae): an alternative route to genome tetraploidization in unisexuals. Genetics 151 227–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, M. L., C. J. Biggers and C. J. Barker, 1984. Chromosomal and electrophoretic analyses of hybrids between grass carp and bighead carp. Copeia 81 337–342. [Google Scholar]
- Cherfas, N. B., B. L. Gomelsky, O. V. Emelyanova and A. V. Recoubratsky, 1994. Induced diploid gynogenesis and polyploidy in crucian carp, Carassius auratus gibelio (Bloch) × common carp, Cyprinus carpio L., hybrids. Aquac. Fish. Manage. 25 943–945. [Google Scholar]
- Guo, X. H., S. J. Liu and Y. Liu, 2006. Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics 172 1745–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan, A., K. Fredga and A. Sandburg, 1964. Nomenclature for centromeric positions on chromosomes. Hereditas 52 201–220. [Google Scholar]
- Liu, S. J., Y. Liu, G. J. Zhou, X. J. Zhang, C. Luo et al., 2001. The formation of tetraploid stocks of red crucian carp × common carp hybrids as an effect of interspecific hybridization. Aquaculture 192 172–186. [Google Scholar]
- Liu, S. J, Y. D. Sun, G. J. Zhou, X. J. Zhang and Y. Liu, 2003. The ultra-microstructure of the mature gonads and erythrocytes in allotetraploids. Prog. Nat. Sci. 13 194–197 (in Chinese). [Google Scholar]
- Liu, S. J, Y. D. Sun, C. Zhang, K. K. Luo and Y. Liu, 2004. Production of gynogenetic progeny from allotetraploid hybrids red crucian carp × common carp. Aquaculture 236 193–200. [Google Scholar]
- Marian, T., and Z. Kraszai, 1978. Karyological investigation on Ctenopharyngodon idella and Hypophthalmichthys nobilis and their crossing-breeding. Aquac. Hungarica 1 44–50. [Google Scholar]
- Sambrook, J., and D. W. Russell, 1989. Molecular Cloning: A Laboratory Manual, Ed. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shimizu, Y., N. Shibata, M. Sakaizumi and M. Yamashite, 2000. Production of diploid eggs through premeiotic endomitosis in the hybrid medaka between Oryzias latipes and O. ourvinatus. Zool. Sci. 17 951–958. [Google Scholar]
- Sun, Y. D., S. J. Liu, C. Zhang, J. Z. Li, W. R. Wang et al., 2003. The chromosome number and gonadal structure of F9–F11 allotetraploid crucian carp. Chin. J. Genet. 30 37–41 (in Chinese). [PubMed] [Google Scholar]
- Yu, X. J., 1989. China Freshwater Fisheries Chromosome, pp. 44–75. Science Publishing House, Beijing (in Chinese).