Abstract
Asymmetric postmating isolation, where reciprocal interspecific crosses produce different levels of fertilization success or hybrid sterility/inviability, is very common. Darwin emphasized its pervasiveness in plants, but it occurs in all taxa assayed. This asymmetry often results from Dobzhansky–Muller incompatibilities (DMIs) involving uniparentally inherited genetic factors (e.g., gametophyte–sporophyte interactions in plants or cytoplasmic–nuclear interactions). Typically, unidirectional (U) DMIs act simultaneously with bidirectional (B) DMIs between autosomal loci that affect reciprocal crosses equally. We model both classes of two-locus DMIs to make quantitative and qualitative predictions concerning patterns of isolation asymmetry in parental species crosses and in the hybrid F1 generation. First, we find conditions that produce expected differences. Second, we present a stochastic analysis of DMI accumulation to predict probable levels of asymmetry as divergence time increases. We find that systematic interspecific differences in relative rates of evolution for autosomal vs. nonautosomal loci can lead to different expected F1 fitnesses from reciprocal crosses, but asymmetries are more simply explained by stochastic differences in the accumulation of U DMIs. The magnitude of asymmetry depends primarily on the cumulative effects of U vs. B DMIs (which depend on heterozygous effects of DMIs), the average number of DMIs required to produce complete reproductive isolation (more asymmetry occurs when fewer DMIs are required), and the shape of the function describing how fitness declines as DMIs accumulate. Comparing our predictions to data from diverse taxa indicates that unidirectional DMIs, specifically involving sex chromosomes, cytoplasmic elements, and maternal effects, are likely to play an important role in postmating isolation.
The degree of sterility does not strictly follow systematic affinity, but is governed by several curious and complex laws. It is generally different, and sometimes wildly different, in reciprocal crosses between the same two species.
ISOLATION asymmetry occurs when the strength of reproductive isolation between taxa differs significantly between reciprocal crosses. While interest in asymmetric reproductive isolation has often focused on behavioral (sexual) isolation between animal species (e.g., Kaneshiro 1980; Kawanishi and Watanabe 1981; Arnold et al. 1996), postmating isolation asymmetry, expressed as reciprocal-cross differences in F1 viability or fertility or in postmating, prezygotic isolation, is also common. It was originally reported in plants by J. G. Kölreuter in 1761, 1763, 1764, and 1766 (cited and partially translated in Mayr 1986), the first researcher to systematically create interspecific hybrids (his key work is reviewed in Roberts 1929, Chap. II; in Olby 1966a, Chap. 1, 1966b; and, especially, in Mayr 1986). Isolation asymmetry was emphasized by Darwin (1859, Chap. 8, esp. pp. 258–261), who noted, “… hybrids raised from reciprocal crosses … generally differ in sterility in a small, and occasionally in a high degree,” citing Kölreuter and Gärtner, the same plant hybridizers whose hundreds of intra- and interspecific crosses inspired Mendel in 1865 (translated in Bateson 1901). Asymmetry was subsequently found in essentially all systems subject to systematic hybridization experiments, including many invertebrates (e.g., Muller 1942; Oliver 1978; Harrison 1983; Coyne and Orr 1989a; Gallant and Fairbairn 1997; Presgraves and Orr 1998; Navajas et al. 2000; Willett and Burton 2001; Presgraves 2002), vertebrates (e.g., Thornton 1955; Rakocinski 1984; Bolnick and Near 2005), and fungi (e.g., Dettman et al. 2003). A recent analysis of reciprocal species crosses within 14 diverse angiosperm genera found significant isolation asymmetry in 35–45% of all species pairings, evaluated at three different postmating stages of reproductive isolation (Tiffin et al. 2001). Similarly, Muller (1942, p. 101) noted that the viability and fertility of Drosophila F1 males derived from reciprocal crosses “… are so often very different …” and Turelli and Orr (1995) estimated that ∼15% of the cases of Haldane's rule in Drosophila show qualitative asymmetry, with males being sterile or inviable in one cross but not in the reciprocal cross. Despite its ubiquity, however, reciprocal isolation asymmetry, especially asymmetry that occurs after the interspecific transfer of gametes, has received very little theoretical attention.
Because Darwin (1859, Chap. 8) first drew attention to both the generality and the evolutionary significance of asymmetric postmating isolation, we propose “Darwin's corollary” as a name for this phenomenon. It joins Haldane's (1922) rule, concerning the preferential inviability/sterility of the heterogametic sex of interspecific F1 hybrids, and “Coyne's rule” (also known as the “large X effect”; Coyne and Orr 1989b), the disproportionate contribution of the X chromosome to heterogametic F1 inviability/sterility, as a third widespread pattern concerning intrinsic postmating isolation. We describe this reciprocal-cross asymmetry as a “corollary,” both because it is less common than the two “rules” (for reasons that are elucidated by our theoretical analysis) and because it is often produced by the same genetic mechanism that explains the other two, namely interspecific epistatic incompatibilities (Dobzhansky 1936, 1937, p. 256; Muller 1940, 1942; Turelli and Orr 2000). Although our quote from Darwin (1859) suggests that he was concerned only with hybrid sterility, he emphasizes in the opening pages of Chapter 8 that he is discussing two different classes of “sterility”: “sterility of the species when first crossed,” meaning an absence of progeny, which can arise from barriers to fertilization or F1 inviability, and “sterility of the hybrids produced from them,” namely F1 sterility. Indeed, the asymmetry Darwin describes includes both pre- and postmating isolation. We emphasize the latter because of its genetic implications and close connection to previous analyses of the genetics of intrinsic postzygotic isolation.
As summarized by Coyne and Orr (2004, Chaps. 8 and 9), inviability and sterility of species hybrids can often be explained by between-locus “Dobzhansky–Muller incompatibilities” (DMIs)—inappropriate epistatic interactions between alleles that characterize independently evolving lineages. Many DMIs involve interactions between autosomal loci and affect both reciprocal crosses identically. In contrast, DMIs between autosomal loci and uniparentally inherited factors, including mitochondria (mtDNA), chloroplasts, maternal transcripts, and sex chromosomes in heterogametic hybrids, are specific to a particular direction of hybridization and can therefore contribute to asymmetric reproductive isolation. [Note that genetic imprinting has been implicated as a possible source of asymmetric postmating isolation in mammals (Vrana et al. 2000) and angiosperms (Bushell et al. 2003). Given that it effectively corresponds to uniparental inheritance, it can be also be included in our theoretical framework; see discussion.] Orr (1993) and Turelli and Orr (1995, 2000) described between-sex asymmetries associated with X–autosome interactions, cytoplasmic–nuclear interactions, and maternal effects. Here we generalize those analyses by elaborating the dynamic model of Orr and Turelli (2001) to quantitatively analyze how same-sex (including hermaphrodite) asymmetry between reciprocal crosses is expected to vary with divergence time. We provide an idealized quantitative treatment that contrasts symmetrically acting incompatibilities with asymmetrically acting ones.
In addition, because isolation asymmetry appears to be particularly common and taxonomically widespread in angiosperms (Darwin 1859, Chap. 8; Tiffin et al. 2001), we consider in some detail asymmetric genetic interactions that are common in angiosperms: nuclear–cytoplasmic interactions, gametophyte–sporophyte interactions, and triploid endosperm interactions (see Table 1 and Figure 1). Each involves asymmetric interactions, and only the first has been treated previously (Turelli and Orr 2000).
TABLE 1.
Summary of phenomena analyzed that contribute to asymmetric postzygotic isolation
| Focal incompatibilities | Maternal contributions | Paternal contributions | Types of DMIs |
|---|---|---|---|
| X–autosomea | X, autosomes | Autosomes | Ub and Bc |
| Cytonuclear | Cytoplasmic organelles, nuclear genes | Nuclear genes | U and B |
| Maternal effects | Maternal transcripts and proteins, nuclear genes | Nuclear genes | U (and B)d |
| Triploid endosperm | Diploid genome (doubled haploid) | Haploid genome | U (and B)e |
| Gametophytic–sporophyticf | Diploid sporophyte | Haploid gametophyte | U |
Our analysis considers males in male-heterogametic species. In the text, we discuss the loci that make maternal (paternal) contributions to U DMIs as “female acting” (male acting).
Unidirectional: These are the DMIs responsible for asymmetric postzygotic isolation.
Bidirectional: These DMIs contribute equally to postzygotic isolation in both reciprocal crosses.
Depending on the phenotype observed, B DMIs may or may not act simultaneously with U DMIs. For early embryo lethality, U DMIs may act alone. When considering embryo-to-adult viability, both act together.
Depending on the phenotype observed, B DMIs may or may not act simultaneously with U DMIs. Triploid endosperm DMIs can be experimentally distinguished from zygotic DMIs; but if seed viability is assayed directly, both U and B DMIs will contribute.
This includes pollen–style interactions.
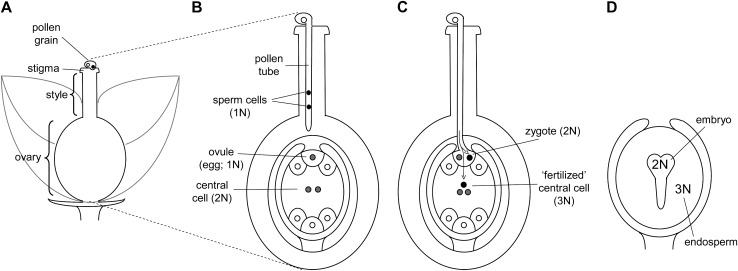
Figure 1.—
Generalized angiosperm gametogenesis and double fertilization. (A) During pollination pollen is transferred to the stigma of the recipient flower. (B) During fertilization the pollen tube germinates and travels through the female stigmatic tissue into the ovary. The mature male gamete (pollen or “microgametophyte”) comprises two genetically identical haploid sperm cells that result from the mitotic division of a single meiotic product. The mature female gametophyte comprises eight genetically identical haploid nuclei resulting from mitotic division of a single meiotic product. The “central cell” differs from the haploid ovule (1N) and other cells in that it is binucleate (2N). (C) Double fertilization: One haploid sperm cell fertilizes the ovule, while the other sperm cell fuses with the diploid central cell to form a triploid endosperm. (D) Postfertilization development: The triploid endosperm functions as a primary storage and nutritive tissue for the developing embryo.
CLASSES OF ASYMMETRIC GENETIC INTERACTIONS
Nuclear–cytoplasmic interactions:
Nuclear–cytoplasmic (“cytonuclear”) interactions occur in all organisms where the function of haploid cytoplasmic organelles (including mitochondria, chloroplasts, and plastids) depends on coordinated expression with the diploid nuclear genome. In angiosperms, hybrid male sterility is thought to result frequently from negative epistatic interactions between cytoplasmic (most probably mitochondrial) and nuclear genes in interspecific hybrids (Frank 1989; Schnable and Wise 1998). Negative cytonuclear interactions are also known to contribute to reproductive isolation between animal species and even populations, where they have been identified as the genetic basis of both hybrid inviability and infertility [e.g., Tigriopus (Willett and Burton 2001; Harrison and Burton 2006) and Drosophila (Rand et al. 2001; Sackton et al. 2003)]. Assuming uniparental inheritance of the relevant organelle, cytonuclear interactions involve interactions between a cytoplasmic genome from one parent (usually maternal, Grun 1976) and the genes in the hybrid nuclear genome derived from the second parent. For specificity, we assume maternal inheritance of cytoplasmic genomes.
X–autosome interactions:
In species with sex chromosomes, heterogametic hybrids often experience asymmetric incompatibilities between sex-chromosome alleles from one parent and autosomal alleles from the other parent. In the F1 generation, these incompatibilities are analogous to cytonuclear interactions (as no recombination of the X chromosome has occurred). For specificity, we consider male hybrids in male-heterogametic species. Obviously, asymmetries will also arise from Y–autosome and/or X–Y interactions.
Genetic maternal effects:
In all metazoans, embryonic development begins under the control of maternal mRNAs and proteins. Early in development, control shifts from these maternal factors to zygotic transcripts (often referred to as the “maternal–zygotic transition”; Wang and Dey 2006). Incompatibilities can occur between paternally inherited alleles and the maternal factors that initiate development. Indeed, genetic analyses reveal that these incompatibilities underlie most exceptions to Haldane's rule in Drosophila (Sawamura 1996; Turelli and Orr 2000). Unlike the previous two classes of incompatibilities, in which asymmetric DMIs always act simultaneously with symmetric DMIs (e.g., autosome–autosome incompatibilities that are identical in reciprocal crosses), all maternal–zygotic incompatibilities are expected to be asymmetric. However, given the gradual turnover of control from maternal factors to zygotic, these DMIs can also be expressed simultaneously with the symmetric DMIs in the hybrid nuclear genome.
Asymmetric incompatibilities in plants:
Gametophytic–sporophytic (GS) (including pollen–style) and triploid endosperm (TRE) interactions are restricted to flowering plants. Like DMIs involving early maternal effects, all GS and TRE DMIs are expected to be asymmetric. Modeling them requires a basic understanding of postpollination and early fertilization processes in angiosperms (see Figure 1). Following transfer of pollen to the stigma of a flower, each pollen grain germinates to produce a tube—containing two genetically identical haploid sperm cells—that grows down the maternal style and into the ovary (Mascarenhas 1989; Figure 1, A and B). Importantly, angiosperm pollen is known to express many genes during this haploid phase (estimated in some cases as >70% of the total haploid genome; Ottaviano and Mulcahy 1989; Frova and Pe 1997), rather than solely expressing paternal gene products. GS interactions therefore occur when genes expressed by the haploid pollen (male gametophyte) interact with genes expressed in the stigma and style of the diploid (sporophyte) maternal parent (Figure 1B). For simplicity, we generally refer to these as pollen–style interactions although they can also operate between pollen and stigmatic and ovary tissues (Willemse and van Lammeren 2002). In interspecific crosses, dysfunctional GS interactions frequently cause postpollination prezygotic cross failure, including failure of pollen to germinate, retardation or rupture of pollen tubes in foreign styles, and failure of pollen tubes to penetrate ovules (de Nettancourt 2001 and references therein).
In contrast to GS interactions, TRE interactions predominate after fertilization, during formation and development of the new zygote. During angiosperm “double fertilization” within a single ovule, one of the pollen haploid sperm cells fertilizes a haploid egg cell to produce a diploid embryo while the remaining sperm cell fuses with a binucleate (doubled haploid) maternal “central cell” to produce a 3N endosperm (Cresti and Tiezzi 1997) (Figure 1C). This triploid endosperm develops rapidly and sequesters maternal resources to act as the primary nutritive body for the developing embryo (Figure 1D). In crosses among angiosperms, hybrid seed failure frequently results from abnormal development of hybrid endosperm (rather than the embryo itself), likely due to dysfunctional interactions between the haploid male and doubled haploid female genetic components within the triploid endosperm (e.g., Lin 1984; Katsiotis et al. 1995; Gutierrez-Marcos et al. 2003). Experiments can determine whether a defect in hybrid seed development is caused by TRE DMIs or symmetric DMIs acting within the diploid embryo (e.g., by assessing viability of the embryo when cultured independently of the endosperm; Bridgen 1994). However, when experiments simply score overall seed development, inviability can be caused by a combination of the asymmetric TRE interactions and symmetric embryonic DMIs. Hence, it is important to consider TRE interactions both in isolation and in conjunction with symmetric DMIs.
We present a theoretical analysis that encompasses all of the asymmetric DMIs discussed above. First, we generate expected fitnesses of reciprocal F1 hybrid genotypes, on the basis of the expected number and relative effects of different classes of DMIs. Second, extending the analytical approximations for the time-dependent distribution of the cumulative effects of DMIs developed in Orr and Turelli (2001), we examine the transient dynamics of asymmetric reproductive isolation expected during allopatric speciation. We use this first to make quantitative predictions about the magnitude and divergence-time dependence of fitness differences between reciprocal crosses, in particular to identify biological factors likely to contribute most to asymmetric isolation. Then we consider the probability that complete reproductive isolation will be seen in one cross, but incomplete isolation in the reciprocal cross (e.g., asymmetric Haldane's rule). Finally, we review data relevant to estimating the parameter values critical to our predictions and then examine plant and animal data on asymmetry to assess its likely causes. We encourage readers who are primarily interested in our predictions to read the Introduction describing our model and its parameters, look at the figures that present our numerical results, and then skip to the discussion of data.
MODELS AND ANALYSES
We present a model of two-locus DMIs that distinguishes different classes of interactions in hybrids, with respect to both the magnitude and the symmetry of their expected effects. Because the bulk of available data that demonstrate asymmetry comes from initial hybridizations rather than backcrosses or the F2 generation, our treatment focuses on isolation expressed during parental hybridization and in the resulting F1. Our model incorporates symmetric DMIs (e.g., between autosomal loci or between sex-linked and autosomal loci in the homogametic sex) that affect reciprocal crosses equally (“bidirectional,” B) and asymmetric DMIs (as described above) that differ between reciprocal crosses (“unidirectional,” U). We use this framework to discuss all of the sources of asymmetry identified above. Note that, throughout our treatment, reproductive isolation due to DMIs falls into two cases: those involving only U DMIs, and those involving both U and B DMIs. For instance, GS interactions are exclusively unidirectional and occur before other interactions that affect zygote viability can act. Hence, only the U DMIs they produce need be considered to understand the resulting asymmetric failure of hybridization. Similarly, TRE effects can be experimentally isolated from embryonic B DMIs that may simultaneously affect seed development. Incompatibilities involving GS and TRE interactions act sequentially, so when we consider only one of them, we implicitly assume that viability is assayed from the beginning to the end of the relevant stage of fertilization/development. In contrast, cytonuclear DMIs (U) act simultaneously with nuclear genome DMIs (B); so both must be considered simultaneously. Similarly, in heterogametic males, X–autosome interactions (U) act simultaneously with autosomal–autosomal interactions (B). These cases are clear cut, but in others there may be no clear expectation about the relative importance or frequency of U vs. B DMIs. For instance, although maternal-effect DMIs act early in embryogenesis, some zygotic transcripts appear extremely early (e.g., Tadros and Lipshitz 2005; Wang and Dey 2006). As soon as zygotic transcripts are active, U maternal-effect DMIs act simultaneously with B nuclear DMIs to determine viability. Similarly, asymmetric TRE interactions will act simultaneously with symmetric embryonic incompatibilities to determine seed viability. Our idealized analyses below include parameters to weight the relative importance of U vs. B DMIs. In some cases, like X–autosome vs. autosome–autosome DMIs, we have simple a priori predictions; but in cases like maternal effects vs. zygotic effects, the biology is much less well understood and the relevant weighting will depend on the timing of expression of the loci involved. Many of these details are irrelevant to our predictions, which depend only on composite parameters that describe the cumulative consequences of U vs. B DMIs.
In general, F1 hybrids experience only one form of B DMIs, namely those involving heterozygous loci on biparentally inherited chromosomes. However, F1 individuals can be afflicted by several types of U DMIs simultaneously. For instance, heterogametic males may experience at least four types: those involving cytonuclear interactions, maternal effects, X-linked DMIs, and Y-linked DMIs. It is usually difficult to know the relative contributions of these to a phenotype like F1 inviability without detailed developmental and genetic analyses. We simplify our mathematical treatment by considering only one type of U DMI. Our analysis can be easily generalized to consider both sequential and simultaneous effects of different DMIs, but we concentrate on the simplest cases to illustrate some central principles.
Basic model:
Following Turelli and Orr (1995, 2000) and Orr and Turelli (2001), we assume that individual DMIs contribute additively to a hybrid breakdown score, S, which maps onto fitness in a simple way. To have a single framework, we assume that U and B DMIs act simultaneously, so that situations in which only U DMIs occur (like GS and TRE) appear as special cases. Let the random variable eU denote the effect of a specific U incompatibility in a parental cross or resulting F1, and let eB denote the effect of a specific B incompatibility. In general, the hybrid breakdown score after a divergence time of t depends on both the number and the kind of DMIs that have accumulated. We denote the number of B DMIs by 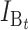 , the number of U DMIs by
, the number of U DMIs by 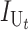 , and the total number by It (we analyze their accumulation below). Table 2 provides a summary of repeatedly used notation. We assume that
, and the total number by It (we analyze their accumulation below). Table 2 provides a summary of repeatedly used notation. We assume that
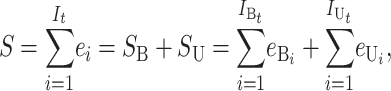 |
(1) |
where by definition 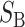 is identical for the reciprocal F1. The ei are all assumed to be independent, and the
is identical for the reciprocal F1. The ei are all assumed to be independent, and the 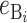 (
(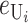 ) are assumed to be identically distributed (the latter assumption can be easily relaxed to allow for different types of U DMIs acting simultaneously, e.g., X–autosome and X–Y). We assume that hybrid fitness is a decreasing function of the breakdown score, v(S), that gives a relative fitness of 1 when S = 0 and declines to 0 when S reaches a threshold value C for complete sterility/inviability; i.e.,
) are assumed to be identically distributed (the latter assumption can be easily relaxed to allow for different types of U DMIs acting simultaneously, e.g., X–autosome and X–Y). We assume that hybrid fitness is a decreasing function of the breakdown score, v(S), that gives a relative fitness of 1 when S = 0 and declines to 0 when S reaches a threshold value C for complete sterility/inviability; i.e.,
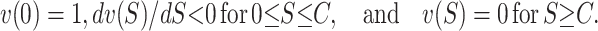 |
(2) |
These general conditions suffice to analyze qualitative asymmetry (see Equations 27–30); but to predict quantitative asymmetry, a particular function v(S) must be chosen (see Equation 10). Let Sij denote the breakdown score produced with a mother from taxon i and a father from taxon j. When considering X–autosome incompatibilities in heterogametic individuals, we assume for definiteness that males are heterogametic. To understand the forces that lead to systematic differences between reciprocal crosses, we first seek conditions under which E(S12) ≠ E(S21).
TABLE 2.
Glossary of repeatedly used notation
| Symbol | Usage (relevant equation in the text) |
|---|---|
| A | Measure of postmating asymmetry, measured as a difference between the more successful minus the less successful direction of hybridization (11) |
| C | Threshold value of the hybrid breakdown score that leads to complete postmating isolation; the value is scaled as a multiple of the average effect of the most deleterious DMI (44) |
| CV | Coefficient of variation of effects of both B and U DMIs (13) and (24) |
| gB | 1 − gU, fraction of substitutions that can contribute to B DMIs (23) |
| gU | Fraction of substitutions relevant to U DMIs that occur at “female-acting” loci (17) |
| h0 | Average contribution of B DMIs to the hybrid breakdown score, assuming that the U DMIs have been scaled to have an average effect of 1 (21) |
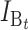 |
No. of bidirectional (B) DMIs accumulated after divergence time t (1) |
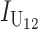 |
No. of unidirectional (U) DMIs experienced in a cross with a taxon 1 maternal parent |
| p | Probability that an allelic difference at two B loci produces a B DMI (22) |
| pU | Probability that allelic differences at one “male-acting” locus and one “female-acting” locus produce a U DMI (5) |
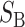 |
Contribution to the hybrid breakdown scores (both 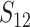 and and 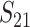 ) from B DMIs (1) and (3) ) from B DMIs (1) and (3) |
| Sij | Hybrid breakdown score produced with a taxon i maternal parent (1) |
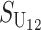 |
Contribution to the hybrid breakdown score 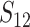 from U DMIs from U DMIs |
| TC | Geometric mean of 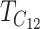 and and 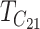 (9) (9) |
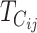 |
Divergence time at which E(Sij) = C, (9), i.e., the time at which the expected breakdown score corresponds to complete postmating isolation |
| v(S) | Function describing how fitness declines as the hybrid breakdown score, S, increases (10) |
| α | Exponent in the function describing how fitness declines as DMIs accumulate (10) |
| β | p(1 − gU)/(pUgU), ratio of the expected no. of B DMIs to the expected no. of U DMIs (26) |
| δ1 | υ1 − 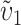 , difference in the relative taxon-specific rates of evolution of female-acting vs. male-acting loci that contribute to U DMIs (4b) , difference in the relative taxon-specific rates of evolution of female-acting vs. male-acting loci that contribute to U DMIs (4b) |
| η | h0β, ratio of the expected contribution to the hybrid breakdown scores caused by B DMIs (which have average effect h0) to the expected contribution from U DMIs (which are assumed to have average effect 1) (28) |
| τ | t/TC, scaled divergence time between the taxa hybridized (19) and (25) |
Expected differences between reciprocal crosses:
From (1), the breakdown scores from reciprocal crosses are
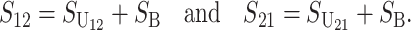 |
(3) |
Thus, E(S12) ≠ E(S21) if and only if 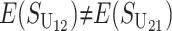 . Because each U DMI is assumed to follow the same distribution of effects, (1) and (3) imply that E(S12) ≠ E(S21) if and only if
. Because each U DMI is assumed to follow the same distribution of effects, (1) and (3) imply that E(S12) ≠ E(S21) if and only if 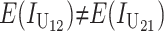 , where
, where 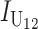 and
and 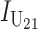 denote the number of U DMIs afflicting the cross or the resulting F1 from the reciprocal combinations. A detailed derivation of the stochastic accumulation of DMIs is presented in appendix a. Here we describe only the assumptions and parameters necessary to explain our biological conclusions. Following the model of Orr (1995) as elaborated by Orr and Turelli (2001), we assume that each pairwise interlocus allelic difference between the diverging lineages can potentially produce a DMI. Each such pair is viewed as an independent “Bernoulli trial.” For pairs that may produce B DMIs, the probability of “success,” i.e., the probability that the difference yields a DMI, is denoted p, as in Orr and Turelli (2001). For pairs of loci that may produce U DMIs, the probability that a pairwise allelic difference yields a DMI is denoted pU. From (3), only pU enters the conditions for E(S12) ≠ E(S21).
denote the number of U DMIs afflicting the cross or the resulting F1 from the reciprocal combinations. A detailed derivation of the stochastic accumulation of DMIs is presented in appendix a. Here we describe only the assumptions and parameters necessary to explain our biological conclusions. Following the model of Orr (1995) as elaborated by Orr and Turelli (2001), we assume that each pairwise interlocus allelic difference between the diverging lineages can potentially produce a DMI. Each such pair is viewed as an independent “Bernoulli trial.” For pairs that may produce B DMIs, the probability of “success,” i.e., the probability that the difference yields a DMI, is denoted p, as in Orr and Turelli (2001). For pairs of loci that may produce U DMIs, the probability that a pairwise allelic difference yields a DMI is denoted pU. From (3), only pU enters the conditions for E(S12) ≠ E(S21).
To calculate the expected number of U DMIs, we must consider substitutions differentiating the diverging lineages at two sets of loci, characterized by whether their DMI effects involve maternally or paternally inherited alleles. Let KU denote the number of substitutions in either lineage at loci that can produce maternally inherited alleles involved in the U DMIs being considered. For X–autosome incompatibilities expressed in males, KU would be the number of X-linked substitutions. For cytonuclear incompatibilities, KU would denote the number of substitutions in organelle genomes; whereas for pollen–style interactions, KU would denote the number of substitutions affecting style function. We assume that KU1 of these substitutions occurred in lineage 1 and KU2 in lineage 2. We assume that the relevant changes are sufficiently rare that all such substitutions have occurred in only one of the two lineages (i.e., only one lineage contains a derived allele at each locus), so that the total number of substitutions differentiating the taxa at these loci is KU = KU1 + KU2. Similarly, we let 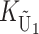 (
(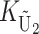 ) denote the number of substitutions in either lineage at loci whose paternally inherited alleles can participate in the F1 U DMIs being considered. For X–autosome incompatibilities expressed in males,
) denote the number of substitutions in either lineage at loci whose paternally inherited alleles can participate in the F1 U DMIs being considered. For X–autosome incompatibilities expressed in males, 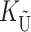 would be the number of autosomal substitutions. For cytonuclear incompatibilities,
would be the number of autosomal substitutions. For cytonuclear incompatibilities, 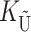 would denote the number of substitutions in the nuclear genomes; whereas for pollen–style interactions,
would denote the number of substitutions in the nuclear genomes; whereas for pollen–style interactions, 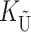 would denote the number of substitutions affecting pollen function. For brevity, we refer to the loci that contribute to KU (
would denote the number of substitutions affecting pollen function. For brevity, we refer to the loci that contribute to KU (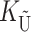 ) as “female-acting” (male-acting) loci.
) as “female-acting” (male-acting) loci.
The expected number of U DMIs experienced by reciprocal crosses can differ because of differences in the rates of molecular evolution of the relevant loci. Although the female-acting and male-acting loci may overlap (for instance, some nuclear loci may contribute to both pollen and style function), we simplify our analyses by assuming that 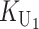 ,
, 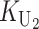 ,
, 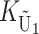 , and
, and 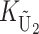 are independent Poisson processes (Orr and Turelli 2001). (Overlap in these sets of loci can be handled more formally by ignoring products of the small parameters p and pU, as discussed below, but the conclusions are unchanged.) The conditions for asymmetry depend on the fraction of the expected number of substitutions of each type in each lineage. Let
are independent Poisson processes (Orr and Turelli 2001). (Overlap in these sets of loci can be handled more formally by ignoring products of the small parameters p and pU, as discussed below, but the conclusions are unchanged.) The conditions for asymmetry depend on the fraction of the expected number of substitutions of each type in each lineage. Let
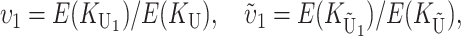 |
(4a) |
and
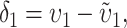 |
(4b) |
so that υ1 (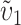 ) is the fraction of the expected female-acting (male-acting) substitutions that occur in lineage 1 and δ1 measures the difference in the relative rates of evolution of these two sets of loci in the taxa being hybridized. In a parental cross with a taxon 1 mother, the expected number of U DMIs conditional on the number of substitutions of each type in each lineage [i.e., conditional on
) is the fraction of the expected female-acting (male-acting) substitutions that occur in lineage 1 and δ1 measures the difference in the relative rates of evolution of these two sets of loci in the taxa being hybridized. In a parental cross with a taxon 1 mother, the expected number of U DMIs conditional on the number of substitutions of each type in each lineage [i.e., conditional on 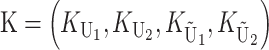 ] is
] is
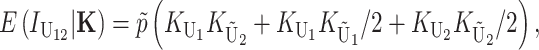 |
(5) |
where the first, second, and third terms in parentheses describe derived–derived, derived–ancestral, and ancestral–derived U interactions, respectively (see appendix a). Using our assumption that the components of K are independent, we have
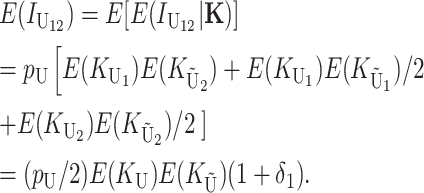 |
(6) |
For the reciprocal cross,
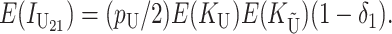 |
(7) |
Equation 5 shows that reciprocal crosses can differ only because of U DMIs between derived alleles in the two lineages. In general, the parameter pU in (6) and (7) as well as the expected effect of each U DMI will differ among alternative types of U incompatibilities. For instance, cytonuclear DMIs involving mismatches between mitochondrial and nuclear loci that disrupt ATP production may have systematically larger effects than typical X–autosome DMIs. The implications of such systematic differences are considered below.
From (6) and (7), we see that the expected number of DMIs differs between reciprocal crosses [i.e., 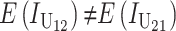 ] only when
] only when
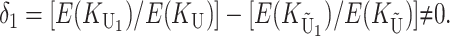 |
(8) |
That is, we expect systematic asymmetries when two diverging lineages show different relative rates of evolution for the female-acting and male-acting loci 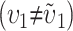 . Note that it is relative rates, not absolute rates, that matter. If taxon 1 evolves uniformly twice as fast as taxon 2 at both sets of loci, we have
. Note that it is relative rates, not absolute rates, that matter. If taxon 1 evolves uniformly twice as fast as taxon 2 at both sets of loci, we have 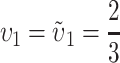 and equal expected breakdown scores from the reciprocal crosses. In contrast, if taxon 1 exhibits a faster relative rate of evolution for female-acting loci than for male-acting loci (i.e., υ1 >
and equal expected breakdown scores from the reciprocal crosses. In contrast, if taxon 1 exhibits a faster relative rate of evolution for female-acting loci than for male-acting loci (i.e., υ1 > 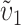 ), crosses using taxon 1 females are expected to produce systematically less-fit F1 (or lower probability of fertilization success) than the reciprocal cross, even if the overall substitution rate for taxon 1 is lower than that for taxon 2 (e.g.,
), crosses using taxon 1 females are expected to produce systematically less-fit F1 (or lower probability of fertilization success) than the reciprocal cross, even if the overall substitution rate for taxon 1 is lower than that for taxon 2 (e.g., 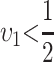 and
and 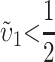 ).
).
Stochastic dynamics of asymmetric sterility/inviability—quantitative asymmetry:
Even without lineage-specific differences in rates of accumulation of the female- vs. male-acting loci [i.e., δ1 = 0 so that E(S12) = E(S21)], reciprocal crosses can produce different hybrid fitnesses, i.e., v(S12) ≠ v(S21), because of chance differences in the numbers and effects of the separate DMIs that contribute to 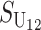 and
and 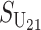 (see Equation 3). To quantify the fitness asymmetry expected under allopatric divergence, we develop a time-dependent probabilistic description of intrinsic postmating isolation by extending the treatment of Orr and Turelli (2001) to U DMIs.
(see Equation 3). To quantify the fitness asymmetry expected under allopatric divergence, we develop a time-dependent probabilistic description of intrinsic postmating isolation by extending the treatment of Orr and Turelli (2001) to U DMIs.
Without any calculations, it is apparent that divergence time, t, must affect asymmetry. For any model of accumulating DMIs, there will be a divergence time, denoted 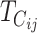 , at which E(Sij) = C, the value that produces complete postmating isolation. As noted above, if δ1 ≠ 0,
, at which E(Sij) = C, the value that produces complete postmating isolation. As noted above, if δ1 ≠ 0, 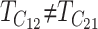 . As shown below, a mathematically convenient reference timescale for postmating asymmetry is the geometric mean
. As shown below, a mathematically convenient reference timescale for postmating asymmetry is the geometric mean
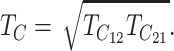 |
(9) |
Divergence time, t, affects asymmetry, because early in divergence (i.e., t ≪ TC) few DMIs have accumulated and we expect v(S12) ≈ v(S21) ≈ 1; whereas after extensive divergence (i.e., t ≫ TC), we expect v(S12) ≈ v(S21) ≈ 0. Thus, asymmetry must be maximal for intermediate values of t (0 < t < TC).
Some recent studies on isolation asymmetry report quantitative differences between the fitnesses of reciprocal F1 (e.g., Tiffin et al. 2001; Bolnick and Near 2005). Moreover, some of those data also show how asymmetry changes with divergence time (e.g., Figure 4 of Bolnick and Near 2005). To make quantitative asymmetry predictions, we need an explicit fitness function, v(S), and a model from which we can derive the time-dependent bivariate distribution of the hybrid breakdown scores (S12, S21). As demonstrated below, the shape of v(S) significantly affects expected levels of asymmetry. To illustrate this, we consider a family of fitness functions that satisfy (2),
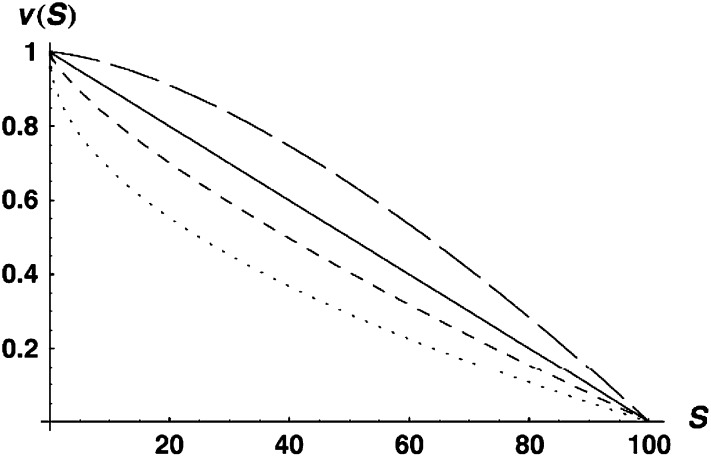
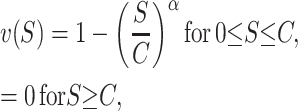 |
(10) |
with α > 0. This function is displayed in Figure 2 for α = 0.5, 0.75, 1, and 1.5. Because S is expected to increase roughly quadratically (Orr 1995; Orr and Turelli 2001) as two-locus DMIs accumulate (and even faster for multilocus DMIs), α = 0.5 would produce a roughly linear decline of hybrid fitness with divergence time, whereas α = 1 and α = 1.5 would produce a roughly quadratic or cubic decline, respectively. Given that meta-analyses reveal at most a slightly faster than linear decline of hybrid fitness with divergence (e.g., Lijtmaer et al. 2003; Bolnick and Near 2005), we focus our numerical examples on an intermediate case, α = 0.75.
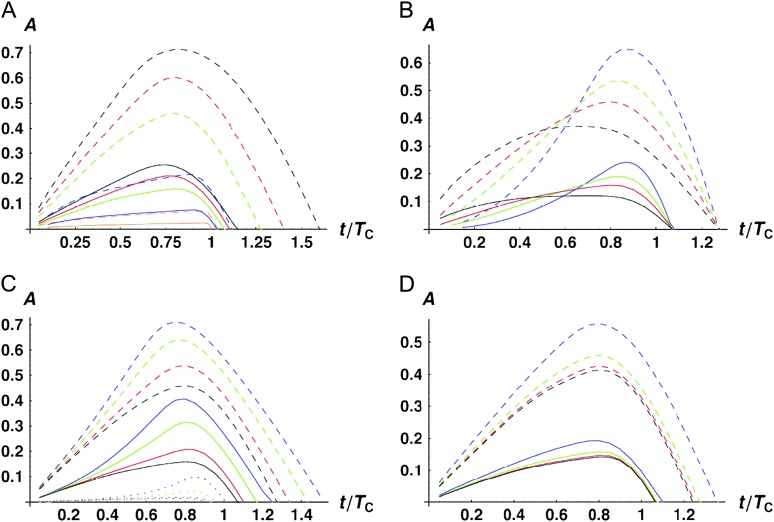
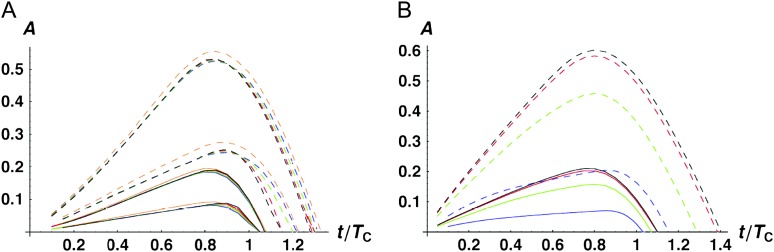
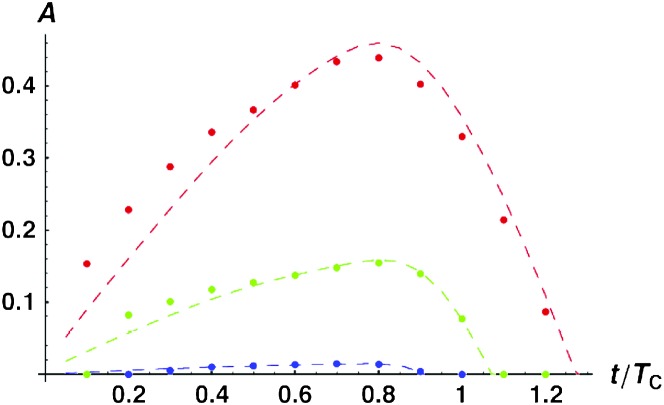
Figure 4.—
Time-dependent medians (solid curves) and 95th percentiles (dashed curves) of asymmetry values A [i.e., P(A ≤ a) = 0.5 vs. 0.95] when only U DMIs contribute to reproductive isolation between lineages. (A) The effects of varying C with α = 0.75, δ1 = 0, and CV = 0.5. The curves are C = 5 (black), 10 (red), 20 (green), 100 (blue), and 1000 (orange). (B) The effects of varying α with C = 20, CV = 0.5, and δ1 = 0. The curves are α = 0.5 (black), 0.75 (red), 1 (green), and 1.5 (blue). (C) The effects of varying δ1 (which controls expected differences between reciprocal breakdown scores) with C = 20, α = 0.75, and CV = 0.5. The curves are δ1 = 0 [E(S12) = E(S21), black], δ1 = 0.166667 (υ1 = 2υ2, red), δ1 = 0.3 (υ1 = 4υ2, green), and δ1 = 0.4 (υ1 = 9υ2, blue). In addition to the median and the 95th percentiles, the 5th percentiles are shown as dotted curves. (D) The effects of varying CV with C = 20, α = 0.75, and δ1 = 0. The curves are CV = 0 (black), 0.25 (red), 0.5 (green), and 1.0 (blue).
Figure 2.—
The fitness function v(S) described by (10) with C = 100 and α = 0.5 (dotted curve), 0.75 (short-dashed curve), 1.0 (solid curve), and 1.5 (long-dashed curve).
To quantify asymmetry, we define
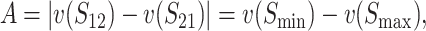 |
(11) |
where Smax = max(S12, S21) and Smin = min(S12, S21). The index A ranges from 0 (no asymmetry) to 1 (complete isolation observed in one cross and none in the reciprocal cross). Its distribution depends on divergence time, which we measure in units of TC, defined in (9). Hence τ = t/TC = 1 corresponds to the time (averaged over the two reciprocal crosses) at which their expected breakdown scores reach C, the value that produces complete postmating isolation.
To determine probable values of A, we need an approximation for the bivariate distribution of (S12, S21). Once that is specified, we can obtain the quantiles of A from the identity
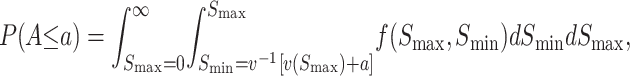 |
(12) |
where f(Smax, Smin) denotes the bivariate distribution of the order statistics (Smax, Smin) and v−1(x) = C(1 − x)1/α for 0 < x < 1, C for x ≤ 0, and 0 for x ≥ 1. Let g(S12, S21) denote the joint distribution of the reciprocal incompatibility scores (S12, S21). Then for Smax ≥ Smin, f(Smax, Smin) = g(Smax, Smin) + g(Smin, Smax). To apply (12), we must approximate g(S12, S21), the time-dependent bivariate distribution of (S12, S21).
Assuming that at least a moderate number of DMIs (on the order of 10) contribute to the incompatibility score, S, Orr and Turelli (2001, Appendix 1) gave a heuristic analytical argument for approximate normality of S. They supported this approximation with numerical simulations of the underlying stochastic processes. We extend this Gaussian approximation to the bivariate distribution of (S12, S21) but must also condition the distribution so that the breakdown scores remain nonnegative. This additional approximation is inconsequential when the means of the breakdown scores are several standard deviations from 0. The adequacy of this approximation for f(Smax, Smin), which involves both truncation and applying a Gaussian approximation even when S is small, is supported by numerical results in appendix b. There we show reasonable agreement between the percentiles of A obtained from (12) and the percentiles obtained from simulating an explicit stochastic model of accumulating DMIs with random effects. Using the (conditional) Gaussian approximation for (S12, S21), we can apply (12) once we have approximated the means, variances, and covariances of S12 and S21. For simplicity, we first assume that all DMIs are asymmetric.
All asymmetric (U) DMIs:
In this case, (1) implies that
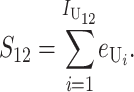 |
(13) |
We can assume without loss of generality that E(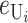 ) = 1, which is equivalent to measuring C in units of the expected number of DMIs required to produce complete postmating isolation. For consistency with the more general calculations below, we assume that Var(
) = 1, which is equivalent to measuring C in units of the expected number of DMIs required to produce complete postmating isolation. For consistency with the more general calculations below, we assume that Var(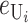 ) = CV2. Even though the
) = CV2. Even though the 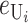 that contribute to S12 and S21 are assumed to be independent, S12 and S21 covary because of their shared dependence on K = (
that contribute to S12 and S21 are assumed to be independent, S12 and S21 covary because of their shared dependence on K = (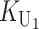 ,
, 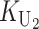 ,
, 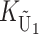 ,
, 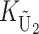 ). However, as shown in appendix c, this covariance is proportional to
). However, as shown in appendix c, this covariance is proportional to 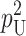 , which is negligible in comparison to the dominant terms in the means and variances, which are proportional to pU. (Orr and Turelli (2001) and Presgraves (2003) estimate p to be <10−5; and even if pU is much larger, it is unlikely to exceed 10−2.) Using expressions (6) and (7) for E(
, which is negligible in comparison to the dominant terms in the means and variances, which are proportional to pU. (Orr and Turelli (2001) and Presgraves (2003) estimate p to be <10−5; and even if pU is much larger, it is unlikely to exceed 10−2.) Using expressions (6) and (7) for E(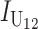 ) and E(
) and E(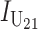 ), our Gaussian approximation for (S12, S21) is completely specified by the moments
), our Gaussian approximation for (S12, S21) is completely specified by the moments
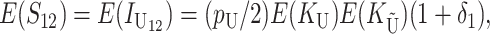 |
(14a) |
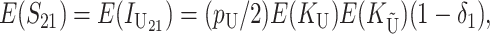 |
(14b) |
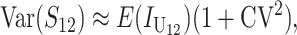 |
(14c) |
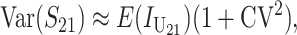 |
(14d) |
and
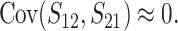 |
(14e) |
The variances and covariance are approximate because they ignore terms proportional to 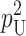 .
.
Our analyses require two additional parameters that describe the overall rate of molecular evolution and how it is apportioned between 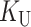 and
and 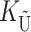 . Let
. Let
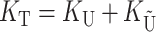 |
(15) |
denote the total number of substitutions. For each of the four independent Poisson processes, 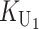 ,
, 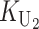 ,
,
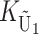 , and
, and 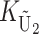 , we denote the corresponding rate parameter by kX, with X = U1,U2, etc. So, for instance, after a divergence time of t,
, we denote the corresponding rate parameter by kX, with X = U1,U2, etc. So, for instance, after a divergence time of t,
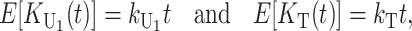 |
(16) |
where 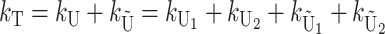 . In terms of these parameters, we have υ1 =
. In terms of these parameters, we have υ1 = 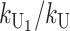 and
and 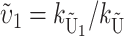 . We introduce the new parameter
. We introduce the new parameter
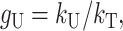 |
(17) |
which is the fraction of substitutions relevant to U DMIs that occur at female-acting loci. For example, for nuclear–cytoplasmic (or X–autosome) interactions, gU describes the fraction of cytoplasmic (or X chromosome) substitutions that contribute to U DMIs. Substituting into (14a), we obtain
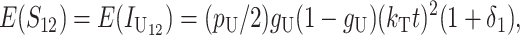 |
(18a) |
which implies that
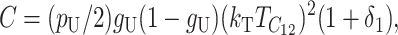 |
(18b) |
by the definition of 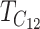 . The other moments can be expressed similarly as explicit functions of the parameters and divergence time.
. The other moments can be expressed similarly as explicit functions of the parameters and divergence time.
Equation 18b implies that if we measure time in units of 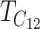 by setting
by setting 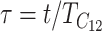 , at time
, at time 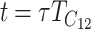 , E(S12) = τ2C. This scaling leads to a major simplification of the expressions for the first- and second-order moments of (S12, S21), which demonstrates that the levels of asymmetry expected in the F1 generation depend on C, α, CV, and δ1 but do not depend on the values of
, E(S12) = τ2C. This scaling leads to a major simplification of the expressions for the first- and second-order moments of (S12, S21), which demonstrates that the levels of asymmetry expected in the F1 generation depend on C, α, CV, and δ1 but do not depend on the values of 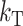 , gU, and pU. This is easiest to see when δ1 = 0, so that E(S12) = E(S21) and
, gU, and pU. This is easiest to see when δ1 = 0, so that E(S12) = E(S21) and 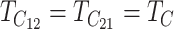 . In this case, when t/TC = τ, (14) and (18) imply that E(S12) = E(S21) = τ2C, Var(S12) = Var(S21) ≈
. In this case, when t/TC = τ, (14) and (18) imply that E(S12) = E(S21) = τ2C, Var(S12) = Var(S21) ≈ 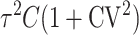 , and Cov(S12, S21) ≈ 0, irrespective of pU, gU, and
, and Cov(S12, S21) ≈ 0, irrespective of pU, gU, and 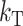 . In contrast, the cumulative distribution defined by (12) clearly depends on C, which is proportional to the means and variances, CV, which inflates the variances, and the shape of v(S). If δ1 ≠ 0, it also affects the results. When t/TC = τ, (14) and (18) imply
. In contrast, the cumulative distribution defined by (12) clearly depends on C, which is proportional to the means and variances, CV, which inflates the variances, and the shape of v(S). If δ1 ≠ 0, it also affects the results. When t/TC = τ, (14) and (18) imply
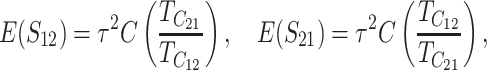 |
(19a) |
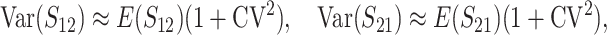 |
(19b) |
and
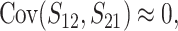 |
(19c) |
where
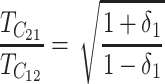 |
(20) |
depends only on δ1. Hence, we see that only C (the number of DMIs required to produce complete postmating isolation), α (the shape of the fitness function, Equation 10), CV (the coefficient of variation of DMI effects), and δ1 [the parameter that determines whether E(S12) = E(S21)] can affect asymmetry when only U DMIs act.
Numerical results:
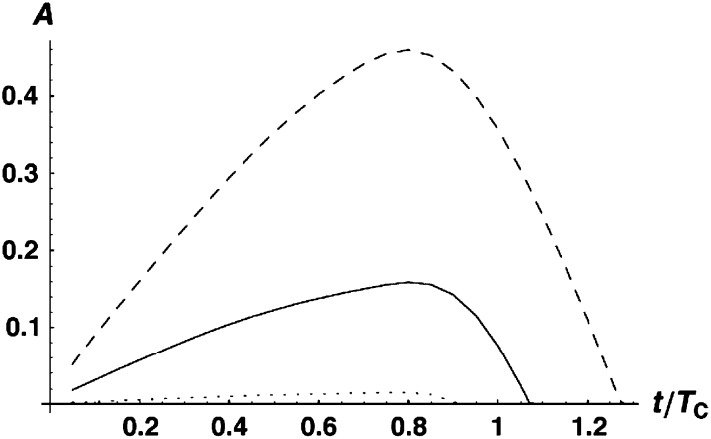
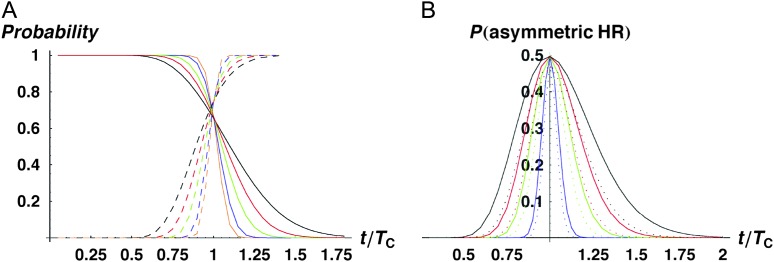
To understand the levels of asymmetry expected as the parameters vary, we used (12) to approximate the quantiles of A by numerically solving the equation P(A ≤ a) = P for a at various values of P, such as 0.05, 0.5, and 0.95. This was done for a range of times, resulting in plots that display expected levels of asymmetry as a function of t/TC. The numerical analysis was performed in Mathematica 5.2 (Wolfram 2003). As noted above, the quantiles of A depend only on C, α, CV, and δ1. Figure 3 illustrates the time-dependent quantiles with C = 20, α = 0.75, CV = 0.5, and δ1 = 0. Note that for these parameters, maximal asymmetry is observed near 0.8TC, and the distribution of A tends to be quite broad (at each time, the dotted curves define the 90% confidence interval for A). Because the 5th percentile is generally very near zero, it is uninformative and is not displayed in most of the figures below, except when δ1 ≠ 0 produces nonnegligible values. When the lower quantile is not shown, asymmetry values indistinguishable from zero even in large experiments (e.g., A ≤ 0.05) would generally be statistically consistent with the parameter values considered.
Figure 3.—
Time-dependent quantiles of A, our measure of quantitative asymmetry defined in Equation 11 [i.e., P(A ≤ a) = P] for C = 20, α = 0.75, δ1 = 0, and CV = 0.5, with P = 0.05 (dotted curve), 0.5 (solid curve), and 0.95 (dashed curve).
Figure 4, A–D, shows how the percentiles of A change as C, α, δ1, and CV vary around base values of C = 20, α = 0.75, δ1 = 0, and CV = 0.5. Figure 4A shows that C has a major effect on the quantiles, with lower asymmetry expected as C increases. This supports Muller's (1942) intuition that greater asymmetry is expected when fewer DMIs are required to produce complete postmating isolation. However, even when C = 100, moderate levels of asymmetry are produced, with the 95th percentile of A near 0.2 for t/TC between ∼0.65 and 0.95. Likely levels of asymmetry are roughly doubled when C is reduced to 20 and roughly tripled (relative to C = 100) when C = 10. Figure 4B addresses the robustness of this pattern to different shapes of the fitness function v(S), with C = 20. As α decreases from 0.75 to 0.5 (which produces a roughly linear decrease of hybrid fitness with divergence time), maximal asymmetry is reduced but significant asymmetry is seen over a larger range of divergence times. The intuitive explanation is that if fitness declines more quickly initially, stochastic differences in breakdown scores lead to significant asymmetry more quickly. Conversely, as α increases from 0.75 to 1 (which produces a roughly quadratic decline of hybrid fitness with time) or 1.5 (roughly cubic decline), maximal asymmetry increases sharply and is markedly peaked for t/TC near 1. The areas under these curves (corresponding to the average asymmetry) are far more consistent for different values of α than the maxima. Comparing the most extreme cases, the area under the 95th percentile curve is ∼0.33 for α = 0.5 vs. 0.39 for α = 1.5.
Figure 4C examines the effect of unequal relative rates of evolution that produce E(S12) ≠ E(S21). Holding C = 20, α = 0.75, CV = 0.5, and 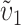 = 0.5, we increased υ1 from 0.5 (δ1 = 0) until the rate of evolution of the female-acting loci in taxon 1 is nine times that in taxon 2 (δ1 = 0.4). The qualitative result is that even when taxon 1 evolves twice as fast as taxon 2 at the female-acting loci, so that δ1 = 0.166667, probable levels of asymmetry rise only slightly—and primarily for t/TC near 1. However, extreme relative rate differences, producing δ1 ≥ 0.3, have an appreciable effect on asymmetry. This is most clearly expressed by the fact that the 5th percentile of A rises to near 0.1 for t/TC between 0.8 and 0.9. These analyses suggest that unless relative rate differences are extreme (e.g., female-acting loci evolve at least four times as fast in one lineage as in the other, while male-acting loci evolve at similar rates in both lineages), stochastic effects are more likely to explain observed levels of postmating asymmetry than systematic interspecific differences in the relative rates of molecular evolution.
= 0.5, we increased υ1 from 0.5 (δ1 = 0) until the rate of evolution of the female-acting loci in taxon 1 is nine times that in taxon 2 (δ1 = 0.4). The qualitative result is that even when taxon 1 evolves twice as fast as taxon 2 at the female-acting loci, so that δ1 = 0.166667, probable levels of asymmetry rise only slightly—and primarily for t/TC near 1. However, extreme relative rate differences, producing δ1 ≥ 0.3, have an appreciable effect on asymmetry. This is most clearly expressed by the fact that the 5th percentile of A rises to near 0.1 for t/TC between 0.8 and 0.9. These analyses suggest that unless relative rate differences are extreme (e.g., female-acting loci evolve at least four times as fast in one lineage as in the other, while male-acting loci evolve at similar rates in both lineages), stochastic effects are more likely to explain observed levels of postmating asymmetry than systematic interspecific differences in the relative rates of molecular evolution.
Figure 4D considers the influence of varying the fitness effects of individual DMIs. It shows that, as expected, asymmetry increases as CV increases, corresponding to increasing variance among the effects of individual DMIs. This qualitative effect is easy to understand because as CV increases, the variances of the breakdown scores increase while their means remain fixed. However, values of CV up to 0.5 have relatively little effect on probable asymmetry values.
Both asymmetric (U) and symmetric (B) DMIs:
More generally, hybrid fitness is determined by both B and U DMIs (see Table 1), and we must consider their relative contributions to the hybrid breakdown score. In Turelli and Orr's (2000) analysis of two-locus X–X, X–autosome, and autosome–autosome DMIs, they described three categories of two-locus DMIs. In increasing order of expected severity, they are: H0 DMIs that involve heterozygous incompatible alleles at both loci, H1 DMIs that involve heterozygous alleles at one locus and either a homozygous or a hemizygous incompatible allele at a second locus, and H2 incompatibilities in which the incompatible alleles at both loci are either hemizygous or homozygous. In general, hemizygosity and homozygosity may lead to different distributions of effects; but we restrict our F1 analyses to a single class of U DMIs and hence a single parameter will suffice to describe the relative effects of the U DMIs (which are either H1 or H2) and the B DMIs (which are all H0). The three classes of DMIs are assumed to contribute on average h0, h1, and h2, respectively, to the hybrid breakdown score, S, with h0 < h1 < h2.
All B DMIs in the F1 involve interactions between heterozygous nuclear loci. Hence, all are H0 incompatibilities, each of which contributes h0 on average to both S12 and S21 [see (1) and (3)]. In contrast, the U DMIs we consider may be either type H1 (e.g., X–autosome in the heterogametic sex, cytonuclear, or maternal effects) or type H2 (e.g., triploid endosperm). To unify our notation and facilitate comparison with the results in the previous section, we assume that the U DMIs have average effect 1, whether they are type H1 or H2, whereas we denote the average effect of the B DMIs by h0 < 1. With this simplification, we can model the U DMI contributions to S12 and S21 by using the approximations described in the previous section. Similarly, we can model the B DMI contributions using the framework developed in Orr and Turelli (2001). Overall, we have
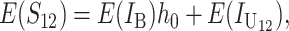 |
(21) |
where (18) provides an explicit time-dependent expression for E(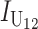 ). A comparable expression for E(IB) is given by Equation 6 of Orr and Turelli (2001). If we let p denote the probability that an allelic difference at two B loci leads to a DMI, the expected number of B DMIs in an F1 is
). A comparable expression for E(IB) is given by Equation 6 of Orr and Turelli (2001). If we let p denote the probability that an allelic difference at two B loci leads to a DMI, the expected number of B DMIs in an F1 is
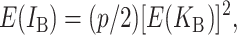 |
(22) |
where E(KB) denotes the number of substitutions at nuclear loci (summed across both lineages) contributing to B DMIs. As in (16), we have 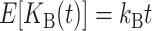 . Unlike (6) and (7) for U DMIs, this result is independent of the expected fraction of these substitutions that occur in each lineage (Orr 1995; appendix a).
. Unlike (6) and (7) for U DMIs, this result is independent of the expected fraction of these substitutions that occur in each lineage (Orr 1995; appendix a).
To apply our bivariate Gaussian approximation and identity (12) for determining the quantiles of the asymmetry index A, we must first clarify the relationship between KB and the stochastic process K = (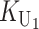 ,
, 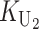 ,
, 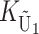 ,
, 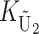 ) that entered our analysis of U DMIs and then approximate the variances and covariance of S12 and S21. From Table 1, we see that for the cases we consider, the loci contributing to KB are either all nuclear loci or, in the case of X–autosome U DMIs, all autosomal loci. In these cases, the loci that potentially contribute to KB are identical with the loci that potentially contribute to
) that entered our analysis of U DMIs and then approximate the variances and covariance of S12 and S21. From Table 1, we see that for the cases we consider, the loci contributing to KB are either all nuclear loci or, in the case of X–autosome U DMIs, all autosomal loci. In these cases, the loci that potentially contribute to KB are identical with the loci that potentially contribute to 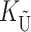 (namely the male-acting loci in U DMIs). Hence, the parameter gU =
(namely the male-acting loci in U DMIs). Hence, the parameter gU = 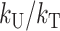 introduced in (17) also can be used to differentiate the loci contributing to U vs. B DMIs (with gB = 1 − gU). In particular, for X–autosome DMIs in heterogametic males, gU is the parameter gX of Turelli and Orr (2000), the fraction of nuclear substitutions that are X-linked. For cytonuclear interactions, gU describes the fraction of substitutions that occur in the relevant cytoplasmic organelles. When TRE incompatibilities interact with zygotic incompatibilities to determine seed development, all nuclear DMIs in the F1 (which are B) can act effectively simultaneously with TRE incompatibilities (which are U). In contrast, the interaction between maternal effects and the zygotic genome is much less clear cut, because the relevant zygotic loci are those expressed earliest in development. These could well be only a very small subset of the nuclear loci. However, in our idealized model of DMI origins, the zygotic incompatibilities that manifest as development moves from maternal to zygotic control can potentially involve the entire nuclear genome. Hence, in the case of simultaneous action of maternal-effect DMIs and zygotic DMIs, it seems unlikely that a single parameter gU can capture both the relative role of male-acting loci involved in maternal–zygotic U DMIs and the relative role of zygotic B DMIs affecting early embryos (because the latter could involve a much smaller subset of loci). As discussed below, this complication can be accommodated by suitable interpretation of composite parameters that emerge in our analysis.
introduced in (17) also can be used to differentiate the loci contributing to U vs. B DMIs (with gB = 1 − gU). In particular, for X–autosome DMIs in heterogametic males, gU is the parameter gX of Turelli and Orr (2000), the fraction of nuclear substitutions that are X-linked. For cytonuclear interactions, gU describes the fraction of substitutions that occur in the relevant cytoplasmic organelles. When TRE incompatibilities interact with zygotic incompatibilities to determine seed development, all nuclear DMIs in the F1 (which are B) can act effectively simultaneously with TRE incompatibilities (which are U). In contrast, the interaction between maternal effects and the zygotic genome is much less clear cut, because the relevant zygotic loci are those expressed earliest in development. These could well be only a very small subset of the nuclear loci. However, in our idealized model of DMI origins, the zygotic incompatibilities that manifest as development moves from maternal to zygotic control can potentially involve the entire nuclear genome. Hence, in the case of simultaneous action of maternal-effect DMIs and zygotic DMIs, it seems unlikely that a single parameter gU can capture both the relative role of male-acting loci involved in maternal–zygotic U DMIs and the relative role of zygotic B DMIs affecting early embryos (because the latter could involve a much smaller subset of loci). As discussed below, this complication can be accommodated by suitable interpretation of composite parameters that emerge in our analysis.
Given normality, the distribution of (S12, S21) depends only on their means, variances, and covariance. These are computed in appendix d, using the simplifying assumption that all of the random variables describing DMI effects have equal coefficients of variation (CV); i.e., Var(eU)/[E(eU)]2 = Var(eB)/[E(eB)]2 = CV2, where E(eU) = 1 and E(eB) = h0. To understand how asymmetry depends on the parameters, it is useful to express the moments given in (D2), (D5), and (D7) as functions of the scaled time, τ = t/TC, with TC defined by (9). After some simplification, we obtain
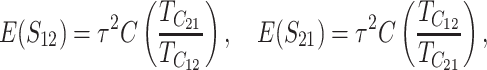 |
(23a) |
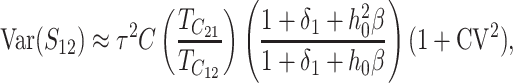 |
(23b) |
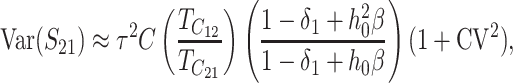 |
(23c) |
and
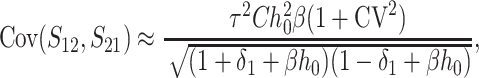 |
(23d) |
where
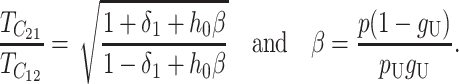 |
(24) |
Thus, asymmetry is independent of 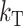 but does depend on C, CV, δ1, v(S) [i.e., α in (10)], and h0; and it depends on p, pU, and gU only through the ratio β defined in (24). Multiplying the numerator and denominator of β by (1 − gU), we see that β is the ratio of the expected number of B DMIs to the expected number of U DMIs, providing a straightforward biological interpretation of this parameter. β can also be expressed as pkB/(pUkU).
but does depend on C, CV, δ1, v(S) [i.e., α in (10)], and h0; and it depends on p, pU, and gU only through the ratio β defined in (24). Multiplying the numerator and denominator of β by (1 − gU), we see that β is the ratio of the expected number of B DMIs to the expected number of U DMIs, providing a straightforward biological interpretation of this parameter. β can also be expressed as pkB/(pUkU).
When the term 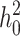 β makes a negligible contribution to the variances and covariance, the implications of analyzing both B and U DMIs become much clearer. Setting
β makes a negligible contribution to the variances and covariance, the implications of analyzing both B and U DMIs become much clearer. Setting 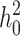 β = 0 in (23), we obtain
β = 0 in (23), we obtain
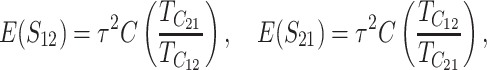 |
(25a) |
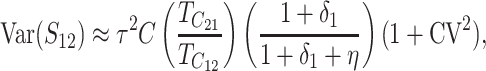 |
(25b) |
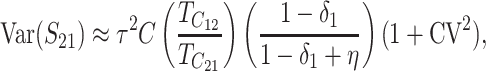 |
(25c) |
and
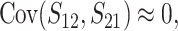 |
(25d) |
with
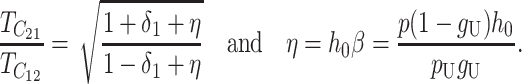 |
(26) |
Hence when 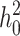 β is negligible (the relevant range of parameters is discussed below), four parameters that are likely to be very difficult to estimate individually, namely p, pU, h0 and gU, enter only as a single composite parameter η defined by (26). Like β, η has a simple biological interpretation. It is the ratio of the expected contribution to the hybrid breakdown scores from B DMIs (which have average effect h0) to the expected contribution from U DMIs (which have average effect 1). This interpretation of η simplifies our analysis of maternal effects for which we have little data to guide our choice of gU.
β is negligible (the relevant range of parameters is discussed below), four parameters that are likely to be very difficult to estimate individually, namely p, pU, h0 and gU, enter only as a single composite parameter η defined by (26). Like β, η has a simple biological interpretation. It is the ratio of the expected contribution to the hybrid breakdown scores from B DMIs (which have average effect h0) to the expected contribution from U DMIs (which have average effect 1). This interpretation of η simplifies our analysis of maternal effects for which we have little data to guide our choice of gU.
Comparing (25) with the expressions that arise with only U DMIs (19), we see that for fixed values of C, CV, and δ1, the reciprocal breakdown scores remain approximately uncorrelated, their means are brought closer to one another by the appearance of the positive term η in the numerator and denominator of the ratio in 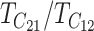 , and their variances are reduced by the appearance of η in the denominators of (25b) and (25c). Hence, as expected, incorporating both B DMIs and U DMIs systematically reduces asymmetry. The same result emerges from the more general approximations (23), which also have a positive covariance contributing to reduced asymmetry. A simple biological interpretation is that when both U and B DMIs act, the U DMIs, which are solely responsible for reproductive asymmetry, account for a smaller fraction of the total isolation. Hence, less asymmetry is expected.
, and their variances are reduced by the appearance of η in the denominators of (25b) and (25c). Hence, as expected, incorporating both B DMIs and U DMIs systematically reduces asymmetry. The same result emerges from the more general approximations (23), which also have a positive covariance contributing to reduced asymmetry. A simple biological interpretation is that when both U and B DMIs act, the U DMIs, which are solely responsible for reproductive asymmetry, account for a smaller fraction of the total isolation. Hence, less asymmetry is expected.
Numerical results:
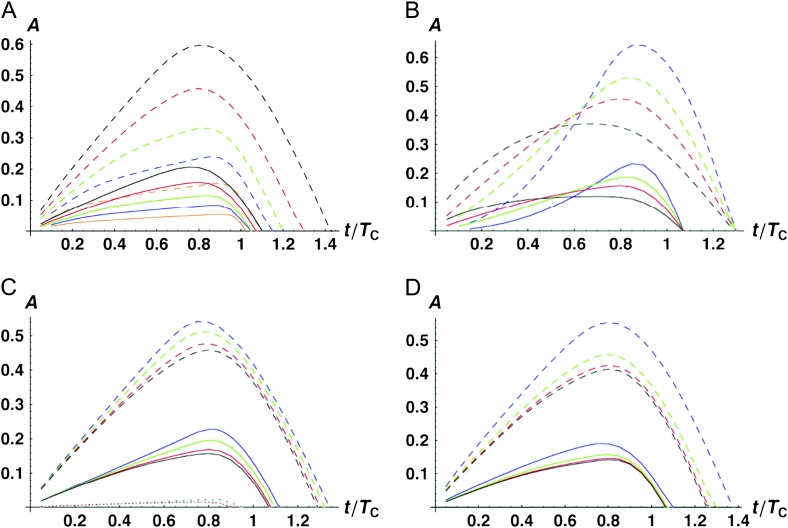
As in the case with only U DMIs, we analyze the levels of asymmetry expected as the parameters vary by using (12) to approximate the quantiles of A. We first consider the range of parameters under which the simpler moment approximations (25), which depend only on η rather than on β and h0 separately (as in 23), are adequate for describing probable values of asymmetry. Figure 5A considers the effect of varying h0 from 0.05 to 0.4 when η is held fixed at either 1 (corresponding to equal average contributions of B and U DMIs to postmating isolation) or 10 (corresponding to 10-fold greater contribution from B DMIs than from U DMIs). As expected, asymmetry is much greater when η is smaller. Less obvious is how little influence h0 has once η is known. As Figure 5A shows, for η = 10, h0 has no appreciable effect on asymmetry until h0 = 0.4, which we consider implausibly high (as discussed below). When η = 1, the value of h0 is essentially irrelevant, even up to h0 = 0.4. Our qualitative conclusion is that although h0 can have a major effect on asymmetry through its role in determining η, we can understand this effect by varying η rather than varying β and h0 separately. Hence, Figure 5B explores the role of varying η while holding h0 fixed at 0.1. [Note that in Figure 5B and all subsequent figures, we use the full approximation (23) for the moments, but hold h0 = 0.1 as we vary η.] It shows that for these parameter values, η on the order of 0.1 produces levels of asymmetry comparable to those seen with only U DMIs (η = 0). Conversely, once η is as large as 10, relatively little asymmetry is expected (but it is likely to still be detectable in moderate-sized experiments when t/TC is near 1).
Figure 5.—
Effects of combining symmetric (B) and asymmetric (U) DMIs on the time-dependent asymmetry values, A. The solid lines are medians, the dashed lines are 95th percentiles. (A) The effect of varying the dominance parameter h0, while holding fixed the parameter η, defined in (26), which quantifies the relative contribution of B vs. U DMIs to hybrid dysfunction. Two sets of results are provided: η = 1 (top) and η = 10 (bottom). The curves are h0 = 0.05 (black), 0.1 (red), 0.2 (green), 0.3 (blue), and 0.4 (orange). The other parameters are C = 10, α = 0.75, δ1 = 0, and CV = 0.5. (B) The effect of varying the mix of symmetric (B) vs. asymmetric (U) DMIs, as measured by η. The curves are η = 0 (all U, black), η = 0.1 (red), η = 1 (green), and η = 10 (blue). The other parameters are h0 = 0.1, C = 10, α = 0.75, δ1 = 0, and CV = 0.5.
Figure 6, A–D, shows how the percentiles of A change as C, α, δ1, and CV vary around base values of C = 10, α = 0.75, δ1 = 0, and CV = 0.5, while holding η = 1 and h0 = 0.1. Figure 6A, like Figure 4A, shows that C has a major effect on the quantiles, with lower asymmetry expected as C increases. However, even when C = 40, detectable levels of asymmetry are produced. High levels of asymmetry are probable when C is on the order of 5 or 10. It is important to realize that C has a very different interpretation in Figure 6A than in Figure 4A. In Figure 4A, C is simply the average number of U DMIs needed to produce complete postmating isolation. When η = 1, we expect U and B DMIs to make equal average contributions to hybrid dysfunction, with each U DMI having average effect 1 and each B DMI having average effect h0. Thus, when C = 10, η = 1, and h0 = 0.1, we expect that complete postmating isolation will be produced by 55 DMIs on average, with 5 U DMIs and 50 B DMIs. These numbers are halved when C = 5. Thus, even when C = 5, “simple” genetics would not underlie complete postmating isolation; yet significant asymmetry would arise by chance without systematic differences producing E(S12) ≠ E(S21). On the other hand, a small number of DMIs (namely the U DMIs) would account for an appreciable fraction of postmating isolation. This can be viewed as a generalization of the large X effect for heterogametic hybrids, in which X-linked DMIs (which would be U DMIs) contribute disproportionately to hybrid inviability/sterility.
Figure 6.—
Time-dependent medians (solid curves) and 95th percentiles (dashed curves) for the asymmetry index A [i.e., P(A ≤ a) = 0.5 vs. 0.95] when both U and B DMIs contribute to reproductive isolation between lineages. (A) The effects of varying C with η = 1, h0 = 0.1, α = 0.75, δ1 = 0, and CV = 0.5. The curves are C = 5 (black), 10 (red), 20 (green), 40 (blue), and 100 (orange). (B) The effects of varying α, the shape of the fitness function, with C =10, η = 1, h0 = 0.1, δ1 = 0, and CV = 0.5. The curves are α = 0.5 (black), 0.75 (red), 1 (green), and 1.5 (blue). (C) The effects of varying δ1 (which controls expected differences between reciprocal breakdown scores) with C =10, η = 1, h0 = 0.1, α = 0.75, and CV = 0.5. The curves are δ1 = 0 [E(S12) = E(S21), black], δ1 = 0.166667 (υ1 = 2υ2, red), δ1 = 0.3 (υ1 = 4υ2, green), and δ1 = 0.4 (υ1 = 9υ2, blue). In addition to the median and the 95th percentiles, the 5th percentiles are shown as dotted curves. (D) The effects of varying CV with C = 20, α = 0.75, and δ1 = 0. The curves are CV = 0 (black), 0.25 (red), 0.5 (green), and 1.0 (blue).
Figure 6B looks at the effects of varying α, with C = 10, η = 1, h0 = 0.1, δ1 = 0, and CV = 0.5. The results are very similar to those in Figure 4B. As α increases, more extreme asymmetry is produced for t/TC near 1, but lower asymmetry is expected at lower (earlier) divergence times. Like Figure 4C, Figure 6C examines the effect of unequal relative rates of evolution that produce E(S12) ≠ E(S21). Holding C = 10, η = 1, h0 = 0.1, α = 0.75, CV = 0.5, and 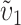 = 0.5, we increased υ1 from 0.5 (δ1 = 0) until the rate of evolution of the female-acting loci in taxon 1 is nine times that in taxon 2 (δ1 = 0.4). The qualitative result is that, in contrast to the case with all U DMIs (η = 0), even when taxon 1 evolves nine times as fast as taxon 2 at the female-acting loci, so that δ1 = 0.4, probable levels of asymmetry rise only slightly. These analyses suggest that when both U and B DMIs act, stochastic effects are more likely to explain postmating asymmetry than systematic interspecific differences in the relative rates of molecular evolution.
= 0.5, we increased υ1 from 0.5 (δ1 = 0) until the rate of evolution of the female-acting loci in taxon 1 is nine times that in taxon 2 (δ1 = 0.4). The qualitative result is that, in contrast to the case with all U DMIs (η = 0), even when taxon 1 evolves nine times as fast as taxon 2 at the female-acting loci, so that δ1 = 0.4, probable levels of asymmetry rise only slightly. These analyses suggest that when both U and B DMIs act, stochastic effects are more likely to explain postmating asymmetry than systematic interspecific differences in the relative rates of molecular evolution.
Figure 6D considers the influence of varying the fitness effects of individual DMIs. It shows that, as expected, asymmetry increases as CV increases, corresponding to increasing variance among the effects of individual DMIs. This qualitative effect is easy to understand because as CV increases, the variances of the breakdown scores increase while their means remain fixed. However, values of CV up to 0.5 have relatively little effect. The implications of these numerical results are discussed further when we discuss our predictions in light of estimates of the critical parameters and observed patterns of asymmetry.
Stochastic dynamics of asymmetric sterility/inviability— qualitative asymmetry:
Muller (1942, p. 101) argued that the extent of asymmetry in the viability and fertility of Drosophila F1 males derived from reciprocal crosses suggested that relatively few DMIs must be involved in postzygotic isolation. Muller (1942) provided no quantitative data on asymmetry, but Turelli and Orr (1995) estimated that ∼15% of the cases of Haldane's rule in Drosophila show qualitative asymmetry, with males being sterile or inviable in one cross but not in the reciprocal cross. In the context of our parameterization, Muller's (1942) conjecture depends on the values of the threshold C and the parameter η that measures the relative contribution of B vs. U DMIs that contribute to postzygotic isolation (and hence dominance). Turelli and Orr (1995) discussed numerical simulations that seemed to refute Muller's conjecture. However, our analytical approximation for the bivariate distribution of the hybrid breakdown scores permits a more thorough analysis.
A complete treatment of qualitative asymmetry for Haldane's rule must simultaneously treat male and female hybrid-breakdown scores. However, motivated by Coyne and Orr's (1989a, 1997) Drosophila data, which show a long time lag between the onset of male vs. female sterility/inviability, we present a simplified analysis that considers only the reciprocal male incompatibility scores. Thus, for any fixed divergence time, we approximate the probability of observing an asymmetric example of Haldane's rule as the probability that Smin < C conditional on Smax > C, (i.e., isolation is complete in one direction of the cross but only partial isolation in the other direction, conditioned on Haldane's rule being observed). This probability is
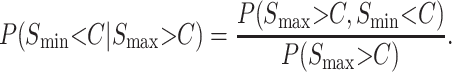 |
(27) |
The qualitative behavior of (27) can be understood without calculations. Relatively early in divergence, when t ≪ TC, this probability must be one (because if there are any U DMIs, one of the two reciprocal scores must reach C first); while late in divergence, when 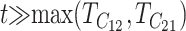 , this probability must be zero. Hence, detailed models can produce only the precise shape of the decline. The problem with using this conditional probability to examine Muller's conjecture is that it fails to weight the conditional probabilities by the probability that complete postmating isolation is observed in a least one of the two reciprocal crosses. If we knew the distribution of divergence times at which Haldane's rule was observed in either reciprocal cross, we could calculate the probability of observing qualitative asymmetry by averaging the time-dependent probability (27) over the distribution of observation times.
, this probability must be zero. Hence, detailed models can produce only the precise shape of the decline. The problem with using this conditional probability to examine Muller's conjecture is that it fails to weight the conditional probabilities by the probability that complete postmating isolation is observed in a least one of the two reciprocal crosses. If we knew the distribution of divergence times at which Haldane's rule was observed in either reciprocal cross, we could calculate the probability of observing qualitative asymmetry by averaging the time-dependent probability (27) over the distribution of observation times.
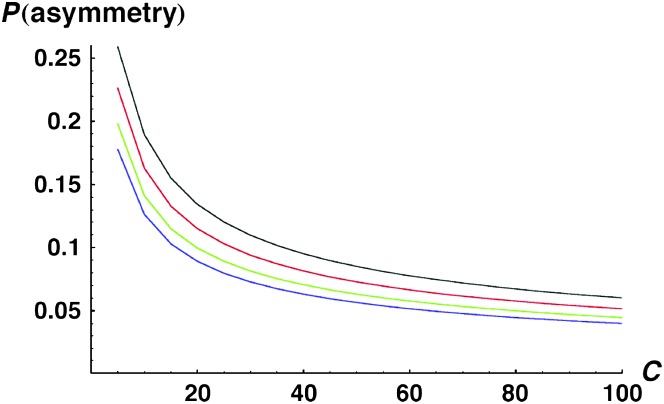
Without information on the distribution of observation times, we can understand more about the probability of observing qualitative asymmetry by examining the numerator and denominator of (27) separately, i.e., calculating
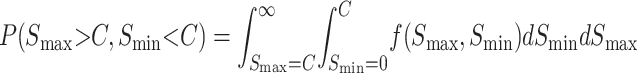 |
(28) |
and
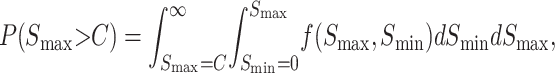 |
(29) |
where, as in (12), f(Smax, Smin) denotes the bivariate distribution of the order statistics (Smax, Smin) derived from the reciprocal incompatibility scores (S12, S21). These integrals were evaluated using Mathematica 5.2. Expression (28) is probably most informative because it describes the probability of observing qualitative asymmetry at any specific divergence time. In particular, if we postulate that hybridizations are observed at divergence times uniformly distributed between T1 and T2, we can approximate the frequency of qualitative asymmetry as
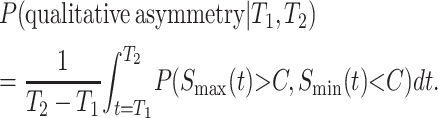 |
(30) |
Given that Muller's (1942) conjecture was motivated by Drosophila data, we focus on this case. At least four sources of asymmetry are relevant: X-linked incompatibilities, Y-linked incompatibilities, cytonuclear incompatibilities, and maternal effects. As discussed in Turelli and Orr (2000), all four have been repeatedly demonstrated. Hence, we ask what parameter values would allow these U DMIs to account for the observed frequency of quantitative asymmetry. First consider the simplest case of X–autosome incompatibilities. For most Drosophila, ∼20% of the genome is X-linked (for the minority, the X chromosome includes ∼40% of the nuclear genome). Assuming roughly equal rates of X and autosome evolution, we have gU = 0.2. If the large-X effect in Drosophila males is attributable to dominance (Turelli and Orr 2000), we expect p = pU. With these assumptions, η = 4h0 (in contrast, for “large-X” species, with 40% of the nuclear genome X-linked, we expect η = 3h0/2). Hence, for h0 between 0.05 and 0.4, we expect η between 0.2 and 1.6 (for large-X species, the corresponding range is 0.075–0.6). “Faster-X” evolution (Charlesworth et al. 1987) or an inherent tendency of the X to accumulate male-specific DMIs (i.e., pU > p) would simply lower η.
The X–autosome and autosome–autosome DMIs discussed above will generally act in conjunction with X–Y and cytonuclear DMIs and will possibly also interact with maternal effects. Although the sizes of the coding regions on the Y and mitochondria are very small relative to the autosomes, we know that DMIs involving the Y and mtDNA can have large effects on hybrid inviability/sterility. These DMIs, as well as maternal effects, will effectively act to decrease η, making U DMIs relatively more important in producing postmating isolation and increasing the likelihood of qualitative asymmetry. We illustrate this below with some plausible parameter values.
Numerical results:
From our analyses of quantitative asymmetry, we expect that the likelihood of qualitative asymmetry will be largely determined by C and η. We explore the consequences of varying both below. Figure 7A shows how the monotone functions of divergence time, 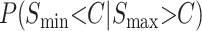 and
and 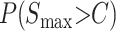 , change with C assuming that η = 0.5. As expected, as C increases from 5 to 100, these curves become increasingly steep, showing that the transition from neither reciprocal cross producing complete isolation to both producing complete isolation becomes increasingly deterministic. Figure 7B shows how the dependence of
, change with C assuming that η = 0.5. As expected, as C increases from 5 to 100, these curves become increasingly steep, showing that the transition from neither reciprocal cross producing complete isolation to both producing complete isolation becomes increasingly deterministic. Figure 7B shows how the dependence of 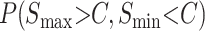 on divergence time changes with both C and η. The most striking feature of this graph is the similarity of the curves corresponding to η = 0.1 and η = 1.0, suggesting that for a wide range of plausible values of η, the probability of observing asymmetry depends primarily on C, unless C is fairly small (on the order of ≤10). This point is made more explicitly in Figure 8, which uses (30) to approximate the probability of observing asymmetry as a function of C, with η varying from 0.1 to 1.5, assuming that hybridization data are uniformly distributed between divergence times T1 = 0.5TC and T2 = 1.5TC. The conclusion is that a 15% frequency of asymmetric Haldane's rule (as seen in Drosophila) is consistent with a value of C as high as 25 if η = 0.1 or a value of C near 10 if η is as large as 1.5 (which seems implausible). We discuss this further below, after reviewing the data concerning the dominance parameter h0.
on divergence time changes with both C and η. The most striking feature of this graph is the similarity of the curves corresponding to η = 0.1 and η = 1.0, suggesting that for a wide range of plausible values of η, the probability of observing asymmetry depends primarily on C, unless C is fairly small (on the order of ≤10). This point is made more explicitly in Figure 8, which uses (30) to approximate the probability of observing asymmetry as a function of C, with η varying from 0.1 to 1.5, assuming that hybridization data are uniformly distributed between divergence times T1 = 0.5TC and T2 = 1.5TC. The conclusion is that a 15% frequency of asymmetric Haldane's rule (as seen in Drosophila) is consistent with a value of C as high as 25 if η = 0.1 or a value of C near 10 if η is as large as 1.5 (which seems implausible). We discuss this further below, after reviewing the data concerning the dominance parameter h0.
Figure 7.—
Time-dependent values of probabilities related to qualitative asymmetry between reciprocal crosses. (A and B) The effect of varying C with gU = 0.2, h0 = 0.1, α = 0.75, δ1 = 0, CV = 0.5, and p/pU adjusted to produce the desired value of η. The colors indicate C = 5 (black), 10 (red), 20 (green), 50 (blue), and 100 (orange). (A) Assumes η = 0.5 and shows the effect of varying C on the conditional probability of qualitative asymmetry given that postmating isolation is complete in one of the two reciprocal crosses (P[Smin < C | Smax > C], solid curves) and the probability that postmating isolation is complete in at least one direction (P[Smax > C], dashed curves). (B) The joint probability that Smax > C and Smin < C as t varies with η = 0.1 (solid curves) and η = 1.0 (dotted curves).
Figure 8.—
Approximate probabilities of qualitative asymmetry as a function of C, calculated using (30) with T1 = 0.5TC and T2 = 1.5TC. The parameters are the same as in Figure 7, and the curves correspond to η = 0.1 (black), η = 0.5 (red), η = 1.0 (green), and η = 1.5 (blue).
COMPARISON TO DATA
We next consider data relevant to estimating the parameters that determine predicted levels of asymmetry and then compare our theoretical predictions to various data. Our theoretical analyses suggest that the critical parameters determining postmating asymmetry are: C (the number of DMIs required to produce complete postmating isolation), α (the shape of the fitness function, see Equation 10), CV (the coefficient of variation of DMI effects), h0/h1 and h0/h2 (the relative magnitude of B vs. U DMIs affecting F1), δ1 (differences in the relative rates of evolution between lineages for loci whose maternally vs. paternally inherited alleles contribute to U DMIs, see Equation 8), and η (the relative contribution of B vs. U DMIs to hybrid dysfunction, see Equation 26). We first consider what is known about individual parameter values and then ask how the data on asymmetry constrain the parameters about which little is known. Although the available data are undoubtedly sparse and, for qualitative patterns, potentially subject to high uncertainty, this examination represents a first-pass comparison of our model parameters and predictions to what is currently known. Ideally, this assessment will become more nuanced—and statistically rigorous—as more data are collected.
Data concerning individual parameters:
Number of DMIs contributing to postmating isolation, C:
Several recent studies have focused on loci involved in DMIs that have apparently large effects on hybrid fitness (e.g., Barbash et al. 2003; Matsubara et al. 2003; Presgraves et al. 2003; reviewed in Coyne and Orr 2004, Chap. 8; Brideau et al. 2006; Sweigart et al. 2006). These studies might suggest that relatively few DMIs are needed to produce complete postmating isolation, i.e., that C is on the order of five or less. However, all of these studies involve homozygous introgressions that produce H2 incompatibilities (Turelli and Orr 2000), i.e., interactions between loci that are both hemizygous or homozygous for incompatible alleles. In contrast, in F1 individuals, these loci would produce either H1 or H0 incompatibilities; i.e., one or both loci would be heterozygous for compatible alleles. Given the generality of partially recessive DMI effects (e.g., Presgraves 2003; Tao and Hartl 2003; and our discussion below), these “large-effect” DMIs would be expected to make much smaller contributions to F1 postmating isolation than indicated by the introgression results.
Nonetheless, there are cases in which F1 reproductive isolation has a simple genetic basis. For example, a single genetic interaction acts as a dominant semilethal in hybrids between Crepis tectorum and C. capiliaris, resulting in death of the seedling carrier (Hollingshead 1930; see Gerstel 1954 for other classic examples of two-locus nuclear interactions that result in F1 inviability). An intraspecific example is the two-locus interaction between genes from copper-tolerant and copper-intolerant forms of Mimulus guttatus, which causes F1 inviability (Macnair and Christie 1983). In addition to these pairwise nuclear interactions, one of the best examples of large-effect DMIs that act in the F1 generation is cytoplasmic male sterility (CMS), commonly found in interspecific plant crosses. The nuclear–mitochondrial interactions underlying CMS are directly responsible for sterility (Schnable and Wise 1998); but they are atypical DMIs for two reasons. First, the mitochondrial mutations that confer CMS and the nuclear genes with which they interact are thought to arise and fix sequentially within the same population. Second, and more importantly, the male sterility phenotype is thought to be the direct target of selection (albeit to favor organelle transmission via female function; Frank 1989; Burt and Trivers 2006, Chap. 5), rather than the inadvertent byproduct of population divergence, as under the classical Dobzhansky–Muller model. These empirical examples show that some cases of asymmetric isolation are due to very few (i.e., less than five) DMIs.
These cases, which would be expected to produce greater levels of asymmetry, fall outside our theoretical treatment, which emphasizes normally distributed hybrid-breakdown scores produced by the cumulative effects of multiple DMIs. However, following Orr and Turelli (2001), our analyses could be easily redone assuming that only a single DMI is needed to produce complete isolation (i.e., C = 1). We have emphasized C ≫ 1 because empirical observations in many systems indicate that hybrids typically show quantitative declines in viability and sterility (Coyne and Orr 2004, Chap. 2), including those apparently due to U DMIs (e.g., multiple cases of cytonuclear interactions cited in Levin 2003). In addition, mapping experiments in Drosophila (Coyne and Orr 2004, Chap. 8), Heliconius butterflies (Salazar et al. 2005), and the plant group Lycopersicon (Moyle and Graham 2005) also generally implicate several to many loci in hybrid sterility and/or inviability. Most of these data (e.g., the Drosophila and Heliconius examples) provide only upper bounds on the parameter C that enters our analyses, as they are performed between taxa for which sex-limited hybrid inviability or sterility is already complete. Because DMIs are expected to accumulate in proportion to at least the square of divergence time, the number of DMIs observed after postmating isolation is complete can greatly exceed C (Orr and Turelli 2001). Nevertheless, both these mapping data and the qualitative observations outlined above are consistent with at least moderate numbers of loci underlying postmating isolation between species (with the number of DMIs contributing to hybrid inviability and sterility typically on the order of ≥10). Translating this into a value for C requires understanding the mix of U vs. B DMIs and their relative effects, which depend on dominance relations, considered below.
Interactions among DMI effects, α:
To our knowledge there have been no published experimental studies of how fitness declines with increasing numbers of DMIs, analogous to the many intraspecific studies of fitness decline with accumulating deleterious alleles (e.g., Charlesworth et al. 2004). Instead, there are comparative data that deal with divergence time rather than DMI number. Lijtmaer et al. (2003) and Bolnick and Near (2005) present the most detailed quantitative analyses of the decline in hybrid viability with evolutionary divergence time, t; and their viability data seem more consistent with a linear than a quadratic decline. A quadratic relation would be expected if DMIs accumulate in proportion to t2 and hybrid fitness falls linearly with cumulative DMI effects (i.e., α = 1 in Equation 10). The apparent linearity motivated our focus on numerical examples assuming α < 1. Although Mendelson et al. (2004) appear to find evidence for a faster-than-linear decline in postmating isolation in anurans and Lepidoptera, their evidence is compromised by the fact that they combine data on F1 viability and fecundity from both males and females; but we expect viability and male and female fecundity to be affected by different DMIs (Orr 1989). Mendelson et al. (2004) also use the quantized scores (0, 0.25, 0.5, 0.75, and 1) of postzygotic isolation introduced by Coyne and Orr (1989) that rely on categorical data describing complete inviability/sterility rather than quantitative measures of hybrid performance. Our quantitative analysis focuses on a single form of postmating isolation in a particular sex and assumes a continuous (rather than categorical) performance measure. A complete treatment of viability and fecundity for male and female hybrids requires considering the multivariate distribution of hybrid breakdown scores relevant to each fitness component in each sex.
Our conjecture that α < 1 seems difficult to reconcile with intraspecific data, suggesting that deleterious alleles interact roughly multiplicatively (e.g., Charlesworth
et al. 2004). For alleles of small effect, multiplicative effects would translate into an approximately linear decline of fitness; i.e., α ≈ 1. There are at least three ways to reconcile this apparent contradiction. First, contrary to theoretical expectations, DMIs may not accumulate faster than linearly with divergence time (Welch 2004). For instance, Kondrashov (2003) has suggested that with gene flow, two-locus DMIs may accumulate linearly rather than quadratically with time. Second, the experiments examining the interactions of deleterious alleles, which generally deal with relatively small fitness effects of alleles segregating within species, may not be informative about the shape of the fitness function that describes the progress to complete postmating isolation as DMIs accumulate between species. In particular, for alleles underpinning DMIs, it is generally assumed that they have no appreciable deleterious effects on their native genetic background; therefore it might be inappropriate to consider these analogous to intraspecific deleterious loci in terms of their expected epistatic interactions. Third, the relatively meager quantitative data (as opposed to the discrete, categorical data of Coyne and Orr 1989) on hybrid viability/sterility may be insufficient to understand how hybrid fitness falls with divergence time. We tentatively conclude that current data indicate α between  and 1.
and 1.
Dominance, h0/h1:
There are two classes of data to consider: qualitative data that indicate that H0 and H1 DMIs are much more difficult to detect and hence tend to have much smaller effects than H2 DMIs (i.e., h0, h1 ≪ h2) and quantitative data that attempt to estimate the relative magnitudes of these effects. The qualitative data are reviewed in Coyne and Orr (2004, Chap. 8). They are typified by the comparison between Coyne et al. (1998), which used deficiencies to find H1 DMIs affecting viability in F1 females with Drosophila melanogaster mothers and D. simulans fathers, and Presgraves (2003), which used a hybrid male rescue mutation and deficiencies to find H2 DMIs affecting viability in F1 males from the same cross. In contrast to the paucity of H1 DMIs with large effect uncovered by Coyne et al. (1998), Presgraves (2003) found many more lethality-inducing H2 DMIs, and he showed that the lethality was due to epistatic interactions between X-linked and autosomal loci. This suggests that h1 ≪ h2.
The best quantitative estimates of d1 ≡ h1/h2 come from Tao and Hartl (2003), who estimated this ratio separately for DMIs affecting hybrid lethality vs. hybrid male sterility. Their analysis is instructive because it reveals how estimates of dominance are affected by the shape of the fitness function. Indeed, we chose fitness function (10) because it makes this point simply. The essence of Tao and Hartl's (2003) procedure is to compare the fitness reductions produced by small homozygous vs. heterozygous introgressions of the D. mauritiana genome into D. simulans. Homozygous introgressions produce H2 DMIs, whereas the heterozygous introgressions produce mainly H1 DMIs (as well as a much smaller expected number of H0 DMIs, whose cumulative effects should be negligible). The calculations are simplest for inviability, but the same logic applies to their analysis of fecundity.
As Tao and Hartl (2003) argue, the expected breakdown scores associated with small homozygous vs. heterozygous introgressions are
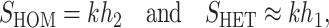 |
(31) |
where the same constant k appears in both expressions. Tao and Hartl (2003) equate these expectations to the actual fitness reductions observed [i.e., to 1 − v(S) in the notation of Equation 10]. This implicitly assumes that α = 1, i.e., that fitness falls linearly as S increases. Thus, they obtain
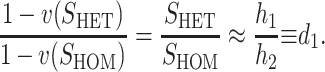 |
(32) |
However, with fitness function (10), this ratio becomes
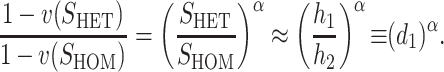 |
(33) |
Assuming α = 1 (i.e., applying Equation 34), Tao and Hartl (2003) estimate d1 = h1/h2 = 0.23–0.29 for hybrid male sterility (HMS) and d1 = 0.33–0.39 for hybrid lethality (HL). If instead α = 0.75 (or α = 0.5), their data would produce approximate estimates of h1/h2 in the range 0.14–0.19 (0.05–0.08) for HMS and 0.23–0.28 (0.11–0.15) for HL, corresponding to greater recessivity. The individual introgressions of Tao and Hartl (2003) (or the near-isogenic lines of Moyle and Graham 2005) could be experimentally combined to estimate α directly (cf. Szafraniec et al. 2003).
These are the best quantitative estimates of DMI dominance, but they deal with h1/h2 rather than the ratio h0/h1 (or h0/h2) that enters our analysis of the combined effects of B and U DMIs. Nevertheless, qualitative and quantitative data suggest that h0/h1 is < (as postulated by the “dominance theory” of Haldane's rule for hybrid inviability; Orr 1993; Turelli and Orr 1995, 2000; Turelli and Begun 1997), and values in the range 0.1–0.4 are plausible. The ratio h0/h2 for nuclear genes should be at most 0.2. The ratio involved in our joint treatment of B and U DMIs may be even smaller as noted below, because the U DMIs may have larger average effects.
(as postulated by the “dominance theory” of Haldane's rule for hybrid inviability; Orr 1993; Turelli and Orr 1995, 2000; Turelli and Begun 1997), and values in the range 0.1–0.4 are plausible. The ratio h0/h2 for nuclear genes should be at most 0.2. The ratio involved in our joint treatment of B and U DMIs may be even smaller as noted below, because the U DMIs may have larger average effects.
Relative rates of molecular evolution, δ1:
Different chromosome types and different genomes routinely evolve at different rates; for instance, animal mitochondria tend to evolve faster than nuclear genomes, whereas in plants nuclear genes tend to evolve faster than chloroplasts, which evolve faster than mitochondria (Wolfe et al. 1987). However, the relevant rate differences for our model concern not whether these different genomic components evolve at different rates, but whether the ratios of their evolutionary rates differ among closely related lineages. Empirical data on the relative rates of evolution of “male-acting” vs. “female-acting” loci in different lineages are sparse, but some information suggests that these could be large, even among closely related taxa. For example, Cho et al. (2004) document up to a 4000-fold difference among lineages in their relative rates of mitochondrial evolution across a range of plant genera, given a standardized rate of substitution at nuclear loci. More relevant to our analyses are within-genus estimates of rate heterogeneity. For example, within the genus Plantago, mtDNA rate variation (standardized to nuclear rates) varies from 1.5- to 50-fold between sister taxa (see Table 1 in Cho et al. 2004). These, along with other reports of mtDNA rate variation (Whittle and Johnston 2002; see other references in Cho et al. 2004), indicate that plant mtDNA rate heterogeneity is likely to be widespread, even among very closely related species. Other relevant data are from studies of evolutionary rate differences for sex-specific loci. Several recent comparative studies suggest that sex-chromosome-specific rates can be quite different among closely related taxa. For example, heterogeneity in evolutionary rates among five Galliformes bird species approaches 2-fold at W-linked loci and 1.5-fold at Z-linked loci (Berlin and Ellegren 2006). Comparisons among 11 Mus species at Y- and X-linked loci showed rate differences of >5-fold and >4-fold, respectively, among sister taxa (see Figure 1 and Table 2 in Sandstedt and Tucker 2005).
Interpreting the significance of these empirical data for the parameters used in our model requires caution. In particular, our δ1 term refers to rates of evolution at loci that specifically affect DMIs, rather than genome- or chromosomewide rates of molecular evolution. In addition, most cases of rate heterogeneity assess variation at silent sites only, whereas most DMIs presumably result from nonsynonymous (phenotype-affecting) substitutions. Nonetheless, there is reason to expect that nonsynonymous rates may be similarly different between sister taxa. For example, a recent analysis indicates a more than twofold difference in rates of nonsynonymous substitution in male-acting accessory gland proteins (Acp's) among sister species of Drosophila, although average rates of nonsynonymous substitution in nuclear loci do not differ (Wagstaff and Begun 2005). Acp's are known to mediate postcopulatory female responses including sperm storage, ovulation, egg laying, and receptivity (see Wagstaff and Begun 2005 for references) and could therefore have substantial effects on postmating, prezygotic isolation between species or on male fertility of F1 hybrids. Given these data, and in the absence of direct data (from any system) on the relative evolutionary rates of loci involved in U DMIs, the parameter values we have explored—relative rate differences between lineages of 0- to 10-fold—appear to span a range that is biologically plausible.
Variation in effects of individual DMIs, CV:
Currently there are no direct estimates of the coefficient of variation (CV) of individual DMIs. However, data from QTL mapping experiments of hybrid incompatibility, and from within-species estimates for relevant quantitative genetic traits, suggest that CV ≤ 1. CV can be estimated from published data on hybrid incompatibility QTL by assuming that each QTL is underpinned by a single locus. (This assumption is conservative in that it overestimates the CV of single-locus effects if several loci typically contribute to individual QTL.) Using data from hybrid incompatibility QTL between tomato species (Moyle and Graham 2005), we calculate an average CV of between 0.1 and 0.4 for a range of hybrid incompatibility traits. Given our lack of information on the H0 and H1 DMIs expected to act in hybrids, we assume that these values obtained from H2 DMIs are representative. They are consistent with those for quantitative genetic traits including viability, early and late fecundity, longevity, productivity, and fitness in Drosophila, where the maximum estimated CV is ∼0.5, but is generally <0.1 for phenotypic effects of spontaneous mutations or standing genetic variation (see Table 2 in Houle 1998). Values of CV > 0.5 for DMI effects would make it easier to explain high levels of asymmetry with larger values of C.
Relative contributions of B vs. U DMIs, η:
Different types of DMIs will make systematically different contributions to postmating isolation. F1 individuals experience only one form of B DMIs, namely H0 DMIs between nuclear loci. In contrast, they can experience several classes of U DMIs simultaneously that are either H1 or H2. As discussed above, several lines of evidence suggest that for nuclear loci, h0 ≪ h1 ≪ h2. Our composite parameter η is defined by (26) with the assumption that only one class of U DMIs is considered and each contributes 1 to the hybrid breakdown (whether they are H1 or H2), while the H0 B DMIs each contribute h0 < 1. Thus, in many cases η conflates the relative effects of different classes of DMIs (e.g., H0 vs. H1) with the relative effects of DMIs involving different classes of loci, e.g., nuclear–nuclear vs. cytonuclear. Anecdotal evidence suggests that the U DMIs, especially those involving cytoplasmic and maternal effects, tend to make relatively large contributions to postmating isolation. This may be primarily a consequence of them being H1 or H2 rather than H0. Although our analyses have focused on modeling a single class of U DMIs, it is important to understand differences among different classes of U DMIs when comparing the levels of asymmetry seen at different developmental stages and among different groups of taxa.
For example, in contrast to the generally small to moderate effects of the H0 and H1 nuclear DMIs experienced by F1 hybrids, there are many cases in which heterospecific organelles (or even those from differentiated populations) have readily detectable effects, despite the fact that organelle genomes are three orders of magnitude smaller than nuclear genomes (e.g., Levin 2003; Rand et al. 2004; Harrison and Burton 2006). Such data suggest both relatively large individual effects and a higher relative probability for such incompatibilities arising; i.e., pU ≫ p. Cytoplasmic male sterility is one extreme but common example where individual nuclear–cytoplasmic interactions can confer complete male sterility. The commonality of cytonuclear incompatibilities probably reflects the close coevolution of organelle and nuclear genomes (Rand et al. 2004; Linke and Borner 2005), their potential for intragenomic conflicts (Burt and Trivers 2006), and the centrality to cellular metabolism of intergenome interactions (e.g., the synthesis of ATP). Close coevolution presumably also underlies the high frequency and easily detectable consequences of maternal effects in animals (Parker et al. 1985; Sawamura 1996; Turelli and Orr 2000, Table 1; Tadros and Lipshitz 2005) and postzygotic parent-of-origin effects in plants (Tiffin et al. 2001). Despite uncertainty concerning the number of loci contributing to maternal-effect incompatibilities, we know at least for Drosophila that they tend to be relatively common and have large effects. As Sawamura (1996) has argued, most of the exceptions to Haldane's rule in Drosophila occur with respect to viability and seem to be caused by maternal-effect loci interacting with the male-derived X-linked loci. Not infrequently, these maternal-effect-X H1 incompatibilities that afflict only females have a greater cumulative effect than the H1 X–autosome incompatibilities that afflict only males and generally lead to Haldane's rule for viability.
Similarly, individual effects of U DMIs acting prezygotically can also apparently be large. For example, “unilateral incompatibility” (complete GS isolation in one direction, but no GS isolation in the reciprocal one, presumed to be due to changes in pollen–stigma signaling) is regularly observed, even among very closely related species (Lewis and Crowe 1958; de Nettancourt 2001; see also Figure 10A). These effects appear to be mediated by few loci of relatively large effect in several cases (e.g., Rashid and Peterson 1992; Bernacchi and Tanksley 1997; reviewed in de Nettancourt 2001). TRE and maternal–zygotic interactions that act immediately after fertilization in plants are also likely to have comparatively large effects. For example, interactions between three loci appear to account for nearly complete isolation due to endosperm failure in one direction of a cross among rice subspecies (Matsubara et al. 2003). Other studies have similarly indicated the involvement of few loci of large effect in endosperm failure of interspecific crosses (e.g., Ehlenfeldt and Hanneman 1988; Camadro and Masuelli 1995; Johnston and Hanneman 1999). The comparative sizes of GS and TRE interactions are harder to judge directly against X–autosome U DMIs, as there are few heterogametic plant systems in which the necessary hybridizations have been described. Nonetheless, because total isolation can potentially be conferred by relatively few genetic changes affecting GS and TRE interactions, these individual U DMIs are also presumably larger in effect than typical X–autosome interactions. (Note that, because of these apparent differences in the size of individual effects, systematic differences between biological groups in the relative frequencies of different kinds of U incompatibilities will obviously affect their propensity for isolation asymmetry—see below).
Figure 10.—
Changes in isolation asymmetry over time in Silene species crosses at the stage of pollen–stigma (GS) interactions (A) and interactions during endosperm (TRE) and seed development (B).
Our conclusion is that the parameter η (defined in Equation 26) that measures the relative contribution of B vs. U DMIs to postmating isolation is likely to be far smaller than expected on the basis of gene counting for two reasons: (1) U DMIs seem to have a far higher probability of occurring than B DMIs (pU ≫ p), and (2) U DMIs seem to have systematically larger effects (h0 ≪ 1). Observational biases may contribute to both of these conjectures, but the asymmetry data seem difficult to explain otherwise.
Data on quantitative asymmetry:
Inviability in centrarchids:
The most extensive data relating divergence time to quantitative asymmetry come from Bolnick and Near's (2005) analysis of inviability between centrarchid fishes with a fossil-calibrated phylogeny. Figure 9 presents a corrected version of the data from their Figure 4 (kindly provided by D. I. Bolnick). These data show the pattern of rising and then falling asymmetry expected. The level of quantitative asymmetry is quite high, with A on the order of 0.2 early in divergence and A > 0.4 later. Comparing these empirical results to our theoretical predictions, as illustrated in Figures 4–6, three principal conclusions emerge. To explain such high levels of asymmetry, our theoretical analyses suggest, first, that U DMIs must dominate hybrid inviability (i.e., η must be small, certainly <1 and probably closer to 0) and, second, that C must be fairly small, on the order of ≤5. A third, more tentative, inference follows from the marked rightward skew of the time course of asymmetry. Comparing this with Figures 4B and 6B suggests that α is unlikely to be as small as 0.5, which produces more symmetric trajectories for the quantiles.
Figure 9.—
Quantitative asymmetry, A, from crosses between centrarchid fishes with a fossil-calibrated phylogeny [adapted from Bolnick and Near's (2005) Figure 4, with corrections provided by D. I. Bolnick]. The data from each cross describe the mean hatch rate for hybrids divided by the mean hatch rate from intraspecific crosses. The y-axis is the difference between reciprocal crosses, and the x-axis is estimated divergence time in millions of years.
Are these inferences consistent with what is known about the causes of hybrid inviability in centrarchids? Bolnick and Near (2005, p. 1763) review data suggesting that maternal–zygotic interactions play a major role in the developmental dysfunction of centrarchid hybrids (cf. Parker et al. 1985). As discussed previously, early development is likely to be dominated by U interactions. Sawamura's (1996) observations concerning the exceptions to Haldane's rule in Drosophila and our discussion below of asymmetry data from Drosophila, mosquitoes, and Lepidoptera suggest that these DMIs often have large effects. Bolnick and Near's (2005) viability data (their Figures 3 and 5) motivated our examination of models in which fitness falls more slowly than linearly with increasing DMIs. However, although there is little statistical power in so few data, the shape of the asymmetry trajectory seems more consistent with linearity (α = 1) or α = 0.75 (the canonical value used in our numerical illustrations) than with α = 0.5, which would produce a roughly linear decline of fitness with time (see Figures 3 and 5 of Bolnick and Near 2005). Of course, our ability to draw definitive conclusions about this and other parameters is limited by the inevitable statistical and biological noise in the asymmetry data. Nonetheless, comparing the general pattern and magnitude of asymmetry over time in the data and in our model seems to strongly indicate a major role for U DMIs of large effect.
Pollen–stigma and early F1 inviability in angiosperms:
Moyle et al. (2004) examined patterns of change in reproductive isolation over increasing genetic distance in three plant genera. In those analyses, reproductive isolation values for reciprocal crosses (if available) were averaged to give a single data point per species pair. Here we have reanalyzed reciprocal-cross data for the genus Silene, to examine patterns of asymmetry through time at two stages of isolation: pollen germination rates following transfer to the female stigma (corresponding to GS interactions) and seed set per successful fertilization (corresponding to TRE interactions, in addition to any B interactions occurring in the F1 hybrid embryo) (Figure 10). As predicted, overall the magnitude of observed asymmetry generally increases with increasing genetic distance between taxa and then falls once at least one reciprocal cross produces complete isolation. (Note that complete isolation in both reciprocal crosses is observed for many species pairs in this group, unlike the extended duration of incomplete isolation observed for centrarchids.)
In Figure 10, A and B, we have divided the data into two categories: crosses involving only hermaphrodite species and crosses that involve at least one dioecious species (i.e., with separate males and females). In Silene these dioecious species have heteromorphic sex chromosomes and heterogametic (XY) males. For GS interactions, it is clear that hermaphrodite crosses show earlier and larger asymmetry than dioecious crosses (Figure 10A). In comparison, the opposite pattern is observed for TRE and early postzygotic interactions. We have little additional data to suggest why these two stages of isolation differ. However, it might suggest that that the sex chromosomes play a small role in mediating GS interactions, in comparison to TRE and early embryonic interactions. Maternal effects play a strong role at this later stage, as do unequal parental contributions of cytoplasmic factors, perhaps indicating a larger role for X–cytoplasmic interactions and/or endosperm-based X–autosomal interactions in early postzygotic development. Regardless, given the extreme levels of quantitative asymmetry observed in Figure 10, A and B, it seems clear that C must be very small for many species pairs at both of these developmental stages, indicating that individual GS and TRE DMIs can produce nearly complete isolation. Accordingly, our detailed quantitative predictions are likely less relevant to these species pairs.
Data on qualitative asymmetry:
Drosophila data on male and female asymmetry:
Muller (1942, p. 101) was the first to suggest that genetic inferences might be made from the frequency with which F1 males from reciprocal crosses differ in fertility and viability. Turelli and Orr (1995) estimated that the frequency of qualitative asymmetry for Haldane's rule in Drosophila is ∼15% (this is consistent with data subsequently tabulated by Coyne and Orr 1997 and Turelli and Begun 1997). Assuming η ≤ 1, our results in Figure 8 suggest that this level of qualitative asymmetry is consistent with C on the order of 10–20. Hence, if h0 is no more than 0.2, the number of loci involved in male hybrid dysfunction would be at least 20 and possibly closer to 100.
More surprising is the apparently even higher frequency of qualitative asymmetry observed for F1 females. Taking the data from Coyne and Orr's (1989a, 1997) postzygotic isolation classes 0.75 and 1.0, >25% of the cases of F1 female sterility/inviability are asymmetric. The frequency is particularly high in the repleta and virilis species groups. This suggests that the sex-specific U DMIs that specifically affect F1 females, for instance, interactions between the paternally inherited X and cytoplasmic factors (including maternal effects), can often have individually large effects, consistent with the pattern of exceptions to Haldane's rule for inviability in Drosophila (Sawamura 1996). This pattern merits a more careful treatment, using quantitative data on hybrid fitness that partitions viability vs. fertility effects. For example, because the interactions between the maternal cytoplasm and the paternal X in F1 females are likely to act relatively early in development, we might expect that more female asymmetry will be observed in hybrid viability than sterility (Turelli and Orr 2000). Nevertheless, the mosquito data discussed below suggest that both viability and fecundity can be affected by such interactions.
Mosquito data on male and female asymmetry:
Asymmetric postzygotic isolation is well documented in mosquitoes, and in principle these patterns could be used to assess the relative importance of different mechanisms for this asymmetry. In particular, Presgraves and Orr (1998) compared patterns of isolation between species of Anopheles (which exhibit male heterogamety) and Aedes (with sex determined by a small chromosomal region—possibly a single locus—rather than a sex chromosome); all else being equal, taxa with sex chromosomes are expected to experience more U DMIs and hence produce more asymmetric results. Unfortunately, we cannot test this hypothesis statistically using the available Aedes–Anopheles data because of the relatively small number of crosses involving Aedes and the phylogenetic nonindependence of many of the crosses in both groups. Nonetheless, it is possible to compare patterns of male vs. female asymmetry in mosquitoes. It is interesting that of the 25 crosses that show Haldane's rule for sterility in Anopheles (i.e., males are sterile in one or both directions of a cross), four are asymmetric (16%, see Table 2 of Presgraves and Orr 1998), in line with the results for qualitative asymmetry in Drosophila. In contrast, 4 of 15 crosses produce asymmetric female inviability and 4 of 10 crosses produce asymmetric female sterility. These nominally higher levels of female asymmetry suggest that the female-specific U DMIs, namely those between the paternally inherited X and the cytoplasm (including maternal effects), tend to have relatively larger effects than the male-specific U DMIs (acting between the maternally derived X and the paternal autosomes). This conclusion is reinforced by Lepidoptera data.
Lepidoptera data on female asymmetry:
Lepidoptera, which have heterogametic (ZW) females, provide an interesting contrast to male-heterogametic Drosophila and Anopheles. To simplify the comparison to other taxa, we refer to the female karyotype in Lepidoptera as XY. Presgraves (2002) examined patterns of reproductive isolation, and evidence for Haldane's rule, in Lepidoptera. Using data on reciprocal crosses reported in Appendix 2 of Presgraves (2002), we find that 60% (15/25) of crosses that show female-limited inviability are asymmetric. (One pair may be an exception to Haldane's rule, but this would reduce the fraction only slightly to 58%.) While these sample sizes are small, this extraordinarily high level of female isolation asymmetry suggests that despite the small size of the X chromosome in Lepidoptera, the genetic basis of postzygotic isolation in this group involves a major role for X-linked DMIs, as seen in male-heterogametic groups like Drosophila and Anopheles (and possibly Silene). As discussed by Turelli and Orr (2000) and Presgraves (2002), such interactions contribute to (or oppose) Haldane's rule depending on the nature of sex determination. In male-heterogametic clades, F1 males potentially experience Y–cytoplasmic and Y–maternal-effect DMIs, but the X chromosome is always on its native cytoplasmic/maternal background. In contrast, in all groups with heteromorphic sex chromosomes, F1 females can suffer from unique X–cytoplasmic and X–maternal-effect interactions. In male-heterogametic clades, these female-specific H1 DMIs can produce exceptions to Haldane's rule, as emphasized by Sawamura (1996). In contrast, there should be many fewer exceptions to Haldane's rule in female-heterogametic clades, because in these cases F1 females potentially experience more severe H2 X–cytoplasmic or X–maternal-effect DMIs, in addition to the hemizygous X–autosome interactions that can underpin Haldane's rule. Note that the “faster-male” theory (Wu and Davis 1993) predicts the opposite—that exceptions to Haldane's rule should be greater in female heterogametic species groups. Future data will help illuminate which prediction is more frequently observed. It is possible that our prediction will ultimately be more relevant to patterns of hybrid inviability than sterility if, as some data suggest (e.g., Presgraves and Orr 1998), faster-male effects are more important with respect to the development of hybrid sterility.
In general, we expect the X-mediated DMIs to be substantially more frequent and/or stronger than Y-mediated DMIs because Y chromosomes are generally heterochromatic with far fewer active genes. This is consistent with both Lepidoptera and Drosophila data, where female asymmetry appears to be more exaggerated than male asymmetry. Asymmetry in Lepidoptera females is expected to be even more severe because not only do they experience H2 (rather than H1) X–cytoplasmic or X–maternal-effect DMIs, but also they may suffer more H2 X–Y DMIs than Drosophila do, if the Y chromosome in Lepidoptera is less degenerate, as some data suggest (Presgraves 2002, p. 1175). Whatever the cause, such a large apparent sex-chromosome effect may be surprising in Lepidoptera because the X (W) chromosome contains only ∼3–5% of the nuclear genome (in terms of both estimated gene number and DNA content); yet this exaggeration is consistent with other data supporting a large-X effect for both morphological differences and postmating isolation in Lepidoptera (Presgraves 2002).
DISCUSSION
Orr (1993, 1995) pioneered mathematical analyses of two-locus DMIs to understand the accumulation of postzygotic isolation and the resulting patterns of reproductive isolation. These analyses were extended and generalized by Turelli and Orr (1995, 2000), Orr and Orr (1996), and Orr and Turelli (2001). Here we extend those analyses to understand one of the oldest and most common patterns in the hybridization literature, fitness differences emerging from reciprocal crosses. In doing so, we provide a probabilistic framework for understanding the fitness consequences of DMI accumulation that can produce testable genetic inferences from readily obtained phenotypic data.
We sought to infer the most likely causes of isolation asymmetry in different empirical systems. Several general principles are evident, even without theoretical development. The first is that postmating isolation asymmetry requires genetic interactions involving genes that are asymmetrically transmitted (i.e., U DMIs). Incompatibilities between genes that are symmetrically transmitted and expressed (i.e., B DMIs) cannot generate isolation differences between reciprocal crosses, regardless of relative rates of evolution in the two diverging lineages. Nonetheless, to understand the quantitative implications of U DMIs, B DMIs must generally also be considered because these two classes of DMI often act simultaneously (Table 1), and less asymmetry is expected with increasing contributions to postmating isolation from B DMIs. Thus, asymmetry is most likely, and most extreme, at reproductive stages where U DMIs predominate, e.g., early embryogenesis in animals when maternal effects act and early stages of plant fertilization dominated by GS interactions and TRE effects. In addition to these general principles, several less obvious generalizations emerge from our analysis.
Factors that increase predicted levels of postmating isolation asymmetry:
Although our treatment is necessarily idealized, our model indicates regions of parameter space where isolation asymmetry is expected to be either very large or very small. Using readily available phenotypic data from both model and nonmodel organisms, we can therefore make inferences and generate hypotheses that will be testable as genetic tools become available. Of particular interest is the shape of the fitness function (10) that translates DMI effects into hybrid fitness and indirect estimates of the numbers and types of DMIs that contribute to postmating isolation [i.e., the parameters α and C in (10) and η in (26)]. Specifically, Figures 4 and 6 suggest that asymmetry is relatively large when fewer DMIs are needed to produce complete postmating isolation (low C), when U DMIs make relatively large contributions to postmating isolation (η small), when there are large differences in the relative rates of evolution of loci contributing to U vs. B DMIs in diverging lineages (large δ1), and when there is a large variance in the fitness effects of individual DMIs (large CV). More subtly, we expect a more even distribution of asymmetry (and a lower maximum) over a wide range of divergence times if the fitness function that transforms DMI effects into fitness falls faster than linearly (α < 1).
Our analyses also make predictions about the relative importance of these parameters in contributing to observed asymmetry. For example, the effects of increasing CV over a plausible range of values appear to be negligible in comparison to the effect of reducing the number of DMIs (C) required to produce complete isolation. Muller (1942, p. 101) originally suggested that asymmetry data could be used to make inferences about the genetics of hybrid sterility/inviability. He reasoned that because there were so many cases in which F1 males from reciprocal crosses differed (often substantially) in fertility and viability, hybrid dysfunction was “… in the main dependent on only a very few loci each of which had a considerable influence on viability or fertility, while the great majority of the gene differences between species or subspecies had very little effect …” Muller was surely correct in hypothesizing that only a tiny fraction of interspecific genetic differences lead to DMIs (as supported by estimates suggesting that our parameter p is on the order of 10−5; Orr and Turelli 2001; Presgraves 2003). Similarly, Muller correctly understood that smaller values of C (corresponding to few DMIs of large effect) would lead to greater asymmetry. Nevertheless, our analysis of qualitative asymmetry indicates that for plausible parameter values, the levels of asymmetry observed for Haldane's rule in Drosophila are consistent with scores of loci contributing to hybrid male sterility/inviability.
An unexpected outcome of our analysis is the surprisingly small relative effect that deterministic rate differences between lineages (δ1 > 0) have on the size and duration of asymmetry, over a wide range of parameter values (see Figures 4C and 6C). One of our initial goals was to assess whether observed levels of asymmetry could be explained by stochastic effects or whether the magnitude or duration of asymmetry implied that deterministic differences (in particular, rate variation among lineages in genes contributing to U vs. B DMIs) must be involved. Our analysis indicates that empirical patterns of postmating asymmetry are generally consistent with a stochastic explanation. In addition, patterns emerging from our comparative analyses suggest that, in cases where extreme levels of asymmetry are observed (for instance, the Silene data shown in Figure 10), postmating isolation is more likely attributable to a small number of DMIs of large effect than to deterministic rate differences between lineages. Nonetheless, our analysis also indicates that if systematic rate differences (i.e., δ1 ≠ 0) do play a significant role in postmating asymmetry, then the direction of asymmetry should be predictable from the relative rates of evolution for the loci that contribute to U DMIs. For instance, if asymmetry is caused by U DMIs involving maternally inherited cytoplasmic genomes (either mitochondria or chloroplasts), and evolutionary rate differences play a decisive role, we expect that crosses will be systematically less successful when the lineage showing faster plastid evolution (relative to nuclear evolution) is used as the maternal parent. This prediction can be easily tested.
Systematic differences among groups in the predicted frequency and magnitude of isolation asymmetry:
An interesting generalization from our analyses is that various groups should differ systematically in their propensity for postmating isolation asymmetry, depending on biological features such as the types of asymmetric interactions that can act, the nature of those interactions (especially the predicted severity of the interactions, i.e., H0 vs. H1 or H2 DMIs), and specific features of reproductive biology. For example, angiosperms can potentially experience a unique and larger range of potential U DMIs than animals, because they exhibit haploid gene expression in male gametes during pollination and fertilization (enabling GS interactions) and because of early postzygotic asymmetric interactions in the endosperm (TRE interactions), on top of more “universal” asymmetries like maternal effects and cytonuclear interactions. In addition to the larger range of potential U DMIs, the magnitude of effects of these additional U DMIs is likely to be larger in the initial parental cross and resulting F1's, primarily because GS and TRE interactions involve homozygous–hemizygous (H2) interactions, whereas X–autosome and cytonuclear interactions involve heterozygous–hemizygous (H1) interactions in the F1. Assuming roughly equal numbers of factors are involved in each type of interaction (and equivalent sizes of effect—see above), if DMIs are largely recessive, then H2 GS and TRE interactions are expected to produce much stronger and more asymmetric reproductive barriers in parental crosses in comparison to DMIs acting later in development. In fact, although the sample sizes vary substantially, statistically significant asymmetric postmating isolation appears to be observed more frequently at stages where GS and TRE incompatibilities predominate. For instance, failure to set seed or fruit following pollination was found to be significantly asymmetric in 132 of 293 (45%) reciprocal species crosses in 13 plant genera, and F1 seed viability was found to be significantly asymmetric between reciprocal crosses in 60 of 132 crosses (45%) performed in 6 plant genera. In contrast, F1 male fertility, where H1 cytonuclear interactions may predominate was significantly asymmetric in only 8 of 23 (35%) of reciprocal crosses in 4 plant genera (Tiffin et al. 2001).
Differences in the developmental biology and sex determination of groups should also systematically affect levels of asymmetry. For example, it is plausible to expect differences between phylogenetic groups on the basis of systematic differences in the nature and timing of the maternal–zygotic transition during embryogenesis, especially the length of time over which maternal effects direct embryonic development. Species in which there is an extended phase of maternal control and/or simultaneous maternal and embryonic developmental control are expected to be more prone to isolation asymmetry at the stage of early postzygotic embryonic development. Accordingly, frequent observations of isolation asymmetry at these developmental stages might suggest maternal–zygotic interactions as specific candidates for DMIs. Similarly, in animals with sex chromosomes, only the heterogametic sex can experience H2 DMIs between the sex chromosomes, whereas females experience interactions between the sex chromosome and cytoplasmic factors (including maternal effects) that are not experienced by males. Comparative analyses of asymmetry between the sexes, and across taxa with different forms of sex determination, can therefore indicate the relative importance of different types of DMIs. The empirical asymmetry data currently available in animals point to a major role for cytoplasmic and maternal effects in both postmating isolation and isolation asymmetry. In particular, the Drosophila, Anopheles, and Lepidoptera data all point to the specific importance of X–cytoplasmic and X–maternal-effect DMIs. Our analyses suggest that patterns of postmating asymmetry can help us understand the genetics of hybrid dysfunction. Further, detailed, quantitative data on isolation asymmetry will facilitate evaluating our conclusions and predictions.
Finally, it is worth noting that it is straightforward to apply our general model to instances where parent-of-origin (female- or male-acting) effects on reciprocal reproductive isolation are due to “epigenetic” rather than genetic effects. Epigenetic modifications (i.e., heritable changes in gene expression or function due to extra-DNA modifications, such as methylation or chromatin remodeling) are increasingly recognized as a potentially common mechanism of differentiation between species (e.g., Rapp and Wendel 2005). Just as standard DMIs are due to inappropriate interactions between lineage-specific sequence-based differences, dysfunctional F1 hybrid interactions could be due to extra-DNA-based differences that characterize diverged lineages. (All that is required are divergent lineage-specific heritable differences that interact to produce hybrid dysfunction.) Indeed, it may eventuate that epigenetic effects are even more likely to be associated with asymmetric isolation, as the current (albeit limited) data suggest that they are frequently associated both with modified gene expression early in embryogenesis and with meiotic processes during gametogenesis (Rapp and Wendel 2005; Dawe and Henikoff 2006). This prediction awaits future investigation of the “epigenetics of speciation.”
Acknowledgments
We are grateful to N. H. Barton, D. I. Bolnick, J. A. Coyne, H. A. Orr, M. W. Hahn, A. R. Lemmon, B. C. O'Meara, P. C. Phillips, D. C. Presgraves, M. Slatkin, and two anonymous reviewers for comments and discussions that improved earlier versions of the manuscript. M.T. thanks A. A. Hoffmann, M. Slatkin, C. Moritz, Centre for Environmental Stress and Adaptation Research, the University of Melbourne, and the University of California (UC) Berkeley Museum of Vertebrate Zoology for hospitality. L.C.M. was supported by a postdoctoral fellowship from the Center for Population Biology, UC Davis, the Indiana University Department of Biology, and National Science Foundation (NSF) grant DEB 0532097. M.T. was supported by NSF grant DEB 0089716 and UC Berkeley's Miller Institute for Basic Research in Science.
APPENDIX A: STOCHASTIC ACCUMULATION OF TWO-LOCUS INCOMPATIBILITIES AND EXPECTED DIFFERENCES IN RECIPROCAL BREAKDOWN SCORES
We first calculate the potential number of incompatibilities between diverged genomes, noting that these incompatibilities can occur between derived alleles in each taxon (DD) or between derived and ancestral alleles (DA). Incompatibilities between two ancestral alleles (AA) are impossible as this genotype represents a fit wild-type state, as generally assumed under the Dobzhansky–Muller model. We assume that at any locus, at most one lineage has a derived allele. As explained by Orr (1995), two-locus DA incompatibilities involve interactions between loci that have both undergone substitutions in one of the lineages. Orr (1995) analyzed the accumulation of B DMIs in F1 individuals given some number of substitutions in each lineage. Orr and Turelli (2001) generalized his treatment to allow for the random (Poisson) accumulation of substitutions in each lineage and random contributions of DMIs to the hybrid breakdown score S. We first derive the relevant result from Orr and Turelli (2001) and then consider the accumulation of U DMIs that are the focus of this article.
First consider only loci that produce B DMIs and let p denote the probability that an allelic difference at two such loci leads to a DMI. As noted in Equation 6 of Orr and Turelli (2001), the expected number of B DMIs in an F1 is
 |
(A1) |
where E(KB) denotes the number of substitutions at these loci, summed across both lineages. To understand our analysis of U DMIs, it is useful to explain how (A1) arises from an explicit consideration of DD vs. DA incompatibilities. Note that if lineage 1 undergoes successive substitutions at loci A and B, the derived allele at locus B may be incompatible with the ancestral allele at locus A, which remains fixed in lineage 2. The order of the substitutions is critical. If the B locus substitution occurred first, there could be no DMI involving the ancestral allele at locus A and the derived allele at B, because the ancestral allele at locus A was part of the genetic background in which the B substitution occurred, and we assume that DMI-producing substitutions are unopposed by selection. Following Orr (1995), we use D1A2 to denote DMIs between a derived allele in lineage 1 and an ancestral allele in lineage 2. If lineage 1 has undergone K substitutions, the number of possible D1A2 incompatibilities is 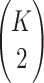 (namely half of the number of ordered pairs of loci), because for any pair of loci, only the locus that experiences the second substitution can produce a D1A2 DMI.
(namely half of the number of ordered pairs of loci), because for any pair of loci, only the locus that experiences the second substitution can produce a D1A2 DMI.
To derive (A1), let 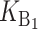 (
(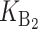 ) denote the number of autosomal substitutions in lineage 1 (lineage 2) and assume that each substitution in each lineage has probability p of producing a DMI with any locus in the other lineage that harbors a different allele. We have
) denote the number of autosomal substitutions in lineage 1 (lineage 2) and assume that each substitution in each lineage has probability p of producing a DMI with any locus in the other lineage that harbors a different allele. We have
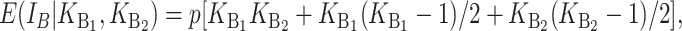 |
(A2) |
where the first, second, and third terms in the brackets describe DD, D1A2, and D2A1 interactions, respectively. Assuming that 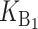 and
and 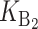 follow Poisson distributions, so that
follow Poisson distributions, so that 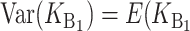 ) and
) and 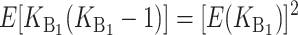 , and defining
, and defining 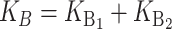 , we use
, we use
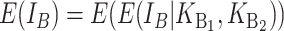 |
(A3) |
to obtain (A1) after algebraic simplification. Note that the expected number of B DMIs is unaffected by the relative rates of molecular evolution in the two lineages—it depends only on the total number of substitutions separating the taxa, E(KB).
To understand the accumulation of U DMIs, we follow the terminology and notation introduced between Equations 3 and 4 in the text. In particular, for pairs of loci that can produce U DMIs, the probability that a difference yields a DMI is denoted pU. Now there are four categories of substitutions to be considered, K = (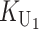 ,
, 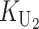 ,
, 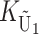 ,
, 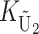 ), where
), where 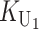 and
and 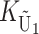 denote the number of “female-acting” and “male-acting” substitutions in taxon 1, respectively. Because U DMIs are intrinsically asymmetric, we must specify the direction of the cross to enumerate them. We consider taxon 1 females mated to taxon 2 males and count the potential DD, D1A2, and D2A1 interactions. Following the logic above, we see that
denote the number of “female-acting” and “male-acting” substitutions in taxon 1, respectively. Because U DMIs are intrinsically asymmetric, we must specify the direction of the cross to enumerate them. We consider taxon 1 females mated to taxon 2 males and count the potential DD, D1A2, and D2A1 interactions. Following the logic above, we see that
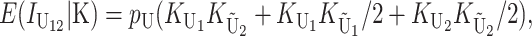 |
(A4) |
where the first, second, and third terms in parentheses describe DD, D1A2, and D2A1 U interactions, respectively. The first term is obvious. The only subtlety is the appearance of the factors of  in the second and third terms. As in the derivation of (A2) for B DMIs, the order of substitutions determines whether loci that have both experienced substitutions in one lineage can produce D1A2 or D2A1 U DMIs. For U DMIs as for B DMIs, any ordered pair of relevant loci that have both experienced substitutions in maternal lineage 1 will have either probability pU of producing a D1A2 DMI or probability 0 of producing a D1A2 DMI, depending on whether the first locus considered underwent the first or second substitution, respectively. Thus, conditional on K, the number of D1A2 U DMIs is a binomial random variable, with parameters
in the second and third terms. As in the derivation of (A2) for B DMIs, the order of substitutions determines whether loci that have both experienced substitutions in one lineage can produce D1A2 or D2A1 U DMIs. For U DMIs as for B DMIs, any ordered pair of relevant loci that have both experienced substitutions in maternal lineage 1 will have either probability pU of producing a D1A2 DMI or probability 0 of producing a D1A2 DMI, depending on whether the first locus considered underwent the first or second substitution, respectively. Thus, conditional on K, the number of D1A2 U DMIs is a binomial random variable, with parameters 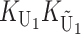 , the number of ordered pairs of the relevant loci, and pU/2, with the
, the number of ordered pairs of the relevant loci, and pU/2, with the  arising because only one temporal ordering of the substitutions can produce a D1A2 U DMI.
arising because only one temporal ordering of the substitutions can produce a D1A2 U DMI.
APPENDIX B: ACCURACY OF THE BIVARIATE GAUSSIAN APPROXIMATION
As discussed in Appendix 1 of Orr and Turelli (2001), our Gaussian approximation for the hybrid breakdown score S requires that a moderate number of DMIs contribute to S. We showed very close numerical agreement between simulation results and our analytical approximations for the mean and variance of waiting times to achieve complete postzygotic isolation when C ≥ 20 and p < 10−4. We conjecture that the approximation will remain adequate for C on the order of 5 or 10. The question we first address is how the moments of S are biased by conditioning on S > 0. Because this conditioning increases the mean and decreases the variance, it will lower the predicted level of asymmetry. The issue is complicated by the fact that we are concerned with the time course of postmating isolation, including times when the mean of S is only a fraction of C. We consider only the simplest case with all U DMIs and equal relative rates of molecular evolution in the two lineages (δ1 = 0), so that the moments of S are
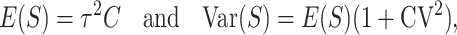 |
(B1) |
where τ = t/TC and TC is the time at which E(S) = C (see Equation 19).
Numerical calculations were performed using Mathematica to determine the percentage of relative error of the mean and standard deviation of S obtained with truncation. The percentage of relative error of the mean is computed as
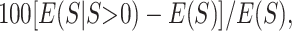 |
(B2) |
assuming that S has a Gaussian distribution with moments given by (B1). For τ ≥  , C ≥ 20, and CV ≤
, C ≥ 20, and CV ≤  , the maximum relative error in the mean is <3% (results not shown). Note that τ =
, the maximum relative error in the mean is <3% (results not shown). Note that τ =  and C = 20 correspond to an average of only five DMIs contributing to S. In contrast, when τ ≥
and C = 20 correspond to an average of only five DMIs contributing to S. In contrast, when τ ≥  , C ≥ 20, and CV ≤
, C ≥ 20, and CV ≤  , the standard deviation of S is underestimated by ∼6%, but this falls to <4% with C ≥ 25. We conclude that truncating our Gaussian approximation for S to allow only positive values is likely to have a minimal effect on our predicted levels of asymmetry for τ ≥
, the standard deviation of S is underestimated by ∼6%, but this falls to <4% with C ≥ 25. We conclude that truncating our Gaussian approximation for S to allow only positive values is likely to have a minimal effect on our predicted levels of asymmetry for τ ≥  , C ≥ 20, and CV ≤
, C ≥ 20, and CV ≤  and that near τ =
and that near τ =  and C = 20, this approximation is likely to underestimate the predicted level of asymmetry. Although the relative errors quickly become quite large when the mean of S is small, our Gaussian approximation is itself suspect for these values, because so few DMIs are contributing. Thus, even for C ≥ 20 and CV ≤
and C = 20, this approximation is likely to underestimate the predicted level of asymmetry. Although the relative errors quickly become quite large when the mean of S is small, our Gaussian approximation is itself suspect for these values, because so few DMIs are contributing. Thus, even for C ≥ 20 and CV ≤  , the predicted levels of asymmetry for τ ≤
, the predicted levels of asymmetry for τ ≤  are best viewed as interpolations between the predicted value at τ =
are best viewed as interpolations between the predicted value at τ =  and the complete symmetry that must hold for τ = 0.
and the complete symmetry that must hold for τ = 0.
To test the adequacy of these interpolations, we performed replicated stochastic simulations to model the accumulation of DMIs. Orr and Turelli (2001) showed that under our model, the number of DMIs accumulated by time t satisfies Var(It) ≈ E(It), so it is reasonable to approximate It by a Poisson distribution. Hence, we can model reciprocal breakdown scores with all U DMIs by applying (13) assuming that It is Poisson, with the individual DMI effects chosen as independently and identically distributed observations. We used a gamma distribution for DMI effects with mean 1 and variance equal to CV2. For a range of times, we generated independent values of S12 and S21 and then calculated A = |S12 − S21|. This process was replicated 10,000 times for each time point and quantiles were computed and compared to the results of (12) with our Gaussian approximation. Figure A1 shows the results with C = 20, α = 0.75, δ1 = 0, and CV = 0.5. This example shows that even for small values of E(Sij), our approximations for the percentiles of A are sufficiently accurate to interpret the data on quantitative asymmetry now available. The complications associated with approximating the distribution of A for relatively short divergence times, e.g., τ <  , are irrelevant to our analyses of qualitative asymmetry.
, are irrelevant to our analyses of qualitative asymmetry.
Figure A1.—
Comparison between numerical predictions for quantitative asymmetry, A, based on our bivariate Gaussian approximation and Equation 12 (dashed curves) and simulation results of a compound Poisson process (dots) as described in appendix b. The colors correspond to 95th (red), 50th (green), and 5th (blue) percentiles of A. Our calculations assume only U DMIs with C = 20, α = 0.75, δ1 = 0 and CV = 0.5.
APPENDIX C: MOMENTS FOR THE RECIPROCAL BREAKDOWN SCORES WITH ONLY U INCOMPATIBILITIES
We derive the moments for the reciprocal breakdown scores (S12, S21) given in (14). We begin with our definition
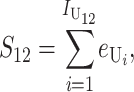 |
(C1) |
where 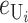 denotes the effect of the ith DMI, and recall our assumptions that these random variables are independent and identically distributed with
denotes the effect of the ith DMI, and recall our assumptions that these random variables are independent and identically distributed with 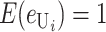 and
and 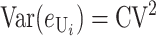 . Using the properties of conditional expectations, we have
. Using the properties of conditional expectations, we have
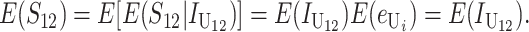 |
(C2) |
Similarly,
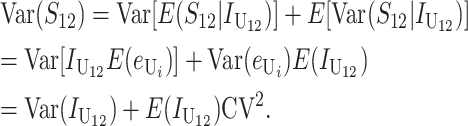 |
(C3) |
To calculate Var(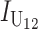 ), we must, as in appendix a, consider the fact that there are three types of DMIs, denoted DD, D1A2, and D2A1, and the number of each follows a binomial distribution. We first consider the distribution of the numbers of each type conditional on K = (
), we must, as in appendix a, consider the fact that there are three types of DMIs, denoted DD, D1A2, and D2A1, and the number of each follows a binomial distribution. We first consider the distribution of the numbers of each type conditional on K = (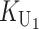 ,
, 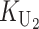 ,
, 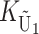 ,
, 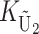 ) and then consider the (negligible) covariance between these random variables induced by their shared dependence on K. Conditional on K, the number of DD DMIs is binomial with parameters
) and then consider the (negligible) covariance between these random variables induced by their shared dependence on K. Conditional on K, the number of DD DMIs is binomial with parameters 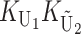 and pU (because each ordered pair of substitutions, corresponding to either possible temporal ordering of the two substitutions, can produce a DMI). Hence, conditional on K, DD DMIs contribute
and pU (because each ordered pair of substitutions, corresponding to either possible temporal ordering of the two substitutions, can produce a DMI). Hence, conditional on K, DD DMIs contribute 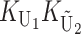 pU(1 − pU) to Var(
pU(1 − pU) to Var(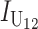 ). In contrast, as explained in appendix a, the number of D1A2 DMIs is binomial with parameters
). In contrast, as explained in appendix a, the number of D1A2 DMIs is binomial with parameters 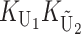 and pU/2. These DMIs contribute
and pU/2. These DMIs contribute 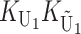 (pU/2)[1 − (pU/2)] to
(pU/2)[1 − (pU/2)] to  ). Adding these and the analogous result for D2A1 DMIs and comparing the result to (A4) for E(
). Adding these and the analogous result for D2A1 DMIs and comparing the result to (A4) for E( ), we have
), we have
 |
(C4) |
Next, note that
 |
(C5) |
Combining (C3) and (C5), we see that
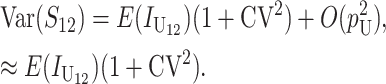 |
(C6) |
Similarly, we have
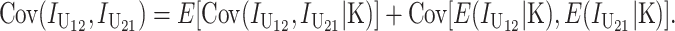 |
(C7) |
The first term is 0 because each DMI is assumed to arise independently, given the sequence of substitutions. The second term is the covariance between two conditional expectations, each of which is proportional to pU (see Equation A4); hence, this covariance is O( ). Overall, we have
). Overall, we have
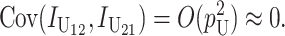 |
(C8) |
The final step in our derivation of (14) is to note that
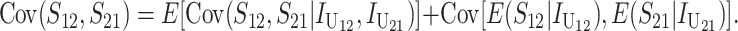 |
(C9) |
The first term in (C9) is zero because conditioned on the number of DMIs, S12 and S21 are just sums of independent random variables. The second term is O(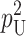 ) because it is a covariance between terms of the form
) because it is a covariance between terms of the form 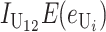 and hence differs from
and hence differs from 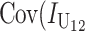 ,
, 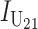 ), which is O(
), which is O(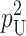 ), only by a constant.
), only by a constant.
APPENDIX D: MOMENTS FOR THE RECIPROCAL BREAKDOWN SCORES WITH BOTH U AND B INCOMPATIBILITIES
Here we derive the means, variances, and covariance for (S12, S21). We begin with our definitions,
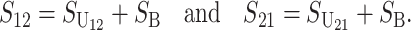 |
(D1) |
The means can be assembled from (18), (21), and (22) to produce
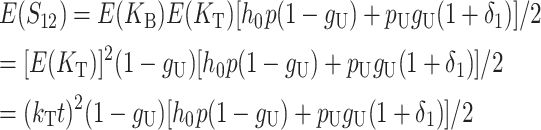 |
(D2a) |
and
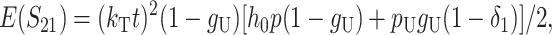 |
(D2b) |
where we have used our convention that the single class of U DMIs being considered has average effect 1.
Next we approximate the variances and covariance of S12 and S21. When both B and U DMIs are considered, S12 and S21 are correlated for two reasons. The major contribution comes from the fact that both S12 and S21 include  (see Equation D1). This contributes Var(
(see Equation D1). This contributes Var( ) to Cov(S12, S21). As noted in appendix c, additional negligible contributions arise from correlations between
) to Cov(S12, S21). As noted in appendix c, additional negligible contributions arise from correlations between 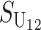 and
and 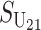 and between
and between 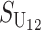 and
and 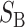 caused by their shared dependence on K = (KU1, KU2,
KB1, KB2). As in appendix c, we show that these covariance terms are second order in the small parameters p and pU and hence are ignored in our approximations. To calculate the variances and covariances, we must specify the variances of the individual DMI effects
caused by their shared dependence on K = (KU1, KU2,
KB1, KB2). As in appendix c, we show that these covariance terms are second order in the small parameters p and pU and hence are ignored in our approximations. To calculate the variances and covariances, we must specify the variances of the individual DMI effects 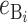 and
and 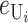 in (1). For simplicity, we assume that all of the random variables describing DMI effects have equal coefficients of variation (CV); i.e., Var(eU)/[E(eU)]2 = Var(eB)/[E(eB)]2 = CV2, where E(eU) = 1 and E(eB) = h0.
in (1). For simplicity, we assume that all of the random variables describing DMI effects have equal coefficients of variation (CV); i.e., Var(eU)/[E(eU)]2 = Var(eB)/[E(eB)]2 = CV2, where E(eU) = 1 and E(eB) = h0.
From (D1) we have
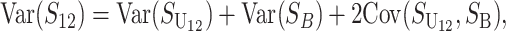 |
(D3) |
where Var( ) is given by (C6) and Var(SB) is given by Equation 21 of Orr and Turelli (2001). Following the derivation for (C8) above, we see that
) is given by (C6) and Var(SB) is given by Equation 21 of Orr and Turelli (2001). Following the derivation for (C8) above, we see that
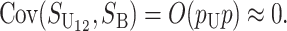 |
(D4) |
Assembling the pieces, we find that to leading order in p and pU (i.e., ignoring products of these small terms),
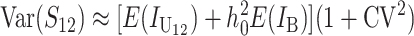 |
(D5a) |
and
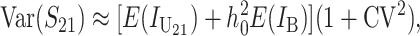 |
(D5b) |
where the expectations are given by (18) and (22).
Finally, we have
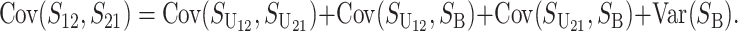 |
(D6) |
As shown in appendix c and above, each of the first three terms is proportional to products of the small parameters p and pU; hence to leading order (i.e., ignoring products of small terms),
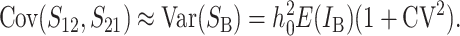 |
(D7) |
Note that if we consider only B DMIs, we can set gU = 0 [which implies that 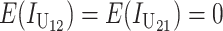 ], h0 = 1, and identify
], h0 = 1, and identify 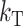 (which sums the substitutions over both taxa) with 2k in Orr and Turelli (2001) to show that (D2a) and (D5a) reproduce the results in Orr and Turelli (2001). Conversely, with only U DMIs, (D2), (D5), and (D6) reduce to (14) in the text.
(which sums the substitutions over both taxa) with 2k in Orr and Turelli (2001) to show that (D2a) and (D5a) reproduce the results in Orr and Turelli (2001). Conversely, with only U DMIs, (D2), (D5), and (D6) reduce to (14) in the text.
We dedicate this article to H. Allen Orr, whose pioneering theoretical work on DMIs made our analyses possible.
References
- Arnold, S. J., P. A. Verrell and S. G. Tilley, 1996. The evolution of asymmetry in sexual isolation: a model and test case. Evolution 50 1024–1033. [DOI] [PubMed] [Google Scholar]
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolating in Drosophila. Proc. Natl. Acad. Sci. USA 100 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson, W., 1901. Experiments in plant hybridization. J. R. Hortic. Soc. 26 1–32. [Google Scholar]
- Berlin, S., and H. Ellegren, 2006. Fast accumulation of nonsynonymous mutations on the female-specific W chromosome in birds. J. Mol. Evol. 62 66–72. [DOI] [PubMed] [Google Scholar]
- Bernacchi, D., and S. D. Tanksley, 1997. An interspecific backcross of Lycopersicon esculentum × L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147 861–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick, D. I., and T. J. Near, 2005. Tempo of hybrid inviability in centrarchid fishes (Teleostei: Centrarchidae). Evolution 59 1754–1767. [PubMed] [Google Scholar]
- Brideau, N. J., H. A. Flores, J. Wang, S. Maheshwari, X. Wu et al., 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314 1292–1295. [DOI] [PubMed] [Google Scholar]
- Bridgen, M. P., 1994. A review of plant embryo culture. Hortscience 29 1243–1246. [Google Scholar]
- Burt, A., and R. Trivers, 2006. Genes in Conflict. Belknap, Cambridge, MA.
- Bushell, C., M. Spielman and R. J. Scott, 2003. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camadro, E. L., and R. W. Masuelli, 1995. A genetic model for the endosperm balance number (EBN) in the wild potato Solanum Acaule Bitt and 2 related diploid species. Sex. Plant Reprod. 8 283–288. [Google Scholar]
- Charlesworth, B., J. A. Coyne and N. H. Barton, 1987. The relative rates of evolution of sex chromosomes and autosomes. Am. Nat. 130 113–146. [Google Scholar]
- Charlesworth, B., H. Borthwick, C. Bartolomé and P. Pignatelli, 2004. Estimates of the genome mutation rate for detrimental alleles in Drosophila melanogaster. Genetics 167 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y., J. P. Mower, Y. L. Qiu and J. D. Palmer, 2004. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. USA 101 17741–17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. a Patterns of speciation in Drosophila. Evolution 43 362–381. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. b Two rules of speciation, pp. 180–207 in Speciation and Its Consequences, edited by D. Otte and J. A. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., and H. A. Orr, 1997. “Patterns of speciation in Drosophila” revisited. Evolution 51 295–303. [DOI] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 2004. Speciation. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., S. Simeonidis and P. Rooney, 1998. Relative paucity of genes causing inviability in hybrids between Drosophila melanogaster and D. simulans. Genetics 150 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresti, M., and A. Tiezzi, 1997. Sexual Plant Reproduction. Springer, Berlin.
- Darwin, C., 1859. On the Origin of Species. John Murray, London.
- Dawe, R. K., and S. Henikoff, 2006. Centromeres put epigenetics in the driver's seat. Trends Biochem. Sci. 31 662–669. [DOI] [PubMed] [Google Scholar]
- De Nettancourt, D., 2001. Incompatibility and Incongruity in Wild and Cultivated Plants. Springer, Berlin.
- Dettman, J. R., D. J. Jacobson, E. Turner, A. Pringle and J. W. Taylor, 2003. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution 57 2721–2741. [DOI] [PubMed] [Google Scholar]
- Dobzhansky, T., 1936. Studies on hybrid sterility II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T., 1937. Genetics and the Origin of Species. Columbia University Press, New York.
- Ehlenfeldt, M. K., and R. E. Hanneman, 1988. Genetic control of endosperm balance number (EBN)—3 additive loci in a threshold-like system. Theor. Appl. Genet. 75 825–832. [Google Scholar]
- Frank, S. A., 1989. The evolutionary dynamics of cytoplasmic male sterility. Am. Nat. 133 345–376. [Google Scholar]
- Frova, C., and M. E. Pe, 1997. Gene expression during pollen development, pp. 31–48 in Sexual Plant Reproduction, edited by M. Cresti and A. Tiezzi. Springer, Berlin.
- Gallant, S. L., and D. J. Fairbairn, 1997. Patterns of postmating reproductive isolation in a newly discovered species pair, Aquarius remigis and A. remigoides (Hemiptera; Gerridae). Heredity 78 571–577. [DOI] [PubMed] [Google Scholar]
- Gerstel, D. U., 1954. A new lethal combination in interspecific cotton hybrids. Genetics 39 628–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun, P., 1976. Cytoplasmic Genes and Evolution. Columbia University Press, New York.
- Gutierrez-Marcos, J. F., P. D. Pennington, L. M. Costa and H. G. Dickinson, 2003. Imprinting in the endosperm: a possible role in preventing wide hybridization. Philos. Trans. R. Soc. Lond. B 358 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane, J. B. S., 1922. Sex-ratio and unidirectional sterility in hybrid animals. J. Genet. 12 101–109. [Google Scholar]
- Harrison, J. S., and R. S. Burton, 2006. Tracing hybrid incompatibilities to single amino acid substitutions. Mol. Biol. Evol. 23 559–564. [DOI] [PubMed] [Google Scholar]
- Harrison, R. G., 1983. Barriers to gene exchange between closely related cricket species. I. Laboratory hybridization studies. Evolution 37 245–251. [DOI] [PubMed] [Google Scholar]
- Hollingshead, L., 1930. A lethal factor in Crepis effective only in an interspecific hybrid. Genetics 15 114–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle, D., 1998. How should we explain variation in the genetic variance of traits? Genetica 103 241–253. [PubMed] [Google Scholar]
- Johnston, S. A., and R. E. Hanneman, 1999. The nature of the genetic control of endosperm balance number based on aneuploid analysis of Datura. Sex. Plant Reprod. 12 71–75. [Google Scholar]
- Kaneshiro, K. Y., 1980. Sexual isolation, speciation and the direction of evolution. Evolution 34 437–444. [DOI] [PubMed] [Google Scholar]
- Katsiotis, A., R. E. Hanneman and R. A. Forsberg, 1995. Endosperm balance number and the polar-nuclear activation hypothesis for endosperm development in interspecific crosses of Solanaceae and Gramineae respectively. Theor. Appl. Genet. 91 848–855. [DOI] [PubMed] [Google Scholar]
- Kawanishi, M., and T. K. Watanabe, 1981. Genes affecting courtship song and mating preference in Drosophila melanogaster, Drosophila simulans and their hybrids. Evolution 35 1128–1133. [DOI] [PubMed] [Google Scholar]
- Kondrashov, A. S., 2003. Accumulation of Dobzhansky-Muller incompatibilities within a spatially structured population. Evolution 57 151–153. [DOI] [PubMed] [Google Scholar]
- Levin, D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28 5–11. [Google Scholar]
- Lewis, D., and L. K. Crowe, 1958. Unilateral interspecific incompatibility in flowering plants. Heredity 12 233–256. [Google Scholar]
- Lijtmaer, D. A., B. Mahler and P. L. Tubaro, 2003. Hybridization and postzygotic isolation patterns in pigeons and doves. Evolution 57 1411–1418. [DOI] [PubMed] [Google Scholar]
- Lin, B. Y., 1984. Ploidy barrier to endosperm development in maize. Genetics 107 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke, B., and T. Borner, 2005. Mitochondrial effects on flower and pollen development. Mitochondrion 5 389–402. [DOI] [PubMed] [Google Scholar]
- Macnair, M. R., and P. Christie, 1983. Reproductive isolation as a pleiotropic effect of copper tolerance in Mimulus guttatus. Heredity 50 295–302. [Google Scholar]
- Mascarenhas, J. P., 1989. The male gametophyte of flowering plants. Plant Cell 1 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, K., T. Khin and Y. Sano, 2003. A gene block causing cross-incompatibility hidden in wild and cultivated rice. Genetics 165 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr, E., 1986. Joseph Gottlieb Kölreuter's contributions to biology. Osiris 2 135–176. [Google Scholar]
- Mendelson, T. C., B. D. Inouye and M. D. Rausher, 2004. Quantifying patterns in the evolution of reproductive isolation. Evolution 58 1424–1433. [DOI] [PubMed] [Google Scholar]
- Moyle, L. C., and E. B. Graham, 2005. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle, L. C., M. S. Olson and P. Tiffin, 2004. Patterns of reproductive isolation in three angiosperm genera. Evolution 58 1195–1208. [DOI] [PubMed] [Google Scholar]
- Muller, H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by J. Huxley. Clarendon Press, Oxford.
- Muller, H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6 71–125. [Google Scholar]
- Navajas, M., A. Tsagkarakov, J. Lagnel and M. J. Perrot-Minnot, 2000. Genetic differentiation in Tetranychus urticae (Acari: Tetranychidae): Polymorphism, host races or sibling species? Exp. Appl. Acarol. 24 365–376. [DOI] [PubMed] [Google Scholar]
- Olby, R. C., 1966. a Origins of Mendelism. Schocken Books, New York.
- Olby, R. C., 1966. b Joseph Koelreuter, 1733–1806, pp. 33–65 in Late Eighteenth Century European Scientists, edited by R. C. Olby. Pergamon Press, Oxford.
- Oliver, C. G., 1978. Experimental hybridization between the nymphalid butterflies Phyciodes tharos and P. campestris montana. Evolution 32 594–601. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1989. Localization of genes causing postzygotic isolation in two hybridizations involving Drosophila pseudoobscura. Heredity 63 231–237. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1993. A mathematical model of Haldane's rule. Evolution 47 1606–1611. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1995. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics 139 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr, H. A., and L. H. Orr, 1996. Waiting for speciation: the effect of population subdivision on the time to speciation. Evolution 50 1742–1749. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., and M. Turelli, 2001. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution 55 1085–1094. [DOI] [PubMed] [Google Scholar]
- Ottaviano, E., and D. L. Mulcahy, 1989. Genetics of angiosperm pollen. Adv. Genet. 26 1–64. [Google Scholar]
- Parker, H. R., D. P. Philipp and G. S. Whitt, 1985. Gene regulatory divergence among species estimated by altered developmental patterns in interspecific hybrids. Mol. Biol. Evol. 56 217–250. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2002. Patterns of postzygotic isolation in Lepidoptera. Evolution 56 1168–1183. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., and H. A. Orr, 1998. Haldane's rule in taxa lacking hemizygous X. Science 282 952–954. [DOI] [PubMed] [Google Scholar]
- Presgraves, D. C., L. Balagopalan, S. M. Abmayr and H. A. Orr, 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423 715–719. [DOI] [PubMed] [Google Scholar]
- Rakocinski, C. F., 1984. Aspects of reproductive isolation between Campostoma oligolepis and Campostoma anomalum pullum (Cypriniformes: Cyprinidae) in northern Illinois. Am. Midl. Nat. 112 138–145. [Google Scholar]
- Rand, D. M., A. G. Clark and L. M. Kann, 2001. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics 159 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., R. A. Haney and A. J. Fry, 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19 645–653. [DOI] [PubMed] [Google Scholar]
- Rapp, R. A., and J. F. Wendel, 2005. Epigenetics and plant evolution. New Phytol. 168 81–91. [DOI] [PubMed] [Google Scholar]
- Rashid, A., and P. A. Peterson, 1992. The Rss system of unidirectional cross-incompatibility in maize. 1. Genetics. J. Hered. 83 130–134. [Google Scholar]
- Roberts, H. F., 1929. Plant Hybridization Before Mendel. Princeton University Press, Princeton, NJ.
- Sackton, T. B., R. A. Haney and D. M. Rand, 2003. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution 57 2315–2325. [DOI] [PubMed] [Google Scholar]
- Salazar, C. A., C. D. Jiggins, C. F. Arias, A. Tobler, E. Berminghan et al., 2005. Hybrid incompatibility is consistent with a hybrid origin of Heliconius heurippa Hewiston from its close relatives, Heliconius cydno Doubleday and Heliconius melpomene Linnaeus. J. Evol. Biol. 18 247–256. [DOI] [PubMed] [Google Scholar]
- Sandstedt, S. A., and P. K. Tucker, 2005. Male-driven evolution in closely related species of the mouse genus Mus. J. Mol. Evol. 61 138–144. [DOI] [PubMed] [Google Scholar]
- Sawamura, K., 1996. Maternal effect as a cause of exceptions for Haldane's rule. Genetics 143 609–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable, P. S., and R. P. Wise, 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3 175–180. [Google Scholar]
- Sweigart, A. L., L. Fishman and J. H. Willis, 2006. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics 172 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafraniec, K., D. M. Wloch, P. Sliwa, R. Borts and R. Korona, 2003. Small fitness effects and weak genetic interactions between deleterious mutations in heterozygous loci of the yeast Saccharomyces cerevisiae. Genet. Res. 82 19–31. [DOI] [PubMed] [Google Scholar]
- Tadros, W., and H. D. Lipshitz, 2005. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232 593–608. [DOI] [PubMed] [Google Scholar]
- Tao, Y., and D. L. Hartl, 2003. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. III. Heterogeneous accumulation of hybrid incompatibilities, degree of dominance, and implications for Haldane's rule. Evolution 57 2580–2598. [DOI] [PubMed] [Google Scholar]
- Thornton, W. A., 1955. Interspecific hybridization in Bufo woodhousei and Bufo valliceps. Evolution 9 455–468. [Google Scholar]
- Tiffin, P., M. S. Olson and L. C. Moyle, 2001. Asymmetric crossing barriers in angiosperms. Proc. R. Soc. Lond. Ser. B 268 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and D. J. Begun, 1997. Haldane's rule and X chromosome size in Drosophila. Genetics 147 1799–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 1995. The dominance theory of Haldane's rule. Genetics 140 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli, M., and H. A. Orr, 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrana, P. B., J. A. Fossella, P. Matteson, T. del Rio, M. J. O'Neill et al., 2000. Genetic and epigenetic incompatibilities underlie hybrid dysgenesis in Peromyscus. Nat. Genet. 25 120–124. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., and S. K. Dey, 2006. Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 7 185–199. [DOI] [PubMed] [Google Scholar]
- Welch, J. J., 2004. Accumulating Dobzhansky-Muller incompatibilities: reconciling theory and data. Evolution 58 1145–1156. [DOI] [PubMed] [Google Scholar]
- Whittle, C. A., and M. O. Johnston, 2002. Male-driven evolution of mitochondrial and chloroplastidial DNA sequences in plants. Mol. Biol. Evol. 19 938–949. [DOI] [PubMed] [Google Scholar]
- Willemse, M., and A. Van Lammeren, 2002. Fertilization, from attraction to embryo. Biologia 57 13–22. [Google Scholar]
- Willett, C. S., and R. S. Burton, 2001. Viability of cytochrome C genotypes depends on cytoplasmic backgrounds in Tigriopus californicus. Evolution 55 1592–1599. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., W. H. Li and P. M. Sharp, 1987. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 84 9054–9058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram, S., 2003. The Mathematica Book, Ed. 5. Wolfram Media, Champaign, IL.
- Wu, C.-I, and A. W. Davis, 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane's rule and its genetic bases. Am. Nat. 142 187–212. [DOI] [PubMed] [Google Scholar]