Abstract
Characterization of mitochondrial genomes from individual Thaumamermis cosgrovei nematodes, obligate parasites of the isopod Armadillidium vulgare, revealed that numerous mtDNA haplotypes, ranging in size from 19 to 34 kb, are maintained in several spatially separated isopod populations. The magnitude and frequency of conspecific mtDNA size variation is unprecedented among all studied size-polymorphic metazoan mitochondrial genomes. To understand the molecular basis of this hypervariation, complete nucleotide sequences of two T. cosgrovei mtDNA haplotypes were determined. A hypervariable segment, residing between the atp6 and rrnL genes, contributes exclusively to T. cosgrovei mtDNA size variation. Within this region, mtDNA coding genes and putative nonfunctional sequences have accumulated substitutions and are duplicated and rearranged to varying extents. Hypervariation at this level has enabled a first insight into the life history of T. cosgrovei. In five A. vulgare hosts infected with multiple nematodes, four carried nematodes with identical mtDNA haplotypes, suggesting that hosts may become infected by ingesting a recently hatched egg clutch or become parasitized by individuals from the same brood prior to dispersal of siblings within the soil.
METAZOAN mitochondrial genomes are generally small in size (12–20 kb) and encode 36–37 genes (Boore 1999) whose products contribute to the processes of electron transport, oxidative phosphorylation, and mitochondrial protein synthesis. Much larger genomes (up to 43 kb) have occasionally been described and their expanded sizes are usually the result of duplications of mitochondrial DNA (mtDNA) sequences, rather than increased gene content (Fuller and Zouros 1993; Boore et al. 2005; Dellaporta et al. 2006). A comprehensive survey of animal mitochondrial DNA size polymorphism revealed that small, incremental mtDNA length variation is common among conspecifics; much of this variation is caused by copy number variation of short, noncoding tandem repeat arrays, often confined to the main replication control region of the molecule (Lunt et al. 1998). Larger size variations also occur among animal mtDNAs, but at much lower frequencies (Gach and Brown 1997).
We report here an atypically large and hypervariable mtDNA found in the nematode Thaumamermis cosgrovei, an obligate parasite of the terrestrial isopod Armadillidium vulgare (Poinar 1981). In a study of >100 T. cosgrovei individuals from several geographically isolated populations, we found that this mitochondrial genome is strikingly variable in size (Tang and Hyman 2005); individual nematodes maintain haplotypes ranging from 19 to 34 kb. Surprisingly, no two individuals appear to share the same haplotype, and we have employed the term “hypervariation” to describe this observation. Mitochondrial genome size heteroplasmy within some individuals was also observed. To understand the molecular basis of hypervariation among T. cosgrovei mitochondrial genomes, complete nucleotide sequences of two T. cosgrovei mtDNA haplotypes have been determined. Our data revealed that T. cosgrovei mtDNA can be divided into a constant region and a hypervariable region. It is the hypervariable region that contributes exclusively to T. cosgrovei mtDNA size variation, as within this locus, mtDNA coding and putative nonfunctional sequences have been duplicated and rearranged to varying extents. Different copy numbers, sequence rearrangement, and extensive nucleotide substitutions within these repeated elements give rise to size polymorphism and sequence divergence among T. cosgrovei mtDNAs.
The extensive and frequent mtDNA size variation in T. cosgrovei may also provide the key to a better understanding of its life history. For T. cosgrovei, no information is available as to the mechanism of isopod infection and dispersal. Infrequently, a single isopod host is multiply infected with 2–16 juvenile nematodes. Hypervariable mtDNA haplotypes can be used to test whether individuals parasitizing the same host are the result of spatially or temporally separate infection events.
MATERIALS AND METHODS
Nematodes:
Isopod hosts were collected from the Botanic Garden on the University of California-Riverside campus and from private yards in Riverside, Highland, and Rancho Cucamonga, California. The isopods were bent backward by hand, splitting the exoskeleton, and submerged into 0.15 m NaCl. Slim white J3 stage nematodes emerged from the thoracic cavity of the host and were transferred with a dental pick into fresh saline.
DNA isolation:
A rapid alkaline lysis procedure was employed to prepare an enriched population of circular molecules from nematodes that was suitable template for rolling circle amplification (RCA) reactions, as described previously (Tang and Hyman 2005). T. cosgrovei total genomic DNA was purified from individual nematodes using a mini-lysate procedure (Powers et al. 1986) and used for hybridization analysis and as template in conventional PCR amplifications.
PCR amplification:
Primer pairs were designed on the basis of conserved nematode cytochrome oxidase subunit 1 (cox1), large mitochondrial ribosomal RNA (rrnL), cytochrome b (cob), and ATP synthase F0 subunit 6 (atp6) gene sequences and are listed in Table 1.
TABLE 1.
Primers used in PCR amplification
| Primer | Primer sequence |
|---|---|
| cox1F | 5′-CGGTATGGAGACTACCTAGATTCCTACTTG-3′ |
| cox1R | 5′-GATCGTCTGTCAATATCCAACCCAACTC-3′ |
| cobF | 5′-AATCTGACCTCGATGTTGACTTAA-3′ |
| cobR | 5′-TTAAGTCAACATCGAGGTCAGATT-3′ |
| rrnLF | 5′-CTTCGATCAATTCCTAACAAGCTGGGTGC-3′ |
| rrnLR | 5′-GCACCCAGCTTGTTAGGAATTGATCGAAG-3′ |
| atp6F | 5′-TGTAATATTATAGCAGGACATGTG-3′ |
| atp6R | 5′-CACATGTCCTGCTATAATATTACA -3′ |
Regions between rrnL and cox1, cox1 and cob, and cob and atp6 were amplified by long-distance PCR (Expand 20kbplus kit, Roche, Mannheim, Germany). Long-distance PCR amplifications were performed in 50-μl reactions with 38.5 μl sterilized distilled water, 5 μl 10× Expand PCR reaction buffer (with 22.5 mm MgCl2), 2.5 μl 10 mm dNTP, 1 μl of each primer (10 μm), 1 μl DNA template prepared by a mini-lysate procedure (Powers et al. 1986), and 1 μl Expand Long Template enzyme mix. Long-distance PCR cycling conditions include 92° for 2 min (initial strand denaturation), followed by 35 cycles that included 92° for 10 sec (denaturation), 56° for 40 sec (primer annealing), and 68° for 10 min (extension). A final elongation step was then conducted at 68° for 7 min.
RCA:
RCA reactions were conducted using the TempliPhi kit (Amersham Biosciences, Piscataway, NJ) as previously described (Tang and Hyman 2005).
Restriction enzyme digestion, gel electrophoresis, and Southern blot analysis:
Total genomic DNA from individual T. cosgrovei nematodes were digested to completion with restriction endonucleases according to manufacturer's instructions. Restriction fragment products were fractionated by agarose gel electrophoresis, visualized by ethidium bromide staining, and then transferred to Immobilon-NY+ membranes (Millipore, Bedford, MA). Digoxigenin-tagged DNA probes were generated by random primed labeling, hybridized to target sequences, immunodetected with antidigoxigenin-AP and Fab fragments and then visualized with the colorimetric substrates nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche, Mannheim, Germany).
Molecular cloning and sequencing:
PCR products were cloned into the pGEM-T EASY vector (Promega, Madison, WI) via TA cloning. Targeted mtDNA restriction fragments were excised from agarose gels using the QIAEXII gel extraction kit (QIAGEN, Valencia, CA) and subcloned into the plasmid pCR2.1 (Invitrogen, Carlsbad, CA). Sequencing reactions for recombinant clones were carried out at the University of California-Riverside Institute for Integrative Genome Biology Core Facility on an Applied Biosystems (Foster City, CA) 3730xl automated sequencer.
Sequence analysis:
DNA sequence assembly and alignment were conducted using the Accelrys (San Diego) SeqWeb platform v.2 of the GCG package (Madison, WI). Protein and rRNA gene sequences were identified by comparing their similarity to published metazoan mtDNA sequences. Some tRNA genes were recognized by tRNAscan-SE (Lowe and Eddy 1997) and DOGMA algorithms (Wyman et al. 2004); others were identified by making visual searches for their anticodon sequences and screened for the potential of adjacent sequences to fold into cloverleaf structures. The secondary structure of rRNA genes was deduced by analogy to other published nematode mitochondrial rRNA gene structures and drawn with the RnaViz program (De Rijk and De Wachter 1997).
Mitochondrial gene rearrangement distance determination:
Mitochondrial gene rearrangement distances among different nematodes were determined by GRIMM (Multiple Genome Rearrangements; Tesler 2002). The rearrangement distance between any two genomes is the minimum number of rearrangement steps (including reversals and translocations) required to transform one genome into the other (Sankoff et al. 1992).
RESULTS
Physical characteristics of T. cosgrovei mtDNA: large size, high frequency of conspecific size variation, and heteroplasmy:
Two approaches were used to estimate the sizes of the mtDNA molecules from different individual T. cosgrovei nematodes. These included transfer-hybridization experiments with total genomic DNA and restriction enzyme digests of RCA-amplified complete mitochondrial genomes.
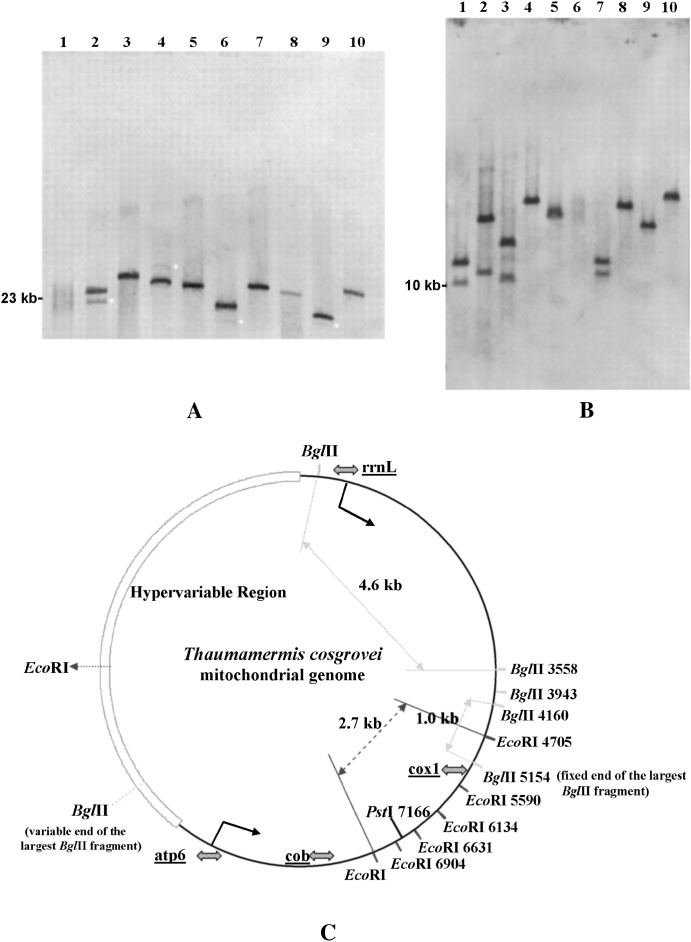
T. cosgrovei mtDNA displays a remarkable frequency and extent of length variation, as demonstrated by Southern blot analysis. Figure 1, A and B, depicts representative examples of mtDNA size polymorphism within nine individual T. cosgrovei nematodes as revealed by transfer hybridization of an 11-kb digoxigenin-labeled mtDNA segment encompassing rrnL to atp6 (Figure 1C) to PstI- and EcoRI-cleaved genomic DNA. PstI is expected to linearize the T. cosgrovei mitochondrial genome on the basis of nucleotide sequence (Figure 1C); thus, the size of the hybridized PstI fragment represents the length of the complete mitochondrial genome in each genomic digest. As shown in Figure 1A, the size of the hybridized PstI fragment varies considerably among individuals, indicating mtDNA length heterogeneity within the population. Four of the nine T. cosgrovei individuals examined here are heteroplasmic for two distinct size classes of mtDNA (Figure 1A, lanes 2, 4, 6, and 9); typically, in these heteroplasmic individuals, one mtDNA size class is present in substoichiometrical yields. Another restriction enzyme, EcoRI, is known to cleave the molecules at six invariant sites located within a 2.7-kb region (between positions 4705 and 7371) (Figure 1C). Comparison of the hybridization profiles of PstI and EcoRI digests from the homoplasmic individuals revealed that EcoRI might also cleave at one additional position in mtDNAs of some individuals. The size of the hybridized EcoRI fragment (Figure 1B, lanes 4, 5, and 8–10) or the sum of the sizes of the two hybridized EcoRI fragments (Figure 1B, lanes 1–3 and 7) represents the length of the complete mtDNA circle minus 2.7 kb of the common sequence (the five small bands that have migrated off the bottom of the gel).
Figure 1.—
Hybridization analysis of restriction-enzyme-digested T. cosgrovei genomic DNAs from 9 individual and 15 pooled nematodes. (A) Genomic DNA cleaved by PstI. Lane 1, pooled nematodes; lanes 2–10, individuals I–X. (B) Genomic DNA cleaved by EcoRI. Lanes 1–5, individuals I–V; lane 6, total DNA from pooled nematodes; lanes 7–10, individuals VI–IX. (C) Restriction map of the T. cosgrovei mitochondrial genome based on the complete nucleotide sequence. The mitochondrial segment used as probe in hybridization (rrnL–atp6) is delimited by angled arrows; shaded arrows show positions of primers used in long-distance PCR amplification; light arrows delimit the 4.6-and 1.0-kb BglII fragments shared by all the T. cosgrovei individuals in the local University of California-Riverside population; dashed arrow defines a 2.7-kb region that is not expected to be visualized by hybridization in B (see text); small light dots indicate the substoichiometric haplotype in heteroplasmic individuals. All nematodes were obtained from A. vulgare hosts collected on the University of California-Riverside campus.
The hybridization profiles in Figure 1, A and B, revealed the presence of mtDNA size polymorphism and the numerous haplotypes maintained in the T. cosgrovei isolate found on the University of California-Riverside campus. When total cellular DNA prepared from 15 pooled nematodes was cleaved by PstI or EcoRI (Figure 1A, lane 1, and Figure 1B, lane 6), distinct bands were not detected; instead, a smear was observed, suggestive of a continuum of mtDNA size variants within the population. Data from PstI and EcoRI cleavage were used to estimate the mtDNA size carried within individual nematodes. This analysis revealed that the sizes of their mitochondrial genomes range from 19 to 34 kb within this sampling.
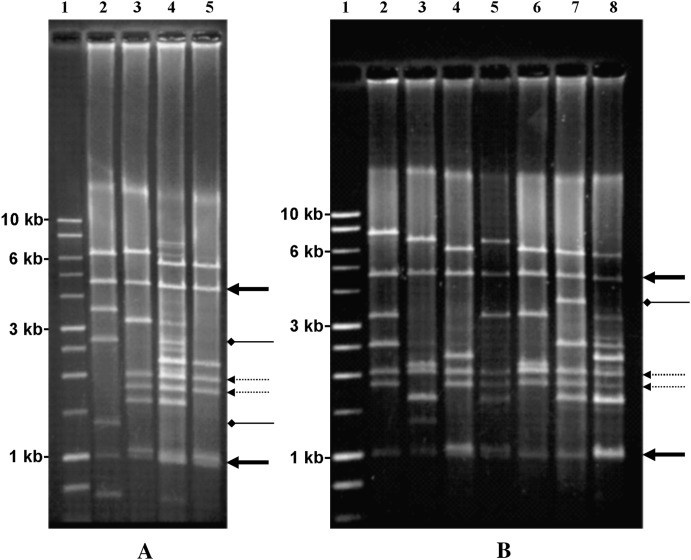
Rolling circle amplification of complete T. cosgrovei mitochondrial genomes (Tang and Hyman 2005) provides a more vivid demonstration of mtDNA size variation (Figure 2, A and B). A complex array of restriction products was obtained when the RCA products amplified from the pooled T. cosgrovei template were digested with BglII (Figure 2A, lane 4, and Figure 2B, lane 8). BglII digestion of RCA products from the eight individual nematodes gave simpler patterns composed of seven to nine visible bands that often represent a subset of the fragments observed when pooled template was employed. On occasion, bands derived from individual haplotypes do not match fragments found in the pool. This is likely the result of sampling a limited number of nematodes to create the mixture. There are only two BglII restriction products shared by all the individuals sampled, sized at 4.6 and 1 kb. Determination of the complete nucleotide sequence of two haplotypes reveals two additional shared bands of 385 and 217 bp, positioned between rrnL and cox1 (Figure 1C; not visible on the gels displayed in Figure 2). The size and distribution of other BglII bands differ among individuals. Some additional bands are shared by many of the individuals (Figure 2, dashed arrows), while others may be unique or infrequently shared among individuals (Figure 2, diamond arrowheads).
Figure 2.—
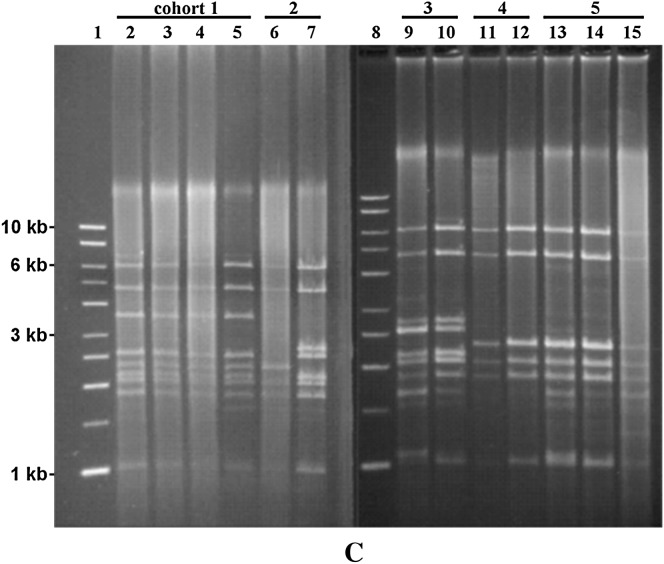
Rolling circle amplification of T. cosgrovei mtDNA from pooled and individual nematodes. Rolling circle amplification reaction products were digested with BglII and fractionated on a 1.0% agarose gel. Solid arrows designate the 4.6- and 1.0-kb BglII fragments shared by all T. cosgrovei individuals in our local population; dashed arrows indicate BglII bands that are shared by the majority of individuals; diamond-headed arrows indicate BglII bands that are infrequently shared among individuals. (A) Lane 1, 1-kb DNA molecular weight marker (Promega); lane 4, BglII-cleaved RCA product using template from eight pooled nematodes; lanes 2, 3, and 5, BglII-cleaved RCA product using template from a single nematode; lanes 2 and 5 correspond with mtDNA haplotypes II and I, respectively. (B) Lane 1, 1-kb DNA molecular weight marker; lane 8, BglII-cleaved RCA product using template from eight pooled nematodes; lanes 2–7, BglII-cleaved RCA product using template from a single nematode. (C) Characterization of mtDNA structures of nematodes from multiply infected isopod hosts. Lanes 1 and 8, 1-kb DNA molecular weight marker; lanes 2–5 (cohort 1), lanes 9 and 10 (cohort 3), lanes 11 and 12 (cohort 4), and lanes 13–15 (cohort 5) represent four hosts containing cohorts with identical mtDNA structures; lanes 6 and 7 (cohort 2) revealed different mtDNA structures. All nematodes were obtained from A. vulgare hosts collected on the University of California-Riverside campus.
mtDNA haplotypes are shared by T. cosgrovei individuals in the same A. vulgare host:
Occasionally, individual isopod hosts are multiply infected with J2 stage nematodes. Our surveys have indicated that A. vulgare hosts can be infected with 2–16 nematodes. Hypervariation of mtDNA haplotypes allows us to test whether multiple T. cosgrovei individuals developing within a single host are the result of temporally or spatially independent infections. Nematodes from multiply infected, individual A. vulgare hosts were dissected as a cohort, and the mtDNA haplotype from each individual nematode was determined using RCA and BglII cleavage. Four of five cohorts (Figure 2C, cohorts 1 and 3–5) reveal identical mtDNA structure whereas differences in mtDNA structure are observed in a fifth group (cohort 2, Figure 2C). Only in multiply infected A. vulgare hosts have we found nematodes with shared mtDNA haplotypes.
Two completely characterized T. cosgrovei mtDNA haplotypes: genome size and general features:
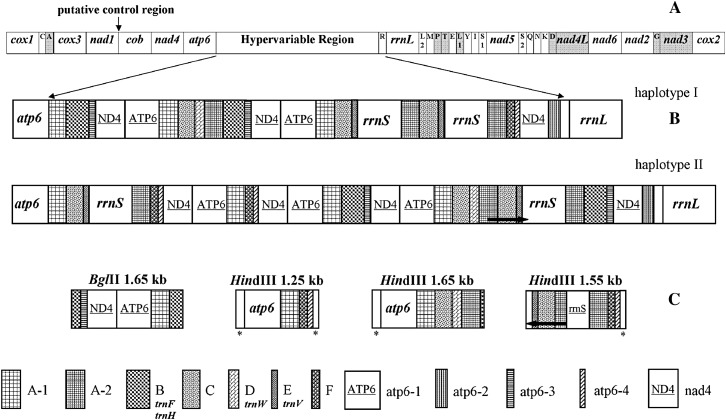
To further understand the molecular basis of hypervariation in T. cosgrovei mitochondrial genome size, complete nucleotide sequence was determined for two mtDNA haplotypes. Haplotype I (Figure 2A, lane 5) was chosen because it displays a rather simple BglII restriction product pattern relative to other haplotypes and all BglII fragments can be found within the mtDNAs of a majority of the individuals sampled. Haplotype II (Figure 2A, lane 2), in contrast, represents a rather unique architecture; except for the 4.6- and 1-kb shared bands, all other BglII fragments derived from haplotype II are present infrequently within the population. Both of the chosen haplotypes are also relatively small in size, facilitating their sequencing and assembly. T. cosgrovei mitochondrial genome haplotype I is 20,013 bp; haplotype II mtDNA is 21,508 bp in size.
Both T. cosgrovei mtDNA haplotypes encode the same set of 36 genes that are typically found in metazoan mtDNA: 12 protein coding genes, two rRNA genes, and 22 tRNA genes. The 36 genes are encoded on both strands (Figure 3A). The ATPase subunit 8 gene is absent from T. cosgrovei mitochondrial genomes, as in all other nematode mtDNAs except that of Trichinella spiralis (Lavrov and Brown 2001). The mitochondrial gene order of T. cosgrovei is unique among known metazoan mitochondrial genomes.
Figure 3.—
Transcription organization of two T. cosgrovei mtDNA haplotypes. (A) Transcriptional map of T. cosgrovei mtDNA common skeleton. (B) Hypervariable regions of haplotypes I and II. (C) Unique segments derived from other hypervariable regions within the haplotype pool. These mtDNA segments were obtained after restriction enzyme digestion and molecular cloning of RCA-amplified mtDNAs using total cellular DNAs from pooled nematodes as template. These fragments did not conform to restriction fragments from the hypervariable region from haplotypes I and II. Although circular, the maps (A) have been displayed in a linear form for ease of comparison. Italized gene names indicate functional, intact gene copies.The tRNA genes are abbreviated using the one-letter amino acid code. L1, L2, S1, and S2 indicate trnL(UAA), trnL(UAG), trnS(UGA), and trnS(UCU), respectively. In A, open boxes are genes transcribed from left to right while shaded boxes are genes transcribed from right to left. Repeated units marked with an asterisk in C are partial nad4 repeats. Thick arrows delimit regions with identical nucleotide sequences in direct (→) or reversed (←) orientations.
On the basis of the restriction enzyme digestion patterns of RCA products from different T. cosgrovei individual nematodes and the complete sequence of the two mtDNA haplotypes, the mtDNA of T. cosgrovei can be divided into a constant region and a hypervariable region (Figures 1C and 3). The constant region includes most of the coding genes and is shared among all the haplotypes. The hypervariable region is located between the atp6 and rrnL genes (Figures 1C and 3). The hypervariable segment contains complete rrnS gene copies (the only copies of this gene in the mtDNA) and partial, likely nonfunctional copies of some mitochondrial genes (nad4, atp6, and sometimes rrnS), duplicated to different copy numbers, resulting in mtDNA size variation and occasional heteroplasmy.
Nucleotide composition:
The A+T content of T. cosgrovei mtDNA is 71.36 and 71.33% for haplotypes I and II, respectively. A+T composition is evenly distributed along the genome. Sequence of the constant mtDNA region is composed of 71.72% A+T. The A+T content decreases slightly to 70.79 and 70.81% in the hypervariable regions of haplotypes I and II.
Protein-coding genes:
Most mitochondrial protein genes in T. cosgrovei are modestly shorter than orthologs in other nematodes except when compared to those of Xiphinema americanum, which maintains the smallest nematode mtDNA described to date (He et al. 2005). Eleven protein-coding genes use the standard start codons typical of invertebrate mitochondrial genes (ATT, ATA, ATG, and TTG) as their translation initiation codons, while an ATC codon was predicted to occur at the beginning of the T. cosgrovei cox2 gene. While not used as an initiation codon in other nematodes studied to date, ATC has been predicted as the start codon for some mitochondrial protein-coding genes in arthropods (Stanton et al. 1997 and Kim et al. 2006), which are considered close relatives of the nematodes as both are pseudocoelomates (Adoutte et al. 1999).
Of the 12 protein coding genes, 10 (atp6, cob, cox1, cox2, cox3, nad1, nad4, nad4L, nad5, and nad6) employ a complete translation termination triplet codon (either TAA or TAG). Two others (nad2 and nad3) appear to use only a template encoded “T,” which can be converted to a complete stop codon TAA by polyadenylation (Ojala et al. 1981).
Each amino acid in T. cosgrovei mtDNA is represented as either a two- or a four-codon family, except for leucine and serine, which are specified by a combination of two families. Within each codon family, codons with either A or T in their third position are used more frequently. The most abundant acids encoded by the T. cosgrovei mitochondrial genome (supplemental Table 1 at http://www.genetics.org/supplemental/) are leucine, serine, phenylalanine, and isoleucine, all of which except serine are nonpolar amino acids. Arginine, cysteine, aspartic acid, and glutamine, the least frequently represented amino acids, are all hydrophilic. These observations are consistent with the fact that the mtDNA-encoded proteins are inner-membrane enzymes involved in oxidative phosphorylation.
Protein-coding gene families:
Both atp6 and nad4 genes are duplicated in the hypervariable region as non-identical, putative, nonfunctional, partial copies. Reiterated atp6 copies are not organized as tandem repeats, but are interspersed between other sequences in the hypervariable region. There are four types of partial atp6 genes (hereafter designated as atp6-1–atp6-4; intact, functional gene designations are italized while possible pseudogenes are in regular type). The functional atp6 gene is 534 bp in length and is located just upstream from the hypervariable region in haplotypes I and II (Figure 3). atp6-1 is 529 bp and differs from the intact atp6 gene at 10 positions, including five nucleotide deletions (supplemental Figure 1A at http://www.genetics.org/supplemental/). Nucleotide sequence divergence between the functional, intact atp6 gene and the putative, nonfunctional atp6-1 repeat mostly resides in the first 24 bp of the functional atp6 gene copy. Encoded amino acids conserved among mermithid nematode ATP6 proteins (SPF at positions 7–9, W at position 76) are not found in atp6-1 copies (supplemental Figure 1B at http://www.genetics.org/supplemental/); therefore, atp6-1 is considered a pseudogene. There are two copies of atp6-1 in haplotype I and three copies in haplotype II (Figure 3B). atp6-2 is composed of 217 bp and its sequence is identical to that of the 5′ 217 bp of atp6-1. Only one copy of atp6-2 is found in the hypervariable region of either haplotype I or haplotype II. atp6-3 is 106 bp in length. Four insertions and 18 substitutions (10 transitions and 8 transversions) can be identified when its sequence is aligned with that of the 5′ portion of the intact atp6 gene, rendering this copy a pseudogene as well. atp6-4, 65 bp in size, presents a further degenerated copy of the atp6 gene. Its 3′ 32-bp nucleotide sequence is the same as that of atp6-3 while the 5′ 33 bp have undergone substantial substitution (supplemental Figure 1A at http://www.genetics.org/supplemental/).
The intact, functional nad4 gene copy is located 5′ to the intact atp6 gene copy within the common skeleton (Figure 3). The nad4 gene is also duplicated multiple times in the hypervariable region (three times in haplotype I and four times in haplotype II). The duplicated copies are direct, interspersed copies as with the atp6 gene family copies (Figure 3). The putative, nonfunctional nad4 pseudogene copies, 432 bp in length, are truncated and represent only ∼36% of the complete nad4 gene, which is 1197 bp in size (supplemental Figure 1C at http://www.genetics.org/supplemental/). Several major deletions have occurred in the nad4 copies. Nucleotide sequences of the nad4 pseudogene copies are almost identical to each other and have not undergone copy-specific changes, as observed among the atp6 pseudogene families.
Ribosomal RNA genes:
The relative positions of the two ribosomal RNA genes (rrnL, rrnS) are not spaced similarly in different T. cosgrovei mtDNA haplotypes. There is one large ribosomal RNA gene (rrnL) located in the constant region. Two copies of the small ribosomal RNA gene (rrnS) reside in the hypervariable region in both haplotypes. In another T. cosgrovei mtDNA haplotype analyzed by restriction enzyme mapping, only one copy of rrnS resides within the hypervariable region (data not shown).
The predicted T. cosgrovei rrnS is 667 bp with a nucleotide composition of T(37.3%) > A(36.9%) > G(14.1%) > C(11.7%) (supplemental Figure 2 at http://www.genetics.org/supplemental/). Its size is similar to that of mitochondrial rrnS genes in other nematodes. The rrnL gene is predicted to be 821 bp (supplemental Figure 3 at http://www.genetics.org/supplemental/) and the nucleotide composition of this gene is T(39.1%) > A(38.6%) > G(12.2%) > C(10.1%). It is the second smallest of all available nematode rrnL gene sequences after that of X. americanum (729 bp; He et al. 2005).
Transfer RNA genes:
Encoded within the T. cosgrovei mtDNA is a set of 22 tRNA genes (Figure 3) typical of metazoan mitochondrial genomes. The mt-tRNA genes vary in size from 52 bp (trnS-UCU, trnS-UGA, trnQ, and trnR) to 62 bp (trnH and trnV). Predicted secondary structures of the mt-tRNA genes (supplemental Figure 4 at http://www.genetics.org/supplemental/) are similar in topology to those found in other nematode mitochondrial genomes.
There are two intact copies of trnF, trnV, and trnH in both mtDNA haplotypes of T. cosgrovei, located within the hypervariable region. Interestingly, phenylalanine, valine, and histidine are not the most frequently used amino acids in T. cosgrovei mitochondrial proteome (9.53, 6.12, and 1.89%; supplemental Table 1 at http://www.genetics.org/supplemental/). The presence of multiple copies of the three tRNA genes appears to be a by-product of duplication events, and not due to selective pressure to increase gene numbers to meet the needs of mitochondrial translation.
Noncoding regions in the constant region:
There are 749 bp in the constant region (6.1%) unassigned to genes. The longest noncoding region, between nad1 and cob genes and 401 bp in length, is possibly the T. cosgrovei mtDNA control region, regulating replication and transcription. It is 73% A+T, slightly elevated relative to the remainder of the mitochondrial genome. This large noncoding region also has the potential to form elaborate stem-loop structures, a common characteristic of control regions (supplemental Figure 5 at http://www.genetics.org/supplemental/).
Hypervariable region in the two fully characterized haplotypes:
The hypervariable region reveals a complicated organization; segments within this expanse can be cataloged into nine elements (gene families atp6, nad4, and rrnS as described earlier and the apparent noncoding elements A, B, C, D, E, and F), some of which can be further divided into subtypes (as with atp6-1–atp6-4) (Figure 3 and Table 2). All repeated sequences are arranged as direct repeats, but not positioned as tandem arrays.
TABLE 2.
Comparisons among repeated elements (gene families and repeated elements A–F)
| Copy no. |
Variation among copies (bp)b |
||||||
|---|---|---|---|---|---|---|---|
| Type | Length (bp) | A+T content (%) | Ia | II | I | II | I and II |
| atp6 | |||||||
| atp6-1 | 529 | 72 | 2 | 3 | 1 | 1 | 2 |
| atp6-2 | 217 | 74 | 1 | 1 | — | — | 0 |
| atp6-3 | 106 | 69 | 2 | 2 | 0 | 0 | 0 |
| atp6-4 | 65 | 68 | 1 | 2 | — | 0 | 0 |
| nad4 | 432 | 73 | 3 | 4 | 4 | 1 | 5 |
| rrnS | 673 | 74 | 2 | 2 | 0 | 3 | 3 |
| A | |||||||
| A-1 | 293 | 68 | 3 | 4 | 1 | 1 | 2 |
| A-2 | 278 | 69 | 3 | 3 | 0 | 0 | 0 |
| B | 291 | 64 | 2 | 2 | 0 | 1 | 1 |
| C | 258 | 69 | 3 | 3 | 1 | 1 | 1 |
| D | 130 | 72 | 1 | 1 | — | — | 0 |
| E | 91 | 74 | 2 | 2 | 0 | 0 | 0 |
| F | 174 | 68 | 1 | 2. | — | 0 | 0 |
Roman numerals I and II refer to haplotypes I and II.
Number of nucleotide differences in base pairs.
Repeated elements A, B, C, D, E, and F vary between 91 to 293 bp in size and range from 64 to 74% in A+T content (Table 2). No significant nucleotide sequence similarities have been detected among these repeated elements nor among any known sequences deposited in the public databases.
There are two subtypes of repeated element A. A-1 is 293 bp in length and is usually adjacent to the 3′-end of the atp6 gene or its pseudogenes (Figure 3). A-2 is 278 bp and is a truncated form of A-1 in that it does not contain the first 5′ 15 bp present in A-1. Copies of repeated element A exist as direct repeats in the hypervariable region and are separated by other types of repeated sequences. Six and seven copies of repeated element A are identified in T. cosgrovei mtDNA haplotypes I and II, respectively.
Repeated element B is 291 bp with 64% A+T content, representing the region of lowest A+T content in the T. cosgrovei mitochondrial genome. Functional trnF and trnH tRNA genes are located in repeated element B. Repeated element C is 258 bp in length and has the potential to encode a polypeptide of 74 amino acids. The element D is 136 bp. The 21 bp at the 3′-end of element D is a duplicated copy of the 3′-end of rrnS. trnW is encoded within the type D component. The trnV gene is situated within element E, which is 91 bp in length. There are two copies of the element E in each haplotype. The element F, 174 bp in length, is present once in haplotype I and twice in haplotype II.
Size variation between haplotypes I and II can be ascribed to the difference in the number of copies of these repeated elements. There is one extra copy of each of the five repeated elements (atp6-1, atp6-4, nad4, type A-1, and F) in haplotype II and the sum of their size (1593 bp) exactly represents the size difference between mtDNA haplotype I and II.
Other characterized fragments from the hypervariable region:
Four additional segments within the hypervariable region have been identified on the basis of restriction enzyme analysis after amplification from template obtained from pooled T. cosgrovei nematodes; these cannot be assigned to either haplotype I or haplotype II (Figure 3C). These fragments include 1.65-kb BglII, 1.65-kb HindIII, and 1.25-kb HindIII fragments that are composed of the same set of direct repeated elements detailed above but not syntenic with those in haplotypes I and II. Typically, there is only one copy of the functional atp6 gene, located in the common skeleton, 5′ of the hypervariable region as found in the two fully characterized haplotypes (Figure 3A). However, a complete atp6 gene copy can also be embedded within the hypervariable region (as in 1.25- and 1.65-kb HindIII fragments) of some T. cosgrovei mtDNAs. The 1555-bp HindIII fragment (Figure 3C) contains complete or deleted rrnS copies in both direct and inverted orientations relative to element F. The region from nucleotide 628 to 969 is identical to the last 342 bp of rrnS 3′ and therefore represents a pseudo-rrnS. DNA sequence from position 449 to 627 in this 1.5-kb element is the 3′ 179 bp of repeated element A and is in inverted direction compared to pseudo-rrnS. Repeated elements C and E are also found in a reverse orientation within this fragment. These observations indicate that additional rearrangements, including inversions, have occurred within the hypervariable region of some mtDNA molecules and contribute to T. cosgrovei mtDNA hypervariation.
DISCUSSION
Hypervariability:
The magnitude of mtDNA haplotype size variation (19–34 kb) in T. cosgrovei is unprecedented among all studied size-polymorphic metazoan mitochondrial genomes. The frequency of intraspecific mtDNA size variation in T. cosgrovei individuals is also unique among animal mitochondrial genomes; virtually every T. cosgrovei individual present as a single parasite within its A. vulgare host carries its own mtDNA haplotype. Only nematodes present as a cohort in multiply infected hosts share a common mtDNA haplotype. We have extended the investigation to three other populations in reproductively isolated locations, including the cities of Riverside, California (∼2 km from the University of California campus), Highland, California (∼24 km north of Riverside), and Rancho Cucamonga, California (∼40 km northwest of the University of California-Riverside campus), and obtained similar results. No common haplotypes among locations were identified; this result stands in stark contrast to what is described for the sea scallop Placopecten magellanicus, where ∼50% of the scallops surveyed maintained the most common type in a single population (La Roche et al. 1990). Since a common T. cosgrovei haplotype was never observed, it is inferred that T. cosgrovei mtDNA molecules undergo frequent rearrangement. In another mermithid nematode, Romanomermis culicivorax, at least three mtDNA size variants (26, 29, and 32 kb) have been fixed (Powers et al. 1986), indicating that extensive mtDNA size polymorphism might be a common trait within this nematode lineage.
It has been proposed that segregation of mitochondrial genome haplotypes is governed by a balance of several factors, including genetic drift during germline vegetative segregation, selection of smaller mtDNA molecules with replicative advantages, and mutation to different mtDNA sizes due to replication errors (Rand and Harrison 1989; Rand 1993). If smaller mtDNAs exhibit prominent selective advantage in replication, it is expected that mitochondrial genomes of shorter contour length would be the most frequent haplotypes in the population and the most prevalent forms within a heteroplasmic individual (Solignac et al. 1984; Rand and Harrison 1986). In T. cosgrovei, smaller mtDNA haplotypes are often present in substoichiometric yields in most heteroplasmic individuals (Figure 1A, lanes 2, 6, and 9). Moreover, in population surveys cataloguing homoplasmic individuals, intermediate-sized mtDNA haplotypes (Figure 1A, lanes 3–5, 7, 8, and 10) are often observed. This result is consistent with earlier observations in crickets and in P. magellanicus, where the most common size class is the one with an intermediate copy number of repeated sequences (Rand and Harrison 1989; Zouros et al. 1992). The abundance of these intermediate-sized molecules may represent a balance between replication errors generating expansion of repeats and recombination or slipped-strand mispairing excising some repeat units.
Nematode gene order:
Nematode mitochondrial protein-coding and rRNA gene orders are remarkably diverse, and especially so within the Enoplea (supplemental Figure 6 at http://www.genetics.org/supplemental/). The rearrangement distance between any two Enoplean nematodes is no smaller than that between any Enoplean nematode and a Chromadorean nematode, including comparisons within the Mermithidae (supplemental Table 2 at http://www.genetics.org/supplemental/). Nematode phylogenetic affinities based on mitochondrial gene order are misleading because of the high frequencies of gene rearrangement; it is unlikely that gene order can be used as a phylogenetic character with reliable signal for Enoplean nematodes, although further sampling among the Enoplea may be required to verify this premise. Similar conclusions regarding the utility of mtDNA gene order as characters in phylogenetic reconstructions were drawn from mollusk mtDNA studies (Le et al. 2000; Dreyer and Steiner 2004).
Molecular evolution of the hypervariable region:
The hypervariable region in T. cosgrovei mtDNA is of unparalleled complexity among metazoan mtDNAs, resulting in length polymorphism generating haplotypes that are not shared between individuals and are not duplicated in reproductively isolated populations. The repeated region contains several active and putative pseudogene copies that vary in copy number among different mtDNA haplotypes. These gene copies are not arranged in tandem, a departure from the most common form of animal mtDNA size polymorphism, which is often related to a variable number of tandem repeats (Lunt et al. 1998). Interestingly, repeats arranged in an inverted orientation can coexist with direct repeats in the same haplotype. The combination of these features is novel for animal mtDNAs. The brachiopod Lingula anatine also has an elaborate mtDNA repeat sequence structure. Nevertheless, the size of repeated segments in L. anatine is constant and all repeated elements are organized in the same orientation (Endo et al. 2005).
Possible molecular mechanisms involved in generating the repeat region:
The most commonly invoked mechanism to explain mtDNA size variation and gene rearrangement is the duplication–random loss model (Moritz et al. 1987; Mueller and Boore 2005). In this model, a portion of the genome is duplicated due to replication errors. Those errors may include slipped-strand mispairing (Levinson and Gutman 1987), imprecise replication initiation or termination (Boore and Brown 1998), or illegitimate priming of DNA synthesis by a tRNA at the replication origin (Jacobs et al. 1989). After duplication, and depending on the patterns of random repeat copy loss due to mutation in the absence of selective pressure, the original mtDNA gene order is either restored or altered. However, this duplication–random loss model cannot easily explain repeated elements positioned in a nontandem fashion nor in inverted orientation.
Within T. cosgrovei mtDNA, repeated sequences are organized as nontandem arrays in the two fully characterized haplotypes and some repeated segments are separated by as much as 7.6 kb (two A elements in haplotype II). Although it is possible for the duplication–random loss model to explain this complicated architecture, this mechanism would require a large number of intermediates after extensive duplication. Moreover, the duplication–random loss model cannot easily account for the presence of inverted repeated units observed in the 1555-bp HindIII fragment identified from the haplotype pool. Mitochondrial DNA recombination can explain such arrangements if mtDNA is broken and the excised region then is reinserted in the opposite orientation; mtDNA sequence translocation accompanied by inversion could be the end product.
Occurrence of recombination in animal mtDNAs was controversial (Moritz et al. 1987). However, a growing literature from physical evidence (Lunt and Hyman 1997; Kajander et al. 2000; Ladoukakis and Zouros 2001a; Hoarau et al. 2002; Passamonti et al. 2003; D'Aurelio et al. 2004; Kraytsberg et al. 2004; Guo et al. 2006), biochemical studies (Thyagarajan et al. 1996), and quantitative genetics analysis (Awadalla et al. 1999; Ladoukakis and Zouros 2001b; Piganeau et al. 2004; Gantenbein et al. 2005; Tsaousis et al. 2005) from metazoa spanning large taxonomic distances suggests that intramolecular and/or intermolecular recombination can operate on animal mtDNAs. Recombination is now proposed to be a major underlying mechanism that generates highly rearranged mitochondrial genomes in numerous animal lineages (Dowton et al. 2003; Miller et al. 2004; Mundy and Helbig 2004; Endo et al. 2005; Nohara et al. 2005; Shao et al. 2005). Size polymorphism, heteroplasmy, and mitochondrial genome rearrangement is a common feature among available Enoplean nematode mtDNA complete sequences, including the family Mermithidae, and stands in contrast to the conserved economization and stable gene orders that typify the Chromadorean nematodes (He et al. 2005). Duplication, coupled with intra- or intermolecular recombination, would be a plausible and parsimonious explanation for the complicated structure of the hypervariable region and occasional heteroplasmy in T. cosgrovei mtDNA and other mermithid nematodes. Such processes must occur frequently to generate and maintain such a rich polymorphism of mtDNAs of vastly different sizes within different T. cosgrovei individuals.
Origins and degeneration of repeated elements:
Generally, when a gene is duplicated and multiple copies of the same gene coexist in one mitochondrial genome, one or more copies of the repeated gene are expected to evolve under relaxed selection pressure, retaining at least one functional copy. Therefore, sequence divergence among members in the same gene family can suggest the relative ages of the corresponding events.
In T. cosgrovei mtDNAs, several gene families and repeated elements within the hypervariable region that contribute to haplotype variation have been defined. For example, atp6 genes are repeated in four different forms. atp6-1 loci differ from the functional atp6 copy in only 10 positions (∼1.9%), indicating atp6-1 repeats may be products of a recent duplication of authentic atp6; atp6-2 repeats match precisely the first 217 bp of atp6-1, suggestive of a recent incomplete duplication of atp6-1. However, considerable degeneration must have occurred to convert the authentic atp6 gene, or atp6-1, into atp6-3 and atp6-4 repeats (79 and 83% similar to corresponding sites in atp6; supplemental Figure 1A at http://www.genetics.org/supplemental/). The nad4 gene and nad4 repeat copies display an even lower level of nucleotide similarity at 68.3% (supplemental Figure 1C at http://www.genetics.org/supplemental/). With such substantial divergences, we may infer that the very first duplication of nad4 and atp6 genes to yield the nad4 and the atp6-3 and atp6-4 pseudogene copies are earlier events. atp6-4 is more similar to atp6-3 than to atp6, atp6-1, or atp6-2, implying that atp6-4 might be the consequence of amplification of atp6-3 and further deterioration. A+T content of other repeated elements is similar to that of the constant region (64–74% vs. 72%), suggesting that these repeats might also have originated from within the mitochondrial genome but decayed beyond recognition. If so, the degree of substitution indicates that these duplication events were also quite early.
Low-level divergence between copies of the same repeated element in current haplotypes:
Although substantial divergences have been identified between functional atp6, nad4 genes, and their putative pseudogene forms, sequence conservation is found at levels >99% among different copies of the same gene family members (e.g., all atp6-1 and nad4 copies) in T. cosgrovei mtDNA (Table 2). This is true for both coding and noncoding regions of the hypervariable segment. Such a high level of nucleotide conservation among repeats could be explained by two possible mechanisms: either these major duplications have occurred recently or concerted evolution mechanisms may be active to homogenize repeats, as suggested for R. culicivorax mtDNA (Hyman and Azevedo 1996). In T. cosgrovei, because virtually every individual nematode carries a different mtDNA haplotype and new haplotypes are generated frequently, recent duplications appear to be the more plausible explanation for the low-level divergence between copies of the same repeat unit.
In addition, we have sequenced 4-kb PCR products from the common skeleton using templates derived from both pooled nematodes and from T. cosgrovei individuals. This expanse encompasses five protein-coding genes. Greater than 99.5% sequence identity was found among all these segments (data not shown). This evidence indicates that strong selection is acting on the common skeleton to maintain the integrity and function of major coding genes.
Factors that may contribute to hypervariation and unique gene order:
Previous research suggests that there may be a link between compact mitochondrial genomes and metabolic efficiency (Selosse et al. 2001). In human cell lines, high levels of mitochondrial duplications lead to measurable reduction in respiratory chain efficiency (Holt et al. 1997). T. cosgrovei, as well as other mermithid nematodes, are obligate parasites of invertebrates and have low metabolic requirements in their postparasitic stage (Nickle 1972). Such life-history traits may be a factor in the evolution of the enlarged and hypervariable T. cosgrovei mitochondrial genome.
An accelerated rate of mtDNA rearrangement associated with a parasitic life style has been proposed (Dowton and Austin 1995; Castro et al. 2002). Factors suggested to impact rearrangements within parasite mitochondrial genomes include elevated speciation rates, increased rates of mutagen flux, and deficiency in mtDNA repair (Castro et al. 2002).
Another possible factor in generating unique parasite mitochondrial genome architectures could involve horizontal gene transfer between the host and the parasite organelle genomes (Davis and Wurdack 2004; Andersson 2005). A. vulgare, the host of T. cosgrovei, also maintains an extremely unusual mitochondrial genome; the host mtDNA is structurally polymorphic, composed of 14-kb monomer and 28-kb dimeric circular molecules (Raimond et al. 1999). However, mitochondrial gene order is not available for A. vulgare and it is not yet possible to determine if there were gene transfers between mitochondrial genomes in this host–parasite interface.
Mitochondrial DNA strand nucleotide composition bias (Crozier and Crozier 1993) can influence the success of rearrangement by placing genes in an unfamiliar, A+T-rich context. Analysis of nucleotide composition in the T. cosgrovei mitochondrial genome reveals no strand bias (data not presented), indicating that this mtDNA may be tolerant of a wide ensemble of events.
Using mtDNA haplotypes to study the mode of isopod infection by T. cosgrovei:
The life history of T. cosgrovei has not been studied and little is known about how A. vulgare becomes infected by nematodes. There are two general routes for multiple nematode infection. One possible mechanism involves host ingestion of an egg clutch containing J1 stage juvenile nematodes (passive infection, as was described for two other mermithid nematodes, Mermis and Pheromermis; Kaiser 1991); a second pathway to parasitism suggests that hatched, multiple J2 stage infectious juveniles independently infect a single host, perhaps separated in a temporal and spatial sense. The second mode is termed active infection and is characteristic of all other mermithid genera with which infectivity has been studied (Kaiser 1991). The extensive hypervariation of mtDNA haplotypes in T. cosgrovei now leads to an answer to the question of whether multiple T. cosgrovei individuals developing within a single host are the result of independent parasitism by infectious juveniles that may represent different maternal lineages. Identical mtDNA haplotypes in four of five cohorts (Figure 2C) suggest that most often (i) isopod hosts ingest T. cosgrovei eggs carrying juveniles from a single maternal source and genetically related T. cosgrovei individuals develop within the host or (ii) these parasites, derived from the same maternal lineage, enter the isopod at an early preparasitic stage just after hatching but before they disperse to different localities in the environment. The difference in mtDNA haplotypes among individuals of one cohort suggests that, on less-frequent occasions, (i) mixed cohorts can be the result of parasitism by a single, heteroplasmic matrilineal lineage or (ii) juvenile T. cosgrovei nematodes derived from different lineages may independently infect isopods by direct penetration. However, the fluidity of T. cosgrovei mtDNA architecture may suggest that the mixed haplotype cohort could instead be due to de novo changes after infection.
Conclusions:
The T. cosgrovei mitochondrial genome displays several features that are unusual among metazoan mtDNA architectures. These distinctive features include (i) frequent and large-scale intraspecific mtDNA size variation (hypervariation); (ii) extensive duplication of coding genes (atp6, nad4, rrnS, trnF, trnV, and trnH); (iii) remnants of ancient duplications (e.g., nad4 and atp6 genes and their pseudogene copies), as well as indications of recent DNA amplifications (e.g., low-level divergence among nad4 pseudogene copies); and (iv) a unique metazoan mtDNA gene order. The two T. cosgrovei mtDNA mitochondrial genomes analyzed in this study provide the first record of multiple intraspecific mtDNA haplotypes with large-scale size variation that has been fully characterized at the nucleotide level. The nontandem arrangement of repeated units in these two haplotypes and the discovery of occasional inversion of some duplicated segments in the haplotype pool suggest that duplication, coupled with recombination, could be an underlying mechanism to generate T. cosgrovei mtDNA size polymorphism and occasional heteroplasmy. These processes must occur very frequently, possibly in real time, to generate and maintain such a rich polymorphism. Full characterization of additional T. cosgrovei mtDNA haplotypes may be helpful in providing additional insights into the molecular evolutionary history of T. cosgrovei mitochondrial genomes.
The high frequency of size variation makes T. cosgrovei an excellent model system for categorizing genetic variation among intraspecific individuals, for investigating the nature and frequencies of mtDNA mutations, for studying life histories and evolutionary origins of mermithid species, and for examining the mechanisms for mitochondrial gene rearrangements and patterns of non-Mendelian inheritance. This system may also offer the opportunity for “real time” observation of sequence trafficking that contributes to hypervariation while maintaining the integrity of the common skeleton.
Acknowledgments
The authors thank Edward Platzer and Paul De Ley for critical reading of the manuscript. We also thank Diana Linares for assistance with analysis of multiply infected nematodes. This work was supported in part by awards from the University of California-Riverside Agricultural Experiment Station, the Committee on Research, University of California Pacific Rim Program, and the Institute for Integrative Genome Biology.
References
- Adoutte, A., G. Balavoine, N. Lartillot and R. De Rosa, 1999. Animal evolution: The end of the intermediate taxa? Trends Genet. 15 104–108. [DOI] [PubMed] [Google Scholar]
- Andersson, J. O., 2005. Lateral gene transfers in eukaryotes. Cell Mol. Life Sci. 62 1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla, P., A. Eyre-Walker and J. M. Smith, 1999. Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286 2524–2525. [DOI] [PubMed] [Google Scholar]
- Boore, J. L., 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore, J. L., and W. M. Brown, 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 8 668–674. [DOI] [PubMed] [Google Scholar]
- Boore, J. L., J. R. Macey and M. Medina, 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 395 311–348. [DOI] [PubMed] [Google Scholar]
- Castro, L. R., A. D. Austin and M. Dowton, 2002. Contrasting rates of mitochondrial molecular evolution in parasitic diptera and hymenoptera. Mol. Biol. Evol. 19 1100–1113. [DOI] [PubMed] [Google Scholar]
- Crozier, R. H., and Y. C. Crozier, 1993. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics 133 97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aurelio, M., C. D. Gajewski, M. T. Lin, W. M. Mauck, L. Z. Shao et al., 2004. Heterologous mitochondrial DNA recombination in human cells. Hum. Mol. Genet. 13 3171–3179. [DOI] [PubMed] [Google Scholar]
- Davis, C. C., and K. J. Wurdak, 2004. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science 305 676–678. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S. L., A. Xu, S. Sagasser, W. Jakob, M. A. Moreno et al., 2006. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc. Natl. Acad. Sci. USA 103 8751–8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijk, P., and R. De Wachter, 1997. RnaViz, a program for the visualisation of RNA secondary structure. Nucleic Acids Res. 25 4679–4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowton, M., and A. D. Austin, 1995. Increased genetic diversity in mitochondrial genes is correlated with the evolution of parasitism in the hymenoptera. J. Mol. Evol. 41 958–965. [DOI] [PubMed] [Google Scholar]
- Dowton, M., L. R. Castro, S. L. Campbell, S. D. Bargon and A. D. Austin, 2003. Frequent mitochondrial gene rearrangements at the hymenopteran nad3-nad5 junction. J. Mol. Evol. 56 517–526. [DOI] [PubMed] [Google Scholar]
- Dreyer, H., and G. Steiner, 2004. The complete sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca). Mol. Phylogenet. Evol. 31 605–617. [DOI] [PubMed] [Google Scholar]
- Endo, K., Y. Noguchi, R. Ueshima and H. T. Jacobs, 2005. Novel repetitive structures, deviant protein-encoding sequences and unidentified ORFs in the mitochondrial genome of the brachiopod Lingula anatina. J. Mol. Evol. 61 36–53. [DOI] [PubMed] [Google Scholar]
- Fuller, K. M., and E. Zouros, 1993. Dispersed discrete length polymorphism of mitochondrial DNA in the scallop Placopecten magellanicus (Gmelin). Curr. Genet. 23 365–369. [DOI] [PubMed] [Google Scholar]
- Gach, M. H., and W. M. Brown, 1997. Characterization and distribution of long tandem duplications in brook stickleback (Culea inconstans) mitochondrial DNA. Genetics 145 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantenbein, B., V. Fet, I. A. Gantenbein-Ritter and F. Balloux, 2005. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae). Proc. Biol. Sci. 272 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. H., S. J. Liu and Y. Liu, 2006. Evidence for recombination of mitochondrial DNA in triploid crucian carp. Genetics 172 1745–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., J. Jones, M. Armstrong, F. Lamberti and M. Moens, 2005. The mitochondrial genome of Xiphinema americanum sensu stricto (Nematoda: Enoplea): considerable economization in the length and structural features of encoded genes. J. Mol. Evol. 61 819–833. [DOI] [PubMed] [Google Scholar]
- Hoarau, G., S. Holla, R. Lescasse, W. T. Stam and J. L. Olsen, 2002. Heteroplasmy and evidence for recombination in the mitochondrial control region of the flatfish Platichthys flesus. Mol. Biol. Evol. 19 2261–2264. [DOI] [PubMed] [Google Scholar]
- Holt, I. J., D. R. Dunbar and H. T. Jacobs, 1997. Behaviour of a population of partially duplicated mitochondrial DNA molecules in cell culture: segregation, maintenance and recombination dependent upon nuclear background. Hum. Mol. Genet. 6 1251–1260. [DOI] [PubMed] [Google Scholar]
- Hyman, B. C., and J. L. B. Azevedo, 1996. Similar evolutionary patterning among repeated and single copy nematode mitochondrial genes. Mol. Biol. Evol. 13 221–232. [DOI] [PubMed] [Google Scholar]
- Jacobs, H. T., S. Asakawa, T. Araki, K. Miura, M. J. Smith et al., 1989. Conserved transfer RNA gene cluster in starfish mitochondrial DNA. Curr. Genet. 15 193–206. [DOI] [PubMed] [Google Scholar]
- Kaiser, H., 1991. Terrestrial and semiterrestrial Mermithidae, pp. 899–965 in Manual of Agricultural Nematology, edited by W. R. Nickle. Marcel Dekker, New York.
- Kajander, O. A., A. T. Rovio, K. Majamaa, J. Poulton, J. N. Spelbrink et al. 2000. Human mtDNA sublimons resemble rearranged mitochondrial genoms found in pathological states. Hum. Mol. Genet. 9 2821–2835. [DOI] [PubMed] [Google Scholar]
- Kim, I., E. M. Lee, K. Y. Seol, E. Y. Yun, Y. B. Lee et al., 2006. The mitochondrial genome of the Korean hairstreak, Coreana raphaelis (Lepidoptera: Lycaenidae). Insect Mol. Biol. 15 217–225. [DOI] [PubMed] [Google Scholar]
- Kraytsberg, Y., M. Schwartz, T. A. Brown, K. Ebralidse, W. S. Kunz et al., 2004. Recombination of human mitochondrial DNA. Science 304 981. [DOI] [PubMed] [Google Scholar]
- Ladoukakis, E. D., and E. Zouros, 2001. a Direct evidence for homologous recombination in mussel (Mytilus galloprovincialis) mitochondrial DNA. Mol. Biol. Evol. 18 1168–1175. [DOI] [PubMed] [Google Scholar]
- Ladoukakis, E. D., and E. Zouros, 2001. b Recombination in animal mitochondrial DNA: evidence from published sequences. Mol. Biol. Evol. 18 2127–2131. [DOI] [PubMed] [Google Scholar]
- LaRoche, J., M. Snyder, D. I. Cook, K. Fuller and E. Zouros, 1990. Molecular characterization of a repeat element causing large-scale size variation in mitochondrial DNA of the sea scallop Plactopecten magellanicus. Mol. Biol. Evol. 7 45–54. [DOI] [PubMed] [Google Scholar]
- Lavrov, D. V., and W. M. Brown, 2001. Trichinella spiralis mtDNA: a nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics 157 621–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, T. H., D. Blair, T. Agatsuma, P. Humair, N. J. H. Campbell et al., 2000. Phylogenies inferred from mitochondrial gene orders: a cautionary tale from the parasitic flatworms. Mol. Biol. Evol. 17 1123–1125. [DOI] [PubMed] [Google Scholar]
- Levinson, G., and G. A. Gutman, 1987. Slipped-strand mispairing: a major mechanism for DNA-sequence evolution. Mol. Biol. Evol. 4 203–221. [DOI] [PubMed] [Google Scholar]
- Lowe, T. M., and S. R. Eddy, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt, D. H., and B. C. Hyman, 1997. Animal mitochondrial DNA recombination. Nature 387 247. [DOI] [PubMed] [Google Scholar]
- Lunt, D. H., L. E. Whipple and B. C. Hyman, 1998. Mitochondrial DNA variable number tandem repeats (VNTRs): utility and problems in molecular ecology. Mol. Ecol. 7 1441–1455. [DOI] [PubMed] [Google Scholar]
- Miller, A. D., T. T. T. Nguyen, C. P. Burridge and C. M. Austin, 2004. Complete mitochondrial DNA sequence of the Australian freshwater crayfish, Cherax destructor (Crustacea: Decapoda: Parastacidae): a novel gene order revealed. Gene 331 65–72. [DOI] [PubMed] [Google Scholar]
- Moritz, C., T. E. Dowling and W. M. Brown, 1987. Evolution of animal mitochondrial DNA: relevance for population biology and systematics. Annu. Rev. Ecol. Evol. Syst. 18 269–292. [Google Scholar]
- Mueller, R. L., and J. L. Boore, 2005. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol. Biol. Evol. 22 2104–2112. [DOI] [PubMed] [Google Scholar]
- Mundy, N. I., and A. J. Helbig, 2004. Origin and evolution of tandem repeats in the mitochondrial DNA control region of shrikes (Lanius spp.). J. Mol. Evol. 59 250–257. [DOI] [PubMed] [Google Scholar]
- Nickle, W. R., 1972. Contribution to our knowledge of Mermithidae. J. Nematol. 4 113–146. [PMC free article] [PubMed] [Google Scholar]
- Nohara, M., M. Nishida, M. Miya and T. Nishikawa, 2005. Evolution of the mitochondrial genome in Cephalochordata as inferred from complete nucleotide sequences from two Epigonichthys species. J. Mol. Evol. 60 526–537. [DOI] [PubMed] [Google Scholar]
- Ojala, D., J. Montoya and G. Attardi, 1981. Transfer RNA punctuation model of RNA processing in human mitochondria. Nature 290 470–474. [DOI] [PubMed] [Google Scholar]
- Passamonti, M., J. L. Boore and V. Scali, 2003. Molecular evolution and recombination in gender-associated mitochondrial DNAs of the Manila clam Tapes philippinarum. Genetics 164 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piganeau, G., M. Gardner and A. Eyre-Walker, 2004. A broad survey of recombination in animal mitochondria. Mol. Biol. Evol. 21 2319–2325. [DOI] [PubMed] [Google Scholar]
- Poinar, G. O., 1981. Thaumamermis cosgrovei N-Gen, N-Sp (Mermithidae, Nematoda) parasitizing terrestrial isopods (Isopoda, Oniscoidea). Syst. Parasitol. 2 261–266. [Google Scholar]
- Powers, T. O., E. G. Platzer and B. C. Hyman, 1986. Large mitochondrial genome and mitochondrial DNA size polymorphism in the mosquito parasite, Romanomermis culicivorax. Curr. Genet. 11 71–77. [DOI] [PubMed] [Google Scholar]
- Raimond, R., I. Marcade, D. Bouchon, T. Rigaud, J. P. Bossy et al., 1999. Organization of the large mitochondrial genome in the isopod Armadillidium vulgare. Genetics 151 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., 1993. Endotherms, ectotherms, and mitochondrial genome-size variation. J. Mol. Evol. 37 281–295. [DOI] [PubMed] [Google Scholar]
- Rand, D. M., and R. G. Harrison, 1986. Mitochondrial DNA transmission genetics in crickets. Genetics 114 955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., and R. G. Harrison, 1989. Molecular population-genetics of mtDNA size variation in crickets. Genetics 121 551–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankoff, D., G. Leduc, N. Antoine, B. Paquin, B. F. Lang et al., 1992. Gene order comparisons for phylogenetic inference: evolution of the mitochondrial genome. Proc. Natl. Acad. Sci. USA 89 6575–6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse, M. A., B. R. Albert and B. Godelle, 2001. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol. Evol. 16 135–141. [DOI] [PubMed] [Google Scholar]
- Shao, R. F., H. Mitani, S. C. Barker, M. Takahashi and M. Fukunaga, 2005. Novel mitochondrial gene content and gene arrangement indicate illegitimate inter-mtDNA recombination in the chigger mite, Leptotrombidium pallidum. J. Mol. Evol. 60 764–773. [DOI] [PubMed] [Google Scholar]
- Solignac, M., J. Genermont, M. Monnerot and J. C. Mounolou, 1984. Genetics of mitochondria in Drosophila: mtDNA inheritance in heteroplasmic strains of Drosophila mauritiana. Mol. Gen. Genet. 197 183–188. [Google Scholar]
- Stanton, J. L., L. L. Daehler and W. M. Brown, 1997. Mitochondrial gene arrangement of the horseshoe crab Limulus polyphemus L.: conservation of major features among arthropod classes. Mol. Biol. Evol. 14 867–874. [DOI] [PubMed] [Google Scholar]
- Tang, S., and B. C. Hyman, 2005. Rolling circle amplification of complete nematode mitochondrial genomes. J. Nematol. 37 236–241. [PMC free article] [PubMed] [Google Scholar]
- Tesler, G., 2002. GRIMM: genome rearrangements web server. Bioinformatics 18 492–493. [DOI] [PubMed] [Google Scholar]
- Thyagarajan, B., R. A. Padua and C. Campbell, 1996. Mammalian mitochondria possess homologous DNA recombination activity. J. Biol. Chem. 271 27536–27543. [DOI] [PubMed] [Google Scholar]
- Tsaousis, A. D., D. P. Martin, E. D. Ladoukakis, D. Posada and E. Zouros, 2005. Widespread recombination in published animal mtDNA sequences. Mol. Biol. Evol. 22 925–933. [DOI] [PubMed] [Google Scholar]
- Wyman, S. K., R. K. Jansen and J. L. Boore, 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20 3252–3255. [DOI] [PubMed] [Google Scholar]
- Zouros, E., G. H. Pogson, D. I. Cook and M. J. Dadswell, 1992. Apparent selective neutrality of mitochondrial DNA size variation: a test in the deep-sea scallop Placopecten magellanicus. Evolution 46 1466–1476. [DOI] [PubMed] [Google Scholar]