Abstract
We applied association analysis to elucidate the genetic basis for variation in phenotypes affecting postcopulatory sexual selection in a natural population of Drosophila melanogaster. We scored 96 third chromosome substitution lines for nine phenotypes affecting sperm competitive ability and genotyped them at 72 polymorphisms in 13 male reproductive genes. Significant heterogeneity among lines (P < 0.01) was detected for all phenotypes except male-induced refractoriness (P = 0.053). We identified 24 associations (8 single-marker associations, 12 three-marker haplotype associations, and 4 cases of epistasis revealed by single-marker interactions). Fewer than 9 of these associations are likely to be false positives. Several associations were consistent with previous findings [Acp70A with the male's influence on the female's refractoriness to remating (refractory), Esterase-6 with a male's remating probability (remating) and a measure of female offspring production (fecundity)], but many are novel associations with uncharacterized seminal fluid proteins. Four genes showed evidence for pleiotropic effects [CG6168 with a measure of sperm competition (P2′) and refractory, CG14560 with a defensive measure of sperm competition (P1′) and a measure of female fecundity, Acp62F with P2′ and a measure of female fecundity, and Esterase-6 with remating and a measure of female fecundity]. Our findings provide evidence that pleiotropy and epistasis are important factors in the genetic architecture of male reproductive success and show that haplotype analyses can identify associations missed in the single-marker approach.
IN species with polygamous mating systems, male success at gaining copulations may not be a reliable predictor of reproductive fitness, especially when sperm from multiple males are concurrently present in the reproductive tract of a single female (Parker 1970). Multiple mating by females establishes the opportunity for postcopulatory sexual selection through either cryptic female choice or sperm competition (Eberhard and Cordero 2003; Wigby and Chapman 2004). Postcopulatory sexual selection can be an important determinant of male reproductive fitness and studies from a variety of taxa consistently reveal marked differences among males in their ability to outcompete rival sperm for access to fertilizations (e.g., Preston et al. 2003; Konior et al. 2005; Malo et al. 2005).
Sperm competitive ability is a complex trait that is likely influenced by a number of variables, including ejaculate volume (Harcourt et al. 1981; Preston et al. 2003; Dixson and Anderson 2004), sperm motility (Gage et al. 2004), sperm morphology (Oppliger et al. 2003; Dixson and Anderson 2004), and seminal fluid proteins (reviewed in Poiani 2006). In the genus Drosophila, male accessory gland proteins (Acp's) are components of the seminal fluid, and there is abundant evidence that they are important determinants of phenotypes affecting postcopulatory sexual selection (Clark et al. 1995; Wolfner 2002; Chapman and Davies 2004; Fiumera et al. 2005). For example, both RNAi and mutational analyses have shown that Acp70A increases the rate of oviposition and decreases female receptivity to remating (Chen et al. 1988; Chapman et al. 2003; Liu and Kubli 2003). Acp26Aa increases egg-laying rate (Herndon and Wolfner 1995; Heifetz et al. 2000; Chapman et al. 2001) and Acp36DE is necessary for sperm storage (Neubaum and Wolfner 1999; Bloch Qazi and Wolfner 2003). PEB-me, a gelatinous protein product of the ejaculatory bulb, is responsible for the formation the female mating plug, which presumably acts to concentrate sperm near female storage organs (Lung and Wolfner 2001).
Many Acp's exhibit nonneutral patterns of genetic variation, suggesting they are under strong selective pressures (Aguadé et al. 1992; Aguadé 1998, 1999; Begun et al. 2000; Swanson et al. 2001; Tsaur et al. 2001; Kern et al. 2004). Postcopulatory sexual selection through sperm competition, cryptic female choice, or sexually antagonistic coevolution has often been proposed to account for the rapid evolution of male reproductive genes (see Rice 1996; Parker and Partridge 1998; Swanson and Vacquier 2002). The maintenance of persistently high levels of polymorphism within populations for some Acp's creates a paradox, especially when that polymorphism has been demonstrated to be associated with large differences in sperm competitive ability, an important contributor to net fitness (Clark et al. 1995; Fiumera et al. 2005). It appears, however, that nontransitivity of sperm precedence (Clark et al. 2000), male-by-female interactions (Clark et al. 1999), and antagonistic pleiotropy (Fiumera et al. 2005) could account for the preservation of at least some of the intraspecific variation observed in nature. Deciphering the functional links between polymorphisms in male reproductive genes and variation in sperm competitive ability will provide insight into the potential selective pressures affecting patterns of genetic variation within and between species.
Association testing is a powerful approach to screen a large number of candidate genes for natural variation affecting complex phenotypes (Long and Langley 1999). Commonly applied to study human genetic diseases (Hirschhorn and Daly 2005), this approach has also been used by a variety of researchers to identify natural polymorphisms in Drosophila associated with phenotypes. For example, single-nucleotide polymorphisms (SNPs) and transposable element insertions at the achaete-schute complex are associated with variation in bristle number (Mackay and Langley 1990; Long et al. 2000), variation in Egfr associates with wing shape (Palsson and Gibson 2004; Dworkin et al. 2005), and polymorphisms in male reproductive genes associate with traits affecting sperm competitive ability (Clark et al. 1995; Fiumera et al. 2005, 2006). Clark et al. (1995) investigated seven genes on the second and third chromosomes using single-strand conformation polymorphisms, while Fiumera et al. (2005, 2006) focused their analysis on second chromosome loci and typed single-nucleotide polymorphisms in 10 male reproductive genes. Both studies found associations between second chromosome loci and phenotypes affecting sperm competition, but to date no association studies have evidence linking segregating polymorphisms on the third chromosome to natural variation in sperm competitive ability. Here we identify associations between polymorphisms in male reproductive genes on the third chromosome of Drosophila melanogaster and phenotypes affecting sperm competitive ability, with the goal to further characterize the genetic architecture of this important component of reproductive fitness.
METHODS
Drosophila melanogaster fly cultures:
Ninety-six chromosome 3 substitution lines were generated from a natural population of D. melanogaster from State College, Pennsylvania. Each line is homozygous for an individual third chromosome segregating in this natural population and, on average, should be >99% coisogenic for loci on the third chromosome. The second, fourth, and sex chromosomes are derived from the homozygous TM3/TM6 balancer stock and form an identical genetic background across all lines. To generate the lines, single wild females caught between 1998 and 1999 were placed in vials and allowed to oviposit. F1 or F2 males from each vial were mass mated to TM3/TM6 females. Male offspring from the wild × TM3/TM6 matings were backcrossed to females of the same balancer stock. The resulting progeny were selectively intercrossed to eliminate balancers and isolate independent wild third chromosomes in a homozygous state. Individual males were backcrossed to TM3/TM6 for eight generations to isogenize the second, fourth, and X chromosomes. Finally, females were mated to TM3/TM6 males to introduce an identical Y chromosome across all lines. Progeny from these crosses were sib-mated and any offspring that showed balancer phenotypes were eliminated.
Third chromosome extraction lines (red eyes) were mated to the same stock of homozygous cinnabar brown (cn bw, white eyes) females and competed against the same homozygous cn bw males in trials of sperm competitive ability used in Fiumera et al. (2005) and Civetta and Clark (2000). All fly cultures were maintained on standard agar–dextrose–yeast media and housed at 24° on a 12-hr light/dark cycle.
Measuring sperm competition phenotypes:
The 96 chromosome 3 substitution lines were assayed for phenotypes affecting sperm competitive ability using protocols similar to those described in Clark et al. (1995) and Fiumera et al. (2005). Both the defense and the offense components of sperm competition were measured. “Defense” and “offense” refer to scenarios when the experimental male is either the first male or the second male to mate to a given female, respectively. The defense components of sperm competitive ability include male-induced female refractoriness to remating (refractory), the proportion of offspring sired by the experimental male when he is the first male to mate to a doubly mated female (P1′), and fecundity of doubly mated females (fec-def, fec-V1). The offense components include the ability of the experimental male to encourage remating by an already mated female (remating), the proportion of offspring sired by the experimental male when he is the second male to mate to a doubly mated female (P2′), and fecundity of doubly mated females (fec-off, fec-V2).
To estimate defensive metrics of sperm competitive ability, we sequentially mated virgin cn bw females to experimental males and then to cn bw males. All flies were virgins collected under CO2 and aged 4–7 days. For each chromosome 3 substitution line, 10 females were mass mated to 10 experimental males for 12 hr starting at ∼8:00 pm (1 hr after sundown on the light/dark cycle). Males were then discarded and females were transferred, without anesthesia, to individual vials (vial 1) and allowed to oviposit for 2 days. Two virgin cn bw males (tester males) were introduced into each vial and left to mate for 12 hr starting at ∼8:00 pm. Females were then aspirated without CO2 to new vials (vial 2) and males were discarded. After 3 days, females were transferred without anesthesia to a new vial (vial 3) and discarded 5 days later. Live progeny from each vial were counted and eye color was used as marker of paternity. The entire procedure was repeated in a new generation for a total of 20 replicates from each experimental line for each of two different generations (experimental blocks). The offensive metrics were measured similarly except that the cn bw (tester) males were the first males to mate and the chromosome 3 substitution (experimental) males were the second males to mate. We estimated male-induced cost of mating (COM) as the proportion of females that died after mating to both males in the defense experiment. Only those females that survived the entire experiment, had no missing data, and produced at least five progeny were used to estimate phenotypes affecting sperm competitive ability.
Male-induced female refractoriness (refractory) is the proportion of females that do not remate to a tester male after mating with an experimental male and was estimated on the basis of the presence of progeny from the two different males. Only those females who were deduced to have mated to both males were used to determine the proportion of offspring in vials 2 and 3 sired by an experimental male when he is first to mate (P1′). This estimate excludes cases where a female mates to a given male but that male fails to sire any progeny. Defensive fecundity was estimated as the total number of offspring produced by a doubly mated female across all three vials (fec-def). To enable discovery of short-term effects, we also calculated fecundity using only the progeny from vial 1 that were produced immediately after mating to the experimental males (fec-V1). Remating rate was estimated as the proportion of already mated females that remate with an experimental male, again inferred from the presence of progeny from both males (remating). Only those females that remated were used to calculate the proportion of offspring from vials 2 and 3 sired by an experimental male when he is the second male to mate (P2′). Offensive fecundity was estimated as the total number of offspring produced by a doubly mated female across all three vials (fec-off). To enable discovery of short-term effects, we also calculated fecundity using only the progeny from vial 2 that were produced immediately after mating to the experimental male (fec-V2).
Statistical analyses were used to test for significant LINE effects for each of the different phenotypes scored. All the fecundity measures presented adequate fits to the normal distribution. P1′ was arcsine square-root transformed to improve the fit to normality. A general linear model was used to test for significance of these phenotypes as follows: Pijk = Li + Bj + εijk, where Pijk is the trait of interest, Li is the effect of the ith LINE, and Bj is the effect of the jth BLOCK (random factor). The distribution of P2′ was highly skewed, and arcsine square-root transformation did not improve the fit to normality, so a Kruskall–Wallis nonparametric test of line medians was applied. Refractory, remating, and cost of mating were estimated as proportions and permutation tests based on chi-square statistics were conducted using MATLAB (Fiumera et al. 2005, 2006). Line means (or medians) were estimated for each of the phenotypes scored and used in the association testing (see below).
Polymorphism identification:
We used a candidate gene approach to identify natural polymorphisms in male reproductive genes associated with sperm competition phenotypes. Thirteen third chromosome male reproductive genes were selected for investigation in this study (Acp62F, Acp63F, CG6168, Esterase-6, Acp70A, Acp76A, CG14560, Acp95EF, BG642167, Mst57Dc, Mst57Db, Mst57Da, and Acp98A). BG642167 was identified in D. simulans from Swanson et al. (2001) but is unannotated in D. melanogaster Release 4. Some proteins, like Acp70A and Esterase-6, are well characterized seminal fluid proteins (reviewed in Chapman and Davies 2004), while others have only recently been identified from an EST screen (Swanson et al. 2001).
We aimed to identify polymorphisms from entire gene regions, starting ∼1 kb upstream of the transcription start site and ending ∼500 bp downstream of the stop codon. For Acp62F, however, adjacent exons were interrupted by an ∼6.6-kb intron and much of this intergenic region was disregarded in our analysis. Polymorphic sites, mainly SNPs, were identified either from published sequences [Acp70A (Cirera and Aguadé 1997), Esterase-6 (Odgers et al. 2002; Balakirev and Ayala 2003), Acp76A (Begun et al. 2000), and Acp62F (Begun et al. 2000)] or from novel resequencing of a sample of 8–12 of the chromosome 3 substitution lines. In our SNP numbering system, the transcription start site is base pair 1500 except for Mst57Da–Dc, which were not numbered individually given their proximity to each other (coding regions Mst57Da, 565–792; Mst57Db, 1547–1669; and Mst57Dc, 2687–2998).
For resequencing, primers were designed using FlyBase sequences as templates. DNA was extracted from ∼50 whole flies by performing standard phenol/chloroform extractions. We amplified ∼1-kb fragments and visualized products on 1.5% agarose gels to verify primer specificity. Initial amplification products were purified with shrimp alkaline phosphatase and exonuclease I (Promega, Madison, WI) and then prepared for automated sequencing with a BigDye Termination kit (Applied Biosystems, Foster City, CA) and internally located sequencing primers. Dye terminators were removed by filtration though Sephadex columns (Amersham Biosciences, Piscataway, NJ) and prepared samples were loaded onto ABI 3730xl capillary DNA sequencing machines for sequence analysis. Raw sequencing traces were manually assembled using BioEdit V 7.0.5 (written by Tom Hall, available at http://www.mbio.ncsu.edu/BioEdit/bioedit.html, 5/31/05) and polymorphisms were identified by eye. For each gene, we aimed to genotype at least one polymorphism in both the upstream and the downstream regions, as well as one in each intron and exon across the entire set of chromosome 3 substitution lines. To maximize our power to detect associations, preference was given to polymorphisms segregating at intermediate frequencies in our initial sample, polymorphisms with low levels of linkage disequilibrium, and also nonsynonymous amino acid polymorphisms.
Genotyping:
A total of 72 single-nucleotide polymorphisms or indels in 13 male reproductive genes were genotyped across the chromosome 3 substitution lines (Figure 1; supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). Seventy-one were single-nucleotide polymorphisms and one was a 21-nucleotide insertion–deletion polymorphism. Twenty-seven SNPs were genotyped via Pyrosequencing (Ahmadian et al. 2000) using direct-biotinylated, locus-specific primers (supplemental Table 1). PCR amplifications were carried out in 25-μl volumes with final concentrations 1.5 mm MgCl2, 1× PCR buffer (Promega), 0.25 mm each dNTP, 0.3 μm 5′Bio primer, 0.3 μm primer, and 0.5 unit Taq Polymerase (Promega). Reactions were cycled according to the following program: 95° for 2 min; 40 cycles of 95° for 15 sec, 50° for 30 sec, and 72° for 15 sec; and a final extension time of 5 min at 72°. Single-stranded PCR products were isolated according to manufacturer's protocols using a 96-pin vacuum preparation tool (Pyrosequencing) and added to 0.3 μm sequencing primer in annealing buffer (20 mm Tris-acetate; 2 mm MgAc2, pH 7.6). Sequences were analyzed using 0.5× 96 PSQ SNP reagents on a PSQ 96MA (Pyrosequencing). Thirty-eight SNPs (supplemental Table 2) were genotyped via SNPlex technology (Applied Biosystems) and 4 SNPs were genotyped using the SNPstream approach (Beckman Coulter). An insertion–deletion polymorphism and two closely linked SNPs in CG14560 were resolved via direct sequencing. Linkage disequilibrium between loci was calculated using GENEPOP (Raymond and Rousset 1995), treating our genotype information as a haploid data set.
Figure 1.—
Candidate genes and scored polymorphisms. The approximate location of each gene is shown relative to its cytological position on the third chromosome. Typed polymorphisms are given as asterisks. Protein-coding sequences are shown as open boxes and 5′- and 3′-untranslated regions are shaded gray. Introns and upstream and downstream regions are depicted as solid lines.
Association testing:
We investigated the role of variation in male reproductive genes by testing for associations between sperm competition phenotypes and individual SNPs and also investigated epistasis by testing the interaction term of all pairwise SNP combinations. Because of the short-distance linkage disequilibrium in our lines, we were also able to test for associations between sperm competition phenotypes and three-marker haplotypes. False discovery rate (FDR) calculations were applied to each testing procedure to determine the expected number of false positives using the approach of Storey and Tibshirani (2003) and the pava.fdr implementation (Broberg 2005). The two methods were highly consistent (r2 = 0.98) and we report the results from the approach by Storey and Tibshirani (2003).
To test for single-marker associations, we applied simple linear regression and permutation tests (MATLAB) to identify associations between metrics of sperm competitive ability and natural variation in male reproductive genes (Fiumera et al. 2005). For each phenotype, experimentwise and markerwise P-values (Churchill and Doerge 1994) were calculated by comparing the actual F-value for each marker to the distribution of 10,000 permuted F-values for every marker (experimentwise) or for the focal marker (markerwise). Line means were used for arcsine-square root P1′, refractory, remating, cost of mating, and measures of fecundity (fec-def, fec-V1, fec-off, fec-V2), while line medians were used for P2′. Occasionally, lines were eliminated from individual analyses on account of technical difficulties (e.g., failure to amplify at a given marker or scored as a heterozygote), and thus sample sizes vary slightly. To test for epistasis, we used a general linear model (glm in R) to explicitly test the interaction term for all pairwise combinations of the 72 SNPs. Of the 23,004 possible pairwise tests with 72 SNPs and nine phenotypes, 5663 could not be completed because not all four pairwise SNP combinations were present among the sampled lines (due to linkage disequilibrium among SNPs and some missing genotype data).
It is possible that combinations of SNPs, acting in concert as haplotypes, might more accurately identify associations between genotype and phenotype (Clark 2004). Although a small amount of missing data will have a limited effect on associations with single markers, the probability of having missing data at any one marker increases with the number of markers forming a haplotype and this can dramatically affect the number of lines representing each haplotype category. Even though we were missing only 9% of the single-marker genotypes (see results), we would expect to have full data for only ∼75% of the three-marker haplotypes. To circumvent this difficulty we used fastPHASE (Scheet and Stephens 2006) to impute our missing data. The imputed data were then used to calculate the within-gene, three-marker haplotypes via a sliding-window approach. Mst57Da and Mst57Db had only two markers in each gene and thus only two-marker haplotypes were tested. Three markers were chosen on the basis of the extent of linkage disequilibrium in the sample and because including a greater number of markers resulted in most haplotypes being represented by only a single or a few lines. One-way ANOVA was used to test for significant associations between each of the nine phenotypes and the haplotypes. As a test for spurious associations due to imputation error of fastPHASE, we reran the single-marker associations using the imputed data. P-values calculated from these permutations were virtually identical to those reported (seven of the eight associations were the same, one P-value increased from 0.008 to 0.01, and another dropped from 0.02 to 0.005; data not shown). This suggests that imputing the missing data retained the original signatures of the associations between genotype and phenotype and helps validate its utility for haplotype-based tests.
RESULTS
Variation in sperm competitive ability:
A total of 1920 females (20 replicates from each of the 96 chromosome 3 lines) were set up for both the defense and the offense sperm competition experiments. In the defense experiment, 377 females were excluded from all analyses because they had missing data (131), failed to mate to the first male (25), or produced <5 total offspring (221). Thus, 1543 females from 94 of the chromosome 3 lines were used to estimate cost of mating and of these females 94 died. Refractory was then estimated using the 1449 females that met all above criteria and also survived the entire experiment. Of these females, 176 failed to remate (12%), yielding data from 1273 females for the proportion of offspring sired by the first male to mate (P1′) and fecundity estimates in the defense experiment (fec-def and fec-V1). A total of 172,124 offspring were counted during the defense experiment, and of those, 149,670 offspring were included in the estimates of P1′ and fec-def.
In the offense experiment, 463 of the 1920 females were excluded from all analyses because of missing data (161), failure to survive the entire experiment (173), failure to produce at least 5 total offspring (119), or failure to mate to the first male (10). Thus remating was estimated for 93 of the chromosome 3 extraction lines using data from 1457 females. Of those, 232 females failed to remate (16%), yielding data from 1225 females for estimates of the proportion of offspring sired by the second male to mate (P2′) and fecundity (fec-off and fec-V2) in the offense experiment. A total of 195,114 offspring were counted during the offense experiment, and of those, 166,423 were included in estimates of P2′ and fec-off.
Highly significant LINE effects were detected for all the phenotypes scored (P < 0.01) except for male-induced female refractoriness, which was only marginally significant with a P-value of 0.053 (Figure 2). Overall, 92% of females survived the full experiment and, across all lines, cost of mating from the defense experiment was ∼6%. The majority of females mated to each of the males in both the defense (88%) and the offense (84%) experiments. As expected (Lefevre and Jonsson 1962), the majority of the offspring were sired by the second male to mate; on average experimental males sired 16% of the offspring when they were the first males to mate and almost 94% of the offspring when they were the second males to mate.
Figure 2.—
Variation across lines for sperm competition phenotypes. Rank-order line means (or line medians for P2′) are shown with standard errors (or Q1–Q3 plots for P2′). The test statistics and P-values are shown (see text). The grand mean across lines is shown by an open circle on the y-axis. The x-axis always corresponds to the rank-order lines.
In general, sperm competition phenotypes were positively correlated (Table 1). Of the 36 possible pairwise correlations, 17 (47%) were significant at P < 0.05 and seven correlations (19%) were significant at P < 0.001. The two measures of fecundity within each experiment were extremely highly correlated (fec-def with fec-V1, ρ = 0.831 and fec-off with fec-V2, ρ = 0.834), which is expected given that one is a subset of the other. Cost of mating was not significantly correlated with any other scored phenotype while P1′ was significantly positively correlated with every phenotype except cost of mating. Here several phenotypes were correlated across the offense and defense experiments, including P1′ and P2′, as well as fec-off and fec-def.
TABLE 1.
Pearson's correlation coefficient between line means of sperm competition phenotypes
| Cost of mating | Refractory | P1′ | Fec-def | Fec-V1 | Remating | P2′ | Fec-off | |
|---|---|---|---|---|---|---|---|---|
| Refractory | 0.166 | |||||||
| P1′ | 0.074 | 0.239* | ||||||
| Fec-def | −0.164 | 0.011 | 0.377*** | |||||
| Fec-V1 | −0.071 | 0.029 | 0.362*** | 0.831*** | ||||
| Remating | 0.145 | 0.096 | 0.230* | 0.164 | 0.115 | |||
| P2′ | 0.058 | 0.128 | 0.411*** | 0.309** | 0.513*** | 0.109 | ||
| Fec-off | 0.039 | 0.254* | 0.486*** | 0.313** | 0.346** | 0.152 | 0.306** | |
| Fec-V2 | 0.121 | 0.197 | 0.286** | 0.167 | 0.258* | 0.219* | 0.179 | 0.834*** |
Statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001. P1′ was calculated using least-squares line means after ANOVA with arcsine-square-root transformed P1′ and P2′ was calculated using line medians from the Kruskal–Wallis test. Fec-def, fecundity defense; fec-V1, fecundity-V1; fec-off, fecundity offense; fec-V2, fecundity-V2.
Genetic variation:
We genotyped the 94 chromosome 3 substitution lines at 72 loci in 13 male reproductive genes (Figure 1, supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/). Overall we successfully scored 6131 of the 6768 (91%) genotypes. The average frequency of the common allele across all typed loci was 0.77 and only 7 (10%) of the scored loci had the common allele present at a frequency >0.95. Overall, we observed moderate to high levels of linkage disequilibrium within genes and only occasional disequilibrium between loci in different genes (Figure 3). Significant linkage disequilibrium (P < 0.01) was observed for 35% of comparisons between SNPs within the same gene but for only 1.7% of comparisons between SNPs in different genes.
Figure 3.—
Linkage disequilibrium across the 72 markers. Highly significant linkage disequilibrium is shown with solid circles (P < 0.01), and significant linkage disequilibrium is shown with open circles (P < 0.05). Significance tests that could not be completed are marked with a “+” (see text).
Genotype-to-phenotype associations:
Here we present 8 single-marker associations (Table 2) and 12 three-marker haplotype associations (Table 3; Figure 4) between genotype and phenotype that have a markerwise P < 0.01; no experimentwise single-marker associations were significant at P < 0.05. Four pairwise single-marker interaction terms (P < 5 × 10−5; q < 0.05) are presented that represent likely cases of epistasis between genes (Figure 5). FDR calculations indicate that <9 of the 24 associations are likely to be false positives. An additional 22 single-marker and 21 three-marker haplotype associations (markerwise P < 0.05) are presented in supplemental Table 3 at http://www.genetics.org/supplemental/. An additional 19 pairwise interactions (P < 2 × 10−4) were identified but these were driven by a single line with an extremely low phenotypic value that was the only line representing one of the four pairwise marker combinations (supplemental Table 4 at http://www.genetics.org/supplemental/).
TABLE 2.
Associations between single-marker genotypes at male reproductive genes and sperm competition phenotypes
| Phenotype | Marker | Marker type | Adjusted r2 | Other associationsa |
|---|---|---|---|---|
| Refractory | CG6168snp1536 | Synonymous at codon 13 | 0.07 | None |
| P1′ | CG14560snp1474 | ∼30 bp upstream of START | 0.08 | Remating |
| CG14560snp1173 | ∼325 bp upstream of START | 0.07 | Fec-def | |
| Fecundity-defense | CG14560snp1173 | ∼325 bp upstream of START | 0.12 | P1′ |
| Fecundity-V1 | Acp62Fsnp8748 | ∼100 bp downstream of 3′-UTR | 0.07 | Fec-def |
| Remating | Est6snp978 | ∼525 bp upstream of START | 0.07 | None |
| P2′ | CG6168snp1583 | Synonymous at codon 28 | 0.06 | None |
| Fecundity-offense | Mst57Dsnp2852 | ala/thr at codon 56 | 0.07 | Fec-V2 |
Abbreviations for phenotypes: fec-def, fecundity-defense; fec-V2, fecundity-V2.
TABLE 3.
Associations between three-marker haplotypes at male reproductive genes and sperm competition phenotypes
| Phenotype | Marker | Marker typea | Adjusted r2 | Other associationsb |
|---|---|---|---|---|
| Refractory | Acp70Asnp902:1412:1554 | ∼600 bp, ∼80 bp upstream, ser/ala (18) | 0.14 | None |
| P1′ | CG14560snp1173:1306:1474 | ∼325 bp, ∼200 bp, ∼30 bp upstream | 0.13 | Fec-def |
| Fecundity-defense | CG14560snp1042:1131:1173 | ∼450 bp, ∼370 bp, ∼325 bp upstream | 0.15 | None |
| CG14560snp1131:1173:1306 | ∼370 bp, ∼325 bp, ∼200 bp upstream | 0.14 | P1′, fec-V1 | |
| CG14560snp1173:1306:1474 | ∼325 bp, ∼200 bp, ∼30 bp upstream | 0.13 | P1′ | |
| Remating | EST6snp2090:2204:2348 | syn (197), syn (233), syn (283) | 0.14 | None |
| EST6snp2348:3072:3338 | syn (283), ser/ala (508), ∼150 bp downstream | 0.37c | Fec-off, fec-V2 | |
| P2′ | Acp62Fsnp1552:1822:2188 | syn (10), syn (100), intron | 0.16 | None |
| Acp62Fsnp1822:2188:2880 | syn (100), intron, intron | 0.14 | None | |
| Fecundity-offense | Mst57Dsnp2852:3073:3161 | ala/thr (56), ∼70 bp, ∼160 bp downstream | 0.14c | Fec-V2 |
| Fecundity-V2 | EST6snp2348:3072:3338 | syn (283), ser/ala (508), ∼150 bp downstream | 0.12c | Fec-off, remating |
| Mst57Dsnp2852:3073:3161 | ala/thr (56), ∼70 bp, ∼160 bp downstream | 0.12 | Fec-off |
Codon position for synonymous (syn) or nonsynonymous changes is given in parentheses.
Abbreviations for phenotypes: fec-off, fecundity-offense; fec-def, fecundity-defense; fec-V1, fecundity-V1; fec-V2, fecundity-V2.
Associations driven by a single haplotype of low phenotypic value are represented by a single line.
Figure 4.—
Examples of three-marker haplotype associations. The mean value of each haplotype is shown with the standard errors for Acp70A with refractory (A), CG14560 with P1′ (B), Esterase-6 with remating (C), CG14560 with fecundity-defense (D), Acp62F with P2′ (E), and Esterase-6 with fecundity-V2 (F). Association between haplotypes at Esterase-6 and fecundity-V2 is driven by a single line with low phenotypic value (see text). Plot of P1′ is back calculated from ASP1′.
Figure 5.—
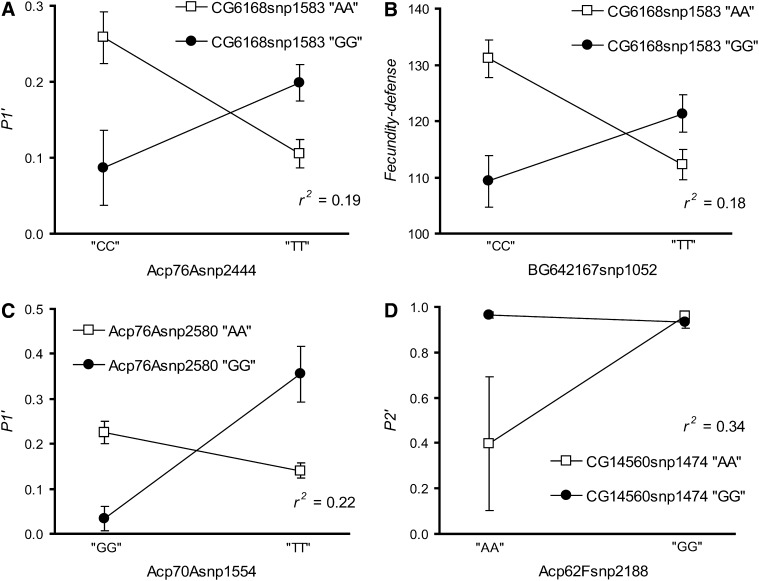
Epistasis at male reproductive genes affecting sperm competitive ability: interaction plots from two-way ANOVA for markers in Acp76A and CG6168 associating with P1′ (A), in CG6168 and BG642167 associating with fecundity-defense (B), in Acp76A and Acp70A associating with P1′ (C), and in Acp62F and CG14560 associating with P2′ (D). Adjusted r2 for each model is shown. Plot of P1′ is back calculated from ASP1′.
The 8 single-marker associations involved polymorphisms in five different genes with seven different sperm competition phenotypes (Table 2). Five of the associations were with markers in noncoding regions, 2 were synonymous changes, and 1, Mst57Dsnp2852 was an alanine-to-threonine amino acid polymorphism. The 12 haplotype associations involved five different genes with seven different phenotypes. Only 7 of these haplotype associations are likely to be independent because several haplotypes share markers across the sliding windows (Table 3). Several of the three-SNP haplotype associations we identified included polymorphisms identified in the single-marker analysis: P1′ with CG14560, fec-def with CG14560, and fec-off with Mst57Dc. The haplotype analysis also revealed several novel associations: refractory with Acp70A, P2′ with Acp62F, fec-off and fec-V2 with Esterase-6, and remate with Esterase-6. Although a single upstream marker, Esterase-6snp978, was associated with remating rate, this marker was not present in either of the Esterase-6 coding sequence haplotypes that associated with male remating rate. Three of the haplotype associations (see Table 3 and Figure 4F) were driven by one haplotype with an extremely low phenotypic value and represented by only a single line. These associations were no longer significant if that single rare haplotype was removed from the analysis.
We also report four likely cases of epistatic interactions among polymorphisms in different male reproductive genes (P < 5 × 10−5, Figure 5); Acp62Fsnp2188 by CG14560snp1474 with P2′, Acp70Asnp1554 by Acp76Asnp2580 with P1′, CG6168snp1583 by Acp76Asnp2444 with P1′, and CG6168snp1583 by BG42167snp1052 with fec-def. A single marker in CG6168 was involved with two different pairwise interactions while two different markers in Acp76A displayed significant interactions for P1′. Interestingly, none of these cases involved two markers in the same gene interacting with each other. An additional 19 epistatic interactions (P < 2 × 10−4) are presented in supplemental Table 4 at http://www.genetics.org/supplemental/. As mentioned in the analysis of haplotypes, these significant interactions were driven by a single line with a very low phenotypic value. Although they may represent biological reality, the results should be viewed with caution.
One-way ANOVA was used to estimate adjusted r2-values for the identified associations. The adjusted r2-values ranged from 0.06 to 0.12 for the single-marker associations (Table 2) and from 0.12 to 0.37 for the three-marker haplotype associations (Table 3). The haplotype-based associations always had a higher adjusted r2-value compared to any of their individual single-marker associations. Several phenotypes were associated with markers in different genes. The combined adjusted r2 was 0.16 for refractory (CG6168 and Acp70A), 0.21 for remating (upstream SNP and coding haplotype for Esterase-6), 0.23 for P2′ (CG6168 and Acp62F), and 0.20 for fec-off (Mst57Dc and Esterase-6). Individually, the models incorporating the interaction terms had adjusted r2-values that ranged from 0.18 to 0.36, suggesting that epistasis may explain a large proportion of the phenotypic variation among lines (Figure 5).
DISCUSSION
To investigate the role of natural third chromosome variation on phenotypes affecting postcopulatory sexual selection, we performed association tests between polymorphisms in 13 male reproductive genes and standard measures of sperm competitive ability in D. melanogaster. Here we report 24 associations (8 single-marker associations, 12 three-marker haplotype associations, and 4 cases of epistasis revealed by single-marker interactions between genes). These associations involved markers in eight male-reproductive genes and eight phenotypes affecting sperm competitive ability. The observed role of natural variation supports the hypothesis that postcopulatory sexual selection is an important evolutionary pressure and helps explain the nonneutral patterns of genetic variation observed in some of these genes. Many of the genes studied in this analysis have not been investigated in detail and our findings provide insight into the complex genetic basis of male reproductive success and highlight excellent candidates for further analysis.
Genotype-to-phenotype associations:
As the name implies, association testing relies on statistical association, and determining an appropriate significance threshold in the face of multiple testing is not trivial. Establishing a stringent experimentwise P-value will decrease the incidence of false positives in the data set, but only at the expense of true associations. On the flip side, the use of a liberal significance threshold will increase the occurrence of both true associations and false positives. The important aspect is to have an estimate of the confidence the study provides for each association because different research projects may have different goals. For example, human geneticists are often interested in using association mapping to define markers that will be valuable diagnostic tools for susceptibility to disease. This goal requires that the association be robust and repeatable in a number of studies and even across populations. With these goals, replication of the association in an independent sample is critical. Association mapping can also be used as a gene discovery technique (and validation tool, see below), particularly in D. melanogaster where the tools for genetic manipulation are available. Association mapping is not the ultimate goal, and reporting a slightly liberal FDR threshold allows subsequent studies to prioritize the candidates while reducing the chances of missing true effects. Knock-down, knock-out, and targeted mutagenesis are more effective experimental approaches for identifying causality, and these studies are being pursued in a variety of laboratories to verify association studies. The power of association tests rests in their capacity to survey a large number of genes, assay a large number of lines to observe the role of natural variation, and identify the function of essential genes when knockouts could be lethal or completely sterile.
With these caveats in mind, false discovery rate calculations (Storey and Tibshirani 2003) indicate that <9 of our presented associations should be false positives, yielding what are likely 15 true positives. Three of these associations validate previous studies and demonstrate the power of our approach to identify genes contributing to natural variation in male reproductive success. Several previous studies have implicated Esterase-6 as an important factor influencing both male mating rate (Gilbert and Richmond 1982; Saad et al. 1994) and female egg-laying rate (Gilbert et al. 1981a; Saad et al. 1994), phenotypes that are very similar to our measures of male remating rate and female fecundity for which we identified associations with polymorphisms in Esterase-6. We also observed a three-marker haplotype in Acp70A associated with male-induced female refractoriness. Experiments using ectopic expression in females (Aigaki et al. 1991), RNAi knockdown in males (Chapman et al. 2003), or knockouts in males (Liu and Kubli 2003) have shown that Acp70A induces female refractoriness and is responsible for the “sperm effect.” Our findings provide additional evidence for these genes' important roles in postcopulatory sexual selection, highlight the role of natural variation, and support our proposition that association testing has yielded true positives, despite the potential for relatively high false discovery rates.
We have also identified many novel associations with uncharacterized male reproductive genes and these now represent strong candidates for directed investigations. CG6168 is a putative protease based on comparative structural modeling (Mueller et al. 2004) and was associated with refractory, P1′, and P2′. Acp62F is a serine protease inhibitor known to enter the hemolymph of the female (Lung and Wolfner 1999) and was associated with fec-V1 and P2′. Markers in CG14560 associate with both P1′ and fec-def. CG14560 contains a 31-amino-acid run of alanines interspersed with proline residues, suggesting that this gene has antimicrobial activity (Mueller et al. 2004). A number of putative antimicrobial peptides are transferred to the female during mating (Lung et al. 2001) and they may be important for guarding the female reproductive tract or male ejaculate against bacterial infection.
Failure to identify a genotype–phenotype association in five of the genes included in this analysis does not eliminate them as putative determinants of sperm competitive ability. For example, we did not identify associations between polymorphisms in Acp70A and measures of female fecundity or cost of mating, despite its known effects on female egg-laying rate (Chen et al. 1988), and male-induced cost of mating (Wigby and Chapman 2005). Our findings imply only that among the lines in this study, there was not variation in these phenotypes ascribable to Acp70A, either because the variation did not exist or because the effects were too small for the power of the test performed.
Using haplotypes for association studies may help overcome some of the limitations of single-marker analyses (see Clark 2004; Schaid 2004). Haplotypes might more accurately define the functional unit of genes if the physical properties such as three-dimensional protein states affect the activity or stability of a protein. Haplotypes may also capture the patterns of population variation more precisely than SNPs, particularly in humans (Wall and Pritchard 2003) where the extent of linkage disequilibrium often extends across whole genes. Finally, haplotypes can reduce the number of tests that are being completed and thus increase the power of the analyses. In our study, we were precluded from using whole-gene haplotypes because linkage disequilibrium was in general so low that whenever more than three SNPs were considered, there were so many distinct haplotypes that the power of the tests was severely compromised. Limiting our attention to three-SNP haplotypes, we find that many of the same associations were identified in both the haplotype tests and the single-marker analysis, but several novel associations were identified using haplotypes. These include Acp70A with refractory, Acp62F with P2′, and Esterase-6 with induced fecundity (fec-V1). The associations with Acp70A and Esterase-6 are well supported by previous experimental studies and suggest that using haplotypes might more accurately capture the true genetic basis for variation in complex traits. It should be noted that some of the haplotype associations were driven by a single line that had a very low phenotypic value and was the only representative of a given haplotype (Figure 4F). Although there may be biological relevance to these cases, as highly deleterious haplotypes are expected to be rare, these findings should be viewed with caution.
Selection on Acp70A and Esterase-6 haplotypes:
Acp70A, arguably the best studied of the accessory gland proteins, is known to affect a variety of phenotypes influencing postcopulatory sexual selection (reviewed in Kubli 2003). Here we have identified an association between male-induced female refractoriness to remating (refractory) and a three-marker haplotype defined by Acp70Asnp902 (∼400 bp upstream), Acp70Asnp1412 (∼90 bp upstream), and Acp70Asnp1554 (serine:alanine at amino acid position 18). Cirera and Aguadé (1997), in a study of the evolutionary history of this gene, noted a strong cluster of linkage disequilibrium in the 5′ region of this gene (near the first marker defining our three-marker haplotype) and another cluster of linkage disequilibrium closer to and including the transcript (containing our next two markers, Figure 6A). The serine:alanine amino acid polymorphism is at the actual cleavage site of the signal peptide. Cirera and Aguadé (1997) observed reduced polymorphism on haplotypes containing the alanine allele, leading them to speculate that this amino acid polymorphism might be under selection.
Figure 6.—
Association between haplotypes at Acp70A and male-induced female refractoriness to remating. (A) The gene region of Acp70A with the two exons (E1 and E2), the two regions of high LD shaded (5′-LD and coding-LD regions identified by Cirera and Aguadé 1997), and the 210-bp promoter region from Styger-Schmucki (1992). The LD regions are open ended, indicating that they may extend further as suggested by Cirera and Aguadé (1997). The asterisks indicate the locations of the three scored markers. (B and C) Associations between male-induced female refractoriness and two-marker haplotypes at Acp70Asnp902:1412 (B) and the serine:alanine amino acid polymorphism (C).
Our single-marker analysis, however, finds no evidence that this site affects refractory (P = 0.50; Figure 6C) and SignalP (Bendtsen et al. 2004) predicts that both amino acids should result in cleavage of the signal sequence. A single marker, Acp70Asnp1412, that is just 142 bp upstream of the amino acid polymorphism is weakly associated with refractory by itself (P = 0.038; supplemental Table 3 at http://www.genetics.org/supplemental/). This marker is in perfect linkage disequilibrium with the serine:alanine polymorphism in the nine lines analyzed by Cirera and Aguadé (1997), but apparent recombinants are detected in our sample. This marker is not solely responsible for the association with refractory; the significance of the three-marker haplotype association requires at least one marker from each of the two high-linkage-disequilibrium (LD) regions described by Cirera and Aguadé (1997). A two-marker haplotype defined by Acp70Asnp902 (near the 5′ LD region) and Acp70Asnp1412 (within the high-LD coding region) is strongly associated with refractory (one-way ANOVA, P = 0.001, Figure 6B). Acp70Asnp1412 is within the 210-bp fragment deemed sufficient for correct tissue- and time-specific expression by Styger-Schmucki (1992), suggesting that this polymorphism might affect transcription. Its effect, however, is realized only in the context of the marker further upstream, suggesting an epistatic interaction among the different markers in the 5′ region of Acp70A. Only three of the four possible haplotypes exist in our sample (Figure 6B), preventing us from explicitly testing for epistasis via the interaction term of a two-way ANOVA. Linkage disequilibrium between these markers (P < 0.01) is likely responsible for the missing haplotype as it should appear in approximately five lines given the allele frequencies in the population. On the basis of unpublished data showing high levels of sequence conservation across species, Cirera and Aguadé (1997) speculated that this region is involved in the regulation of Acp70A and our results are consistent with this region having a functional role in male fitness.
Another well-characterized seminal fluid protein is Esterase-6 (reviewed in Chapman and Davies 2004). A series of experiments show that Esterase-6 affects male mating ability (Gilbert and Richmond 1982; Saad et al. 1994) and female egg-laying rate (Gilbert et al. 1981a; Saad et al. 1994) and influences the likelihood that the female will remate (Richmond et al. 1980; Gilbert et al. 1981b; Scott 1986). Saad et al. (1994) also identified associations between Esterase-6 activity level and variation in male reproductive performance among isofemale lines. In addition to the functional characterization, several studies have identified nonneutral patterns of genetic variation in both the coding (Oakeshott et al. 2001) and the 5′-regulatory regions of Esterase-6 (Balakirev et al. 2002; Odgers et al. 2002). We have identified associations affecting male fitness that map to both the coding region and the 5′-regulatory region of Esterase-6. In fact, the haplotype in the coding region associating with male mating rate (remating) spans the active site (Oakeshott et al. 1987).
The associations with polymorphisms in Acp70A and Esterase-6 are exciting for a variety of reasons. First, they serve to validate the utility of association testing, but more importantly they demonstrate that natural variations in Acp70A and Esterase-6 are important determinants of male reproductive fitness. These findings suggest that postcopulatory sexual selection might be driving the maintenance of nonneutral patterns of genetic variation observed in these genes and that selection might be acting on variation in the coding sequence and operating at the level of transcriptional regulation (see Wray et al. 2003). Clearly the next step is to assess how expression of Acp70A and Esterase-6 varies among the different polymorphisms that we have identified, determine whether natural variation in expression translates into different levels of protein being transferred to the female during mating, and test whether differences in protein level affect reproductive fitness among relevant male genotypes.
Epistasis in male reproductive genes:
The genetic basis for natural variation in male reproductive success is clearly complex even when considering each gene acting only independently. Given the pervasiveness of protein interactions in biological systems, intuition suggests that male reproductive genes do not act independently. Park and Wolfner (1995) demonstrated that the proper cleavage of Acp26Aa requires other accessory gland secretions, providing a clear example of how functional epistasis might occur among different seminal fluid proteins. We tested all pairwise interactions among SNPs and using a conservative cutoff (q < 0.05, P < 5 × 10−5) we have identified four associations that are likely due to epistatic interactions among polymorphisms in male reproductive genes (Figure 5). Two genes, Acp76A and BG642167, that did not show associations in the single-marker or haplotype analysis show strong epistatic interactions affecting sperm competition phenotypes. Interestingly, three of the genes with epistatic interactions are proteases (CG6168) or protease inhibitors (Acp62F, Acp76A). The interactions of proteases and protease inhibitors are known to play important roles in mammalian fertility (Kise et al. 1996), and researchers often speculate that these types of interactions will be important for male reproductive success in Drosophila (see Wolfner 2002).
Only one polymorphism with a significant interaction term was associated with a phenotype in the single-marker tests. It was not, however, with the same phenotype as the interaction term; CG14560snp1474 had a marginal effect on P1′ but interacted with Acp62Fsnp2188 to affect P2′. This is an important observation, as many studies search for epistasis using only polymorphisms that had a significant marginal effect with single-marker tests (see Carlborg and Haley 2004). Our findings, along with others (Carlborg et al. 2003; Montooth et al. 2003), suggest that by focusing only on markers with marginal effects one is likely to miss an important class of genotype–phenotype associations. It is likely that SNPs that lack a marginal effect may be more likely to be maintained in a stable polymorphism through epistatic interactions than would SNPs with large marginal effects as well (Karlin and Carmelli 1975).
The genetic architecture of sperm competitive ability is still not well understood, even in D. melanogaster (but see Hughes 1997; Lew et al. 2006). In particular, little is known about the magnitude of epistatic variance that can have a big impact on the maintenance of variation in the trait as well as the underlying causal genes (Lynch and Walsh 1998). Carlborg et al. (2006) demonstrated that epistasis can “release” genetic variation during long-term directional selection, allowing a greater response to selection than would be expected if gene interactions were excluded. Although it is possible that epistasis could be contributing to the maintenance of genetic variation for phenotypes affecting postcopulatory sexual selection, our study was not designed to address this question. To better understand the role that epistasis might play in maintaining genetic variation for phenotypes affecting postcopulatory sexual selection, it would be valuable to consider how epistasis might influence evolutionary trajectories with male × male (Clark et al. 2000) and male × female interactions (Clark et al. 1999) that are already known to affect sperm competitive ability.
Pleiotropy and male reproductive genes:
Pleiotropy appears to be a common characteristic of many quantitative trait loci (Mackay et al. 2005a,b; Hall et al. 2006). Here we present evidence that four different male reproductive genes have pleiotropic effects on phenotypes affecting sperm competitive ability. In three cases (CG6168 with refractory and P2′; Est-6 with remate and fec-off; Acp62F with fec-V1 and P2′) different markers within a gene associate with different phenotypes. In one case, the same three-marker haplotype upstream of CG14560 associated with both P1′ and fec-def. Here the four observed haplotypes had identical rank-ordered fitness for the two traits (Figure 4, B and D). Interestingly P1′ and fec-def were positively correlated across the lines and our evidence of pleiotropy suggests that a common molecular mechanism might underlie these genetic correlations. On the basis of its consequences for male fitness, this locus should be under directional selection with the high-fitness haplotype moving toward fixation in the population. Perhaps this gene's putative antimicrobial function acts antagonistically on male reproductive success, resulting in antagonistic pleiotropy maintaining genetic variation at this locus.
Although few seminal fluid proteins have been studied in detail, pleiotropy appears to be a common feature of these genes. Fiumera et al. (2005, 2006), using an association approach similar to the one here, found evidence that several male reproductive genes on chromosome 2 have pleiotropic effects on multiple phenotypes, including an example of antagonistic pleiotropy between P2′ and refractory (Fiumera et al. 2005). Acp70A and Esterase-6 are amazing examples of the potential for pleiotropy among seminal fluid proteins (see Kubli 2003; Chapman and Davies 2004). Acp70A is known to affect female egg-laying rate (Chen et al. 1988), male-induced female refractoriness (Chapman et al. 2003; this study), and male-induced cost of mating (Wigby and Chapman 2005) while Esterase-6 affects female productivity (Gilbert et al. 1981a; Saad et al. 1994), male mating success (Richmond et al. 1980; Gilbert and Richmond 1982; Saad et al. 1994), and male-induced female refractoriness (Gilbert et al. 1981b; Scott 1986). Although association testing suggests that male reproductive genes play complex roles in postcopulatory sexual selection, detailed characterization of additional seminal fluid proteins will be required to formally determine their role as multifunctional proteins.
Inbred lines and genetic architecture:
Homozygous chromosome extraction lines provide a powerful means to map the genes underlying complex traits. They allow the role of a single chromosome to be isolated from the rest of the genome and they allow phenotypic measures to be replicated across individuals that are genetically identical. Additionally, they dramatically reduce the samples sizes that are needed to map rare deleterious alleles. There are, however, some limitations to using inbred lines. Some natural variation will be lost when chromosomes contain highly deleterious or lethal alleles (although these should be at very low frequency due to selection). Inbreeding depression may be a concern, and the inability to estimate quantitative genetic parameters is disappointing to evolutionary biologists.
Several metrics of sperm competitive ability were positively correlated across lines in this study (Table 2) and may indicate a shared heritable component (i.e., pleiotropy) but also may be due to inbreeding depression. For example, P1′ and P2′ were positively correlated as were fecundity-defense and fecundity-offense. These observations suggest that there could be shared molecular mechanisms affecting similar phenotypes that are independent of mating order. Several of these correlations are consistent with previous findings. For example, P1′ and refractory were positively correlated in two previous studies (Clark et al. 1995; Fiumera et al. 2005). In addition, P1′ and P2′, as well as P1′ and fec-def were also positively correlated in chromosome 2 extraction lines (Fiumera et al. 2005).
The positive correlations are unlikely to be environmental artifacts, as the sperm competition experiments were conducted over multiple different generations. One does need to consider whether the observed positive correlations could be due to differences in overall line vigor (i.e., inbreeding depression) with healthier lines having higher fitness for many traits. These lines have also been scored for resistance to four bacterial pathogens (T. Sackton, unpublished data) and only 2 of the 36 pairwise correlations (6%) between sperm competition and immunity phenotypes were significant at P < 0.05 compared to 47% within sperm competition phenotypes (Table 2) and 50% within immunity phenotypes (T. Sackton, unpublished data). Therefore, it is unlikely that inbreeding depression is driving these correlations or confounding our association testing because inbreeding is expected to affect all of these traits to some extent. Triple matings, rather than the expected double matings, could also lead to correlations among some phenotypes, but few triple matings resulted when males and females were housed together for the durations used in this study design (Civetta and Clark 2000). Furthermore, this hypothesis would predict that remating and refractory should be positively correlated, and they are not in this experiment or in previous experiments (Clark et al. 1995; Fiumera et al. 2005). The inherent relationship among many of these phenotypes (e.g., P1 and P2) and previous studies showing pleiotropic effects of male reproductive genes (e.g., Acp70A; Chapman et al. 2003; Kubli 2003; Liu and Kubli 2003; Wigby and Chapman 2005) suggest that a shared molecular basis, rather than inbreeding, is a more likely explanation for the observed positive correlations among sperm competition phenotypes.
A number of studies have demonstrated a genetic basis for differences in phenotypes affecting postcopulatory sexual selection in Drosophila, including sperm precedence (Civetta and Clark 2000), male mating success (Hughes and Leips 2006), and female remating rate (Lawniczak and Begun 2005; Lew et al. 2006), just to mention a few. Some studies have even begun to map the variation to chromosomal regions (Lawniczak and Begun 2005; Hughes and Leips 2006) or to candidate genes (Clark et al. 1995; Fiumera et al. 2005). Not only would we want to identify genes and the polymorphism responsible for the phenotypic differences, but we should also strive to characterize the adaptive potential of the focal population, particularly given the nonneutral patterns of genetic diversity seen at male reproductive genes (Swanson and Vacquier 2002).
Ideally the full genetic architecture of sperm competitive phenotypes should be estimated, including additive, dominance, and epistatic components of variance. Given that sperm competitive phenotypes are estimated from large collections of progeny, detailed quantitative genetic analysis is not practical, but some successful efforts have been made. Miller et al. (2001, 2003) demonstrated significant additive variation for sperm length and seminal receptacle length. Lew et al. (2006) found a low but significant coefficient of additive genetic variance for female resistance to male harm, much of which was determined by a female's propensity to remate. Hughes (1997) estimated significant levels of dominance genetic variance for second male sperm precedence but little or no additive genetic variance and inferred that this might be a signature of past directional selection. The results from the association-mapping studies can highlight divergent lines that can be used to create diallel crosses to estimate quantitative genetic parameters while simultaneously testing for the effects of inbreeding. F2 crosses could be created as a genetic test for validation of the association but this would require that additional genotyping be completed. Combining multiple approaches such as association mapping of genes, estimating the quantitative genetic parameters and targeted mutational analysis, will certainly help us understand the evolution of these important fitness-related phenotypes.
Acknowledgments
We thank Phil Olsen, Jenna Rose, and Zoë Weiss for their assistance in scoring progeny. We also thank Xiaoyun Wang for her contributions to the genotyping effort, Bonnie Sceurman and Manolis Dermitzakis for their part in generating the chromosome 3 substitution lines, and Tim Sackton for access to the unpublished data on immunity phenotypes. Two anonymous reviewers and Larry Harshman greatly improved this manuscript. This work was supported by a National Science Foundation grant (DEB-0242987 to A.G.C.), by a National Institutes of Health Ruth L. Kirschstein Postdoctoral Fellowship (NGA 1 F32 GM70300-01 to A.C.F.), and by Cornell University travel funds (to B.L.D.).
References
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., N. Miyashita and C. H. Langley, 1992. Polymorphism and divergence in the Mst26A male accessory gland gene region in Drosophila. Genetics 132 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian, A., B. Gharizadeh, A. C. Gustafsson, F. Sterky, P. Nyren et al., 2000. Single-nucleotide polymorphism analysis by pyrosequencing. Anal. Biochem. 280 103–110. [DOI] [PubMed] [Google Scholar]
- Aigaki, T., I. Fleischmann, P. S. Chen and E. Kubli, 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 7 557–563. [DOI] [PubMed] [Google Scholar]
- Balakirev, E. S., and F. J. Ayala, 2003. Nucleotide variation of the Est-6 gene region in natural populations of Drosophila melanogaster. Genetics 165 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakirev, E. S., E. I. Balakirev and F. J. Ayala, 2002. Molecular evolution of the Est-6 gene in Drosophila melanogaster: contrasting patterns of DNA variability in adjacent functional regions. Gene 288 167–177. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen, J. D., H. Nielsen, G. Von Heijne and S. Brunak, 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. [DOI] [PubMed] [Google Scholar]
- Bloch Qazi, M. C., and M. F. Wolfner, 2003. An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. J. Exp. Biol. 206 3521–3528. [DOI] [PubMed] [Google Scholar]
- Broberg, P., 2005. A comparative review of estimates of the proportion unchanged genes and the false discovery rate. BMC Bioinformatics 6 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg, O., and C. S. Haley, 2004. Epistasis: too often neglected in complex trait studies? Nat. Rev. Genet. 5 618–625. [DOI] [PubMed] [Google Scholar]
- Carlborg, O., S. Kerje, K. Schutz, L. Jacobsson, P. Jensen et al., 2003. A global search reveals epistatic interaction between QTL for early growth in the chicken. Genome Res. 13 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg, O., L. Jacobsson, P. Ahgren, P. Siegel and L. Andersson, 2006. Epistasis and the release of genetic variation during long-term selection. Nat. Genet. 38 418–420. [DOI] [PubMed] [Google Scholar]
- Chapman, T., and S. J. Davies, 2004. Functions and analysis of the seminal fluid proteins of male Drosophila melanogaster fruit flies. Peptides 25 1477–1490. [DOI] [PubMed] [Google Scholar]
- Chapman, T., L. A. Herndon, Y. Heifetz, L. Partridge and M. F. Wolfner, 2001. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proc. R. Soc. Lond. Ser. B Biol. Sci. 268 1647–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. S., E. Stummzollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory-gland peptide that regulates reproductive-behavior of female Drosophila-melanogaster. Cell 54 291–298. [DOI] [PubMed] [Google Scholar]
- Churchill, G. A., and R. W. Doerge, 1994. Empirical threshold values for quantitative trait mapping. Genetics 138 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguadé, 1997. Evolutionary history of the sex-peptide (Acp70A) gene region in Drosophila melanogaster. Genetics 147 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civetta, A., and A. G. Clark, 2000. Chromosomal effects on male and female components of sperm precedence in Drosophila. Genet. Res. 75 143–151. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., 2004. The role of haplotypes in candidate gene studies. Genet. Epidemiol. 27 321–333. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory-gland protein loci in Drosophila melanogaster. Genetics 139 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., D. J. Begun and T. Prout, 1999. Female x male interactions in Drosophila sperm competition. Science 283 217–220. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., E. T. Dermitzakis and A. Civetta, 2000. Nontransitivity of sperm precedence in Drosophila. Evolution 54 1030–1035. [DOI] [PubMed] [Google Scholar]
- Dixson, A. F., and M. J. Anderson, 2004. Sexual behavior, reproductive physiology and sperm competition in male mammals. Physiol. Behav. 83 361–371. [DOI] [PubMed] [Google Scholar]
- Dworkin, I., A. Palsson and G. Gibson, 2005. Replication of an egfr-wing shape association in a wild-caught cohort of Drosophila melanogaster. Genetics 169 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, W. G., and C. Cordero, 2003. Sexual conflict and female choice. Trends Ecol. Evol. 18 438–439. [DOI] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2005. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics 169 243–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera, A. C., B. L. Dumont and A. G. Clark, 2006. Natural variation in male-induced ‘cost-of-mating’ and allele-specific association with male reproductive genes in Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage, M. J. G., C. P. Macfarlane, S. Yeates, R. G. Ward, J. B. Searle et al., 2004. Spermatozoal traits and sperm competition in Atlantic salmon: relative sperm velocity is the primary determinant of fertilization success. Curr. Biol. 14 44–47. [PubMed] [Google Scholar]
- Gilbert, D. G., and R. C. Richmond, 1982. Esterase-6 in Drosophila-melanogaster - reproductive function of active and null males at low-temperature. 10. Proc. Natl. Acad. Sci. USA 79 2962–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, D. G., R. C. Richmond and K. B. Sheehan, 1981. a Studies of Esterase 6 in Drosophila-melanogaster. 5. Progeny production and sperm use in females inseminated by males having active or null alleles. Evolution 35 21–37. [DOI] [PubMed] [Google Scholar]
- Gilbert, D. G., R. C. Richmond and K. B. Sheehan, 1981. b Studies of Esterase-6 in Drosophila-melanogaster. 7. Re-mating times of females inseminated by males having active or null alleles. Behav. Genet. 11 195–208. [DOI] [PubMed] [Google Scholar]
- Hall, M. C., C. J. Basten and J. H. Willis, 2006. Pleiotropic quantitative trait loci contribute to population divergence in traits associated with life-history variation in Mimulus guttatus. Genetics 172 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt, A. H., P. H. Harvey, S. G. Larson and R. V. Short, 1981. Testis weight, body-weight and breeding system in primates. Nature 293 55–57. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., O. Lung, E. A. Frongillo and M. F. Wolfner, 2000. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr. Biol. 10 99–102. [DOI] [PubMed] [Google Scholar]
- Herndon, L. A., and M. F. Wolfner, 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg-laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 92 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn, J. N., and M. J. Daly, 2005. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6 95–108. [DOI] [PubMed] [Google Scholar]
- Hughes, K. A., 1997. Quantitative genetics of sperm precedence in Drosophila melanogaster. Genetics 145 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, K. A., and J. Leips, 2006. Quantitative trait locus analysis of male mating success and sperm competition in Drosophila melanogaster. Evolution 60 1427–1434. [PubMed] [Google Scholar]
- Karlin, S., and D. Carmelli, 1975. Numerical studies on 2-loci selection models with general viabilities. Theor. Popul. Biol. 7 399–421. [DOI] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2004. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics 167 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise, H., J. Nishioka, J. Kawamura and K. Suzuki, 1996. Characterization of semenogelin II and its molecular interaction with prostate-specific antigen and protein C inhibitor. Eur. J. Biochem. 238 88–96. [DOI] [PubMed] [Google Scholar]
- Konior, M., L. Keller and J. Radwan, 2005. Effect of inbreeding and heritability of sperm competition success in the bulb mite Rhizoglyphus robini. Heredity 94 577–581. [DOI] [PubMed] [Google Scholar]
- Kubli, E., 2003. Sex-peptides: seminal peptides of the Drosophila male. Cell. Mol. Life Sci. 60 1689–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak, M. K. N., and D. J. Begun, 2005. A QTL analysis of female variation contributing to refractoriness and sperm competition in Drosophila melanogaster. Genet. Res. 86 107–114. [DOI] [PubMed] [Google Scholar]
- Lefevre, Jr., G., and U. B. Jonsson, 1962. Sperm transfer, storage, displacement and utilization in Drosophila melanogaster. Genetics 47 1719–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew, T. A., E. H. Morrow and W. R. Rice, 2006. Standing genetic variance for female resistance to harm from males and its relationship to intralocus sexual conflict. Evolution 60 97–105. [PubMed] [Google Scholar]
- Liu, H. F., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., and C. H. Langley, 1999. The power of association studies to detect the contribution of candidate genetic loci to variation in complex traits. Genome Res. 9 720–731. [PMC free article] [PubMed] [Google Scholar]
- Long, A. D., R. F. Lyman, A. H. Morgan, C. H. Langley and T. F. C. Mackay, 2000. Both naturally occurring insertions of transposable elements and intermediate frequency polymorphisms at the achaete-scute complex are associated with variation in bristle number in Drosophila melanogaster. Genetics 154 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O., and M. F. Wolfner, 1999. Drosophila seminal fluid proteins enter the circulatory system of the mated female fly by crossing the posterior vaginal wall. Insect Biochem. Mol. Biol. 29 1043–1052. [DOI] [PubMed] [Google Scholar]
- Lung, O., and M. F. Wolfner, 2001. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem. Mol. Biol. 31 543–551. [DOI] [PubMed] [Google Scholar]
- Lung, O., L. Kuo and M. F. Wolfner, 2001. Drosophila males transfer antibacterial proteins from their accessory gland and ejaculatory duct to their mates. J. Insect Physiol. 47 617–622. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and B. Walsh, 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA.
- Mackay, T. F. C., and C. H. Langley, 1990. Molecular and phenotypic variation in the achaete-scute region of Drosophila-melanogaster. Nature 348 64–66. [DOI] [PubMed] [Google Scholar]
- Mackay, T. F. C., S. L. Heinsohn, R. F. Lyman, A. J. Moehring, T. J. Morgan et al., 2005. a Genetics and genomics of Drosophila mating behavior. Proc. Natl. Acad. Sci. USA 102 6622–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, T. F. C., R. F. Lyman and F. Lawrence, 2005. b Polygenic mutation in Drosophila melanogaster: mapping spontaneous mutations affecting sensory bristle number. Genetics 170 1723–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo, A. F., J. J. Garde, A. J. Soler, A. J. Garcia, M. Gomendio et al., 2005. Male fertility in natural populations of red deer is determined by sperm velocity and the proportion of normal spermatozoa. Biol. Reprod. 72 822–829. [DOI] [PubMed] [Google Scholar]
- Miller, G. T., W. T. Starmer and S. Pitnick, 2001. Quantitative genetics of seminal receptacle length in Drosophila melanogaster. Heredity 87 25–32. [DOI] [PubMed] [Google Scholar]
- Miller, G. T., W. T. Starmer and S. Pitnick, 2003. Quantitative genetic analysis of among-population variation in sperm and female sperm-storage organ length in Drosophila mojavensis. Genet. Res. 81 213–220. [DOI] [PubMed] [Google Scholar]
- Montooth, K. L., J. H. Marden and A. G. Clark, 2003. Mapping determinants of variation in energy metabolism, respiration and flight in Drosophila. Genetics 165 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., D. R. Ripoll, C. F. Aquadro and M. F. Wolfner, 2004. Comparative structural modeling and inference of conserved protein classes in Drosophila seminal fluid. Proc. Natl. Acad. Sci. USA 101 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum, D. M., and M. F. Wolfner, 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott, J. G., C. Collet, R. W. Phillis, K. M. Nielsen, R. J. Russell et al., 1987. Molecular-cloning and characterization of Esterase-6, a serine hydrolase of Drosophila. Proc. Natl. Acad. Sci. USA 84 3359–3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott, J. G., E. A. Van Papenrecht, C. Claudianos, B. C. Morrish, C. Coppin et al., 2001. An episode of accelerated amino acid change in Drosophila esterase-6 associated with a change in physiological function. Genetica 110 231–244. [DOI] [PubMed] [Google Scholar]
- Odgers, W. A., C. F. Aquadro, C. W. Coppin, M. J. Healy and J. G. Oakeshott, 2002. Nucleotide polymorphism in the Est6 promoter, which is widespread in derived populations of Drosophila melanogaster, changes the level of Esterase 6 expressed in the male ejaculatory duct. Genetics 162 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppliger, A., Y. Naciri-Graven, G. Ribi and D. J. Hosken, 2003. Sperm length influences fertilization success during sperm competition in the snail Viviparus ater. Mol. Ecol. 12 485–492. [DOI] [PubMed] [Google Scholar]
- Palsson, A., and G. Gibson, 2004. Association between nucleotide variation in Egfr and wing shape in Drosophila melanogaster. Genetics 167 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M., and M. F. Wolfner, 1995. Male and female cooperate in the prohormone-like processing of a Drosophila melanogaster seminal fluid protein. Dev. Biol. 171 694–702. [DOI] [PubMed] [Google Scholar]
- Parker, G. A., 1970. Sperm competition and its evolutionary consequences in insects. Biol. Rev. Camb. Philos. Soc. 45 525–567. [Google Scholar]
- Parker, G. A., and L. Partridge, 1998. Sexual conflict and speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiani, A., 2006. Complexity of seminal fluid: a review. Behav. Ecol. Sociobiol. 60 289–310. [Google Scholar]
- Preston, B. T., I. R. Stevenson and K. Wilson, 2003. Soay rams target reproductive activity towards promiscuous females' optimal insemination period. Proc. R. Soc. Lond. Ser. B Biol. Sci. 270 2073–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, M., and F. Rousset, 1995. Genepop (version-1.2) - population-genetics software for exact tests and ecumenicism. J. Hered. 86 248–249. [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381 232–234. [DOI] [PubMed] [Google Scholar]
- Richmond, R. C., D. G. Gilbert, K. B. Sheehan, M. H. Gromko and F. M. Butterworth, 1980. Esterase-6 and reproduction in Drosophila-melanogaster. Science 207 1483–1485. [DOI] [PubMed] [Google Scholar]
- Saad, M., A. Y. Game, M. J. Healy and J. G. Oakeshott, 1994. Associations of Esterase-6 allozyme and activity variation with reproductive fitness in Drosophila-melanogaster. Genetica 94 43–56. [DOI] [PubMed] [Google Scholar]
- Schaid, D. J., 2004. Evaluating associations of haplotypes with traits. Genet. Epidemiol. 27 348–364. [DOI] [PubMed] [Google Scholar]
- Scheet, P., and M. Stephens, 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78 629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, D., 1986. Inhibition of female Drosophila melanogaster remating by a seminal fluid protein (Esterase-6). Evolution 40 1084–1091. [DOI] [PubMed] [Google Scholar]
- Storey, J. D., and R. Tibshirani, 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styger-Schmucki, D, 1992. Molekulare analyse des sexpeptidgens aus Drosophila melanogaster. Ph.D. Thesis, University of Zürich, Zürich, Switzerland.
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C. I. Wu, 2001. Sex in Drosophila mauritiana: a very high level of amino acid polymorphism in a male reproductive protein gene, Acp26Aa. Mol. Biol. Evol. 18 22–26. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., and J. K. Pritchard, 2003. Haplotype blocks and linkage disequilibrium in the human genome. Nat. Rev. Genet. 4 587–597. [DOI] [PubMed] [Google Scholar]
- Wigby, S., and T. Chapman, 2004. Sperm competition. Curr. Biol. 14 R100–R103. [PubMed] [Google Scholar]
- Wigby, S., and T. Chapman, 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15 316–321. [DOI] [PubMed] [Google Scholar]
- Wolfner, M. F., 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88 85–93. [DOI] [PubMed] [Google Scholar]
- Wray, G. A., M. W. Hahn, E. Abouheif, J. P. Balhoff, M. Pizer et al., 2003. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20 1377–1419. [DOI] [PubMed] [Google Scholar]