Abstract
Defects in lysosomal trafficking pathways lead to decreased cell viability and are associated with progressive disorders in humans. Previously we have found that loss-of-function (LOF) mutations in the Drosophila gene blue cheese (bchs) lead to reduced adult life span, increased neuronal death, and widespread CNS degeneration that is associated with the formation of ubiquitinated-protein aggregates. To identify potential genes that participate in the bchs functional pathway, we conducted a genetic modifier screen based on alterations of an eye phenotype that arises from high-level overexpression of Bchs. We found that mutations in select autophagic and endocytic trafficking genes, defects in cytoskeletal and motor proteins, as well as mutations in the SUMO and ubiquitin signaling pathways behave as modifiers of the Bchs gain-of-function (GOF) eye phenotype. Individual mutant alleles that produced viable adults were further examined for bchs-like phenotypes. Mutations in several lysosomal trafficking genes resulted in significantly decreased adult life spans and several mutants showed changes in ubiquitinated protein profiles as young adults. This work represents a novel approach to examine the role that lysosomal transport and function have on adult viability. The genes characterized in this study have direct human homologs, suggesting that similar defects in lysosomal transport may play a role in human health and age-related processes.
LYSOSOMES are critical organelles for the turnover or degradation of a wide variety of cellular constituents (Dell'Angelica et al. 2000). A complex series of targeting and import pathways direct the flow of material to the lysosome and defects in these pathways are associated with many progressive conditions including lysosomal storage disorders, reduced viability, and neural degeneration (Cataldo et al. 1996; Dell'Angelica et al. 2000; Brunk and Terman 2002; Cuervo 2004). There are three main vesicle-based pathways for transport of material to the lysosome: transport from the trans-Golgi network (TGN), the endocytotic pathway, and macroautophagy (here after called autophagy) (Shih et al. 2002; Klionsky et al. 2003; Raiborg et al. 2003; Luzio et al. 2005). Autophagy involves the sequestration of cytoplasmic material and entire organelles into double-membrane vesicles called autophagosomes, which are transported along microtubules for fusion with lysosomes, generating autolysosomes where the sequestered material is degraded (Klionsky and Emr 2000). Groundbreaking genetic studies in yeast have allowed the identification and characterization of nearly 30 conserved autophagy (atg) genes (Klionsky et al. 2003). Inactivation of key components within the pathway has revealed that autophagy primarily functions as an adaptive response to starvation or cellular stress by recycling nonessential cellular components for nutrition or by clearing old or damaged cytoplasmic material and organelles (Scott et al. 2004; Komatsu et al. 2005). Recent genetic studies in mice have shown that ablation of the atg5 and atg7 genes in the CNS leads to progressive neurological defects, the formation of ubiquitinated inclusion bodies or protein aggregates, and neuronal cell death (Hara et al. 2006; Komatsu et al. 2006).
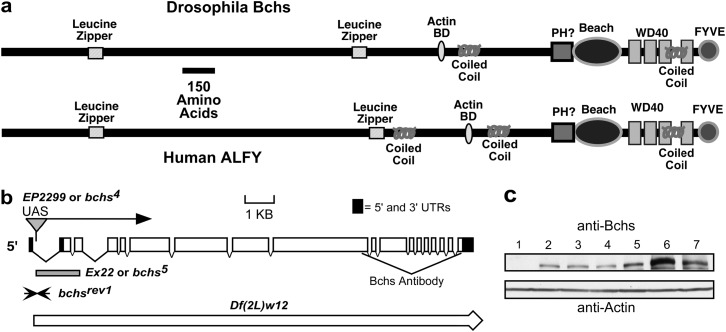
Previously we have shown that mutations in the Drosophila blue cheese (bchs) gene result in a reduced adult life span and age-related neuronal degeneration. These defects include neural atrophy and cell death, preceded by the accumulation of ubiquitin-conjugated protein aggregates throughout the adult CNS (Finley et al. 2003). Consistent with these findings is the recent characterization of Alfy (autophagy-linked FYVE protein) the conserved human bchs homolog (Figure 1a) (Simonsen et al. 2004). Under starvation conditions or following proteasome inhibitor treatment Alfy relocalizes from the nuclear membrane to cytoplasmic structures containing ubiquitin and early autophagic markers (Simonsen et al. 2004). Both Bchs and Alfy proteins are very large, highly conserved proteins that are nearly 400 kDa in size (∼50% identity between fly and human homologs) (Finley et al. 2003; Simonsen et al. 2004). Both proteins contain several conserved protein domains in the C terminus: a BEACH domain followed by a series of WD40 repeats and a PI(3)P-binding FYVE domain (Figure 1a) (Finley et al. 2003; Simonsen et al. 2004). The N-terminal two-thirds of Bchs/Alfy (>2000 amino acids) are also conserved, but do not contain readily identifiable functional domains. However, this region is leucine/isoleucine rich and modeling programs suggest the presence of leucine zippers and coiled-coil domains. On the basis of this sequence analysis it is likely that Bchs/Alfy serve as scaffolding proteins, mediating a diverse series of protein and lipid interactions promoting the recruitment, organization, and transport of vesicles. Taken together, this information suggests that the Bcsh/Alfy family aids in the removal of cytoplasmic ubiquitinated protein aggregates by promoting their autophagic clearance (Bjorkoy et al. 2005). This protein family is not found in yeast and the Bchs/Alfy proteins may have a greater role in multicellular organisms in protein clearance than in starvation-induced autophagy (Finley et al. 2003; Simonsen et al. 2004). However, the cellular pathway(s) that Bchs participates in remains poorly understood.
Figure 1.—
Functional motifs of Bchs/Alfy, mutant alleles, and protein profiles. (a) Members of the bchs/Alfy gene family encode very large, highly conserved proteins with several potential functional protein domains. The C terminus contains a FYVE finger motif [binds PtdIns(3)P lipids], a BEACH domain, and a series of WD40 repeats (protein–protein interaction domain). The remaining N-terminal ∼2000 amino acids of the Bchs and Alfy proteins are also highly conserved and leucine–isoleucine rich. Motif-modeling programs predict several leucine-based motifs including leucine zippers and coiled-coil domains, which primarily facilitate protein interactions and dimerization. Further sequence analysis identifies a second potential phosphatidylinositol interaction motif (PH domain). Sequence analysis of the Bchs/Alfy family indicates that these proteins likely facilitate a diverse series of protein interactions as well having close associations with membrane vesicles. (b) The insertion site of the EP(2L)2299 transposable element is located in the first intron of bchs and allows Gal4-driven expression of the full-length Bchs protein. The Df(2L)w12 deletion was characterized during our the initial characterization and mapping of the bchs and dsf genes and removes nearly 60 kb of genomic sequence containing both genes. Several lines were also isolated that represent individual P-element excisions that generate microexcision alleles of bchs (Ex22 or bchs5) or fully restore the gene (bchsrev1). (c) Western analysis of protein extracts made from adult heads shows that Bchs is a large protein (>250 kDa) that is absent from bchs5 mutants (1) and is expressed at normal levels in bchsrev1 animals (2). Young homozygous EP(2L)2299 flies (3) produce Bchs at levels that are similar to those of Canton-S (4) and w1118 (5) control animals. The Bchs protein is increased fivefold when overexpressed in EP(2L)2299,GMR-Gal4 flies (6) above that of Canton-S flies (4). Using a small deletion that eliminates one bchs allele reduces the total level of Bchs even when it is overexpressed in the eye [EP(2L)2299,GMR-Gal4/Df(2L)W12] (7).
A recent study showed that overexpression of Bchs in the eye using a GMR-Gal4 driver results in a rough eye phenotype (Khodosh et al. 2006). Using a genetic modifier screen, 195 chromosomal deficiency lines were crossed to the Bchs overexpressing line and individual genes uncovered by a single deletion (93B6–7; 93D2) were examined further for Bchs interaction (Khodosh et al. 2006). Individual rab11 mutations were found to significantly enhance the dominant Bchs eye phenotype and additional studies revealed that the Rab11 and Bchs proteins colocalize at the neuromuscular junction and affect bristle development and synaptic function (Khodosh et al. 2006). In this report, we show that overexpressing Bchs in the eye (GMR-Gal4 driver) also results in a rough eye phenotype that is accompanied by the formation of ubiquitin containing varicosities along photoreceptor neural projections. This phenotype is also seen when Bchs is overexpressed in larval motor neurons and is similar to defects associated with perturbations of vesicle transport or fusion pathways (Torroja et al. 1999; Gunawardena and Goldstein 2001; Kraut et al. 2001; Nixon et al. 2005). We further show that young bchs mutants and flies overexpressing Bchs display an altered accumulation profile of ubiquitinated (UB) proteins. Collectively, these findings suggest that Bchs affects protein and vesicle trafficking and is consistent with its role in the transport of lysosomal substrates. To further investigate potential bchs genetic interactions an extensive genetic screen based on alteration of the Bchs eye phenotype was used to identify several unique modifier loci (FlyBase 2007). From this study recessive mutations in lysosomal trafficking genes and cytoskeletal and motor proteins as well as in members of the ubiquitin and SUMO signaling pathways were found to have potential genetic interactions with Bchs (FlyBase 2007). To further characterize mutant phenotypes, mutations in lysosomal genes that produced viable adult flies were examined for UB-protein profiles and changes to adult longevity. As with bchs mutants, the functional losses of several lysosomal transport genes also alter high-molecular-weight UB-protein profiles and reduce adult longevity. Together the genetic analyses of several lysosomal trafficking genes provide a novel mechanistic insight into the requirement of these pathways for the long-term viability of adult Drosophila.
MATERIALS AND METHODS
Protein sequence analysis and motifs:
Identification and characterization of potential functional domains encoded within the Bchs and Alfy protein sequences were performed using online modeling algorithms including http://scansite.mit.edu/motifsca and http://www.ncbi.nlm.nih.gov/Structure/.
Fly culture and stocks:
Flies were cultured and maintained on standard cornmeal–molasses–yeast-based medium. The transgenic line, EP(2L)2299 was originally from the Rörth collection of P-element insertions and allows Gal4-driven expression of the full-length Bchs protein (Rorth 1996; Kraut et al. 2001). The bcsh5 allele and revertant lines that were derived from excision of the EP(2L)2299 P element have been described previously and are outlined in Figure 1b (Finley et al. 2003). The bchs P-element insertion allele, bcsh3 was maintained as a stock over the Df(2L)clot7 chromosome [402-11Cy/Df(2L)clot7] and used for aging and starvation studies (Finley et al. 2003; FlyBase 2007). Drosophila stocks screened in this study were primarily obtained from the Bloomington Stock Center (stock numbers are noted in Figure 2 and Table 1) (FlyBase 2007). The hook11 allele was a gift from H. Kramer (University of Texas Southwestern) (Kramer and Phistry 1999) and the UAS-GFP-CAAX (membrane-targeted GFP) and GMR-Gal4 transgenic lines have been described previously (Finley et al. 1998).
Figure 2.—
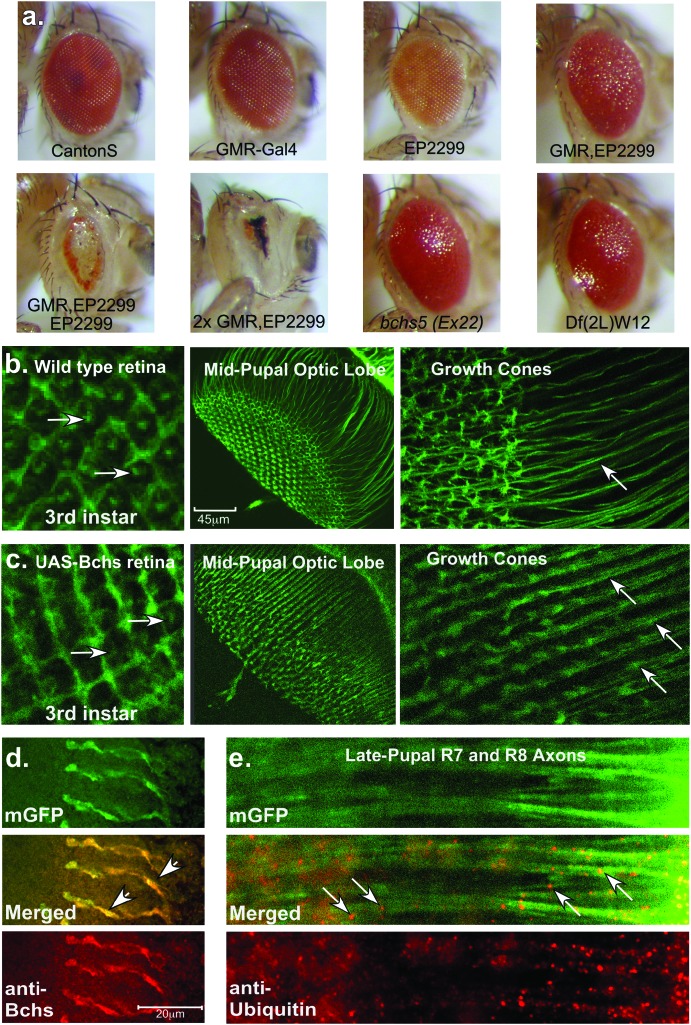
Phenotypes resulting from Bchs overexpression. (a) Characteristic external eye phenotypes of wild type, GMR-Gal4, EP(2L)2299, GMR-Gal4,EP(2L)2299, and 2X GMR-Gal4, EP(2L)2299 flies. (b) Left to right: confocal images from control flies expressing GMR-Gal4/UAS-GFP-CAAX, which express a membrane-targeted green fluorescent protein, show the developing retina (arrows indicate convergence of photoreceptor axons) and neural projections of R7 and R8 photoreceptors (midpupae). (c) Left to right: age-matched tissues from flies coexpressing Bchs [EP(2L)2299]. Retinal development is relatively normal but neural projections into the optic lobe show subtle defects in patterning and growth cone (GC) formation. Arrows indicate the formation of varicosities along the length of axons. (d) In late pupae immunostaining for Bchs shows high levels of Bchs in late pupal axons in EP(2L)2299-overexpressing animals. Bchs colocalizes to terminal synapses and regions of axonal swelling (arrows). (e) In late pupae, ubiquitinated protein aggregates or vesicles (arrows) can be detected along the length of photoreceptor axons in EP(2L)2299-overexpressing animals. These structures are not seen in age-matched wild-type controls (data not shown).
TABLE 1.
Enhancers and suppressors of Bchs overexpression eye phenotype
| Gene and allele | Modification | Bloomington stock no. | Homologs, protein domains, and functions |
|---|---|---|---|
| Vesicle trafficking | |||
| atg100305 | En | 11484 | Serine/threonine kinase, Tor effector |
| atg600096 | En | 11487 | Beclin-1, mitochondrial protein |
| atg18KG03090 | WEn | 13945 | WD40 domains, binds PIP3-5P2 lipid |
| atg2EP3697 | Su | 17156 | Hydrophilic protein, preautophagic structure |
| atg8aEP362 | Su | 14639 | LC3, lipid attachment and microtubule binding |
| orange49h | En | 2385 | ξ-subunit of AP-3 complex |
| ruby1 | En/Su | 88 | β-subunit of AP-3 complex |
| garnet1 | En/Su | 12 | δ-subunit of AP-3 complex |
| carmine1 | En/Su | 21 | μ-subunit of AP-3 complex |
| spinsterk09905 | En | 10948 | Integral membrane protein |
| deep orange1 | En | 3936 | Vps18p, ubiquitin ligase E3 |
| deep orange4 | En | 35 | Vps18p, ubiquitin ligase E3 |
| deep orange8 | En | 28 | Vps18p, ubiquitin ligase E3 |
| carnation1 | En | 19 | Vps 33, sec1 protein |
| ras oppositeG27 | SEn | 4381 | Rop, munc-18, n-Sec1 |
| syntaxin 1AΔ229 | En | 4379 | SNARE, coiled-coil, vesicle targeting |
| syntaxin 1301470 | En | 11536 | SNARE, coiled-coil, vesicle targeting |
| hook1 | En | 306 | Coiled-coiled, MVB interactions |
| rab1193Bi | SEn | 4158 | Small GTPase |
| rab11j2D1 | SEn | 12148 | Small GTPase |
| lightoid1 | En | 338 | Rab-RP1, a GTPase |
| claret1 | En | 459 | Guanine nucleotide exchange factor (GEF) |
| Cytoskeletal and motor proteins | |||
| dlic2G0190 | En | 11951 | Dynein LC, microtubule-based motor activity |
| dlic2G0065 | En | 11696 | Dynein LC, microtubule-based motor activity |
| btvBG01771 | En | 12589 | Dynein HC, microtubule-based motor activity |
| α-tubulin84B5 | En | 2412 | 84B, cytoskeletal and microtubule constituent |
| α-tubulin84B7 | En | 2237 | 84B, cytoskeletal and microtubule constituent |
| β-tubulin D | WEn | 2451 | 85D, cytoskeletal and microtubule constituent |
| Ubiquitin and SUMO pathway | |||
| smt3k06307 | En | 10419 | SUMO, ubiquitin-like protein |
| lesswrightr05486 | En | 11410 | SUMO-conjugating enzyme |
| uba1s3484 | En | 10461 | E1 ubiquitin-activating enzyme |
| effetemer1 | En | 4656 | UbcD1, ubiquitin-conjugating enzyme |
| CG1490 | En | 14505 | Ubiquitin-specific protease 7 |
Modifier genes that also demonstrate a phenotypic change with a corresponding deletion chromosome from the second and third deficiency kit screens are underlined. En, enhancer; WEn, weak enhancer; Su, suppressor; En/Su, complex modifier; SEn, strong enhancer.
Western analysis of the Bchs protein and ubiquitinated conjugated proteins:
Groups of 50 fly heads per genotype were collected and homogenized in 1× PBS, 0.1% Triton-X buffer containing protease inhibitors (4°). After centrifugation the supernatants were collected for each genotype and saved as the protein concentrations were determined for each sample using a Lowry assay (Bio-Rad, Hercules, CA). Twenty micrograms of total protein for each sample were loaded and resolved on 4–20% gradient gels (Bio-Rad) and electroblotted. Western blots were probed sequentially with anti-Bchs (1:4000 dilution, polyclonal rabbit, a gift from Kia Zinn, Division of Biology, California Institute of Technology, Pasadena, CA) or anti-Actin antibodies (1:200 dilution, mouse monoclonal JLA20, developed by Jim Jung-Ching Lin and obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA). For Western analysis of ubiquitinated proteins, 6 heads for each genotype (1-day-old flies) were collected and sonicated in 100 μl protein lysis buffer (2% SDS) and centrifuged. Twenty-five microliters of each sample were run on a 4–20% gradient gel and Western blots were probed sequentially with anti-Ubiquitin (1:1000 dilution, Cell Signaling), anti-Actin, and anti-Bchs antibodies. Following hybridization with appropriate secondary antibodies, immunoreactive bands were detected using standard enhanced chemiluminescence reagents. Autoradiographs were digitally scanned using a GS-800 calibrated desitometer and ImageQuant imaging software and the relative amount of Bchs protein for a given genotype was corrected using actin as a loading control.
Fluorescence confocal microscopy:
Staged third instar larvae and pupae from the UAS-GFP-CAAX,GMR-Gal4 stock line or from the UAS-GFP-CAAX,GMR-Gal4/EP(2)2299 cross were dissected, fixed in 3.5% paraformaldehyde and PBS, and rinsed in PBT (0.05% Triton-X). Tissues were mounted and assayed directly for GFP expression patterns or used for costaining with anti-Bchs or anti-ubiquitin antibodies. For costaining, tissues were fixed in 3.5% paraformaldehyde and PBS for 1 hr at 4° and washed three times in PBT (0.05% Triton-X) at 4°. Following three washes in PBT, tissues were incubated in 5% normal goat serum, PBT, and anti-Bchs (1:1000 dilution; Kia Zinn, Caltech) or with anti-ubiquitin antibodies (1:200 dilution, mouse monoclonal; Cell Signaling) for 2 hr at room temperature, washed three times, and then incubated for 1 hr at room temperature with Cy3-conjugated anti-rabbit or anti-mouse secondary antibodies (1:200 dilution; Jackson Laboratories). Tissues were washed three times in PBT and mounted. Images were collected using a Leica TCS SP2 AOBS confocal microscope.
Design of the Bchs gain-of-function modifier screen:
A Bchs-based gain-of-function (GOF) screen was made possible by a characterization fly line containing an EP-UAS modular expression transposable element insertion, EP(2L)2299, which is located upstream of the bchs coding sequence (Rorth 1996). Previous work reported that this UAS-P element allowed overexpression of full-length Bchs protein in motor neurons and generated a dominant GOF phenotype (Kraut et al. 2001). Overexpressing Bchs in the eye using the GMR-Gal4 driver also produces a highly reproducible dominant phenotype. The GOF phenotype is sensitive to Bchs dosage as shown in flies containing an additional copy of EP(2L)2299 or two copies of both transgenes (Figure 2a). The GMR-Gal4 and EP(2L)2299 P elements were recombined onto a single chromosome and kept as a heterozygous stock by placing it over a double second and third chromosome balancer (CyO:TM6B, Tb1) (FlyBase 2007). Most lines selected for testing in this screen were obtained from the Bloomington Stock Center. Initially we examined stocks representing the majority of the second and third chromosome Bloomington deficiency kits. Subsequent crosses with the GMR-Gal4,EP(2L)2299/CyO:TM6B stock involved lines containing mutations in genes with a wide range of cellular functions. Subsequent mutations were selected that further focused the screen and represented genes involved with lysosomal transport pathways, cytoskeleton or motor proteins, or members of the ubiquitin/SUMO pathways (FlyBase 2007). Individual lines were crossed to the GMR-Gal4,EP(2L)2299/CyO:TM6B stock and grown at 22° and the F1 progeny were scored for modification of eye pigmentation, size, shape, surface texture, and necrosis. On average a size decrease of 30–35% or an increase of 15–20% was set as a lower and an upper significance limit and used to classify a mutation as an enhancer or a suppressor. Representative digital eye images were taken using a Leica MZ6 dissection microscope and Nikon Coolpix 990 camera system. Images were processed and illustrations made using Adobe Photoshop 7.0 and Canvas 8.0 imaging software.
Life span analysis:
Before analyzing mutant lines for changes in adult life span, each stock was first outcrossed into a Canton-S or w1118 background for several generations before reestablishing individual homozygous lines. During the outcrossing process individual mutant lines were assayed for life span and multiple experiments were pooled. For aging analysis at least 100 newly emerged male flies were collected for a given genotype and kept on standard Drosophila culture media (25 per vile) (Finley et al. 2003). Flies were placed at 25° or 29° with a 12-hr light–dark cycle and turned onto fresh food every 2–3 days, and the number of dead flies was counted. The percentage of flies remaining alive for a given time point was calculated from the total starting number of flies aged for a particular genotype. The mean genotype life span and standard deviation were determined using Microsoft Excel and the P-values were determined using GraphPad online software (http://www.graphpad.com).
RESULTS
Characterization of the bchs EP(2L)2299 P element and Bchs expression profiles:
While bchs's loss-of-function (LOF) phenotypes make it an interesting model to study progressive neural degeneration, the subtlety and timing of its adult defects has prevented the efficient design of genetic screens to clarify bchs's functional pathway or to identify potential interacting partners (Finley et al. 2003). We therefore characterized a P-element insertion located in bchs [EP(2L)2299] that could be used to design a dominant GOF genetic modifier screen. Previously we had mapped and sequenced the entire genomic region and cDNA sequence of the bchs gene and carried out a P-element mutagenesis screen on the basis of excision of the EP(2L)2299 construct (Figure 1b). Molecular analysis showed that the EP(2L)2299 P-element line contains a single insertion located within the first bchs intron, upstream of the bchs open reading frame (Figure 1b). Using genetic crosses with a stock containing a constitutively active transposase (Δ2-3), the EP(2L)2299 P element was excised and individual flies lacking the white+ marker were used to generate stocks and characterized for both precise and imprecise removal of the P element. Both types of excisions were identified and characterized for bchs expression and mutant phenotypes (Finley et al. 2003; Simonsen et al. 2004). The bchs5 allele (Ex22) represents an imprecise excision that removes most of the first and second introns and the second and third exons, effectively eliminating the start codon of the message (Figure 1b). Other lines representing precise excisions were identified that restore the original bchs sequence and gene structure (Figure 1b, bchsrev1). Western analysis of proteins made from 1-day-old adult heads revealed that the bchs5 mutation eliminates the production of Bchs, while the bchsrev1 line has normal levels of the protein (Figure 1c, samples 1 and 2). Young EP(2L)2299 flies display a Bchs expression pattern similar to that of our previous findings (becomes absent in older animals) and further phenotypic characterization of this line indicated that it is as a hypomorphic or weak allele of bchs (Figure 1c, sample 3) (Finley et al. 2003). Crossing the EP(2L)2299 line with GMR-Gal4 flies (eye driver) produces F1 offspring that have at least a fivefold increase in the total levels of Bchs within the head (Figure 1c, samples 6 and 4). However, protein levels are substantially decreased when the EP(2L)2299,GMR-Gal4 chromosome is combined with a deletion that removes the entire bchs genomic region (thereby eliminating the wild-type Bchs expression) but remain elevated well above that of controls [Figure 1c, sample 7, Df(2L)W12] (Finley et al. 2003).
Bchs's GOF eye phenotype:
We observed that overexpression of Bchs [GMR-Gal4,EP(2L)2299] led to a morphologically distinct external eye phenotype (Figure 2a). When compared to wild-type controls and the individual parental lines, the GMR-Gal4,EP(2L)2299 combination results in a dark eye pigmentation (similar to GMR-Gal4), a roughened eye surface texture, and a slight decrease in the overall eye size. The dominant phenotype is sensitive to the dosage of Bchs as an additional copy of EP(2L)2299 and two copies of both GMR-Gal4 transgenes further exacerbate the eye phenotype. Excess Bchs in photoreceptor cells (normally expressed at much lower levels) also alters the neural projection patterns. These defects can be visualized by coexpressing bchs with a membrane-targeted GFP (UAS-GFP-CAAX) (Finley et al. 1998). Wild-type eye discs (third instar larvae) display the classic ommatidial organization for this point in retinal development (Figure 2b). In contrast, GMR-Gal4,EP(2L)2299 eye discs have a slight loss in morphology and a decrease in the brightness of the central ommatidial region (Figure 2c, arrows), which marks the confluence of photoreceptor axons before exiting the eye disc.
The innervation patterns of R7 and R8 photoreceptors into the optic lobes can also be visualized in midpupal control and Bchs-overexpressing animals. Wild-type pupae demonstrate the normal stereotypic array of R7 and R8 projections, as well as normal axonal and growth cone morphology for this development stage (Figure 2b) (Finley et al. 1998; Ditch et al. 2005). In the Bchs-overexpressing flies the number and basic array of R7 and R8 axonal projections remain relatively normal (Figure 2c). However, there is a loss of growth cone morphology as well as axonal swelling or varicosities. At higher magnification, control flies show normal axonal projection patterns (Figure 2b, arrow) and growth cone (GC) morphology, while Bchs overexpression results in the loss of growth cone definition and premature termination of developing synapses. These axons also show the formation of varicosities (Figure 2c, arrows). The R7 and R8 terminal synapses are also abnormal and the Bchs protein can be detected near areas of axonal distension (Figure 2d, arrows). These findings are similar to those of previous studies showing that excess Bchs results in synaptic defects and the formation of axonal bulges both in larval motor neurons (Kraut et al. 2001) and in photoreceptors (Khodosh et al. 2006).
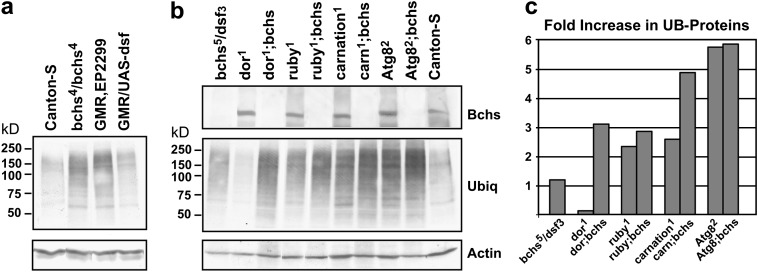
Increased Bchs expression in neural projections results in the formation of varicosities containing ubiquitin-immunopositive inclusions or vesicles (Figure 2e, arrows), which are not detected in wild-type axons (data not shown). Flies with mutations in the bchs gene display a similar phenotype in which ubiquitinated protein profiles are altered in adult neural tissues (Finley et al. 2003; Simonsen et al. 2004). This suggests that either a decrease or overproduction of Bchs can result in the altered accumulation of UB proteins. To quantitatively examine this, Western analysis was performed on protein extracts prepared from heads taken from 1-day-old control flies (Canton-S), homozygous bchs4 mutants [EP(2)2299], and bchs-overexpressing animals [GMR-Gal4,EP(2)2299] as well as from flies expressing the dissatisfaction (dsf) gene (GMR-Gal4/UAS-dsf) (Figure 3a) (Pitman et al. 2002). Although it is a remote possibility the EP(2)2299 P element could potentially increase the expression of a gene located distal to bchs. Therefore Dsf-overexpressing flies (GMR-Gal4/UAS-dsf) were selected as a control since the endogenous dsf gene is located distal to bchs and is the only other gene in the correct orientation for activation by the EP(2)2299 P element. In young EP(2)2299 flies UB proteins are elevated when compared to controls, which is consistent with our previous results showing that loss of bchs alters protein profiles (Finley et al. 2003). In flies overexpressing bchs [GMR-Gal4,EP(2)2299] a significant increase is also seen in the accumulation of high-molecular-weight UB proteins. In contrast, driving dsf expression in the eye (GMR-Gal4,UAS-dsf) does not alter UB-protein levels (Figure 3a) (Pitman et al. 2002). This demonstrates that functional loss and overproduction of bchs both result in the accumulation of UB proteins in neural tissues. In addition, these data show that while activation of the UAS-dsf transgene produces extensive defects in eye development, these defects are distinct from those produced by excess Bchs (Pitman et al. 2002). These data indicate that transcriptional activation by the EP(2)2299 UAS-P element results in an increase only in bchs expression and produces a gene-specific eye phenotype.
Figure 3.—
Ubiquitinated protein profiles. The heads from young adult flies were collected and the ubiquitinated protein profiles (UB proteins) were examined by Western blot analysis. (a) Wild-type flies (Canton-S) show low levels of UB proteins while homozygous bchs4 mutants [EP(2)2299] and flies overexpressing Bchs [GMR-Gal4,EP(2)2299] show enhanced accumulation of high-molecular-weight UB proteins. Flies overexpressing dsf (GMR-Gal4/UAS-dsf), a gene downstream of bchs, do not show an increase in UB proteins. (b) Flies containing the deep orange1, ruby1, carnation1, and Atg8a2 mutations were crossed into the bchs5/Df(2L)dsf3 genetic background. Head homogenates from young male flies were Western blotted and sequentially probed for ubiquitin, actin, and Bchs. Again, bchs mutant animals (bchs5/dsf3) show an early accumulation of UB proteins, as do flies containing single mutations in the ruby1, carnation1, and Atg8a2 genes. Several genotypes with mutations in two lysosomal transport genes saw a synergistic enhancement in UB-protein levels. They include dor1;bchs5/dsf3, ruby1;bchs5/dsf3, carnation1;bchs5/dsf3, and Atg8a2;bchs5/dsf3 mutant combinations. (c) Densitometry values were used to normalize UB proteins against actin levels as loading controls and compared with the corrected Canton-S levels to calculate the fold increase in UB proteins. Flies with the dor1;bchs5/dsf3 and carnation1;bchs5/dsf3 genotypes had the most pronounced synergistic increase while ruby1;bchs5/dsf3 and Atg8a2;bchs5/dsf3 flies show a minor enhancement. In the case of the Atg8a2;bchs5/dsf3 genotype this may be due to the extreme elevation in UB-protein levels seen in Atg8a2 single-mutant flies.
The easy to score dominant eye phenotype from Bchs overexpression provided an opportunity to develop an effective modifier screen. Dominant modifier screens using overexpression of a specific protein have been employed to characterize several diverse genetic pathways including those associated with RAS/MAPK signaling (i.e., GMR-yanact or Sev-yanact) as well as Armadillo and Notch functions (Verheyen et al. 1996; Greaves et al. 1999; Rebay et al. 2000). These types of modifier screens have been used successfully to identify factors that alter polyglutamine aggregation and mutant τ-toxicity (Fernandez-Funez et al. 2000; Kazemi-Esfarjani and Benzer 2000; Shulman and Feany 2003). In the case of Bchs, excessive levels of the protein may deplete interacting cofactors, resulting in the formation of incomplete protein complexes that block axonal transport or prevent the formation or targeting of vesicle subtypes. Eye phenotypes similar to Bchs are observed when SNAP1 (a protein involved with vesicle fusion) is expressed using the same GMR-Gal4 driver (Babcock et al. 2004). UAS-SNAP1 expression in motor neurons disrupts the formation of neuromuscular junctions and in the eye produces a mild rough eye phenotype that is further exacerbated with higher levels of the protein (Babcock et al. 2004). Furthermore, UAS-SNAP1 expressed in combination with mutant alleles of interacting partners (dNSF2155 and syxΔ229) enhances the eye phenotype (including changes to pigmentation, surface texture, and the overall size of the eye), while coexpression of wild-type UAS-dNSF1 or UAS-NSF2 suppresses these defects (Babcock et al. 2004). Therefore we used overexpression of Bchs in the eye to design of a GOF screen. Similar to the previous study by Khodosh et al. (2006) we initially examined a set of chromosomal deletion lines but then primarily focused on individual recessive loss-of-function mutations in a wide range of genes involved with protein turnover, vesicle trafficking, or lysosomal development or function.
Bchs GOF modifier screen:
The similarity between the EP(2L)2299 and the UAS-SNAP1 overexpression eye phenotypes indicated that a modifier screen could be used to detect Bchs interactions using a dominant GOF eye phenotype. For this screen the GMR-Gal4,EP(2L)2299/CyO:TM6B stock line was established and used for individual crosses to mutant lines. Due to the use of the GAL4/UAS system to produce elevated levels of a very large protein (with potentially complex interactions) we were concerned that coexpressing additional proteins could lead to a large number of false positives. Therefore, we primarily examined lines that represented recessive loss-of-function mutations and focused on identifying genetic interactions that enhanced the eye phenotype. This design had the advantage of allowing lethal deletions or mutants to be examined. The GMR-Gal4,EP(2L)2299 stock was initially crossed to lines from the second and third chromosome deficiency kits (Bloomington Stock Center). These results are included in supplemental Tables 1 and 2 at http://www.genetics.org/supplemental/. Most lines do not significantly alter the eye phenotype and those lines that do behaved as enhancers. This initial screen implied that a wide range of genes did not behave as significant modifiers of the GMR-Gal4,EP(2L)2299 phenotype.
Additional lines were screened that represent recessive loss-of-function mutations in specific genes. Initially, mutant lines were selected if their genomic location overlapped with deletion chromosomes that behaved as modifiers. Subsequent lines were chosen that represented divergent cellular pathways affecting lysosomal/endosomal trafficking, neural function or development, synaptic vesicle transport or fusion, and adult life span and cellular metabolism. Later, several UAS lines that affected autophagy gene expression were also included. In all cases F1 progeny were scored for changes in adult eye size, shape, pigmentation, and surface texture. Again, most genetic combinations with GMR-Gal4,EP(2L)2299 did not significantly modify the eye phenotype, indicating a degree of specificity in the detection of potential bchs genetic interactions. These results are outlined in supplemental Table 3 at http://www.genetics.org/supplemental/. Examples of genetic mutations that did behave as modifiers of the Bchs GOF phenotype are illustrated in Figure 4 and are further detailed in Table 1 (individual lines and mutation type). To control for phenotypes resulting from dominant or nonspecific interactions with the GMR-Gal4 driver, lines that were scored as modifiers were crossed individually to the GMR-Gal4 driver and the F1 offspring were independently examined for eye morphology changes. Individual modifier genes that also demonstrated a phenotypic change with a corresponding deletion chromosome from the second and third chromosome deficiency kit screen are noted in Table 1. Of the 20 individual second or third chromosome mutations identified as modifiers, a total of 6 showed an eye phenotype that was similar to the overlapping deletion allele. Significant changes to eye phenotypes were not observed when the GMR-Gal4 driver was used to overexpress independently several other autophagy genes (individual EP-UAS lines) or in combination with recessive loss-of-function mutations (results are shown in supplemental Figure 1 at http://www.genetics.org/supplemental/).
Figure 4.—
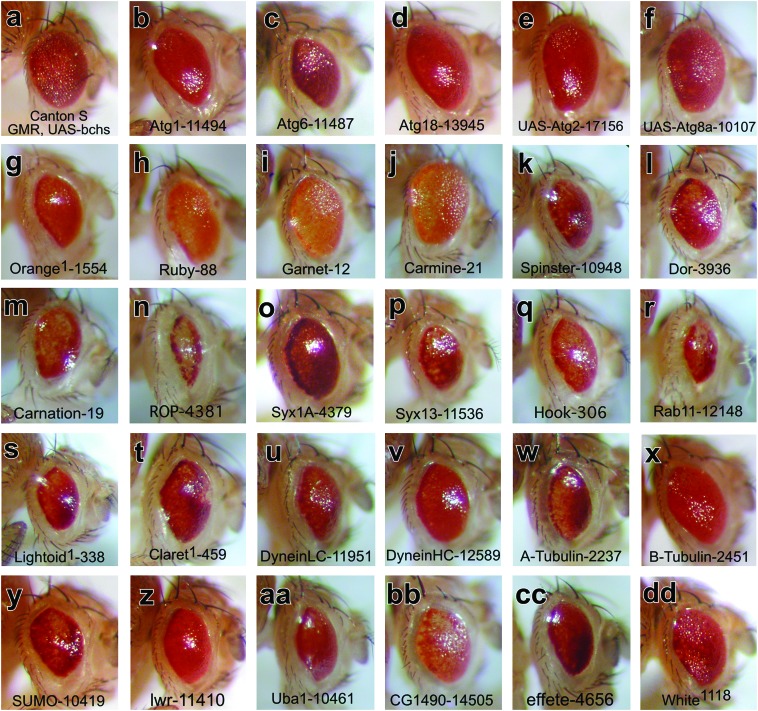
Bchs eye modifiers: fly eyes representing one copy of the GMR-Gal4,EP(2L)2299 expression transgenes in (a) wild-type control (Canton-S) or in heterozygous mutant backgrounds. Mutations in autophagy genes (b) atg1 and (c) atg6 enhance the eye phenotype while (d) a P-element mutation in atg18 has only a slight effect on the Bcsh-GOF phenotype. Coexpression of (e) UAS-atg2 and (f) UAS-atg8a was scored as phenotype suppressors. Mutations in (g) orange (or, adaptor complex AP-3 ξ-subunit) act as phenotype enhancers, while mutations in (h) ruby (rb, AP-3 β-subunit), (i) garnet (g, AP-3 δ-subunit), and (j) carmine (cm, AP-3 μ-subunit) are complex modifiers. Mutations in other vesicle-trafficking genes including (k) spinster (spin), (l) deep orange (dor), (m) carnation (car), (n) ras opposite (rop), (o) syntaxin1A (syx1A), (p) syntaxin13 (syn13), (q) hook (hk), (r) rab-protein 11 (Rab11), (s) lightoid (ltd), and (t) claret (ca) all act as strong Bchs-GOF enhancers. Other proteins involved in lysosomal transport are also Bchs enhancers and include mutations in the (u) dyneinLC and (w) α-tubulin genes and to a much lesser extent (v) dyneinHC and (x) β-tubulin mutations. Mutations in SUMO and ubiquitin pathway members are Bchs-GOF enhancers and include (y) SUMO, (z) lesswright (lwr), (aa) Uba1, (bb) C-terminal ubiquitin hydrolase (CG1490), and (cc) effete mutations. (dd) The GMR-Gal4,EP(2L)2299 transgenes in a white1118 background are shown as a nonmodifying negative control.
Modifying autophagy genes:
Of the six Drosophila autophagy genes examined in this study, mutations in Atg1 (serine/threonine kinase) and Atg6 (Beclin-1) were scored as moderate eye enhancers, while functional loss of the Atg18 gene [PtdIns(3,5)P2 binding protein] only had a minor effect (Figure 4, b–d). In contrast, coexpression of atg2 (EP-UAS line) and atg8a (EP-UAS line) moderately suppressed the Bchs-GOF phenotype (Figure 4, e and f) (FlyBase 2007). Individually these autophagy mutations act as strong homozygous lethal alleles, resulting in death at the late pupal or young adult stage of life. The notable exception is an adult viable mutation in the atg8a gene (data not shown). Their adult survival may in part reflect a partial functional redundancy from a second atg gene, atg8b (third chromosome) (FlyBase 2007). Atg8, also called LC3 (light chain of the microtubule-associated protein complex, MAP), undergoes C-terminal cleavage and covalent linkage to the lipid phosphatidylethanolamine (PE) upon interaction with autophagic membranes (Kouno et al. 2005). Atg8/LC3 remains bound to autophagic membranes until degradation in lysosomes, thus serving as a useful marker of autophagy (Ohsumi 2001). In yeast and flies, Atg1 is required for autophagosome biogenesis and is found on preautophagic and autophagic double-membrane structures together with Atg18 (Reggiori et al. 2004; Scott et al. 2004). Mutations in both Drosophila atg1 and atg18 have been shown to block starvation-induced autophagy in larval fat body tissue (Scott et al. 2004). For our assay, enhancement of the dominant eye phenotype likely indicates that a decreased level of key autophagy components in conjunction with excess Bchs exacerbates the GOF eye defects. At this time the cellular mechanism of this enhancement is unclear but may involve defects with autophagic vesicle formation or global perturbation of microtubule-mediated transport of membrane vesicles.
Modifying mutations in AP-3 complex and late endosomal genes:
Although endogenous Bchs is not expressed in pigment cells and functional loss of Bchs does not alter pigment granule formation (data not shown), mutations that affect eye and body pigmentation were examined because pigment vesicles or granules originate as part of the lysosomal biogenesis pathway. Several genes involved with lysosomal and pigment granule biogenesis acted as strong Bchs-GOF modifiers. They include the four subunits of the Drosophila AP-3 adaptor complex, which mediate trafficking between the trans-Golgi network and lysosomes (Boehm and Bonifacino 2002). Mutations in orange (ξ-subunit, Figure 4g) and ruby (β-subunit, Figure 4h) are strong enhancers while alterations to garnet (δ-subunit, Figure 4i) and carmine (μ-subunit, Figure 4j) behave as complex modifiers by enhancing pigmentation defects and suppressing the eye size reduction (Lloyd et al. 1998). A decrease in pigmentation suggests that expression of Bchs in pigment cells (where it is normally not expressed) has a deleterious effect on pigment granule formation.
Spinster mutations also behave as enhancers of the Bchs-GOF eye phenotype (Figure 4k). Encoding an integral membrane protein, hypomorphic spinster alleles were first identified having adult behavior and neural development defects (Nakano et al. 2001). More recently, spinster (benchwarmer) mutants demonstrated neural degeneration accompanied by the formation of late endosomal inclusions (Dermaut et al. 2005). Localized to lysosomes and late endosomes the vertebrate spinster protein promotes the autophagic cell death pathway in cultured cells (Yanagisawa et al. 2003). Mutations in other genes classified with late endosomal/lysosomal functions also act as Bchs-GOF enhancers. They include deep orange (dor, Vps18p, E3-ligase, Figure 4l) and carnation (Vps33, sec1 protein, Figure 4m) (Sevrioukov et al. 1999). These proteins are members of the homeotypic vacuole fusion and protein sorting (HOPS) trafficking complex and in Drosophila are known to promote late endosomal/lysosomal fusion events (Sriram et al. 2003). Recently we demonstrated that, like its yeast homolog Vps18p, Drosophila dor is required for autophagosome-to-lysosome fusion during programmed autophagy in the larval fat body (Lindmo et al. 2006). Mutant alleles of a second Sec1 protein, ras opposite (rop, munc-18/n-Sec1) enhance the Bchs-GOF phenotype (Figure 4n) as do mutant alleles of syntaxin1A, syntaxin13, and the coiled-coil protein hook (Figure 4, o and p) (FlyBase 2007). Drosophila hook is known to genetically interact with dor during endocytosis by regulating multivesicular endosome (MVE) trafficking (Figure 4q) (Narayanan et al. 2000). Both hook and dor proteins are found in larval neuromuscular synapses and mutations in both alter synaptic size and number (increased in hook and decreased in dor) (Narayanan et al. 2000). Ras opposite, syntaxin1A, and syntaxin 13 are SNARE proteins that are involved in neurotransmitter release (Ciufo et al. 2005).
Another important class of proteins involved in regulating vesicle trafficking and fusion is Rab GTPases. Several independent mutations in Rab11 (Figure 4r) act as very strong enhancers of the Bchs-GOF phenotype. Rab11 is known to associate with multiple vesicle subtypes ranging from recycling endosomes to trans-Golgi vesicles and has important roles in endocytosis (recycling and lysosomal targeting) and membrane targeting during cytokinesis and cellularization (Pelissier et al. 2003). The potential genetic interaction between Rab11 and Bchs was recently confirmed in a similar study that examined their combinatorial effect on synaptic development and morphogenesis (Khodosh et al. 2006). In the Drosophila eye, Rab11 also mediates rhodopsin exocytosis and prevents photoreceptor degeneration (Satoh et al. 2005). Additional genes involved with the biogenesis of lysosomes and related organelles were also identified as having potential interactions with Bchs. They include a second rab gene, lightoid (Figure 4s) and claret, a GTPases guanine nucleotide exchange factor (Figure 4t) (Ma et al. 2004). These two proteins are known to interact and promote the formation of pigment granules during eye development (Ma et al. 2004).
Cytoskeletal/motor proteins and ubiquitin/SUMO pathway member modifiers:
Recent studies examining the removal of aggregate-prone proteins revealed that microtubules and motor proteins are required for appropriate formation and trafficking of autophagosomes (Kamal and Goldstein 2000; Ravikumar et al. 2005) and point mutations in human dynein result in an ALS-like form of neuronal degeneration. Since Bchs contains potential motifs implicated in interactions with motor and cytoskeleton proteins we therefore assayed mutations in this class of genes. The Bchs-GOF phenotype was significantly enhanced by mutations in the dynein light chain (DLC, Figure 4u) and α-tubulin (Figure 4w) genes and to a lesser extent with the dynein heavy chain gene (DHC, Figure 4v) (FlyBase 2007). β-tubulin mutations did not show a significant change in the overall Bchs-GOF phenotype (Figure 4x) as do other functionally related genes (i.e., actin). Since bchs mutant flies demonstrate age-related changes to CNS ubiquitinated protein profiles, genes in the ubiquitin and SUMO signaling pathways were also assayed (Finley et al. 2003). While its unknown if Bchs interacts directly with ubiquitin or SUMO, ubiquitin/SUMO signaling has profound effects on vesicle specification and trafficking to lysosomes and both can be major constituents of cellular inclusions (Steffan et al. 2004; Ciechanover 2005). Mutant alleles of the Drosophila SUMO gene (smt3, Figure 4y) enhance the Bchs-GOF phenotype as do mutations in the SUMO-conjugating enzyme lesswright (lwr, Figure 4z) (FlyBase 2007). Mutations in fly Uba (ubiquitin-activating enzyme 1, Figure 4aa), a C-terminal ubiquitin hydrolase (CG1490, Figure 4bb) and effete (UbcD1, ubiquitin-conjugating enzyme 1, Figure 4cc) also are Bchs-GOF enhancers (Ohlmeyer and Schupbach 2003).
Adult life span profiles of Bchs modifiers:
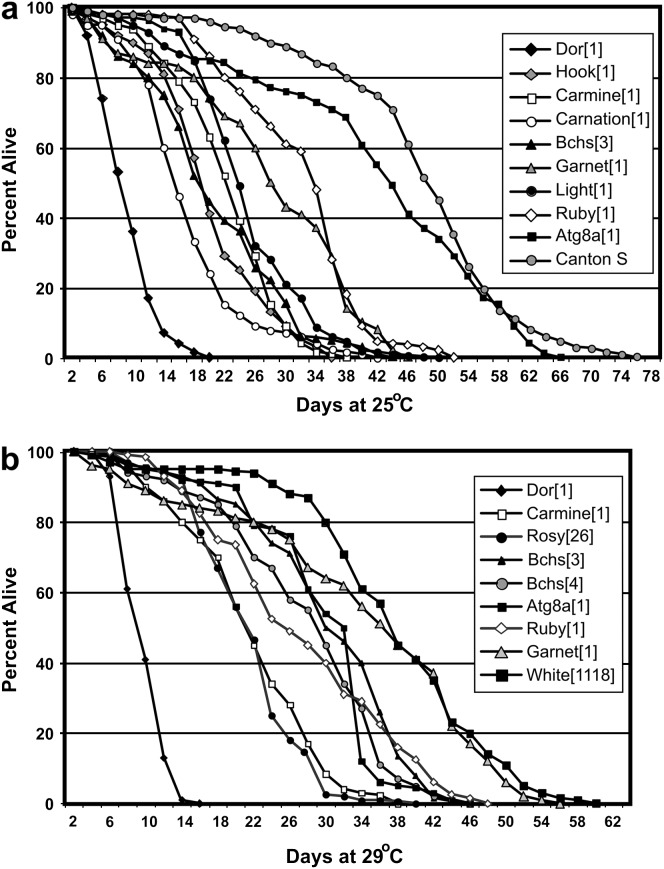
A key phenotype associated with functional loss of bchs in adult Drosophila is a significant reduction in longevity. We therefore asked if genes with potential genetic interactions with bchs demonstrate a similar LOF phenotype. To test this hypothesis, we examined the life span of mutations that were identified by the Bchs modifier screen. Fortunately, several mutant alleles exist that produce homozygous viable adults with normal appearance and activity levels. To control for genetic background effects related to life span, mutant lines were first outcrossed for several generations to wild-type stocks (Canton-S or w1118) and independent lines were reestablished for these studies. The hook1 and hook11 lines were assayed independently from each other and in heterozygous combination (hook1/hook11). Newly emerged adults were collected and aged to determine the life span profiles for deep orange, light, carnation, hook, ruby, garnet, carmine, and atg8a mutants at both 25° and 29° (Figure 5, a and b). The results are summarized in Table 2 (Finley et al. 2003). Canton-S and w1118 and rosy26 strains were used as normal and eye pigment mutant controls (FlyBase 2007).
Figure 5.—
Life span profiles of Bchs modifiers. Newly eclosed males from different genotypes were collected and aged at (a) 25° and (b) 29° for the duration of the experiment. The percentage of individuals for a given genotype that had survived for a given day is calculated from the total number of flies. For each genotype the total number of flies (n) used in the experiment, their mean life spans, standard deviations, and P-values are summarized in Table 2A and 2B.
TABLE 2.
Adult life span profiles
| Genotype | Gender | Total (n) | Mean age (days) | SDa | Maximum age (days) | P-value |
|---|---|---|---|---|---|---|
| A. At 25° | ||||||
| dor1 | Male | 295 | 8.96 | 3.73 | 20 | <0.0001b |
| dor1 | Female | 98 | 10.07 | 3.62 | 15 | <0.0001b |
| dor4 | Male | 106 | 13.94 | 2.95 | 19 | <0.0001b |
| hook1/hook11 | Male | 110 | 18.76 | 8.27 | 58 | <0.0001b |
| hook1 | Male | 338 | 19.861 | 7.26 | 36 | <0.0001b |
| carnation1 | Male | 242 | 17.42 | 8.69 | 48 | <0.0001b |
| bchs3/df clot7 | Male | 330 | 21.69 | 10.54 | 46 | <0.0001b |
| bchs3/bchs5 | Male | 274 | 23.7 | 6.9 | 37 | <0.0001b |
| carmine1 | Male | 351 | 21.97 | 6.96 | 38 | <0.0001b |
| light1 | Male | 247 | 24.92 | 8.8 | 61 | <0.0001b |
| garnet1 | Male | 111 | 27.41 | 11.75 | 45 | <0.0001b |
| ruby1 | Male | 183 | 31.57 | 8.842 | 53 | 0.0001b |
| atg8aep362 | Male | 150 | 41.9 | 15.4 | 66 | 0.0040b |
| Canton-S | Male | 264 | 46.85 | 12.33 | 68 | |
| B. At 29° | ||||||
| deep orange1 | Male | 136 | 8.19 | 2.4 | 14 | <0.0001c |
| rosy26 | Male | 128 | 19.8 | 5.98 | 38 | <0.0001c |
| carmine1 | Male | 122 | 20.69 | 7.73 | 36 | <0.0001c |
| carnation1 | Male | 121 | 23.868 | 8.78 | 42 | <0.0001c |
| bchs3/bchs6 | Male | 156 | 20.08 | 6.74 | 36 | <0.0001c |
| bchs3/Df(2L)clot7 | Male | 80 | 22.23 | 6.3 | 32 | <0.0001c |
| bchs4 (EP2299) | Male | 196 | 25.87 | 9.09 | 44 | <0.0001c |
| agt8a1 | Male | 153 | 27.16 | 7.732 | 45 | <0.0001c |
| garnet1 | Male | 98 | 32.08 | 13.84 | 54 | 0.0194c |
| white1118 | Male | 124 | 36.02 | 10.97 | 58 | |
Standard deviation from the mean.
P-values were calculated using Canton-S as a control.
P-values were calculated using white 1118 as a control.
The mutants that demonstrated the shortest average life span were weak alleles of deep orange (Figure 5a) (FlyBase 2007). Both male and female dor1 flies and dor4 males showed similar aging profiles, with nearly all dead within 16 days (Table 2). Carnation1 and hook1 mutants also have a significant life span reduction when compared to controls. Previous work has found that hook1 mutants develop retinal degeneration (Kramer and Phistry 1996) and this study shows that hook1 and hook1/hook11 trans-heterozygous flies have decreased longevity profiles similar to bchs adults (Table 2). Several mutant lines have average life spans greater than bchs mutants, but still significantly less than control lines. At 25° they include light, carmine, ruby, and garnet mutations, as well as a P-element insertion into the atg8a gene (Figure 5a). Most mutants have similar profiles when compared at 25° and 29°. The exceptions are rosy26, which has a significant decrease in longevity at 29°, and garnet1 mutants that have a nearly normal life span (Figure 5b). While these lysosomal trafficking mutants have been studied extensively in terms of synaptic vesicle function and pigment granule development, to our knowledge this is the first detailed examination of their life span profiles. Our results clearly show that mutations in lysosomal trafficking proteins can reduce adult longevity.
To determine if mutations in bchs and other lysosomal trafficking genes have an additive effect on adult phenotypes we established crosses between different allelic combination of bchs and other lysosomal trafficking mutants. Unfortunately, the combination of strong deletion alleles of bchs [Df(2L)dsf3/Df(2L)clot7 or Df(2L)dsf3/Df(2)w3] with several lysosomal genes produced very few double-mutant adults (Finley et al. 1998, 2003). Using other bchs mutant alleles we were able to generate an occasional double-mutant male fly by combining the bchs5/Df(2L)dsf3 genotype individually with the dor1, ruby1, carnation1, atg8a2, garnet1, and carmine1 mutations (scored for eye color). Again, eclosion rates for the different bchs5/Df(2L)dsf3 genetic combinations were substantially reduced but a few viable flies were produced. Heads were harvested from 1- to 2-day-old animals and protein extracts were examined by Western blot analysis. Since bchs LOF mutations do not produce an easy-to-score phenotype we confirmed functional loss of bchs by Western analysis (anti-Bchs) in conjunction with total UB-protein profiles (Figure 3b). As seen previously, bchs mutants [bchs5/Df(2L)dsf3] have an elevated UB-protein level when compared to age-matched controls (Canton-S). A similar elevation in large UB proteins was also detected for ruby1, carnation1, and atg8a2 single mutants. This phenotype was further exacerbated in double mutants with the bchs5/Df(2L)dsf3 genotype. An increase in UB-protein levels was detected for garnet1 and carmine1 mutant males but was not detected in garnet1;bchs5/Df(2L)dsf3 and carmine1;bchs5/Df(2L)dsf3 double mutants (data not shown). These data indicate that individual mutations in bchs or other genes involved with lysosomal transport/trafficking can alter UB-protein profiles in young adult neural tissues and in several examples double-mutant combinations produce an additive or synergistic phenotype.
DISCUSSION
Lysosomal-autophagic trafficking defects:
There is growing evidence that defects in lysosomal transport or clearance of age-related or disease-associated proteins can play a central role in the etiology of many neural degenerative disorders (Cataldo et al. 1996; Garcia-Mata et al. 2002; Ravikumar et al. 2005; Yu et al. 2005). Previous work on Bchs and Alfy indicated that these proteins are involved with the autophagic clearance of ubiquitinated substrates. Human Alfy colocalizes with cytoplasmic ubiquitin structures and autophagic membranes and older bchs mutant flies accumulate ubiquitinated CNS aggregates (Finley et al. 2003; Simonsen et al. 2004). In this study we show that high levels of Bchs in the Drosophila eye cause a dominant eye phenotype in which external eye structures and neural development are perturbed (Figure 2). Neural defects include alterations to the formation of terminal synapses and axonal varicosities containing ubiquitin. Biochemical analysis reveals that both bchs mutant and overexpressing flies show similar changes in UB-protein profiles (Figure 3). These phenotypes suggest that lysosomal transport defects alter the turnover of UB proteins (Gunawardena and Goldstein 2001; Nixon et al. 2005). A Bchs-GOF screen was designed around this overexpression eye phenotype and used to identify several candidate proteins (modifiers) that potentially have genetic interactions with Bchs.
The role of lysosomal mutations in adult viability:
Several loss-of-function mutations within the autophagic pathway were found to behave as Bchs-GOF enhancers (Atg1 and Atg6), while two transgenic lines that allow GMR-Gal4-driven coexpression with EP(2L)2299 (UAS-atg2 and UAS-atg8a) mildly suppress a dominant eye phenotype. This is similar to previous findings with GMR-Gal4 expression of UAS-SNAP in which the genetic background of known interacting factors enhanced or suppressed the dominant eye phenotype (Babcock et al. 2004). Other classes of proteins that are involved with different aspects of lysosomal transport also act as Bchs-GOF modifiers, including the four subunits of the AP-3 protein complex (orange, ruby, garnet, and carmine). These four protein subunits have primarily been characterized in terms of pigment granule biosynthesis but clearly they have additional cellular functions since all are located within Drosophila nerve terminals and have a subtle role in adult behaviors (Dell'Angelica et al. 2000; Boehm and Bonifacino 2002). Lysosomal trafficking and pigmentation defects are also observed with mutant mammalian AP-3 homologs. Mutant mocha (δ-subunit) and pearl (β3A-subunit) mice have pigmentation defects and lysosomal abnormalities as do humans suffering from Hermansky–Pudlak syndrome (β3A-subunit) (Dell'Angelica et al. 2000; Boehm and Bonifacino 2002). In addition, mocha mice demonstrate progressive inner ear and neurological degeneration.
Several proteins characterized as having roles in the late endosomal pathway were scored as positive Bchs-GOF eye modifiers including mutations in the spinster, hook, deep orange, and carnation genes. Drosophila mutations in these genes have been shown to affect neuronal maintenance and degeneration. Defects in the mouse and human homologs of carnation (Vps33) generate buff mutant mice and the ARC syndrome in humans (Vps33B, lethal multisystem disorder), respectively (Gissen et al. 2004). Previously we have shown that, in addition to the late endosomal–lysosomal fusion events, the Deep orange protein also facilitates the formation of autolysosmes (autophagosome–lysosome vesicles) or amphisomes (autophagosome–late endosome vesicles) (Rieder and Emr 1997; Lindmo et al. 2006). Other known “lysosomal” proteins that mediate a host of trafficking/fusion events in several vesicle subtypes were also scored as Bchs-GOF modifiers. These include ras opposite, syntaxin1A, syntaxin13, Rab11, lightoid, claret, α-tubulin, and the dynein light chain mutations. Defects in dynein-mediated transport are linked with neurodegenerative disorders and recently were shown to impair autophagy-mediated clearance of neuronal protein aggregates (Ravikumar et al. 2005).
The addition of ubiquitin to protein substrates is used by the cell as a signal for proteasome degradation or as a sorting signal for endocytotic trafficking to the lysosome. The accumulation of ubiquitinated-protein aggregates or inclusions is a key feature of many neural degenerative disorders and is a primary phenotype associated with bchs loss-of-function mutations. More recently SUMOylated proteins have also been found in neural inclusions, indicating that multiple changes in protein modification, trafficking, and/or proteolytic turnover pathways can be a central feature of neural degeneration and reduced viability (Steffan et al. 2004). In our study mutations in both the SUMO (SUMO and lesswright) and ubiquitin (uba, CG1490, and effete) pathways behaved as enhancers of the Bchs-GOF phenotype. At this time it remains unclear if this is due to direct interactions between the Bchs protein and ubiqutinated or SUMOylated substrates or to a general blockage in vesicle sorting and transport.
Deletion lines and negative modifiers indicate screen selectivity:
In this study we identified eight regional deletions that behave as Bchs modifiers, which removed individual genes that were also scored as modifiers (Table 1, underlined). The remaining 11 Bchs-modifier genes located on the second and the third chromosome were eliminated by a deletion interval that did not generate a phenotype meeting the criteria set as being sufficiently significant. This discrepancy may be related to the level of modification produced by an individual mutation. In our screen and in a separate study (Khodosh et al. 2006) Rab11 mutations show a pronounced unambiguous eye phenotype that corresponds well with regional deletions. A similar strong phenotype was also seen with the lightoid mutations (Figure 4s) where either a single mutation or elimination of its genomic region resulted in obvious modification of the eye. This implies that an intermediate phenotype may be masked by the removal of other genes in the region that act as even moderate suppressors. A moderate suppression could involve the loss of a direct interaction between a trafficking protein and Bchs or the removal of transcription factors that normally enhance the endogenous bchs gene expression or have an upstream effect on the Gal4/UAS-driven expression. Although the deletion screen was informative the direct testing of individual genes helped to more quickly narrow the focus of this study.
Information regarding Bchs's functional pathway was also obtained from mutations that had little or no effect on the dominant Bchs-GOF eye phenotype. For a complete list of mutations that were tested and determined not to modify the Bchs eye phenotype see supplemental Table 3 at http://www.genetics.org/supplemental/. Eye-pigment mutants that are members of the ABC transporter class of lysosomal proteins (white, brown, and scarlet) did not modify the Bchs-GOF phenotype nor did genes that have early endosomal functions like hrs, clatherin-HC, or lap (AP180-like protein) (FlyBase 2007). Additional genes in vesicle trafficking pathways were also tested and mutations in sec61α, synaptotagmin, doa, rph, rab5, and rab14 genes did not produce significant modification of the Bchs eye phenotype (data not shown) (FlyBase 2007). Since insulin/TOR signaling is known to regulate autophagy, members of this pathway were examined for Bchs interactions (Kapahi et al. 2004; Ravikumar et al. 2004). Mutations in chico (insulin-like receptor), InR, and Tor genes did not have a significant effect on the Bchs-GOF phenotype (Kapahi et al. 2004). This may indicate a lack of direct protein interactions with Bchs or that a second signaling pathway regulates different features of autophagy regarding aggregate clearance in neurons. Kinesin motor proteins play a major role in vesicle trafficking and several mutations in kinesin and kinesin-like genes were examined and none produced a significant change in the Bchs-GOF phenotype. Finally, not all members of the ubiquitin-signaling pathway acted as Bchs modifiers, including mutations in Ubp64E, Uch-L3, fat facets, UbcD 10, and UbcD4. The fact that only certain genes in lysosomal trafficking pathways acted as Bchs-GOF modifiers highlights the interaction specificity of the screen's design. It also indicates that Bchs may preferentially interact with proteins involved in late transport events to the lysosome or the autophagic-to-lysosomal or trans-Golgi/late endosomal-to-lysosomal transport processes.
Decreased longevity and UB-protein accumulation in lysosomal trafficking mutants:
Lysosomal trafficking defects are often associated with progressive disorders and reduced viability (Cataldo et al. 1996; Dell'Angelica et al. 2000; Brunk and Terman 2002; Cuervo 2004). To further clarify the role that lysosomal transport has on the adult Drosophila life span, mutant lines identified from the modifier screen were examined for changes in adult longevity. We found that mutations in several AP-3 genes (ruby, garnet, and carmine), late endosomal pathway genes (i.e., deep orange, carnation, and hook), and the autophagy gene atg8a resulted in reduced average adult life spans. While the decrease in longevity associated with bchs mutations can be attributed to progressive neural defects this may not be the case for mutations in other lysosomal import genes. Bchs is primarily expressed in neurons while the other genes have a broader expression pattern (Finley et al. 2003). For example we have shown that dor (described as an endosomal gene) is required for programmed autophagy in larval fat body cells (Lindmo et al. 2006). This suggests that lysosomal defects in nonneuronal tissues may also contribute to premature adult death associated with these mutants (Lindmo et al. 2006). However, a preliminary examination of UB proteins from mutant flies shows that most genotypes demonstrate a pronounced accumulation of proteins. In addition, several double-mutant combinations with bchs show an additive effect on UB-protein levels, with the most striking effect occurring with dor1 and carnation1 mutations. While both proteins are known to genetically interact and closely associate during late endosomal trafficking we have recently shown that the Dor protein functions during the activation of the programmed autophagic pathway in the larval fat body. The implications are that individual proteins may have dual functions, which reflect tissue-specific or age-dependent requirements coming from different lysosomal trafficking pathways.
Conclusion:
The genetic analysis of bchs interactions further supports the hypothesis that Bchs serves as a large scaffolding protein with multiple interacting partners. These genetic interactions primarily involve genes known to promote the transport, targeting, or fusion of different vesicle subpopulations with the lysosome. Viable loss-of-function mutations in autophagic, trans-Golgi, and late endosomal pathway members produce subtle but progressive defects that decrease adult longevity by altering the transport of substrates to the lysosome. Furthermore, our Drosophila genetic findings have implications for human disease since each gene characterized in this study has a direct human homolog and several are associated with diseases characterized by defects in lysosomal trafficking or function (FlyBase 2007). Many others have not been directly linked to a human disease but several map to loci associated with progressive disorders. Using Drosophila as an efficient model system, these genes and phenotypic defects can become a focus for future studies related to human disorders.
Acknowledgments
We thank Kia Zinn for providing the Bchs antibody, Helmut Kramer for the hook11 allele, and the Bloomington Stock Center for the remainder of the mutant lines examined in this study. This work was supported by grants from the National Institutes of Health, the National Institute on Aging, the National Institute on Neurological Disorders and Stroke (K.F. and R.C.), and the Norwegian Cancer Society (A.S. and K.L.) and by the Research Council of Norway (T.R.).
References
- Babcock, M., G. T. Macleod, J. Leither and L. Pallanck, 2004. Genetic analysis of soluble N-ethylmaleimide-sensitive factor attachment protein function in Drosophila reveals positive and negative secretory roles. J. Neurosci. 24 3964–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy, G., T. Lamark, A. Brech, H. Outzen, M. Perander et al., 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm, M., and J. S. Bonifacino, 2002. Genetic analyses of adaptin function from yeast to mammals. Gene 286 175–186. [DOI] [PubMed] [Google Scholar]
- Brunk, U. T., and A. Terman, 2002. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur. J. Biochem. 269 1996–2002. [DOI] [PubMed] [Google Scholar]
- Cataldo, A. M., D. J. Hamilton, J. L. Barnett, P. A. Paskevich and R. A. Nixon, 1996. Properties of the endosomal-lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer's disease. J. Neurosci. 16 186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover, A., 2005. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell. Biol. 6 79–87. [DOI] [PubMed] [Google Scholar]
- Ciufo, L. F., J. W. Barclay, R. D. Burgoyne and A. Morgan, 2005. Munc18–1 regulates early and late stages of exocytosis via syntaxin-independent protein interactions. Mol. Biol. Cell 16 470–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, A. M., 2004. Autophagy: in sickness and in health. Trends Cell Biol. 14 70–77. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E. C., C. Mullins, S. Caplan and J. S. Bonifacino, 2000. Lysosome-related organelles. FASEB J. 14 1265–1278. [DOI] [PubMed] [Google Scholar]
- Dermaut, B., K. K. Norga, A. Kania, P. Verstreken, H. Pan et al., 2005. Aberrant lysosomal carbohydrate storage accompanies endocytic defects and neurodegeneration in Drosophila benchwarmer. J. Cell Biol. 170 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditch, L. M., T. Shirangi, J. L. Pitman, K. L. Latham, K. D. Finley et al., 2005. Drosophila retained/dead ringer is necessary for neuronal pathfinding, female receptivity and repression of fruitless independent male courtship behaviors. Development 132 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Funez, P., M. L. Nino-Rosales, B. de Gouyon, W. C. She, J. M. Luchak et al., 2000. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408 101–106. [DOI] [PubMed] [Google Scholar]
- Finley, K. D., P. T. Edeen, M. Foss, E. Gross, N. Ghbeish et al., 1998. Dissatisfaction encodes a tailless-like nuclear receptor expressed in a subset of CNS neurons controlling Drosophila sexual behavior. Neuron 21 1363–1374. [DOI] [PubMed] [Google Scholar]
- Finley, K. D., P. T. Edeen, R. C. Cumming, M. D. Mardahl-Dumesnil, B. J. Taylor et al., 2003. blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration. J. Neurosci. 23 1254–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase, 2007. http://flybase.bio.indiana.edu.
- Garcia-Mata, R., Y. S. Gao and E. Sztul, 2002. Hassles with taking out the garbage: aggravating aggresomes. Traffic 3 388–396. [DOI] [PubMed] [Google Scholar]
- Gissen, P., C. A. Johnson, N. V. Morgan, J. M. Stapelbroek, T. Forshew et al., 2004. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat. Genet. 36 400–404. [DOI] [PubMed] [Google Scholar]
- Greaves, S., B. Sanson, P. White and J. P. Vincent, 1999. A screen for identifying genes interacting with armadillo, the Drosophila homolog of β-catenin. Genetics 153 1753–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena, S., and L. S. Goldstein, 2001. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron 32 389–401. [DOI] [PubMed] [Google Scholar]
- Hara, T., K. Nakamura, M. Matsui, A. Yamamoto, Y. Nakahara et al., 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441 885–890. [DOI] [PubMed] [Google Scholar]
- Kamal, A., and L. S. Goldstein, 2000. Connecting vesicle transport to the cytoskeleton. Curr. Opin. Cell Biol. 12 503–508. [DOI] [PubMed] [Google Scholar]
- Kapahi, P., B. M. Zid, T. Harper, D. Koslover, V. Sapin et al., 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani, P., and S. Benzer, 2000. Genetic suppression of polyglutamine toxicity in Drosophila. Science 287 1837–1840. [DOI] [PubMed] [Google Scholar]
- Khodosh, R., A. Augsburger, T. L. Schwarz and P. A. Garrity, 2006. Bchs, a BEACH domain protein, antagonizes Rab11 in synapse morphogenesis and other developmental events. Development 133 4655–4665. [DOI] [PubMed] [Google Scholar]
- Klionsky, D. J., and S. D. Emr, 2000. Autophagy as a regulated pathway of cellular degradation. Science 290 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., J. M. Cregg, W. A. Dunn, Jr., S. D. Emr, Y. Sakai et al., 2003. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5 539–545. [DOI] [PubMed] [Google Scholar]
- Komatsu, M., S. Waguri, T. Ueno, J. Iwata, S. Murata et al., 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu, M., S. Waguri, T. Chiba, S. Murata, J. I. Iwata et al., 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441 880–884. [DOI] [PubMed] [Google Scholar]
- Kouno, T., M. Mizuguchi, I. Tanida, T. Ueno, T. Kanematsu et al., 2005. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 280 24610–24617. [DOI] [PubMed] [Google Scholar]
- Kramer, H., and M. Phistry, 1996. Mutations in the Drosophila hook gene inhibit endocytosis of the boss transmembrane ligand into multivesicular bodies. J. Cell Biol. 133 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer, H., and M. Phistry, 1999. Genetic analysis of hook, a gene required for endocytic trafficking in Drosophila. Genetics 151 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut, R., K. Menon and K. Zinn, 2001. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11 417–430. [DOI] [PubMed] [Google Scholar]
- Lindmo, K., A. Simonsen, A. Brech, K. Finley, T. E. Rusten et al., 2006. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp. Cell Res. 312 2018–2027. [DOI] [PubMed] [Google Scholar]
- Lloyd, V., M. Ramaswami and H. Kramer, 1998. Not just pretty eyes: Drosophila eye-colour mutations and lysosomal delivery. Trends Cell Biol. 8 257–259. [DOI] [PubMed] [Google Scholar]
- Luzio, J. P., P. R. Pryor, S. R. Gray, M. J. Gratian, R. C. Piper et al., 2005. Membrane traffic to and from lysosomes. Biochem. Soc. Symp. No. 72, 77–86. [DOI] [PubMed]
- Ma, J., H. Plesken, J. E. Treisman, I. Edelman-Novemsky and M. Ren, 2004. Lightoid and Claret: a rab GTPase and its putative guanine nucleotide exchange factor in biogenesis of Drosophila eye pigment granules. Proc. Natl. Acad. Sci. USA 101 11652–11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, Y., K. Fujitani, J. Kurihara, J. Ragan, K. Usui-Aoki et al., 2001. Mutations in the novel membrane protein spinster interfere with programmed cell death and cause neural degenerationin Drosophila melanogaster. Mol. Cell. Biol. 21 3775–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, R., H. Kramer and M. Ramaswami, 2000. Drosophila endosomal proteins hook and deep orange regulate synapse size but not synaptic vesicle recycling. J. Neurobiol. 45 105–119. [DOI] [PubMed] [Google Scholar]
- Nixon, R. A., J. Wegiel, A. Kumar, W. H. Yu, C. Peterhoff et al., 2005. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 64 113–122. [DOI] [PubMed] [Google Scholar]
- Ohlmeyer, J. T., and T. Schupbach, 2003. Encore facilitates SCF-Ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development 130 6339–6349. [DOI] [PubMed] [Google Scholar]
- Ohsumi, Y., 2001. Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell. Biol. 2 211–216. [DOI] [PubMed] [Google Scholar]
- Pelissier, A., J. P. Chauvin and T. Lecuit, 2003. Trafficking through Rab11 endosomes is required for cellularization during Drosophila embryogenesis. Curr. Biol. 13 1848–1857. [DOI] [PubMed] [Google Scholar]
- Pitman, J. L., C. C. Tsai, P. T. Edeen, K. D. Finley, R. M. Evans et al., 2002. DSF nuclear receptor acts as a repressor in culture and in vivo. Dev. Biol. 245 315–328. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., T. E. Rusten and H. Stenmark, 2003. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 15 446–455. [DOI] [PubMed] [Google Scholar]
- Ravikumar, B., C. Vacher, Z. Berger, J. E. Davies, S. Luo et al., 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36 585–595. [DOI] [PubMed] [Google Scholar]
- Ravikumar, B., A. Acevedo-Arozena, S. Imarisio, Z. Berger, C. Vacher et al., 2005. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat. Genet. 37 771–776. [DOI] [PubMed] [Google Scholar]
- Rebay, I., F. Chen, F. Hsiao, P. A. Kolodziej, B. H. Kuang et al., 2000. A genetic screen for novel components of the Ras/Mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154 695–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., K. A. Tucker, P. E. Stromhaug and D. J. Klionsky, 2004. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6 79–90. [DOI] [PubMed] [Google Scholar]
- Rieder, S. E., and S. D. Emr, 1997. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 8 2307–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorth, P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, A. K., J. E. O'Tousa, K. Ozaki and D. F. Ready, 2005. Rab11 mediates post-Golgi trafficking of rhodopsin to the photosensitive apical membrane of Drosophila photoreceptors. Development 132 1487–1497. [DOI] [PubMed] [Google Scholar]
- Scott, R. C., O. Schuldiner and T. P. Neufeld, 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7 167–178. [DOI] [PubMed] [Google Scholar]
- Sevrioukov, E. A., J. P. He, N. Moghrabi, A. Sunio and H. Kramer, 1999. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol. Cell 4 479–486. [DOI] [PubMed] [Google Scholar]
- Shih, S. C., D. J. Katzmann, J. D. Schnell, M. Sutanto, S. D. Emr et al., 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell. Biol. 4 389–393. [DOI] [PubMed] [Google Scholar]
- Shulman, J. M., and M. B. Feany, 2003. Genetic modifiers of tauopathy in Drosophila. Genetics 165 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen, A., H. C. Birkeland, D. J. Gillooly, N. Mizushima, A. Kuma et al., 2004. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 117 4239–4251. [DOI] [PubMed] [Google Scholar]
- Sriram, V., K. S. Krishnan and S. Mayor, 2003. deep-orange and carnation define distinct stages in late endosomal biogenesis in Drosophila melanogaster. J. Cell Biol. 161 593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan, J. S., N. Agrawal, J. Pallos, E. Rockabrand, L. C. Trotman et al., 2004. SUMO modification of Huntingtin and Huntington's disease pathology. Science 304 100–104. [DOI] [PubMed] [Google Scholar]
- Torroja, L., H. Chu, I. Kotovsky and K. White, 1999. Neuronal overexpression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Curr. Biol. 9 489–492. [DOI] [PubMed] [Google Scholar]
- Verheyen, E. M., K. J. Purcell, M. E. Fortini and S. Artavanis-Tsakonas, 1996. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics 144 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa, H., T. Miyashita, Y. Nakano and D. Yamamoto, 2003. HSpin1, a transmembrane protein interacting with Bcl-2/Bcl-xL, induces a caspase-independent autophagic cell death. Cell Death Differ. 10 798–807. [DOI] [PubMed] [Google Scholar]
- Yu, W. H., A. M. Cuervo, A. Kumar, C. M. Peterhoff, S. D. Schmidt et al., 2005. Macroautophagy—a novel {beta}-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 171 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]