Abstract
The bursicon gene in Anopheles gambiae is encoded by two loci. Burs124 on chromosome arm 2L contains exons 1, 2, and 4, while burs3 on arm 2R contains exon 3. Exon 3 is efficiently spliced into position in the mature transcript. This unusual gene arrangement is ancient within mosquitoes, being shared by Aedes aegypti and Culex pipiens.
TRANS-SPLICING involves splicing of separate RNA molecules into a single entity (Bonen 1993). It is common in several organisms such as nematodes (e.g., Blumenthal et al. 2002) and kinetoplastids (e.g., Gopal et al. 2005) where a leader sequence is trans-spliced into the front of certain mRNAs, especially those that are downstream in operons. A few other examples of trans-splicing are known, including in the human genome (e.g., Zhang et al. 2003); however, the phenomenon is not common in our genome (Shao et al. 2006). It has also been employed in innovative efforts toward gene therapy involving repair of long transcripts (e.g., Lai et al. 2005; Yang and Walsh 2005).
The bursicon gene CG13419 in Drosophila melanogaster, named burs by Dewey et al. (2004), encodes one-half of the functional heterodimeric insect neurohormone bursicon, which regulates cuticle sclerotization and wing inflation after ecdysis in Drosophila and other insects (Luo et al. 2005; Mendive et al. 2005). This protein, which is also known as the α-chain of bursicon, is conserved throughout insects and related arthropods (Van Loy et al. 2006). In Drosophila, Burs is a 173-amino-acid protein encoded by three exons. Here we show by comparison with all available insect and related arthropod genomes that the ancestral insect gene had four coding exons and in all except mosquitoes it is a conventionally spliced locus. In mosquitoes bursicon is translated from a mRNA derived from four exons (one 5′ UTR and three coding); however, the middle coding exon is on a different chromosome arm (2R) from the first, second, and fourth exons, which are together on chromosome arm 2L. To our knowledge this is the only example of trans-splicing known to involve internal exons of genes on nonhomologous chromosome arms, although this has been demonstrated experimentally for the complicated trans-spliced mod(mdg4) locus in Drosophila (Gabler et al. 2005).
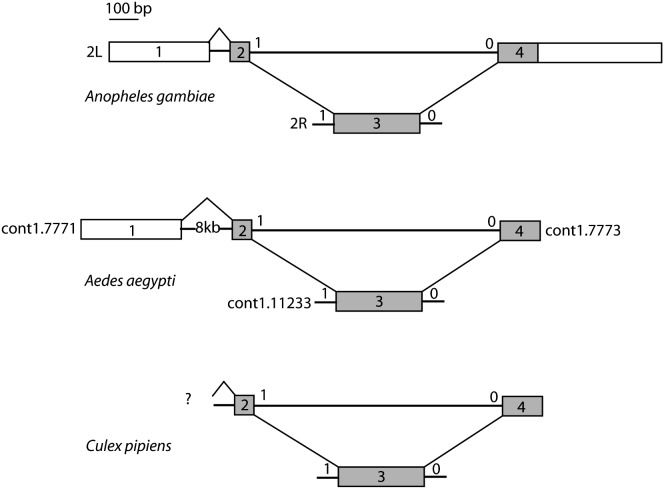
The unusual Anopheles gambiae bursicon gene structure was initially inferred bioinformatically using the Drosophila protein as query in TBLASTN searches of the genome sequences and by comparison of two 5′ ESTs (NAP1-P41-B-11-5 and NAP1-P111-F-12-5) and the available genome sequence (Holt et al. 2002). We obtained the cDNA clones from which these ESTs were generated and completed their sequences (GenBank nos. AY735442 and AY735443). These unambiguously confirm that the first, second, and fourth exons are present contiguously on the same scaffold on chromosome arm 2L (on the reverse strand at ±10.5 Mb on this ±49-Mb chromosome arm), while the third exon is present alone on chromosome arm 2R (on the reverse strand at ±22 Mb on this ±67-Mb chromosome arm) (Figure 1). These regions of the genome are both well assembled for at least 10 kb on each side of the bursicon exons as judged by comparison with the raw traces available in the Trace Archive at NCBI, indicating that this unusual gene structure is not the result of misassembly of the genome sequences. We amplified the locus with exons 1, 2, and 4 from the upstream promoter region to the 3′ UTR, and the single clean PCR product size (1430 bp) and DNA sequence agree fully with the assembled genome sequence.
Figure 1.—
Genomic structures of the burs124 and burs3 loci of three mosquitoes. Boxes represent exons while lines represent introns, approximately to scale. Shaded boxes are coding sequences. The phases of intron splices relative to codons are indicated above and near the intron splices, and the splicing is indicated by the diagonal lines. The 5′ UTR exon for Aedes is ∼8 kb upstream in another scaffolded contig, as indicated by a 5′ EST, while the 5′ UTR exon has not been identified for Culex. The chromosomal locations for the burs124 and burs3 loci are indicated for Anopheles while the contigs containing these are indicated for Aedes (contig1.7772 is entirely within the 8 kb first intron). The Culex loci were assembled from raw reads.
Furthermore, this trans-spliced gene structure encoding bursicon is also present in the Aedes aegypti and Culex pipiens genomes (Figure 1). Although the chromosome locations of the contigs containing the 1, 2, 4, and 3 exons are not known in Ae. aegypti, microsynteny on either side with orthologs of the same genes in An. gambiae and Ae. aegypti implies that they are again on separate chromosome arms (microsynteny cannot be examined easily for C. pipiens until the genome assembly is available, but given its relatively close relationship to Ae. aegypti it is likely to be the same). This unusual gene structure has therefore existed for at least the ±150 million years (MY) of mosquito evolution since the split of the culicid and anophelid family lineages, but is younger than the ±250 MY split of the dipteran suborders Nematocera and Brachycera (containing the Culicidea and Drosophilidae, respectively) (Krzywinski et al. 2006). We propose to call these two complementary mosquito bursicon-encoding loci burs124 and burs3.
This unusual trans-splicing of two separate loci encoding complementary parts of bursicon is restricted to these three mosquito genomes. All other available insect and related arthropod genome sequences contain a single locus encoding bursicon from 3 or 4 coding-exon genes (Figure 2). It appears that the ancestral bursicon locus in insects, at least, consisted of 4 coding exons separated by three introns (Figure 2) (the presence of introns in the 5′ UTRs cannot be conclusively demonstrated or excluded in these species in the absence of cDNA evidence). The drosophilids, Tribolium, and Ixodes lost the last phase 0 intron, the mosquitoes lost the middle phase 0 intron, and the water flea Daphnia pulex and deer tick Ixodes scapularis lost the first phase 1 intron (or this intron is unique to insects). The position of the first phase 1 intron within or near the signal peptide is rather variable, but this kind of intron movement within highly variable coding sequences such as signal sequences is not unusual.
Figure 2.—
Alignment of the Burs proteins encoded by available insect and arthropod genomes. Gene models were built on the basis of TBLASTN searches of the publicly available draft assemblies or the raw reads available at the Trace Archive at NCBI. The signal sequences are shown in italics, as indicated by the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/). The locations and phases of introns are shown; a dot indicates absence of an intron; the dash before the three mosquito proteins indicates the presence of an intron in the 5′ UTR immediately before the start codon. The N-terminal coding exon has not been identified for Nasonia vitripennis. These protein sequences are available in a supplemental FASTA file (at http://www.genetics.org/supplemental/).
We attempted to monitor the trans-splicing of the An. gambiae bursicon mRNA by RT–PCR from the second to fourth exons using primers designed to amplify the entire coding region from start to stop codons (Figure 1). If the bursicon mRNA trans-splicing is a slow inefficient process, then in addition to the expected final product of ∼490 bp we would anticipate obtaining either a relatively short product of ∼230 bp, reflecting the splicing of the second and fourth exons prior to or instead of trans-splicing, or a long product of ∼1300 bp, reflecting the prespliced transcript containing the second and fourth exons connected by the out-of-frame phase1/0 “intron” between them. Instead, the only product we obtain is the final fully trans-spliced product of ∼490 bp (data not shown). Thus this unusual trans-splicing of an internal exon into the middle of a separate transcript is achieved efficiently.
How might this unusual example of trans-splicing have evolved? We hypothesize that it resulted from duplication of the bursicon gene followed by essentially simultaneous inactivation of the third exon in one copy and at least the second or fourth in the other. Association of the two pseudogenic transcripts might have allowed trans-splicing leading to an intact transcript that became indispensable. Subsequent decay and loss of the third exon from the 2L gene copy and the second and fourth exons from the 2R copy would yield the present-day situation. This would be an unusual series of events, accounting perhaps for the rareness of this kind of trans-splicing of an internal exon. If such a duplication involved more than just the bursicon locus we might find hints of genomic similarity on either side of the burs124 and burs3 loci; however, the genes on either side of these loci—in An. gambiae, at least—are unrelated to each other. A duplication might have involved only this locus, or genome flux over the past >150 MY might have removed any remnants of a larger duplication.
How this trans-splicing is now achieved is unclear because we can find no sequence similarity between the burs124 and burs3 loci that might mediate association of the two transcripts. Alignments of the donor and acceptor splice sites involved in the trans-splicing in these three mosquitoes reveal no obvious features of the sites themselves that might predispose them to trans-splicing, and there is no sequence conservation of the intron sequences that might suggest conserved secondary structures mediating the trans-splicing (supplemental Figure 1 at http://www.genetics.org/supplemental/). We also cannot identify a promoter region for the burs3 locus. Isolation of the burs3 transcript, perhaps through temporary inhibition of splicing, might help illuminate how this unusual trans-splicing is achieved today in mosquitoes. An entirely alternative possibility is that the formation of a functional mature mRNA encoding bursicon in mosquitoes involves even more esoteric events, such as genomic rearrangement in the relevant tissues. Our genomic PCR amplification of the burs124 locus revealed no indication of such a larger construct, although it might be present in only a limited set of cells and be out-competed in the PCR reaction by the smaller unrearranged fragment.
Acknowledgments
We thank the various genome sequencing centers for making genome assemblies and raw reads publicly available prior to publication, the Fotis Kafatos laboratory for the two An. gambiae cDNA clones, and two anonymous reviewers for insightful comments on the manuscript.
References
- Blumenthal, T., D. Evans, C. D. Link, A. Guffanti, D. Lawson et al., 2002. A global analysis of Caenorhabditis elegans operons. Nature 417 851–854. [DOI] [PubMed] [Google Scholar]
- Bonen, L., 1993. Trans-splicing of pre-mRNA in plants, animals, and protists. FASEB J. 7 40–46. [DOI] [PubMed] [Google Scholar]
- Dewey, E. M., S. L. McNabb, J. Ewer, G. R. Kuo, C. L. Takanishi et al., 2004. Identification of the gene encoding bursicon, an insect neuropeptide responsible for cuticle sclerotization and wing spreading. Curr. Biol. 14 1208–1213. [DOI] [PubMed] [Google Scholar]
- Gabler, M., M. Volkmar, S. Weinlich, A. Herbst, P. Dobberthien et al., 2005. Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species Drosophila melanogaster and D. virilis. Genetics 169 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, S., S. Awadalla, T. Gaasterland and G. A. Cross, 2005. A computational investigation of kinetoplastid trans-splicing. Genome Biol. 6 R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt, R. A., G. M. Subramanian, A. Halpern, G. G. Sutton, R. Charlab et al., 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298 129–149. [DOI] [PubMed] [Google Scholar]
- Krzywinski, J., O. G. Grushko and N. J. Besansky, 2006. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol. Phylogenet. Evol. 39 417–423. [DOI] [PubMed] [Google Scholar]
- Mendive, F. M., T. Van Loy, S. Claeysen, J. Poels, M. Williamson et al., 2005. Drosophila molting neurohormone bursicon is a heterodimer and the natural agonist of the orphan receptor DLGR2. FEBS Lett. 579 2171–2176. [DOI] [PubMed] [Google Scholar]
- Lai, Y., Y. Yue, M. Liu, A. Ghosh, J. F. Engelhardt et al., 2005. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 23 1435–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C. W., E. M. Dewey, S. Sudo, J. Ewer, S. Y. Hsu et al., 2005. Bursicon, the insect cuticle-hardening hormone, is a heterodimeric cystine knot protein that activates G protein-coupled receptor LGR2. Proc. Natl. Acad. Sci. USA. 102 2820–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, X., V. Shepelev and A. Fedorov, 2006. Bioinformatic analysis of exon repetition, exon scrambling and trans-splicing in humans. Bioinformatics 22 692–698. [DOI] [PubMed] [Google Scholar]
- Van Loy, T., M. B. Van Hiel, H. P. Vandersmissen, J. Poels, F. Mendive et al., 2006. Evolutionary conservation of bursicon in the animal kingdom. Gen. Comp. Endocrinol. (in press). [DOI] [PubMed]
- Yang, Y., and C. E. Walsh, 2005. Spliceosome-mediated RNA trans-splicing. Mol. Ther. 12 1006–1012. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Y. Xie, J. A. Martignetti, T. T. Yeo, S. M. Massa et al., 2003. A candidate chimeric mammalian mRNA transcript is derived from distinct chromosomes and is associated with nonconsensus splice junction motifs. DNA Cell Biol. 22 303–315. [DOI] [PubMed] [Google Scholar]