TO test whether quantitative traits are under directional or homogenizing selection, it is common practice to compare population differentiation estimates at molecular markers (FST) (Wright 1951) and quantitative traits (QST) (Spitze 1993). If the trait is neutral and its determinism additive, then theory predicts that QST = FST, while QST > FST is predicted under directional selection for different local optima, and QST < FST is predicted under homogenizing selection (Merila and Crnokrak 2001). Goudet and Büchi (2006) recently evaluated the effects of dominance, inbreeding, and sampling design on QST for neutral traits. Under dominance, Goudet and Büchi (2006) found that (1) dominance decreases on average the value of QST relative to FST (i.e., QST − FST ≤ 0), (2) the magnitude of the contrast QST − FST increases with population differentiation (i.e., with increasing FST), and (3) dominance is unlikely to lead to QST − FST > 0.
In a recent letter to Genetics, Lopez-Fanjul et al. (2007) questioned the evidence leading to these claims. In particular, they criticized Goudet and Büchi (2006) for using averages over allele frequencies and dominance deviations. Here, taking an analytical approach similar to that used in Lopez-Fanjul et al. (2007), we first show that under an island model, the result QST ≤ FST with dominance obtained by Goudet and Büchi (2006) is strictly true over all allelic frequencies and dominance deviations. We then argue that independently of the underlying population structure, averaging over allele frequencies and dominance deviations is pertinent, since quantitative traits are polygenic, and this is what empiricists study when they estimate QST. We conclude by emphasizing that the major problem faced by empiricists is not the slight negative bias in QST due to nonadditive effects, but the very large variance in this quantity, particularly when the number of samples is small.
As Lopez-Fanjul
et al. (2007) and Goudet and Büchi (2006) used different parameterizations, some clarification might be useful. Goudet and Büchi (2006) used −a, d, and a, while Lopez-Fanjul
et al. (2007) used 1 − s, 1 − hs, and 1 for the genotypic values of AA, AB, and BB, respectively. These two notation schemes are equivalent when a = 1 (and therefore s = 2) and 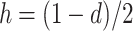 .
.
To obtain the expectation of 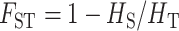 and
and 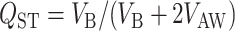 , four quantities are needed: gene diversities within [
, four quantities are needed: gene diversities within [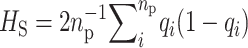 ] and over all [
] and over all [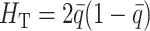 ] populations, where q represents the frequency of the recessive allele and np the number of populations, as well as the additive variance within (VAW) and between (VB) populations. Under strict additivity, these four quantities are functions of the first and second moments of the distribution of allele frequencies only. However, under dominance (when
] populations, where q represents the frequency of the recessive allele and np the number of populations, as well as the additive variance within (VAW) and between (VB) populations. Under strict additivity, these four quantities are functions of the first and second moments of the distribution of allele frequencies only. However, under dominance (when 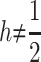 ), VAW and VB also depend on the third and fourth moments of this distribution. As we see below, the difference between Lopez-Fanjul
et al. (2007) results and those of Goudet and Büchi (2006) stems partly from the assumed distribution of allele frequencies.
), VAW and VB also depend on the third and fourth moments of this distribution. As we see below, the difference between Lopez-Fanjul
et al. (2007) results and those of Goudet and Büchi (2006) stems partly from the assumed distribution of allele frequencies.
We consider exactly the same genetical setup as in Lopez-Fanjul
et al. (2007): a biallelic locus with dominance h, additive effect s, and allele frequency q. Using their notation, the mean for the trait, in any given population, is given by M = 1 − 2qhs − q2s(1 − 2h). The variance of trait mean among populations is given by VB = E(M2) − E(M)2, where E denotes expectation with respect to the distribution of q among populations. This turns out to be a polynomial function of the first four moments of allelic frequencies. The additive variance in a given population is given by VAW = 2αq(1 − q) [where α = s(h + (1 − 2h)q is the average effect; Lopez-Fanjul
et al. 2003]. With 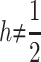 the expectation of this additive variance among populations, E(VAW), is also a polynomial function of the first four moments of allele frequencies.
the expectation of this additive variance among populations, E(VAW), is also a polynomial function of the first four moments of allele frequencies.
The expectations of these quantities are obtained by replacing q, q2, q3, and q4 in their expressions by the first, second, third, and fourth moments of allele frequency distribution (just as in Lopez-Fanjul et al. 2007). In their letter, Lopez-Fanjul et al. (2007) consider a specific model where isolated lines diverge by drift from an infinitely large panmictic population (pure drift model, PDM), while Goudet and Büchi (2006) considered the classical island model (IM) at equilibrium between migration and drift. For the infinite island model at equilibrium, and for a biallelic locus, the distribution of allelic frequencies follows a beta distribution with parameter 4Nm,
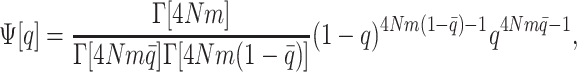 |
(Wright 1937a,b), where 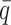 is the average allele frequency among populations (for the multiallelic locus equivalent, the pertinent distribution is a Dirichlet, as used in Goudet and Büchi 2006; see Rannala and Hartigan 1996).
is the average allele frequency among populations (for the multiallelic locus equivalent, the pertinent distribution is a Dirichlet, as used in Goudet and Büchi 2006; see Rannala and Hartigan 1996).
The first four moments of the beta distribution can be expressed in terms of 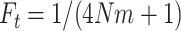 as follows:
as follows:
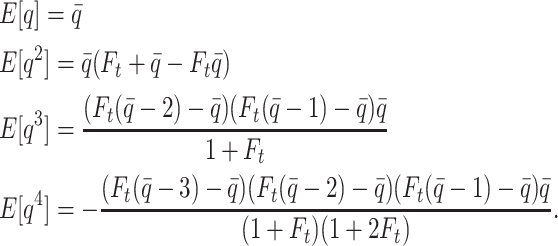 |
(1) |
Under the PDM, the moments of the allele frequency distribution are given by Equations 1–4 in Lopez-Fanjul
et al. (2002). Note that the two models yield the same first and second moments, but differ in their third and fourth moments, which also influence VB and VAW when 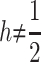 .
.
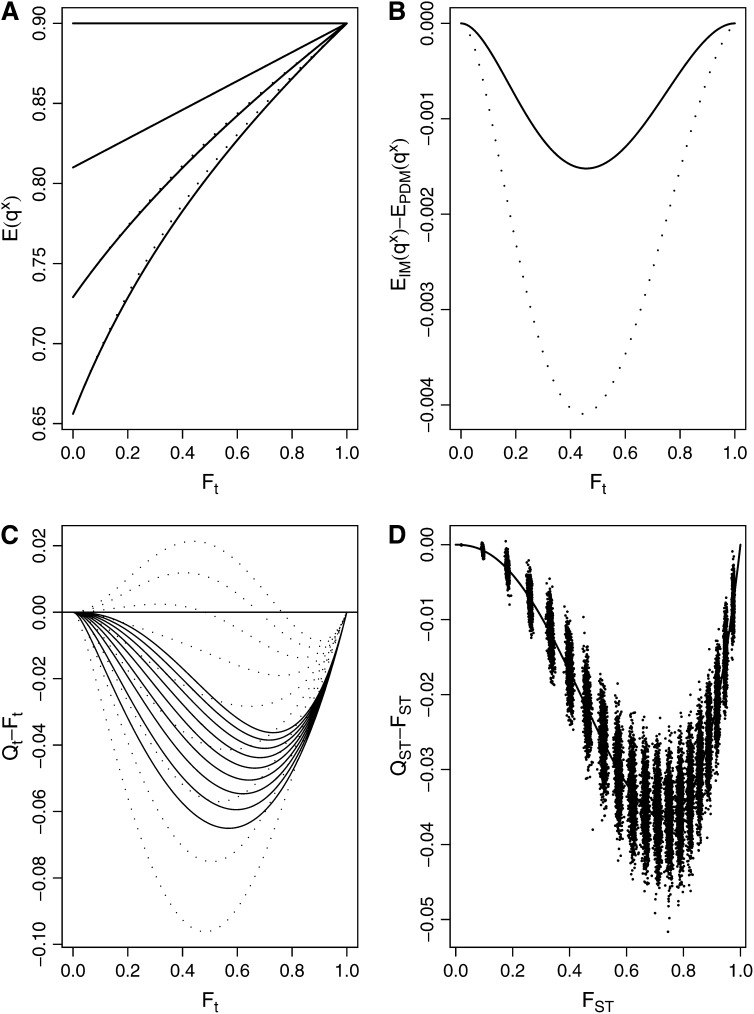
Figure 1A portrays the dynamics of the first four allele frequency moments of the two models, assuming an overall allele frequency of q = 0.9. This frequency was chosen because this is the frequency that gave rise to the largest positive difference between QST and FST in Lopez-Fanjul et al. (2007) (see Figure 1 of their article). The discrepancies between the third and fourth moments are barely notable in Figure 1A, but when the differences between these moments are plotted against Ft (Figure 1B), we can clearly see that the expected third and fourth moments of allelic frequency are smaller in the IM than in the PDM. Figure 1C shows that these tenuous differences lead to a qualitative change in the difference QST − FST. While in the PDM (dotted lines) this difference can be positive or negative, it is always negative in the IM (for any s and any h < 0.5). The formulas used by Lopez-Fanjul et al. (2007) therefore do not apply to an island model (at least at equilibrium), contrary to what they claim. And, since none of the overall allelic frequencies give rise to a positive difference between QST and FST in the IM, it can be concluded that the results of Goudet and Büchi (2006) are true both for a single diallelic locus under the IM and for more realistic multilocus traits, at least under the IM and the PDM. Goudet and Büchi (2006) were therefore correct in identifying the population model as a likely reason for the discrepancies between their results and earlier work by Lopez-Fanjul et al. (2003). Finally, Figure 1D confirms that simulations of a single diallelic locus with overall recessive frequency of 0.9 and h = 0 give results entirely consistent with the theoretical expectations under an IM.
Figure 1.—
Moments of allele frequency for a one-locus, two-allele model, with a = 1, d = 0 (s = 2, h = 0). (A) First four moments for q = 0.9 as a function of Ft. Solid lines are for the island model (IM), and dotted lines are for independent lineages diverging from an ancestral large panmictic population (PDM). First to fourth moments from top to bottom. (B) Difference between IM and PDM for the third (solid line) and fourth (dotted line) moments of allele frequency for q = 0.9 as a function of Ft (the first and second moments for the IM and PDM are identical). (C) Expected value of the difference Qt − Ft as a function of Ft for different frequencies of the recessive allele (0.1–0.9 in steps of 0.1 from bottom to top) for the IM (solid lines) and the PDM (dotted lines; same as Figure 1 in Lopez-Fanjul et al. 2007, with h = 0 instead of h = 0.25; Qt and Ft are used for expectations of QST and FST, respectively). (D) For q = 0.9, simulations of the difference QST − FST in an island model made of 10,000 islands for different FST-values. The solid line is for the expectation of the difference QST − FST in an IM.
Lopez-Fanjul et al. (2007) also assert that contrary to the results of Goudet and Büchi (2006), the difference between QST and FST is largest around FST = 0.5 and decreases thereafter. It is clear that for extremely large population differentiation (e.g., FST ≈ 1) QST and FST should be equal whatever the dominance deviation. This is because when populations are entirely differentiated, no heterozygotes remain, and hence dominance is not expressed. Figure 1, C and D, shows that in the IM the maximum difference between QST and FST due to dominance is obtained for differentiation values of ∼0.7–0.8 rather than 0.5. A cursory survey of the literature shows that FST ≥ 0.7 is seldom reported. Accordingly, the simulation scheme in Goudet and Büchi (2006) did not investigate FST-values much larger than 0.8.
Although it was just shown that under an island model dominance always leads to QST ≤ FST, one might be worried that this result depends on the underlying model of population structure. Indeed, the IM (which assumes an equilibrium between migration and drift) is by no means more realistic than the PDM (which assumes no migration), for which dominance does give rise to the pattern QST > FST for certain allelic frequencies. It is essential to realize that all these results are the expectation of the difference between QST and FST for a single diallelic locus (in the analytical section of Goudet and Büchi 2006, p. 1339 gives the expression of QST for a single diallelic locus independently of the underlying model of population structure, from which it is possible to derive the conditions under which QST > FST). As soon as several loci are considered, and because the parameter space leading to QST > FST is so narrow, the general consequence of dominance deviations is QST ≤ FST. Lopez-Fanjul
et al. (2007) criticized Goudet and Büchi (2006) for using a far too wide parameter space in their simulations. First, as shown above for the IM, QST ≤ FST independently of the distribution of s and h. For the PDM, restricting simulation scenarios within what is known about allelic frequencies and dominance deviations of recessive deleterious alleles (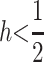 , low q) also leads to QST ≤ FST. The conditions leading to QST > FST for a dominant trait are rather unlikely: over and above it occurring only for a specific population model (PDM), it also requires the frequencies of the recessive alleles at most loci coding for the trait to be large. While this might happen for an isolated locus, it is extremely unlikely that it will occur for any real traits (for which, for a neutral trait, we expect a symmetric distribution of q among loci). Thus, while for a single diallelic locus the relation between QST and FST depends on the underlying model of population structure, for a polygenic trait the different models of population structures consistently lead to QST ≤ FST.
, low q) also leads to QST ≤ FST. The conditions leading to QST > FST for a dominant trait are rather unlikely: over and above it occurring only for a specific population model (PDM), it also requires the frequencies of the recessive alleles at most loci coding for the trait to be large. While this might happen for an isolated locus, it is extremely unlikely that it will occur for any real traits (for which, for a neutral trait, we expect a symmetric distribution of q among loci). Thus, while for a single diallelic locus the relation between QST and FST depends on the underlying model of population structure, for a polygenic trait the different models of population structures consistently lead to QST ≤ FST.
It is perhaps unfortunate that Lopez-Fanjul et al. (2007) drew attention only to one of the conclusions of Goudet and Büchi (2006), namely the slight bias in QST due to dominance, and did not take note of the rest of the article, where Goudet and Büchi (2006) quantified the variance in QST under several experimental designs. From Figure 1 (and Figure 2 of Lopez-Fanjul et al. 2007), we see that the expected bias in QST seldom exceeds 10% of the value of FST. This would matter if the variance of QST was small. But Goudet and Büchi (2006) found that unless the number of populations analyzed to estimate QST is very large (e.g., >20), only extremely large differences between QST and FST (certainly >10% of the value of FST) are likely to be statistically significant.
The slight effect that dominance might have on QST is therefore unlikely to lead to a spurious inference of selection, and the large variance of QST is certainly more worrisome for the prospect of identifying traits under selection.
Acknowledgments
We are grateful to Lucie Büchi, Laurent Lehmann, and particularly François Balloux for discussions and comments on previous versions of this manuscript. This work was carried out in part when J.G. was hosted by François Balloux in the Department of Genetics at Cambridge University. J.G. was under the benefit of a visiting professor Underwood fellowship from the United Kingdom Biotechnology and Biological Sciences Research Council. G.M. was supported by grant 31-108194/1 from the Swiss National Science Foundation to J.G.
References
- Goudet, J., and L. Büchi, 2006. The effects of dominance, regular inbreeding and sampling design on QST, an estimator of population differentiation for quantitative traits. Genetics 172 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanjul, C., A. Fernandez and M. Toro, 2002. The effect of epistasis on the excess of the additive and non-additive variance after population bottlenecks. Evolution 56 865–876. [DOI] [PubMed] [Google Scholar]
- Lopez-Fanjul, C., A. Fernandez and M. Toro, 2003. The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164 1627–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Fanjul, C., A. Fernandez and M. A. Toro, 2007. The effect of dominance on the use of the QST − FST contrast to detect natural selection on quantitative traits. Genetics 176 725–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merila, J., and P. Crnokrak, 2001. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 14 892–903. [Google Scholar]
- Rannala, B., and J. Hartigan, 1996. Estimating gene flow in island populations. Genet. Res. 67 147–158. [DOI] [PubMed] [Google Scholar]
- Spitze, K., 1993. Population structure in Daphnia obtusa: quantitative genetics and allozyme variation. Genetics 135 367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1937. a The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1937. b The distributions of gene frequencies in populations. Science 85 504. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1951. The genetic structure of populations. Ann. Eugen. 15 323–354. [DOI] [PubMed] [Google Scholar]