DISULFIDE bonds in proteins—how these bonds are formed, how they are cleaved, and how they participate in protein folding—have become my passion over the past 25 years. During this period, the discoveries in my lab in this very biochemical realm have come largely from genetic approaches. Many factors have influenced the pace of our progress, including at times ignorance of certain biochemical dogma and at other times unreflective adherence to such dogma. We have been impelled by the conviction that well-thought-out genetic approaches can yield insights into how processes of protein chemistry occur in a living cell. Yet, despite our carefully constructed “rational” approaches, serendipity, a workhorse of science, has consistently led us in unexpected directions.
Disulfide bonds, the covalent bonds between sulfurs of cysteine residues, contribute to the folding, structure, and stability of many proteins. In gram-negative bacteria, structural disulfide bonds are found only among those proteins translocated through the cytoplasmic membrane such as secreted toxins, components of appendages such as flagella, many periplasmic proteins, and the periplasmic domains of some outer membrane and cytoplasmic membrane proteins. In eukaryotic cells, proteins with stable disulfide bonds are among the proteins that pass through the endoplasmic reticulum. They include secreted proteins and the extracytoplasmic domains of plasma membrane proteins. Few, if any, proteins with structural disulfide bonds are located in the cytoplasm, whether in eukaryotes or prokaryotes. However, certain cytoplasmic reductive enzymes that use the redox chemistry of cysteine in their active sites do form disulfide bonds as part of their catalytic cycles, but these bonds are subsequently reduced to regenerate active enzyme.
For many years, the accepted explanation for the specialized subcellular location of proteins with disulfide bonds was based on a simple view: The periplasm of bacteria, because it is exposed to oxygen, and the lumen of the endoplasmic reticulum, perhaps because of the presence of oxidized glutathione, are oxidizing environments. Thus, the formation of disulfide bonds in proteins, an oxidative step, takes place in such environments without need for any enzyme catalysts. In contrast, the cytoplasms of both eukaryotic and prokaryotic cells are reducing environments maintained by electrons transferred from molecules such as NADH, NADPH, and reduced glutathione. Either disulfide bonds cannot form under these reducing conditions or, if they do, they are converted back to free cysteine residues by the reducing environment. It seemed as though any further exploration of these processes was unnecessary; the explanations were at hand.
The assumption that disulfide bonds form spontaneously in an oxidizing compartment derived directly from the important experiments of Anfinsen et al. (1961) on protein folding in the early 1960s. They showed that when bovine pancreatic ribonuclease, which contains four disulfide bonds, was reduced and denatured, it could reassemble into its active structure in the test tube in the presence of oxygen and in the absence of any enzyme catalysts. These findings suggested that no such catalysts for the oxidative folding process should be necessary in vivo. Nevertheless, the kinetics of disulfide bond formation in ribonuclease in these experiments was very slow, incommensurate with the rapid kinetics that we now know occurs in vivo. Furthermore, a significant fraction of ribonuclease folded into a non-native conformation with the “wrong” cysteines joined in disulfide bonds. This latter finding led Anfinsen and co-workers to predict the existence of and then to find the enzyme, protein disulfide isomerase (PDI), that promoted rearrangement of the disulfide bonds of incorrectly folded ribonuclease into the native conformation (Goldberger et al. 1963). They did not see the necessity of looking for an enzyme that catalyzes disulfide bond formation itself.
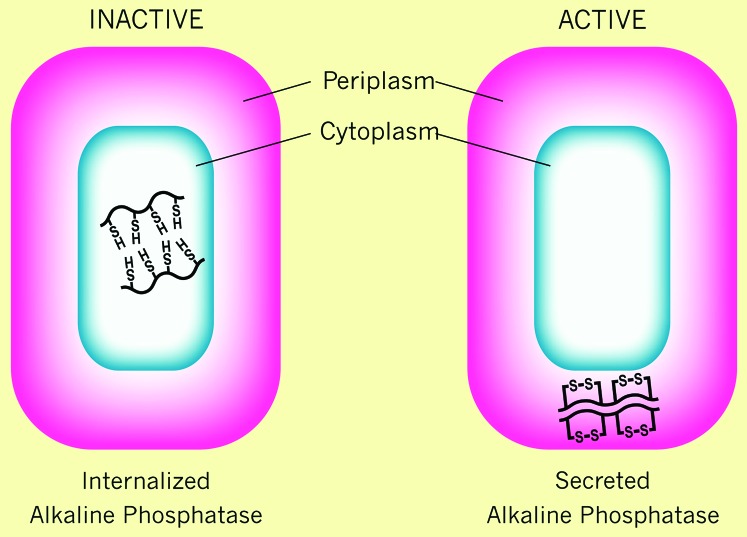
We became interested in the issue of disulfide bond formation during the course of our studies on translocation of proteins across the cytoplasmic membrane of Escherichia coli. We used as one model for the protein secretion process alkaline phosphatase (AP), a homodimeric nonspecific phosphomonoesterase located in the periplasmic space. Each of AP's monomers contains two disulfide bonds, which are essential for its folding into a stable, active enzyme (Sone et al. 1997). To study the signals required for translocation, we used a genetic selection scheme for obtaining mutations that altered the cellular location of AP from the periplasm to the cytoplasm (Michaelis et al. 1983). We then showed that all of these mutations altered the amino-terminal signal sequence of AP. In studying the properties of the signal sequence mutants, we discovered that the AP that was localized to the cytoplasm in these mutants no longer exhibited enzymatic activity (Michaelis et al. 1983, 1986) (Figure 1). We suspected that the lack of activity of cytoplasmically localized AP was due to the absence of the disulfide bonds in the protein, as we knew that the disulfide bonds of AP were required for its functioning and that the reducing environment of the cytoplasm should prevent disulfide bond formation. Subsequently, we showed that, indeed, the four cysteines of cytoplasmic AP were not joined in disulfide linkages, but were each present in reduced form (Derman and Beckwith 1991).
Figure 1.—
Activity of alkaline phosphatase when expressed in the cytoplasm or periplasm of E. coli. The two parts represent the E. coli cell with alkaline phosphatase expressed without its signal sequence in the cytoplasm (internalized alkaline phosphatase) or in the periplasm (secreted alkaline phosphatase). The protein has free cysteines in the cytoplasm and cysteines joined in a disulfide bond in the periplasm. Active alkaline phosphatase is a homodimeric enzyme.
My early training as an undergraduate and graduate student with Lowell Hager during the 1950s was in chemistry and biochemistry. However, during my graduate career I became enchanted with the power of bacterial genetics, inspired particularly by the work of François Jacob, Élie Wollman, Jacques Monod, and their co-workers. I went on to do postdoctoral work, learning bacterial genetics in the laboratories of Arthur Pardee, Bill Hayes, Sydney Brenner, and François Jacob, as I gradually broke away from or even suppressed my earlier training. I saw how much could be learned by taking a genetic approach to fundamental problems, “without touching the biochemistry,” as Sydney Brenner put it. This prelude is to explain how, by the time our work on alkaline phosphatase caused us to come upon this “problem” of disulfide bond formation in the mid-1980s, I had strayed far enough away from the field of biochemistry to be unaware that the problem of disulfide bond formation was considered solved by the Anfinsen experiments. The oxidizing environments of certain compartments were considered sufficient explanation. Thus, out of ignorance or naiveté I chose to initiate studies into the “problem” of disulfide bond formation. I assumed that, like most other chemical processes occurring in biological systems, there must be an enzyme that catalyzes the formation of disulfide bonds.
Our studies on AP led me to formulate what I thought to be two open questions about disulfide bond formation. First, how do disulfide bonds form efficiently in proteins in their specialized compartments? Can we find an enzyme that carries out this process? Second, why do disulfide bonds not form in cytoplasmic proteins? The explanation for the lack of activity of alkaline phosphatase localized to the cytoplasm—“the reducing environment of the cytoplasm”—seemed to me vague and to beg an important question: What are the specific components of the “reducing environment” that prevent disulfide bond formation in the cytoplasm? Is it simply electrons hanging around in the cytoplasm ready to bash any disulfide bond that had the audacity to appear? These lines of questioning caused me to try with my co-workers to devise schemes for isolating mutations that would be defective in the process of disulfide bond formation in the periplasm and mutations that would allow disulfide bond formation in the cytoplasm.
A GENETIC SELECTION FOR MUTANTS DEFECTIVE IN DISULFIDE BOND FORMATION IN THE E. coli PERIPLASM
To find an enzyme responsible for disulfide bond formation initially, I attempted to devise a genetic selection or screen that would provide us with mutants defective in that process. However, the ideas I concocted relied on questionable assumptions and seemed laborious enough that I could not convince anyone in my lab to pursue them. Instead, a few years after I proposed my complicated scheme, we unexpectedly found such mutants using a genetic selection that was designed for totally different purposes (Bardwell et al. 1991). Because of our interest in the process of membrane protein assembly, we had constructed a gene fusion strain in which the cytoplasmic enzyme β-galactosidase was fused to a periplasmic domain of the cytoplasmic membrane protein MalF. The signals in MalF that caused it to insert into the membrane and to translocate its periplasmic domains across the membrane also dragged β-galactosidase into the periplasm where it could not fold into its active conformation. Thus, the fusion strain exhibited a Lac− phenotype due to the absence of β-galactosidase activity. We believed mutations in this strain that were defective in membrane protein insertion would cause the fusion protein to be localized to the cytoplasm where the β-galactosidase would be active. Karen McGovern, a graduate student, began to select mutants of the fusion strain in which the Lac+ phenotype was restored. The first four mutations that she characterized mapped to the same region of the E. coli chromosome. However, examination of the properties of the mutants indicated that they were not defective in membrane protein insertion; they had no effect on the membrane insertion of the MalF protein itself. As Karen was approaching the deadline for finishing her thesis, she moved on to other projects and the four mutant strains were stored in frozen glycerol cultures that remained untouched for another 4 years.
During those 4 years, the frozen mutant strains constantly tantalized me. When new students or postdoctoral fellows arrived in the lab, I offered them, among other projects, a study of these mutants. Since we had not a clue as to what process these mutations might affect, nobody took up the challenge—that is, until Jim Bardwell arrived in the lab as a postdoctoral fellow. Jim's enthusiasm for tackling a project that no one else would touch was consistent with his adventurous character. He took up even more serious challenges in his travels: bicycling muddy trails through the jungles of Madagascar and canoeing down rivers of New Guinea populated by headhunting tribes. Jim began by cloning the gene in which the mutations occurred on the basis of the phenotype of the MalF–LacZ fusion strain. He then determined the DNA sequence of the gene, which tipped us off as to what its function might be. The deduced protein product contained a pair of cysteines separated by two amino acids, a “motif” typical of thiol/disulfide oxidoreductases such as thioredoxin. We postulated that this might be the sought-after protein that catalyzed disulfide bond formation in the periplasm. But, we might not even have thought of this if it had not been for the following factors: (1) I had already been interested in the issue of disulfide bond formation for some time; (2) a graduate student in the lab, Alan Derman, was working on disulfide reduction in the cytoplasm and was focusing on the thioredoxins; and (3) Dana Boyd, a senior researcher in the lab, had noted that there were many cysteines present in β-galactosidase that, in the MalF–LacZ fusion strain, might become disulfide bonded in the periplasm, thus inactivating the enzyme.
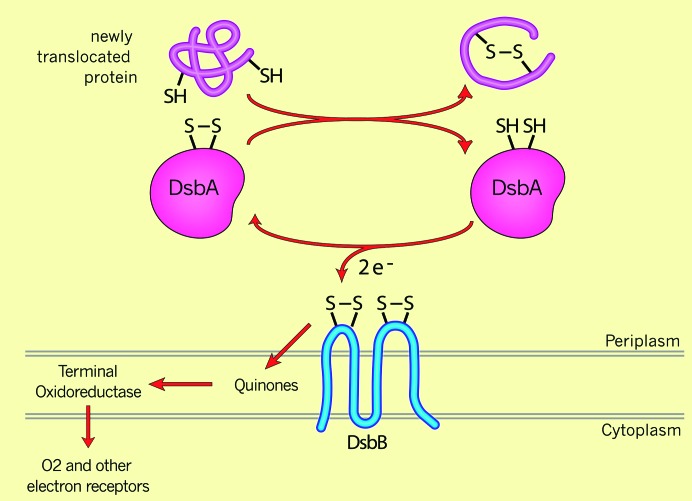
We proceeded to show that the product of this gene was involved in disulfide bond formation by examining the effect of the mutants on formation of disulfide bonds in AP—the protein that had first stimulated our interest in these questions (Bardwell et al. 1991). A 30-sec pulse labeling and immunoprecipitation revealed that AP, which was almost entirely disulfide bonded in the wild-type background, totally lacked disulfide bonds in the pulse labeling of the mutant strain. We named the gene defined by the mutations dsbA for disulfide bond formation. Within the year, other laboratories, equally inadvertently, had detected mutations in the dsbA gene (Kamitani et al. 1992; Peek and Taylor 1992). Subsequently, with these and other selections, a second gene required for disulfide bond formation, dsbB, was discovered (Bardwell et al. 1993; Belin and Boquet 1993; Dailey and Berg 1993; Missiakas et al. 1993). Work over the next few years showed DsbA to be the oxidase that directly catalyzes disulfide bond formation in substrate proteins in the periplasm, utilizing the disulfide bond between the two cysteines of the thioredoxin-like motif. DsbB, a cytoplasmic membrane protein with a pair of redox-active cysteines in each of its periplasmic domains, was necessary for the reoxidation and, thus, the regeneration of DsbA as an active enzyme after it had become reduced during its reaction with substrate proteins (Figure 2).
Figure 2.—
Disulfide bond formation in the periplasm DsbA is a periplasmic enzyme that donates its disulfide bond to substrate proteins. This results in the reduction of the two cysteines of DsbA, which are then reoxidized by the cytoplasmic membrane protein DsbB. Via two pairs of redox-active cysteines, DsbB transfers electrons via quinones and terminal oxidoreductases to oxygen.
Our discovery of DsbA was a surprise to many who had assumed that there was no need for such an enzyme. In fact, subsequent studies have revealed that the PDI originally studied by Anfinsen and colleagues is actually the eukaryotic counterpart of DsbA (Frand and Kaiser 1998; Pollard et al. 1998). Whether it also carries out isomerization of disulfide bonds in vivo is still an open question.
However, it is clear that isomerization is essential as DsbA does not necessarily form the correct disulfide bonds when it first oxidizes its substrates. The appearance of non-native disulfide bonds in these proteins is related, at least in some cases, to the fact that disulfide bond formation takes place during the translocation of proteins into the periplasm where DsbA may simply join the cysteines in order of their appearance in the periplasm. Thus, a protein that in its native form has disulfide bonds between cysteines not appearing in order in the polypeptide chain will end up with the wrong disulfide bonds (Berkmen et al. 2005). This does necessitate a disulfide bond isomerase, which in E. coli is the separate protein DsbC and which, in eukaryotic cells, may reside in PDI, the protein that also oxidizes cysteines. Overall, what we know so far about DsbA suggests that it may not exhibit much specificity in the cysteines that it chooses to join in disulfide bonds.
WHY ARE THERE NO PROTEINS WITH STRUCTURAL DISULFIDE BONDS IN THE CYTOPLASM?
At the same time that we were considering the problem of how disulfide bonds are formed in the periplasm, we were also trying to understand what was responsible for the absence of disulfide bonds in the cytoplasm. We began this study with the assumption that the “reducing” cytoplasm maintained cysteines in the reduced state. We held on to this assumption, even after we had already discovered that DsbA was necessary for disulfide bond formation in the periplasm. This knowledge should have led us to consider the possibility that to make disulfide bonds in the cytoplasm at all, we would need to have a cytoplasmic analog of DsbA. In contrast to the thinking that allowed us to find DsbA, in this case we were strongly influenced by prevailing dogma.
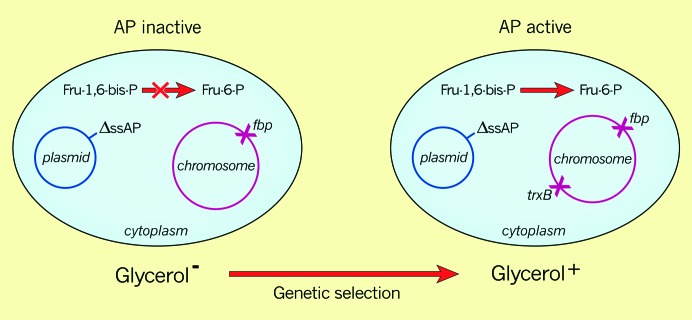
We devised a selection for mutants that would alter the environment of the cytoplasm so that disulfide bonds could form in a cytoplasmically localized protein. We chose AP as the protein since (1) AP localized to the “reducing” cytoplasm is inactive because it lacks disulfide bonds and (2) AP's phophomonoesterase activity is so nonspecific that it seemed likely that, if active in the cytoplasm, it would be able to cleave phosphate from various phosphorylated cytoplasmic metabolic intermediates. The scheme then was to utilize an E. coli mutant that was lacking a phosphatase specific to a particular metabolic pathway, such that the cells were auxotrophic or exhibited another phenotype that could be easily selected against. Specifically, we began with a mutant strain that was lacking the enzyme fructose-1,6-bis-phosphatase (fbp), which is necessary for reverse glycolysis, generating glucose-6-phosphate, which could enter the hexose monophosphate shunt. This strain is unable to use glycerol as a carbon source since glycerol enters metabolism farther down the glycolytic pathway and must be converted to the appropriate hexoses via reverse glycolysis. We introduced into the fbp strain a plasmid expressing a mutant, signal sequenceless AP, which was cytoplasmic but could not restore growth on glycerol, as the AP was inactive. Genetic selection for mutants that allowed growth on glycerol should yield mutations that would express an active AP in the cytoplasm. According to our assumptions, these mutations should define the gene encoding a protein responsible for maintaining AP in a reduced and inactive state (Figure 3).
Figure 3.—
A trxB mutant renders alkaline phosphatase active in the cytoplasm. The genetic selection described in the text utilizes a mutant strain defective in fructose-1,6-bis-phosphatase (fbp) and a plasmid expressing a signal sequenceless alkaline phosphatase from a lac IPTG-inducible promoter.
Alan Derman and Will Prinz carried out this genetic selection and examined 10 mutant derivatives of this strain in which AP had become active in the cytoplasm; all of the mutations mapped to the gene (trxB) for thioredoxin reductase (Derman et al. 1993). We verified that the now-active cytoplasmic AP contained disulfide bonds. The finding of trxB mutants in this selection fit reasonably with our preconceptions, since thioredoxin reductase reduces the disulfide bond of thioredoxin, which, in turn, reduces the disulfide bonds of certain enzymes (e.g., ribonucleotide reductase) that use redox active cysteines to reduce their substrates (Figure 4). According to our preconceptions, thioredoxin in the mutant strains could no longer be reduced and would accumulate in the oxidized form; any disulfide bonds that might form in AP by some unspecified process would no longer have to fear reduction by thioredoxin. Since we were looking for the appropriate reductant, we assumed that thioredoxin was the protein that maintained the cysteines of cytoplasmic proteins (AP in this case) in a reduced state. This thinking led to the obvious prediction that a trxA mutant, lacking thioredoxin itself, should have the same phenotype as the trxB mutant—disulfide bond formation and activity of AP in the cytoplasm. This was not the case; the trxA mutant did not exhibit any cytoplasmic AP activity.
Figure 4.—
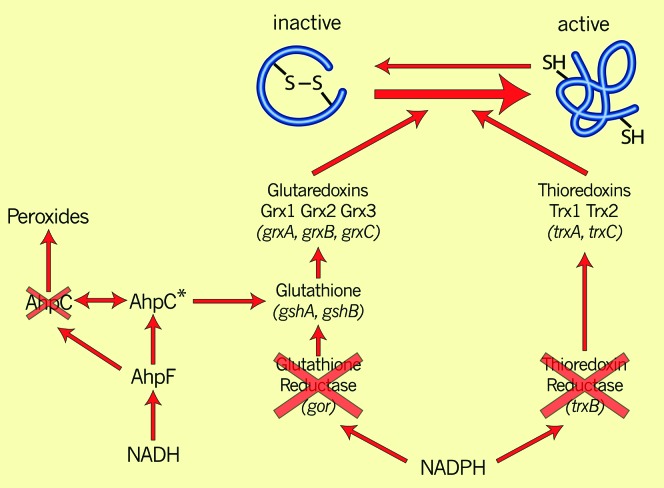
Cytoplasmic disulfide-reducing pathways A double mutant lacking the genes for thioredoxin reductase and glutathione oxidoreductase is unable to grow because cells missing both electron transfer pathways cannot reduce ribonucleotide reductase and regenerate its activity. Selection for suppressor mutations of this growth defect yields mutations in the ahpC gene, which encodes one of the two protein components (AhpC and AhpF) of a pathway for reducing peroxides. The ahpC* suppressor mutation lacks peroxidase activity and confers upon the AhpC protein the ability to generate reduced glutathione.
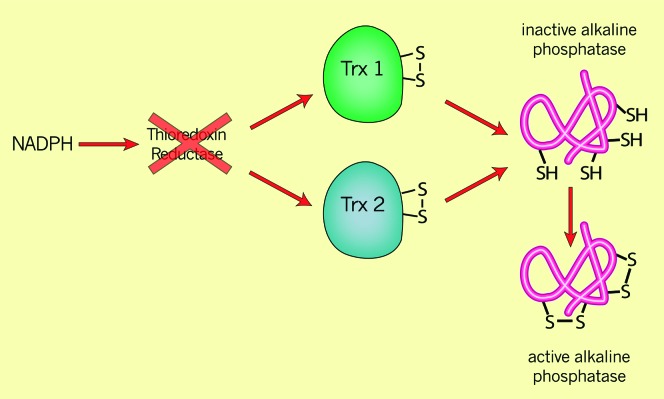
Because of the trxA result, and because the consequences of studies on DsbA for this study were beginning to sink in, we realized that there was an alternative hypothesis to consider. According to this second hypothesis, the trxB mutant accumulated oxidized thioredoxin, a protein closely related to oxidized DsbA. The disulfide bond formation that we observed in the trxB mutant was not due to the absence of a reductant but due to the presence of an oxidant, oxidized thioredoxin. A prediction of this second hypothesis was that in a double mutant, trxB, trxA, where there would be no available oxidized thioredoxin, no disulfide bonds should accumulate in AP. However, in a test of this hypothesis, we found that the introduction of the trxA mutation into the trxB strain only slightly lowered the amount of active AP in the cytoplasm (Derman et al. 1993). This puzzling result led us to propose an auxiliary hypothesis, according to which there were two thioredoxins in E. coli, one of which had not been previously identified. In this modified form of the second hypothesis, both thioredoxin 1 and thioredoxin 2 (the hypothetical one) accumulated in the oxidized form in the trxB mutant and were each capable of introducing disulfide bonds into AP. On the basis of this reasoning, Eric Stewart in my group and the group of Spyrou in Sweden identified a second thioredoxin, thioredoxin 2, the product of the trxC gene (Miranda-Vizuete et al. 1997; Stewart et al. 1998). We could eliminate the “oxidizing” phenotype of the trxB mutants—the activation of AP—by introducing the trxA and trxC mutations together into trxB strains (Stewart et al. 1998). In contrast, simply eliminating thioredoxins 1 and 2 in a trxA, trxC double mutant did not give active cytoplasmic AP, thus neatly distinguishing between the two hypotheses. We concluded that the disulfide bond formation that we observed in the trxB mutant was due to the oxidized thioredoxins acting on the cysteines of AP, not due to the absence of reduction of a disulfide-bonded AP by thioredoxins (Figure 5).
Figure 5.—
Cytoplasmic disulfide bond formation catalyzed by oxidized thioredoxins. Thioredoxins normally act as reductants of many substrates. Mutants lacking thioredoxin reductase, which normally regenerates reduced thioredoxins after their action on substrates, accumulate oxidized (disulfide-bonded) thioredoxins in the cytoplasm. These oxidized thioredoxins can now promote disulfide bond formation in substrates such as alkaline phosphatase to restore the latter protein's enzymatic acivity.
Thus, the explanation for the absence of disulfide bonds in cytoplasmic proteins is not simply “the reducing environment” of the cytoplasm: it is the absence of an oxidant. It is still possible that if a cell contained high concentrations of both oxidized and reduced thioredoxins, the reduced thioredoxins might interfere with the oxidation process. On the other hand, the now “oxidizing” trxB cytoplasm still has the glutathione pathway, which provides both reduced glutathione and reduced glutaredoxins as reductants.
While writing this article, I happened to be reading some history of science in which I came across a well-known example of scientific discovery whose path to discovery seemed analogous to those that led us to finding thioredoxin 2. Newtonian mechanics had been very successful in predicting the orbit of all of the planets except for Uranus. Two 19th century astronomers, John Adams and Urban Leverrier, confronted this problem by suggesting an auxiliary hypothesis. They could predict the correct, known orbit for Uranus if they assumed that there was a hitherto unobserved planet beyond Uranus. Their proposal quickly led to the discovery of the planet Neptune and explained away the apparent violation of Newton's theories. For us, at a much smaller scale both in terms of scientific significance and the actual size of the subject matter, the hiding of thioredoxin 2 behind thioredoxin 1 temporarily made us question our basic assumptions until we conceived our auxiliary hypothesis.
ENHANCING DISULFIDE BOND FORMATION IN THE CYTOPLASM AND ADDING A NEW REDUCTIVE PATHWAY
During the course of these experiments, I hosted a visiting scientist from the University of Geneva, Dominique Belin, a former bacteriophage T4 geneticist who had switched to studying the secretion of mammalian proteins, among them mouse urokinase. Since mouse urokinase contains six disulfide bonds arranged in a complex pattern, Dominique prodded us to ask whether urokinase (lacking its signal sequence) might assemble properly in the oxidizing cytoplasm of the trxB− strain. Indeed, we found a small amount of cytoplasmic urokinase activity in such an experiment (Derman et al. 1993). Perhaps we could enhance this expression even further by mutating other reducing pathways in the cytoplasm. We first constructed a double mutant (trxB,gor) missing both glutathione reductase and thioredoxin reductase, eliminating both major pathways for the reduction of disulfide bonds (Prinz et al. 1997). These mutants grow extremely slowly unless an external reductant such as dithiothreitol (DTT) is added, probably because they lack the major sources of electrons for reducing essential enzymes such as ribonucleotide reductase (Ortenberg et al. 2004). When DTT is removed from the growth medium, the doubly mutant strain exhibits an oxidizing cytoplasm as indicated by the much higher amounts of urokinase expressed compared to the trxB strain (Prinz et al. 1997). We noted that the trxB,gor strain throws off suppressors that restore normal growth at a very high frequency, ∼1 in 1000 bacteria.2
The trxB,gor strains carrying the suppressor mutations grow with near-normal generation times presumably because they have restored ability to reduce proteins such as ribonucleotide reductase. Surprisingly, the strains not only still exhibit a highly oxidizing cytoplasm as indicated by the presence of significant levels of active oxidized AP, but also accumulate much higher amounts of active AP than the original trxB mutant. Furthermore, we showed with our collaborator George Georgiou that these strains express in the cytoplasm much higher amounts of the disulfide-bonded forms of complex eukaryotic proteins with multiple disulfide bonds, such as urokinase, tissue plasminogen activator, immunoglobulins, etc., when these proteins are cloned without signal sequences into the suppressor strains (Bessette et al. 1999; Jurado et al. 2002). The proper assembly of these proteins is further enhanced when a protein disulfide isomerase activity is introduced into the periplasm by expressing a signal sequenceless DsbC. These strains have been widely used for the expression of eukaryotic proteins with disulfide bonds.
Analysis of the suppressors themselves presented some surprises. All of the suppressor mutations obtained at this very high frequency map to a single gene, ahpC (Ritz et al. 2001). The ahpC gene encodes a peroxidase (peroxiredoxin) that is part of a two-protein system for reducing hydrogen peroxide and alkyl hydroperoxides in E. coli and many other organisms (Figure 4) (Poole et al. 2000). In E. coli, this system functions in the following way: peroxides attack one of the two cysteines of AhpC, Cys46, converting it to an oxidized cysteine sulfenic acid form. This oxidized cysteine is then attacked by Cys165 of AhpC to generate a disulfide bond and release water. The Cys46–Cys165 disulfide bond is then reduced to regenerate active AhpC by a second protein component of this pathway, AhpF. AhpF carries out this reduction step using electrons derived from NADH and transferred by several steps between a FAD moiety and domains of the protein.
DNA sequencing revealed that the changes in all the ahpC suppressor mutations obtained from the trxB,gor strain were identical. The mutations, called ahpC*, result from an expansion of four to five copies of a TCT triplet repeat close to the codon for Cys46 of AhpC. The amplification causes a single amino acid addition to AhpC with a remarkable effect on its activity. In collaboration with biochemist Leslie Poole, we have found that this amino acid insertion completely eliminates the peroxidase activity of AhpC and converts it to an enzyme that reduces glutathionylated proteins, thus releasing reduced glutathione (Masip et al. 2006; Y. Yamamoto, D. Ritz, A.-G. Planson, T. J. Jönsson, M. J. Faulkner, D. Boyd, J. Beckwith and L. B. Poole, unpublished results). Generation of a pool of reduced glutathione in these cells appears to explain how the ahpC* mutation restores growth to the trxB,gor strain, which is missing glutathione reductase.
CONCLUSION
I have been stimulated by readings in the philosophy and history of science to try to look back at the scientific projects in my lab and understand how they arose and evolved. Do they fit into the descriptions of science presented by writers such as Thomas Kuhn, Hilary Putnam, Imre Lakatos, and Richard Boyd? Kuhn describes most clearly how dogma (e.g., paradigms), which is important in promoting the genesis of a field, can also have restraining effects on new ways of looking at problems. The concept of the presence or absence of disulfide bonds being simply due to the “reducing” and “oxidizing” environments of different compartments and the interpretation of the Anfinsen experiments limited thinking about the possibility of specific enzyme systems being responsible for these “environments.” As a convert to genetics, no longer sufficiently imbued with or keeping up with all of the assumptions in the field of biochemistry, I approached the problem of disulfide bond formation ignorant of the then-current dogma.
I am not proud of my ignorance; it is just a fact. Certainly there are other, less beneficial consequences of ignorance. Shifting my allegiance to genetics made it more difficult for me to immediately explore issues of enzyme kinetics, electron transfer processes, and protein folding so relevant to the enzymatic processes involved in disulfide bond metabolism. I regretted not having kept up with those fields or even retained the knowledge that had been transmitted to me as a graduate student.
Finally, some remarks about the utility of suppressor analysis in approaching biological problems. Each of the discoveries that I describe in this article were the result of genetic selections for suppressors of cellular defects: suppressors of a nonfunctional β-galactosidase in a gene fusion strain led to the finding of DsbA; suppressors of a defect in growth on glycerol led to the conclusion that oxidized thioredoxins promote disulfide bond formation in the E. coli cytoplasm; suppressors of the growth defect of mutants missing the two disulfide-reducing pathways led to strains that were efficient at producing disulfide bonds in the cytoplasm and to the discovery of an unusual mutant of a hydroperoxidase. In the first case, we remained for many years at a loss in understanding the nature of the suppressors that we had isolated. Yet I was convinced that these suppressors must be interesting because the first four independent mutations that were isolated all mapped to the same genetic locus and they all strongly restored β-galactosidase activity. The frozen cultures sat there for years waiting for the right time and the right person.
The trxB,gor double mutant that we used in these studies contained a leaky gor mutation. In later studies, we used a strain with deletions of both genes, which grew not at all and that threw off suppressor mutations at a lower frequency due to the absence of background growth.
References
- Anfinsen, C. B., E. Haber, M. Sela, and F. H. White, Jr., 1961. The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc. Natl. Acad. Sci. USA 47 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, J. C. A., K. McGovern and J. Beckwith, 1991. Identification of a protein required for disulfide bond formation in vivo. Cell 67 581–589. [DOI] [PubMed] [Google Scholar]
- Bardwell, J. C. A., J.-O. Lee, G. Jander, N. Martin, D. Belin et al., 1993. A pathway for disulfide bond formation in vivo. Proc. Natl. Acad. Sci. USA 90 1038–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin, P., and P.-L. Boquet, 1993. Un second géne impliqué dans la formation des ponts disulfure de protéines localisées dans l'espace périplasmique de Escherichia coli. C. R. Acad. Sci. III Sci. Vie 361 469–473. [PubMed] [Google Scholar]
- Berkmen, M., D. Boyd and J. Beckwith, 2005. The nonconsecutive disulfide bond of Escherichia coli phytase (AppA) renders it dependent on the protein-disulfide isomerase, DsbC. J. Biol. Chem. 280 11387–11394. [DOI] [PubMed] [Google Scholar]
- Bessette, P. H., F. Åslund, J. Beckwith and G. Georgiou, 1999. Efficient folding of proteins with multiple disulfide bonds in the Escherichia coli cytoplasm. Proc. Natl. Acad. Sci. USA 96 13703–13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey, F. E., and H. C. Berg, 1993. Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA 90 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman, A. I., and J. Beckwith, 1991. Escherichia coli alkaline phosphatase fails to acquire disulfide bonds when retained in the cytoplasm. J. Bacteriol. 173 7719–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derman, A. I., W. A. Prinz, D. Belin and J. Beckwith, 1993. Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262 1744–1747. [DOI] [PubMed] [Google Scholar]
- Frand, A. R., and C. A. Kaiser, 1998. The ERO1 gene of yeast is required for the oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell 1 161–170. [DOI] [PubMed] [Google Scholar]
- Goldberger, R. F., C. F. Epstein and C. B. Anfinsen, 1963. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J. Biol. Chem. 238 628–635. [PubMed] [Google Scholar]
- Jurado, P., D. Ritz, J. Beckwith, V. de Lorenzo and L. A. Fernandez, 2002. Production of functional single-chain Fv antibodies in the cytoplasm of Escherichia coli. J. Mol. Biol. 320 1–10. [DOI] [PubMed] [Google Scholar]
- Kamitani, S., Y. Akiyama and K. Ito, 1992. Identification and characterization of an Escherichia coli gene required for the formation of correctly folded alkaline phosphatase, a periplasmic enzyme. EMBO J. 11 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masip, L., K. Veeravalli and G. Georgiou, 2006. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 8 753–762. [DOI] [PubMed] [Google Scholar]
- Michaelis, S., H. Inouye, D. Oliver and J. Beckwith, 1983. Mutations that alter the signal sequence of alkaline phosphatase in Escherichia coli. J. Bacteriol. 154 366–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, S., J. Hunt and J. Beckwith, 1986. Effects of signal sequence mutations on the kinetics of alkaline phosphatase export to the periplasm in Escherichia coli. J. Bacteriol. 167 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Vizuete, A., A. E. Damdimopoulos, J.-A. Gustafsson and G. Spyrou, 1997. Cloning, expression and characterization of a novel Escherichia coli thioredoxin. J. Biol. Chem. 272 30841–30847. [DOI] [PubMed] [Google Scholar]
- Missiakas, D., C. Georgopoulos and S. Raina, 1993. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc. Natl. Acad. Sci. USA 90 7084–7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortenberg, R., S. Gon, A. Porat and J. Beckwith, 2004. Interactions of glutaredoxins, ribonucleotide reductase, and components of the DNA replication system of Escherichia coli. Proc. Natl. Acad. Sci. USA 101 7439–7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek, J. A., and R. K. Taylor, 1992. Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 89 6210–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, M. G., K. J. Travers and J. S. Weissman, 1998. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol. Cell 1 171–182. [DOI] [PubMed] [Google Scholar]
- Poole, L. B., A. Godzik, A. Nayeem and J. D. Schmitt, 2000. AhpF can be dissected into two functional units: tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39 6602–6615. [DOI] [PubMed] [Google Scholar]
- Prinz, W. A., F. Åslund, A. Holmgren and J. Beckwith, 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272 15661–15667. [DOI] [PubMed] [Google Scholar]
- Ritz, D., J. Lim, C. M. Reynolds, L. B. Poole and J. Beckwith, 2001. Conversion of a peroxiredoxin into a disulfide reductase by a triplet repeat expansion. Science 294 158–160. [DOI] [PubMed] [Google Scholar]
- Sone, M., S. Kishigami, T. Yoshihisa and K. Ito, 1997. Roles of disulfide bonds in bacterial alkaline phosphatase. J. Biol. Chem. 272 6174–6178. [DOI] [PubMed] [Google Scholar]
- Stewart, E. J., F. Åslund and J. Beckwith, 1998. Disulfide bond formation in the Escherichia coli cytoplasm: an in vivo role reversal for the thioredoxins. EMBO J. 17 5543–5550. [DOI] [PMC free article] [PubMed] [Google Scholar]