Abstract
The Drosophila BEAF-32A and BEAF-32B proteins bind to the scs′ insulator and to hundreds of other sites on Drosophila chromosomes. These two proteins are encoded by the same gene. We used ends-in homologous recombination to generate the null BEAFAB-KO allele and also isolated the BEAFA-KO allele that eliminates production of only the BEAF-32A protein. We find that the BEAF proteins together are essential, but BEAF-32B alone is sufficient to obtain viable flies. Our results show that BEAF is important for both oogenesis and development. Maternal or zygotic BEAF is sufficient to obtain adults, although having only maternal BEAF impairs female fertility. In the absence of all BEAF, a few fertile but sickly males are obtained. Using both a chromosomal position-effect assay and an enhancer-blocking assay, we find that BEAF is necessary for scs′ insulator function. Lack of BEAF causes a disruption of male X polytene chromosome morphology. However, we did not find evidence that dosage compensation was affected. Position-effect variegation of the wm4h allele and different variegating y transgenes was enhanced by the knockout mutation. Combined with the effects on male X polytene chromosomes, we conclude that BEAF function affects chromatin structure or dynamics.
ENHANCERS can act over large distances and are capable of activating transcription from diverse promoters (Kermekchiev et al. 1991). Chromatin domain insulators are thought to help prevent promiscuous interactions between enhancers and promoters by dividing chromosomes into domains such that interactions can occur within domains but cannot occur between elements located in different domains. Perhaps the best-known example that illustrates the importance of insulators is the imprinted mammalian insulator downstream of the Igf2 gene (Bell and Felsenfeld 2000; Hark et al. 2000). This insulator is not methylated on the maternal chromosome, allowing binding of the CTCF protein, which blocks activation of Igf2 by a downstream enhancer. The insulator is methylated on the paternal chromosome, which prevents binding by CTCF and allows activation of Igf2 by the downstream enhancer. Inactivation of the insulator on both chromosomes can lead to Beckwith–Wiedemann fetal overgrowth syndrome and the development of Wilms' tumor (Reik et al. 1995; Frevel et al. 1999). In Drosophila, deletion of the Fab-7 insulator in the bithorax complex leads to homeotic transformation of adult abdominal segment 6 (AS6) into another copy of the more posterior AS7 (Mihaly et al. 1997).
There are differences between insulators in certain assays, indicating that different molecular mechanisms can result in insulator activity (for examples, see Parnell and Geyer 2000; Hogga et al. 2001). In addition, some insulators are composite elements with separate components responsible for blocking enhancer–promoter communication and for acting as a barrier against chromosomal position effects (Recillas-Targa et al. 2002). It is not clear how any insulator functions at the molecular level. The various models that have been proposed include acting as promoter decoys, influencing chromatin structure or dynamics, and nuclear organization (Labrador and Corces 2002; Kuhn and Geyer 2003; Gaszner and Felsenfeld 2006). These models are not mutually exclusive. To understand how insulators function, it is necessary to study the proteins involved in insulator activity.
We are interested in the two 32-kDa Drosophila boundary element-associated factors, BEAF-32A and BEAF-32B. Throughout this article we refer to these proteins together as “BEAF” and individually as “32A” or “32B.” BEAF binds to the scs′ insulator as well as to hundreds of other sites on chromosomes (Zhao et al. 1995; Hart et al. 1997). A few other genomic BEAF-binding sites have been identified, and they function as insulators in transgenic fly assays (Cuvier et al. 1998, 2002). This suggests that BEAF-dependent insulators are a common class of insulator in Drosophila. 32A and 32B are derived from the same gene. They have unique amino-terminal DNA-binding domains of ∼80 amino acids, but the remaining 200 amino acids are encoded by a shared exon. BEAF forms complexes with itself, and this is mediated by a region near the carboxy-terminus (Hart et al. 1997). Because there were no mutations available in the BEAF gene, we previously designed a transgene under GAL4 UAS control that encodes a dominant-negative BEAF protein (Gilbert et al. 2006). Here we expand on that work by generating and characterizing mutations in the BEAF gene.
We used ends-in homologous recombination (Rong and Golic 2000; Rong et al. 2002) to generate a knockout mutation in the BEAF gene (BEAFAB-KO). In the process, we also isolated an allele that eliminates the ability to produce the 32A protein (BEAFA-KO). We found that the 32B protein is sufficient to obtain healthy, viable flies. In contrast, eliminating both BEAF proteins reveals that BEAF is essential. Both oogenesis and development are affected by a lack of BEAF. We demonstrate that BEAF is required for the insulator activity of scs′, but not for the scs insulator (which binds the Zw5 protein; Gaszner et al. 1999) or the gypsy insulator [which binds the su(Hw) protein; Harrison et al. 1989]. We also provide evidence that BEAF function affects chromatin. This confirms and extends results that we obtained with the dominant-negative BEAF protein and supports the hypothesis that BEAF functions by affecting chromatin structure or dynamics.
MATERIALS AND METHODS
DNA constructions and germline transformation:
Cloning of the BEAF gene as a 5-kb BglII fragment generated from genomic DNA by PCR has been described (Figure 1A), as has generation of transgenic flies containing this gBF rescue transgene (Gilbert et al. 2006). Site-directed mutagenesis was used to introduce mutations into this gene (Figure 1B; Quikchange, Stratagene, La Jolla, CA). One mutation eliminated the ATG start codon of BEAF-32A and destroyed an NsiI site. A second mutation eliminated the ATG start codon of BEAF-32B and created an ApaI site. Alternative ATG codons for both 32A and 32B are in the wrong reading frames. A third mutation introduced two tandem stop codons into the exon shared by both 32A and 32B and destroyed a BamHI site. A fourth mutation introduced an I-SceI site into the intron between the unique 32B exon and the shared exon. The I-SceI site is 3.7 kb downstream of the 5′-end of the cloned sequences and 1.2 kb upstream of the 3′-end. It is also ∼280 bp downstream of the 32B mutation and ∼300 bp upstream of the introduced stop codons. All mutations were confirmed by restriction digestions and sequencing. The resulting mutant BEAF (mBF) gene was cloned into the NotI site of pTV2 (Rong et al. 2002). This plasmid (0.4 μg/μl) was co-injected with the helper plasmid pπ25.7wc (0.1 μg/μl) into preblastoderm y1 w67c23 embryos to generate P[w+ mBF] transgenic flies (Spradling 1986).
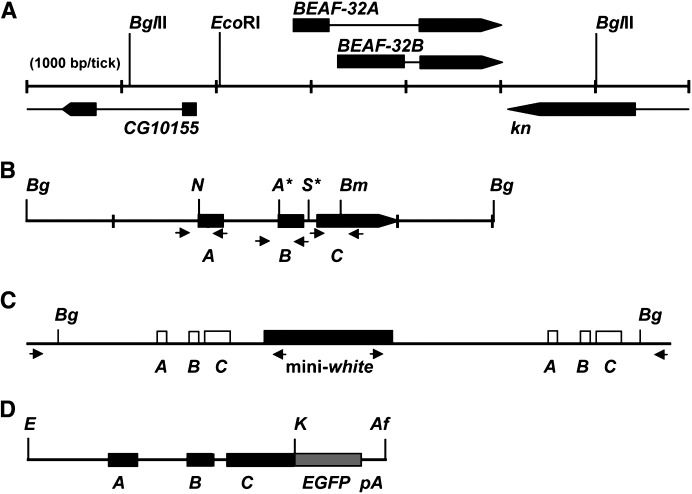
Figure 1.—
Strategy for targeted mutagenesis by homologous recombination. (A) Map of the BEAF gene, showing part of the upstream divergent CG10155 gene and downstream convergent knot (kn) gene. Arrows indicate the direction of transcription, and thin lines represent introns. Note the unique 5′ exons and the shared 3′ exon for BEAF-32A and BEAF-32B. (B) The BEAF gene was cloned as a 4.9-kb BglII fragment (gBF). Mutations were introduced at four locations to make the mutant mBF clone. The mBF gene was used for targeted mutagenesis by ends-in homologous recombination (Rong et al. 2002). Bg, BglII sites; N, destroyed NsiI site; A*, created ApaI site; S*, created I-SceI site; Bm, destroyed BamHI site. A, coding sequences unique to 32A; B, coding sequences unique to 32B. C, coding sequences common to both 32A and 32B. Arrows indicate primer pairs used for PCR. See materials and methods for details. (C) Schematic of the gene duplication expected from ends-in homologous recombination, with the mini-white marker gene between the duplicated BEAF gene. Arrows indicate primer pairs used for gene-specific PCR amplification of the 5′ or 3′ gene copy. (D) Schematic of the GFBF gene. The stop codon of the BEAF gene was converted to a KpnI site. Genomic BEAF sequences on an EcoRI–KpnI fragment were inserted upstream of EGFP sequences in the correct reading frame, with an SV40 polyadenylation sequence downstream of the EGFP sequences. E, EcoRI; K, KpnI; Af, AflII. See materials and methods for details.
A P-element plasmid encoding a BEAF-EGFP fusion gene was also constructed (referred to as GFBF for green fluorescent BEAF; Figure 1D). The stop codon of the BEAF gene was mutated to a KpnI site. pEGFP-N3 (CLONTECH) was modified by deleting a 600-bp AseI–BglII fragment encoding the CMV-IE promoter. A 2.7-kb EcoRI–KpnI BEAF gene fragment was ligated into the modified pEGFP-N3 plasmid to fuse EGFP sequences in frame at the carboxy end of the BEAF sequences. About 900 bp of sequences upstream of the BEAF-32A ATG are present. This likely contains all regulatory elements of the BEAF promoter since a divergent gene, CG10155, is reported to initiate transcription ∼265 bp upstream from the 5′-end of this fragment. An EcoRI–AflII fragment, from the BEAF promoter through the SV40 polyadenylation site, was cloned into pM2 (Cuvier et al. 1998). pM2 is a derivative of pCaSpeR4 with the scs′-derived M2 and scs insulators, so the fusion gene is insulated. This P[w+ GFBF] construct was injected into embryos as described above to generate transgenic fly lines.
Drosophila stocks:
Flies were maintained on standard cornmeal, yeast, and sugar medium with Tegosept. Crosses were performed at 25°. The yellow (y) enhancer blocking lines (2scs′ inserted at 19D; scs inserted at 60A; gypsy inserted at 25C) have been previously described (Kuhn et al. 2004). Generation of the M2 mini-white position-independent expression lines is described in Gilbert et al. (2006). The y variegating lines KV732 (X heterochromatin band 29H), KV600 (X 26H), and KV123 (3L 48H) were kindly provided by G. H. Karpen (University of California at Berkeley). All other fly lines used were from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu).
Isolation of BEAF mutations by ends-in homologous recombination:
Flies with P[w+ mBF] on the X or the CyO balancer chromosome were used to generate mutations in the BEAF gene by homologous recombination (Rong and Golic 2000). BEAF is on the second chromosome. Briefly, P[w+ mBF] females were crossed to 70I-SceI 70FLP/TM6 males. Larvae were given one heat shock at 38° for 1 hr in a water bath. For crosses with P[w+ mBF] CyO, white-eyed female progeny with CyO were crossed to y1 w67c23 males and progeny with red eyes but lacking CyO were crossed to CyO/Sp1 flies to screen for potential homologous recombination events. For crosses with P[w+ mBF] on the X chromosome, white-eyed female progeny from the first cross were crossed to 70FLP/70FLP males and the larvae were given a 1-hr 38° heat shock. This eliminated background in the next generation caused by progeny with the original P[w+ mBF] transposon. Males with red eyes were then crossed to CyO/Sp1 females to screen for potential homologous recombination events. For the P[w+ mBF] CyO strategy, ∼82,500 chromosomes were screened [(1100 vials × 150 flies/vial)/2 because of the CyO chromosome]. Eight mobilizations were recovered, only one of which was due to homologous recombination. For the strategy using P[w+ mBF] on the X chromosome, ∼100,500 chromosomes were screened (670 vials × 150 flies/vial). Three mobilizations were recovered, all of which were due to homologous recombination.
Homologous recombination was confirmed by genomic PCR. Ends-in homologous recombination results in a gene duplication with the mini-white marker gene between the two copies. Primer pairs that would specifically amplify the upstream gene copy, the downstream gene copy, or the original single-copy BEAF gene, all as 5-kb fragments, were used. Amplified DNA was sequenced and analyzed by restriction digestions. We found that one recombination event resulted in both gene copies having a mutated 32A ATG, but the 32B and shared sequences were intact. This is the BEAFA-KO allele, and the chromosome is w+. The other three recombination events had at least one wild-type gene copy. One was determined to have the 32A and 32B ATG mutations as well as the tandem stop codons in the downstream gene copy. This gene duplication was reduced to a single copy by crossing flies to a 70I-CreI Sb/TM6 line and giving the larvae a 1-hr 38° heat shock (Rong et al. 2002). w+ mosaic males were selected and crossed to CyO/Sp1 females. In the following generation, flies with CyO but lacking the 70I-CreI Sb chromosome were selected and individually crossed to CyO/Sp1 flies again. Flies that eclosed and lacked Sp1 were then self-crossed. Flies were screened by PCR and restriction digestion to identify the BEAFAB-KO chromosome, which is w−. Primer sequences used for mutagenesis, PCR, and sequencing are available upon request.
Viability assays:
To examine the effect of the lack of maternal BEAF on female fertility and egg viability, flies of the genotypes indicated in Table 1 were crossed in fly cages sealed with grape juice agar plates smeared with yeast paste. The agar plates were changed every 24 hr and embryos were counted. Hatched larvae were counted and transferred with a brush to vials, and pupae and adults were counted as they appeared. To facilitate collection of BEAFAB-KO female virgins, the BEAFAB-KO chromosome was placed over a CyO GFP w+ balancer chromosome, and third instar larvae were placed in PBS and sorted by fluorescence microscopy. Homozygous BEAFAB-KO larvae were placed in a new vial to pupate and eclose. Surprisingly, this treatment improved the fecundity of the BEAFAB-KO flies and vigor of their progeny.
TABLE 1.
Fertility of BEAFAB-KO females and effect of zygotic BEAF
| ♀ BEAFAB-KO × ♂ BEAFAB-KO |
♀ BEAFAB-KO × ♂ BEAF |
♀ BEAF × ♂ BEAF |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Stage | No./(♀)(day) | Viabilitya | Total | No./(♀)(day) | Viabilitya | Total | No./(♀)(day) | Viabilitya | Total |
| Embryo | 5.07 | 1669 | 4.65 | 897 | 32.0 | 4067 | |||
| Larva | 1.97 | 0.39 | 647 | 1.86 | 0.40 | 359 | 32.0 | 1.0b | 4067 |
| Pupa | 0.81 | 0.41 | 265 | 1.15 | 0.62 | 222 | 26.4 | 0.82c | 3350 |
| Adult | 0.17 | 0.21 | 56 | 0.79 | 0.68 | 152 | 21.8 | 0.83c | 2767 |
A total of 46 BEAFAB-KO females were used in crosses to BEAFAB-KO males, 27 BEAFAB-KO females were used in crosses to BEAF males, and 18 BEAF females were used in crosses to BEAF males.
Viability is the fraction of animals that progress from the previous developmental stage to the indicated stage.
The BEAF females laid a high density of eggs on the collection plates, making it difficult to count embryos. Therefore the number of embryos was estimated to be the same as the number of larvae collected.
The pupal and adult viability from the BEAF females is an underestimate because of mortality caused by crowding in the vials to which the larvae were transferred.
To determine the viability of BEAFAB-KO flies provided with maternal BEAF, six males and six females of the genotype BEAFAB-KO/CyO were placed in a vial for 3 days and then transferred to a new vial for an additional 3 days. The number of BEAFAB-KO and BEAFAB-KO/CyO adults that eclosed was recorded.
Ovary dissection and DAPI staining:
Wild-type or BEAFAB-KO females were mated with wild-type males for 4 days before dissection. Ovaries were dissected in PBS (0.9% NaCl, 14 mm Na2HPO4, 6 mm NaH2PO4, pH 7.3). The dissected ovaries were fixed with 4% formaldehyde in PBS for 15–30 min and then stained with DAPI (250 ng/ml DAPI in PBS/0.1% Triton-X100) or propidium iodide (100 ng/ml) plus RNase A (200 μg/ml) for 30 min. The stained ovaries were transferred to a glass slide with a drop of 60% glycerol in PBS and observed with a Zeiss Axioskop microscope equipped with a SPOT RT Slider CCD camera (Diagnostic Instruments) or a Leica TCS-SP2 confocal microscope.
Insulator and position-effect variegation assays:
All test genes were on the X or third chromosome, and the presence of these chromosomes could be followed by eye pigmentation. The cross strategy took advantage of the normal fertility of BEAFAB-KO males. Males from the test lines were crossed to CyO/Sp1 females. In the next generation, females with the test gene and CyO were selected and crossed to BEAFAB-KO males. Females with the test gene and BEAFAB-KO/CyO were selected and crossed again to BEAFAB-KO males. BEAFAB-KO flies with one copy of the test gene were compared with wild-type flies with one copy of the test gene (generated by crossing test flies to w− y− flies). For position-effect variegation (PEV) assays, phenotypes of BEAFAB-KO/CyO flies with one copy of the test gene were also recorded. Eyes were photographed using dark-field illumination with a ×4 objective on a Zeiss Axioskop microscope equipped with a Spot RT Slider CCD camera (Diagnostic Instruments). Abdomens were photographed at ×50 magnification using fiberoptic illumination on a Zeiss Stemi 2000 stereomicroscope equipped with a Spot RT Slider CCD camera. Eye pigment was quantitated by homogenizing the heads of 20 males in 200 μl 0.1% ammonium hydroxide, extracting once with chloroform, and determining the OD480 of the solution (Ashburner 1989).
Immunostaining polytene chromosomes:
Polytene chromosomes were prepared from salivary glands of healthy, wandering third instar larvae and immunostained as previously described (Gilbert et al. 2006). For this purpose, a fly line with the BEAFAB-KO allele over a w+ CyO GFP balancer was created. Homozygous BEAFAB-KO larvae derived from this line were identified by the lack of green fluorescent protein. Affinity-purified rabbit anti-BEAF antibody was used at a 1:50 dilution. Rabbit antibodies against the X chromosome dosage-compensation complex components MOF, MLE, MSL-1, MSL-2, and MSL-3 were kindly provided by M. I. Kuroda (Howard Hughes Medical Institute and Harvard Medical School) and J. C. Lucchesi (Emory University) and were used at 1:500 dilutions (except MSL-2: 1:250). Rabbit antihistone H4-acetyl-lysine 16 was purchased from Upstate Biotech (07-329) and used at a 1:400 dilution. Texas Red or FITC-conjugated goat anti-rabbit secondary antibodies were used at 1:400 dilutions (Jackson, West Grove, PA). Chromosomes were stained with 100 ng/ml DAPI. Slides were viewed with a Zeiss Axioskop microscope equipped with a Spot RT Slider CCD camera. For viewing GFP fluorescence, salivary glands were fixed for 1 min with 3.7% paraformaldehyde in PBS plus 5% Triton X-100, stained 20 min with 100 ng/ml DAPI in PBS plus 2% Triton X-100, and washed 2 min in 50% glycerol. The chromosomes were then gently spread in a fresh drop of 50% glycerol and viewed immediately.
Scanning electron microscopy:
Flies were prepared and SEM was performed as previously described (Gilbert et al. 2006).
RESULTS
Generation of mutant BEAF alleles by homologous recombination:
The BEAF gene encodes two related 32-kDa proteins, BEAF-32A and BEAF-32B (Figure 1A). These proteins have different amino-terminal DNA-binding domains encoded by unique exons, while the remainder of the proteins are identical and are encoded by a shared exon. We constructed a mutant BEAF transgene (mBF) by introducing point mutations to eliminate the 32A and 32B ATG start codons and insert two tandem stop codons into the shared exon (Figure 1B). Each mutation either created or destroyed a restriction site. Flies containing this mBF transgene were used to generate flies with mutant alleles of BEAF by ends-in homologous recombination (Rong and Golic 2000, 2001; Rong et al. 2002). This commonly results in a gene duplication bracketing the mini-white marker gene (Figure 1C). Using primer pairs anchored in genomic sequences outside of the transgene sequences and in the mini-white sequences (indicated in Figure 1C), we confirmed four such gene duplication events by individual PCR amplification of the BEAF gene upstream and downstream of the mini-white gene. Sequence and restriction digestion analyses found that one gene duplication had the 32A ATG mutation in both gene copies, but lacked the other mutations. We refer to this as the BEAFA-KO allele, and the chromosome is w+. The other three gene duplications had at least one wild-type BEAF allele. One had all three BEAF mutations in the downstream copy. This was reduced to a single copy using I-CreI endonuclease (Rong et al. 2002), and flies retaining all three mutations were identified by PCR analysis. We refer to this as the BEAFAB-KO allele, and the chromosome is w−.
Flies homozygous for both mutant alleles were able to eclose. We analyzed these flies by PCR and Western blotting to confirm that they had the BEAF mutations (Figure 2). For this PCR analysis, gene-specific primers were not used. Instead, primer pairs that generated 500-bp fragments from all BEAF genes present were used (indicated in Figure 1B). Each fragment encompassed a site that was mutated in the mBF transgene to allow detection of the mutations by restriction analysis. The Western analysis of BEAFA-KO flies used antibodies specific for either the 32A or the 32B protein, while the Western analysis of BEAFAB-KO flies used an antibody that recognizes both BEAF proteins. These analyses demonstrated that BEAFA-KO flies only have the 32A ATG mutation and make 32B protein, but no detectable 32A protein (Figure 2, B and C). Similarly, the BEAFAB-KO flies have all three of the BEAF mutations and do not produce any detectable BEAF protein (Figure 2, E and F).
Figure 2.—
Molecular characterization of BEAFA-KO and BEAFAB-KO flies. (A) Schematic of the duplicated BEAFA-KO gene and the mini-white marker gene (shaded box). Solid boxes, mutated BEAF sequences; open boxes, wild-type BEAF sequences. See Figure 1B for details. (B) Restriction analysis of 500-bp PCR products generated from homozygous BEAFA-KO flies (A-KO lanes), the mBF plasmid (Mut lanes), or wild-type flies (Wt lanes). In BEAFA-KO flies, only the 32A ATG mutation is present (NsiI lanes); the 32B ATG is not mutated (ApaI lanes) and the two tandem stop codons are not present (BamHI lanes). (C) Western analysis of BEAFA-KO (A-KO lanes) and wild-type (wt lanes) embryo nuclear extracts with antibodies specific for 32A (anti-32A lanes) or 32B (anti-32B lanes). Antibody specificity is demonstrated by loading 32A protein (Ec A lanes) or 32B protein (Ec B lanes) expressed in Escherichia coli. Note that the anti-32A antibody cross-reacts with another protein, presumably yolk protein. This demonstrates that more total protein was loaded in the A-KO anti-32A lane, yet 32A protein was not detected. Similar amounts of total protein were loaded in the BEAFA-KO and wild-type lanes for the anti-32B blot. (D) Schematic of the single-copy BEAFAB-KO gene. Solid boxes, mutated BEAF sequences. See Figure 1B for details. (E) Restriction analysis of 500-bp PCR products generated from homozygous BEAFAB-KO flies (AB-KO lanes), the mBF plasmid (Mut lanes), or wild-type flies (Wt lanes). The 32A ATG mutation, 32B ATG mutation, and the mutation introducing two tandem stop codons are all present. See B for details. (F) Western analysis of BEAFAB-KO (AB-KO lane) and BEAFA-KO (A-KO lane) adult flies with an antibody that recognizes both forms of BEAF. The cross-reactive band just above BEAF demonstrates that similar amounts of total protein were loaded in both lanes, but no BEAF was detected in the AB-KO lane. See C for details.
Effects of the BEAF mutations on Drosophila viability:
Flies homozygous for the BEAFA-KO allele are viable. They appear healthy, have normal fertility, and can be maintained as a homozygous stock. Thus the 32B protein is sufficient for survival, and the 32A protein is not necessary.
In contrast, flies homozygous for the BEAFAB-KO allele cannot be maintained as a stock. They are weaker than their heterozygous siblings with the CyO balancer chromosome and die within a few days if they are not transferred to a new vial containing a limited number of flies. However, the males have normal fertility when crossed to wild-type females and can live at least 2 weeks if pampered. Females, on the other hand, have reduced fertility and appear to be very sensitive to environmental conditions. When BEAFAB-KO female virgins were collected from their parental vial and crossed to BEAFAB-KO males, they laid few eggs and no larvae were obtained. When crossed to wild-type males, they still laid few eggs but some larvae were obtained. For crosses to wild-type males, counting indicated that BEAFAB-KO females laid <5% the number of eggs laid by wild-type females, the number of larvae obtained per female was <1% of the number from wild-type females, and the number of pupae and adults was <0.2% of the number obtained from wild-type females. As described next, different results were obtained when the experimental protocol was modified. Nevertheless, this result demonstrates that BEAF is important for oogenesis and/or development and that maternal BEAF suffices to obtain adults.
To facilitate the collection of BEAFAB-KO females that had not had an opportunity to mate with males with a wild-type BEAF gene, the mutant chromosome was placed over a CyO GFP w+ balancer. Third instar larvae were placed in PBS and sorted by fluorescence microscopy. BEAFAB-KO larvae lacked GFP and were placed into new vials to pupate and eclose. The resulting BEAFAB-KO females (confirmed by white eyes and lack of curly wings) were used for crosses to BEAFAB-KO or wild-type males. To our surprise, these females laid approximately five times more eggs than their isogenic siblings collected from parental vials. In addition, larvae, pupae, and adults were obtained from inter se crosses. Wild-type females still laid over six times more eggs than these BEAFAB-KO females, confirming that BEAF is important for oogenesis (Table 1). BEAFAB-KO females laid similar numbers of eggs and had similar larval hatch rates of ∼40% whether they were mated with BEAFAB-KO or with wild-type males. This hatch rate was less than half that obtained for wild-type flies, indicating that BEAF is also important for embryonic development. Consistent with this, we have previously shown that expression of a dominant-negative form of BEAF leads to embryonic lethality (Gilbert et al. 2006).
Zygotic BEAF rescued some animals that lacked maternal BEAF, as indicated by the higher proportion of animals from BEAFAB-KO mothers that survived to pupal and adult stages if they had BEAF fathers rather than BEAFAB-KO fathers (Table 1). The adults with zygotic BEAF appeared normal, a roughly equal number of females and males were obtained, and they were fertile. Survival rates to pupal and adult stages remained lower than that obtained for wild-type animals, and the observed wild-type viability was lowered by mortality caused by overcrowding in the wild-type vials. The lowest survival rate for BEAFAB-KO animals was obtained at the pupa-to-adult transition (∼20%). Only ∼5% of these adults were females and these females died shortly after eclosing. At least one-third of the males also died shortly after eclosing, but those that survived were fertile. Thus BEAF is important for postembryonic development, especially of females. Most of the adults, whether or not they survived long, appeared normal. However, a few individuals had various obvious defects in eye, wing, leg, thorax, or abdomen morphology. The BEAFAB-KO genotype was confirmed for six adults by PCR and restriction digestion analysis (data not shown).
To determine if development of homozygous BEAFAB-KO flies that had maternal BEAF is impaired, we recorded the number of BEAFAB-KO and BEAFAB-KO/CyO flies as they eclosed from six lightly populated vials derived from BEAFAB-KO/CyO parents (Table 2). Nearly one-third of the flies were homozygous, with roughly equal numbers of males and females. This indicates that survival to adulthood is not affected when the only BEAF present is maternally provided. However, homozygous flies eclosed 1–2 days later than their heterozygous siblings, indicating a slight developmental delay.
TABLE 2.
Viability of BEAFAB-KO/BEAFAB-KO flies
| No. of flies eclosing of the indicated genotype |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Day: | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | Total |
| BEAFAB-KO/CyO | 43 | 68 | 74 | 46 | 30 | 34 | 18 | 25 | 9 | 8 | 355 |
| BEAFAB-KO/BEAFAB-KO | 2 | 25 | 28 | 19 | 16 | 24 | 18 | 13 | 7 | 8 | 160 |
| % of totala | 4.4 | 19.6 | 22.9 | 24.3 | 25.6 | 27.9 | 29.7 | 30.0 | 30.5 | 31.1 | 31.1 |
Percentage of all eclosed flies that have the genotype BEAFAB-KO/BEAFAB-KO (running total, not daily totals). One-third of the eclosed flies should have this genotype if it does not affect viability because CyO/CyO is embryonic lethal.
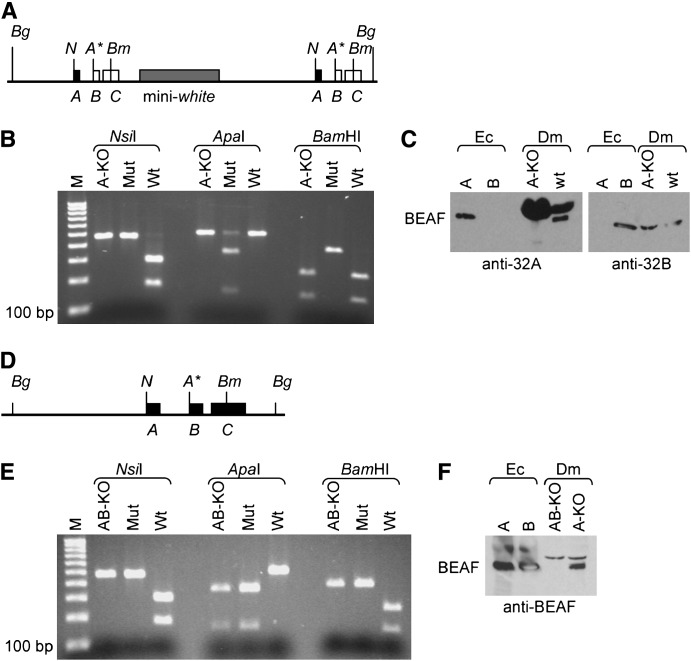
Multiple transgenic fly lines with either the gBF or the GFBF transgene were able to rescue the defects in fertility and vigor. We previously reported that producing a dominant-negative form of BEAF in eye imaginal discs leads to a rough-eye phenotype (Gilbert et al. 2006). Homozygous BEAFAB-KO flies also have a rough-eye phenotype, and this is also rescued by the BEAF transgenes (Figure 3). Hence these defects are due to the lack of BEAF protein and not to an unrelated mutation on the chromosome. The GFBF transgene is driven by a 900-bp BEAF promoter fragment and is insulated from chromosomal position effects. Therefore we expect production of the GFBF proteins to reflect that of the endogenous BEAF proteins. Fluorescence microscopy of homozygous BEAFAB-KO animals rescued by four different GFBF transgenes all gave the same result. Green fluorescent BEAF was observed in every nucleus at every life stage for all tissues examined (data not shown). This is consistent with previous immunolocalization and Western and Northern results that indicated that the BEAF proteins are ubiquitous.
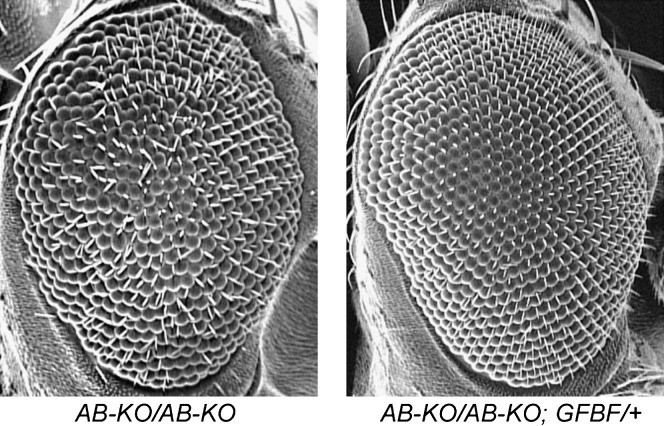
Figure 3.—
BEAFAB-KO flies have a rough-eye phenotype that is rescued by BEAF transgenes. Scanning electron micrograph of a BEAFAB-KO fly (left) shows that it has a rough-eye phenotype. Introducing a single copy of a gBF (not shown) or GFBF transgene (right) rescues this phenotype, resulting in wild-type eye morphology. BEAFA-KO flies do not have a rough-eye phenotype (not shown).
BEAF is required for normal oogenesis:
Because BEAFAB-KO females had low fertility that was rescued by gBF and GFBF transgenes, we decided to examine their ovaries. BEAFAB-KO and wild-type females were mated with wild-type males for 4–7 days prior to dissection. The number of ovarioles per ovary did not appear to differ between BEAFAB-KO and wild-type females, but mutant ovaries were smaller than those from wild type (Figure 4, A–C). This was because most ovarioles from mutant females lacked mature oocytes, whereas most ovarioles from wild-type females ended with a mature oocyte (Figure 4, D and E).
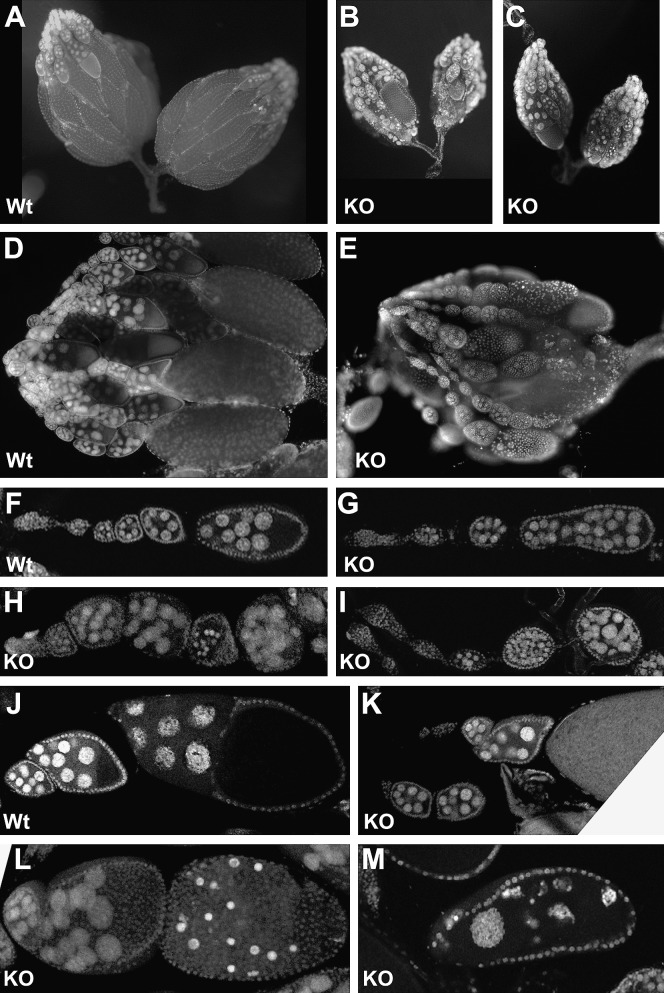
Figure 4.—
Effect of the BEAFAB-KO mutation on oogenesis. Females were mated with wild-type males for 4 days before dissecting out their ovaries and staining DNA with DAPI or propidium iodide plus RNase A. KO, BEAFAB-KO ovaries; Wt, wild-type ovaries. F–M were obtained by confocal microscopy. See text for details.
In normal egg-chamber development, there are 16 interconnected germline cells enveloped by a layer of somatic follicle cells. Of these, 15 become nurse cells with large polyploid nuclei, and the other cell becomes the oocyte. Yolk and the contents of the nurse cells begin to accumulate in the oocyte at stage 8, leading to gradual enlargement of the oocyte (Mahowald and Kambysellis 1980). We did not note any difference between mutant and wild-type germaria, which is where oogenesis initiates. However, we observed a variety of mutant phenotypes that generally became apparent around stage 8 or later. Some egg chambers had too many nurse-cell nuclei, which could be due to the fusion of two egg chambers or to an extra round of cell division (Figure 4, G and I). Others had small, brightly staining nuclei that presumably represented an intermediate step in egg-chamber degeneration. Sometimes such an egg chamber was small like a stage 7 chamber (Figure 4H), and sometimes the chamber was nearly the size of a mature oocyte (Figure 4L). In other cases, the ovariole had a stage 7 or 8 egg chamber adjacent to what appeared to be a mature oocyte, with intermediate stages missing (compare Figure 4, J–K). In yet other cases, an egg chamber the size of a mature oocyte had large, oddly shaped nurse-cell nuclei distributed throughout (Figure 4M). This presumably represents egg-chamber degeneration by a pathway different from that being used in egg chambers with small, brightly staining nuclei. Typically an ovary pair from a female exhibited multiple examples of only one of the phenotypes shown. Thus BEAF is important for oogenesis, particularly at the stages when oocyte size dramatically increases by vitellogenesis and transport of material from the nurse cells. Occasionally, a mature oocyte is formed in the absence of BEAF. If fertilized, some of these oocytes are capable of developing into adults, especially if provided with zygotic BEAF (Table 1).
The BEAFAB-KO mutation affects scs′ insulator function:
Mutating the BEAF-binding sites in scs′ eliminates insulator activity (Cuvier et al. 1998), and expression of a dominant-negative form of BEAF interferes with scs′ insulator activity (Gilbert et al. 2006). To extend these results to the BEAFAB-KO allele, we used two transgene assays. One tested the ability of insulators bracketing the mini-white gene to protect against chromosomal position effects, leading to position-independent expression of mini-white. The other assay tested the ability of insulators to block communication between the wing and body enhancers and the promoter of the y gene.
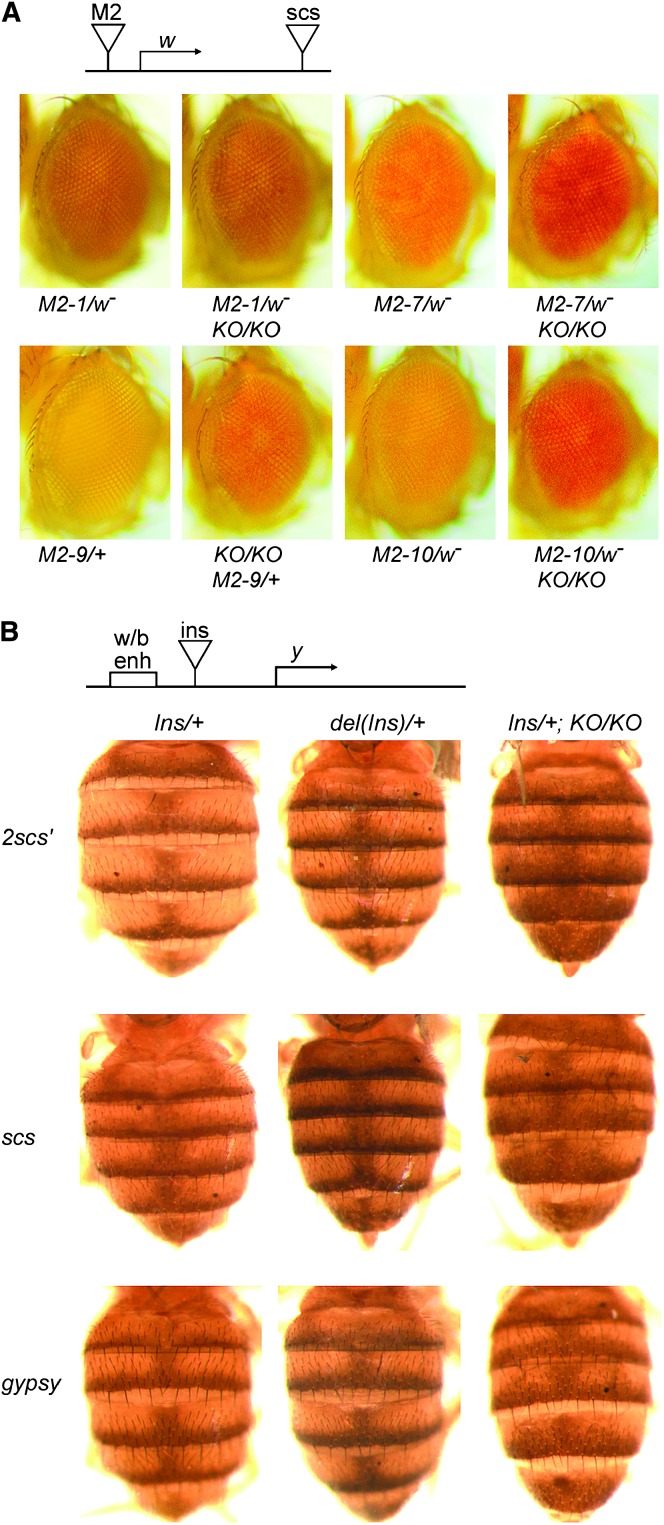
In the position-independent expression assay, mini-white was bracketed by the M2 derivative of scs′ on the 5′ side and by the scs insulator on the 3′ side (Cuvier et al. 1998). The M2 insulator has two copies of the high-affinity BEAF-binding site of scs′, with one copy replacing the low-affinity binding site normally present. Because the mini-white gene lacks an enhancer, bracketing it with insulators should lead to low levels of expression, resulting in flies with yellow or light-orange eyes. In the absence of the 5′ insulator, chromosomal position effects should lead to the activation of mini-white in some fly lines and result in darker eye pigmentation. This was observed: ∼90% of insulated fly lines have yellow or light-orange eyes, while <50% of fly lines insulated only at the 3′-end have such light eye pigmentation (our unpublished results and Cuvier et al. 1998). We had three fly lines with the M2 transposon inserted in the X chromosome, and one line with an insertion in chromosome 3. Females heterozygous for these transposons had yellow or light-orange eyes. Three of these lines had darker pigmentation in a BEAFAB-KO background (Figure 5A), indicating a loss of protection from chromosomal position effects in the absence of BEAF. The insertion in the fourth line is apparently not subject to position effects. Two of these fly lines were also tested in the presence of a dominant-negative BEAF protein (M2-9 and M2-10) and showed similar activation of mini-white (Gilbert et al. 2006).
Figure 5.—
scs′ does not function as an insulator in BEAFAB-KO flies, but the scs and gypsy insulators are still functional. (A) The M2 insulator, an scs′ derivative, does not protect against chromosomal position effects in the absence of BEAF protein. Eyes of 3- to 4-day-old females heterozygous for different M2 transposons and homozygous for BEAF or BEAFAB-KO (KO) are shown. See text for details. (B) A dimer of the scs′ insulator does not block communication between the y wing and body enhancers and the y promoter in the absence of BEAF protein. Lack of BEAF protein does not affect the ability of the scs and gypsy insulators to block this enhancer–promoter communication. Shown are abdomens of 3- to 4-day-old females homozygous for BEAF or BEAFAB-KO (KO) and heterozygous for the indicated transposons, with (Ins) or without [del(Ins)] the indicated insulator between the enhancer and promoter. See text for details.
Three different insulators were tested in the enhancer-blocking assay. An scs′ dimer (2scs′), scs, or gypsy insulator was located between the y wing and body enhancers and the y gene. The scs and gypsy insulators do not have BEAF-binding sites. “Sibling” lines in which the insulators had been removed by the Cre recombinase were also used. This allowed us to compare the level of y-dependent body pigmentation due to the same transposon integration site in the presence and absence of the insulators. Previous studies with these fly lines found that these insulators do not form the boundaries of heat-shock puffs in polytene chromosomes (Kuhn et al. 2004). The level of pigmentation in the dorsal abdomen of 3- to 4-day-old females was recorded for flies heterozygous for the enhancer-blocking transposons with and without the insulators in the presence of BEAF, and with the insulators in the absence of BEAF (Figure 5B). Removal of each of the three insulators resulted in darker pigmentation. In the BEAFAB-KO background, the 2scs′ flies had a level of pigmentation similar to that of their “siblings” lacking the insulator. Enhancer blocking by the scs and gypsy insulators was not affected. We conclude that BEAF is required for the function of 2scs′.
While performing this experiment, we noted that the abdominal pigmentation pattern is altered in all flies lacking BEAF. Pigmentation is concentrated in a thin stripe at the posterior edge of the dorsal side of the abdominal segments, except for the two most-posterior segments in males, which are fully pigmented. The pigment spreads in a diffuse manner to encompass around one-third of each segment. In the BEAFAB-KO background, this diffuse spreading extends further to encompass one-half or more of each segment (Figure 5B). We subsequently found that this spreading occurs in male and female flies. The abdominal pigmentation pattern is visible even in the absence of a functional y gene, although the color is yellow-brown instead of gray-black. The diffuse spreading of the pigmentation also occurs in the absence of a functional y gene and is rescued by gBF and GFBF transgenes (data not shown). Therefore this spreading of the pigmentation is not related to the y transgene used in the enhancer-blocking assay, but is related to the lack of BEAF protein. Perhaps it is due to deregulation of some gene upstream of y that is involved in determining the pigmentation pattern.
BEAF mutations perturb male polytene X chromosome morphology:
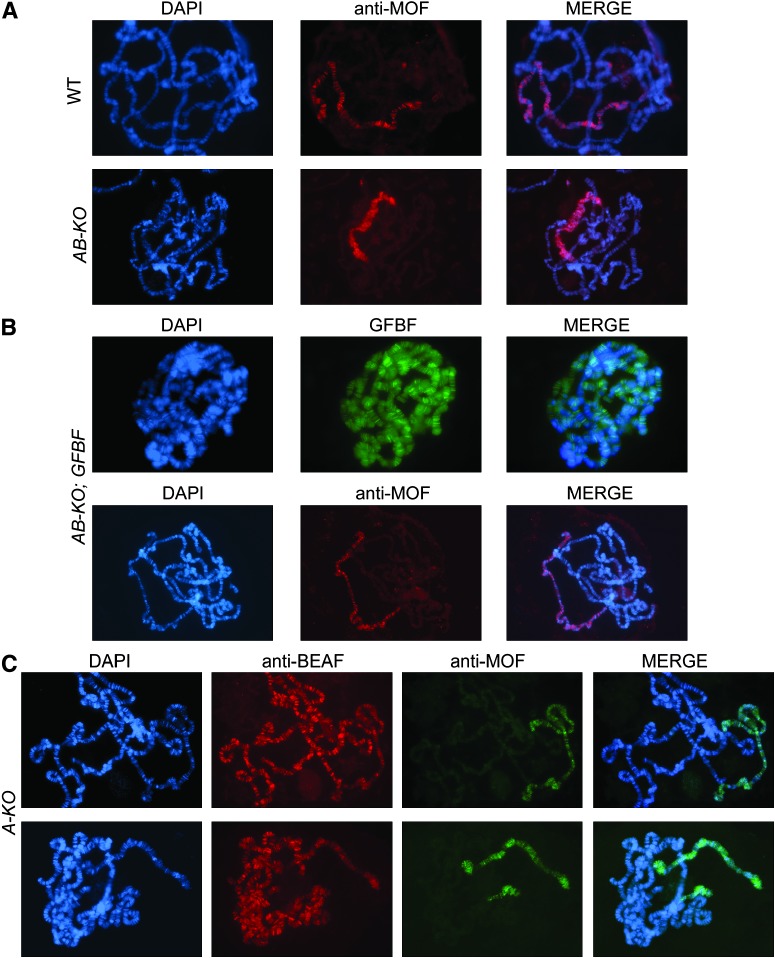
If insulators function by affecting chromatin structure or dynamics, the lack of BEAF could affect chromatin structure. To observe interphase chromatin, polytene chromosome squashes were prepared from salivary glands of third instar larvae. The X chromosome of BEAFAB-KO males from heterozygous mothers exhibited obvious structural defects (Figure 6A). The banding pattern was lost, and the chromosome appeared shorter and puffier. This is less extreme than the results that we previously obtained by producing a dominant-negative form of BEAF in salivary glands, in which the structure of all polytene chromosomes was disrupted in both males and females (Gilbert et al. 2006). Normal X chromosome morphology is restored in the presence of a GFBF transgene, demonstrating that the phenotype is due to a lack of BEAF protein (Figure 6B).
Figure 6.—
BEAF mutations cause a disruption of male X polytene chromosome structure. (A) Salivary gland polytene chromosomes prepared from a wild-type male third instar larvae exhibit a normal banding pattern when the DNA is stained with DAPI. One chromosome arm of polytene chromosomes prepared from a BEAFAB-KO male has lost the banding pattern and appears shorter and broader. Indirect immunostaining with an antibody against MOF shows that it is the X chromosome that appears abnormal. (B) The GFBF transgene rescues the abnormal phenotype of the BEAFAB-KO male polytene X chromosome. (Top) Chromosomes stained with DAPI and gently spread in 50% glycerol without acid treatment to allow direct visualization of green fluorescent BEAF fusion proteins. (Bottom) Chromosomes that have undergone normal fixation, with the X chromosome identified by indirect immunostaining with an antibody against MOF. (C) Polytene chromosomes prepared from BEAFA-KO males show a similar X chromosome phenotype, but it is less extreme and more variable. Note that 32B protein can be detected on these chromosomes by indirect immunofluorescence with an antibody against BEAF. The X chromosome is identified by indirect immunostaining with an antibody against MOF.
Polytene chromosomes were also prepared from larvae from BEAFAB-KO inter se crosses. As for males that had maternal BEAF, the X polytene chromosome from these males showed obvious structural defects but the somatic chromosomes usually appeared normal. There was large variation in polytene chromosome structure from female larvae, ranging from severe disruption of all chromosomes to normal appearance (data not shown). No female larvae survived to become healthy adults, raising the possibility that the variable morphology of their polytene chromosomes represents the variable health of these larvae. Thus the lack of maternal BEAF did not lead to a more severe disruption of polytene chromosome structure except in cases where we believe the health of the larvae was poor.
To positively identify the X chromosome and to determine if the dosage compensation complex (DCC) was affected, we immunostained polytene chromosomes for DCC proteins (Stuckenholz et al. 1999). The DCC associates only with the male X chromosome, where it mediates acetylation of histone H4 on lysine 16. This causes X-linked genes in males to be transcribed at twofold higher rates than in females (Bone et al. 1994; Hamada et al. 2005). Figure 6 shows that association of the DCC protein MOF with the male X chromosome is not affected in the absence of BEAF. The same is true of the DCC proteins MSL-1, MSL-2, MSL-3, and MLE and the dosage-compensation-associated histone modification, acetylation of H4 lysine 16 (data not shown). Therefore, the DCC localizes and functions normally despite the altered morphology of the X chromosome.
We used alleles of the X-linked w gene to further test effects on dosage compensation by examining eye pigmentation. The wa mutation normally shows dosage compensation (males and females have similar eye pigment levels) and the we mutation does not (males have less eye pigment than females) (Lerach et al. 2005). Eye-pigment levels were not affected by the BEAFAB-KO allele, indicating no effect on dosage compensation in this assay (data not shown).
Polytene X chromosomes from BEAFA-KO males also had perturbed morphology (Figure 6C). However, the phenotype was less severe and more variable. The X chromosome morphology ranged from normal or near normal to moderately perturbed. Thus flies lacking 32A protein are not completely normal even though adults have no obvious phenotypes, are healthy, and have normal fertility.
PEV is enhanced in the absence of BEAF:
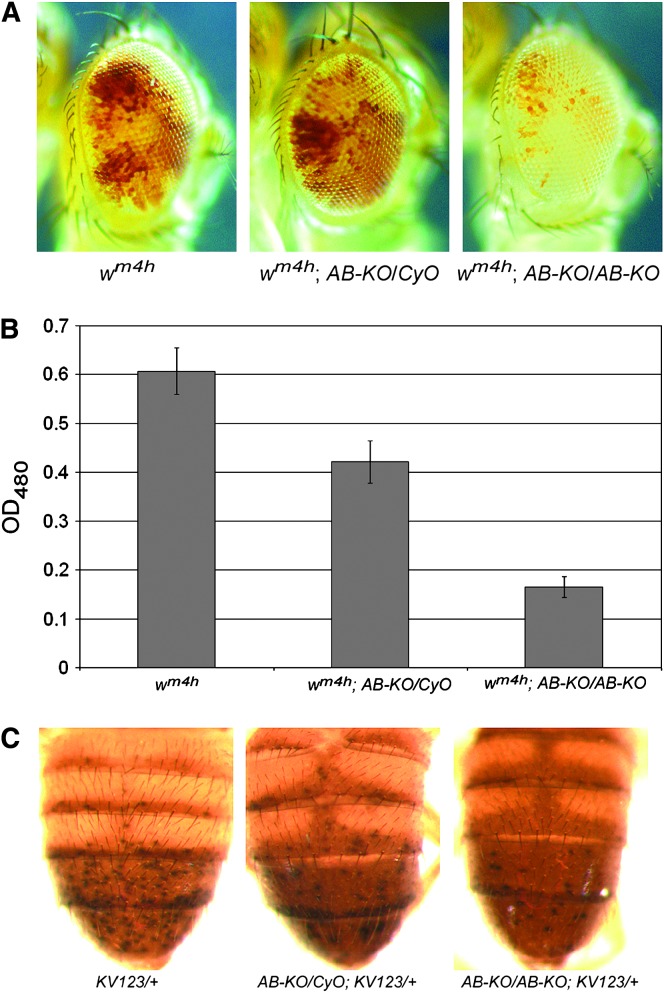
As a second test of the ability of BEAF to affect chromatin organization, we examined the effect of the BEAFAB-KO allele on PEV. The wm4h gene and three different insertions of a transposon carrying a y gene were used. In all four cases, PEV is due to variable spreading of pericentromeric heterochromatin that silences the reporter gene in some cells. The wm4h gene is caused by a chromosomal inversion on the X chromosome (Tartof et al. 1989). The KV732 and KV600 fly lines have the y transgene inserted near the pericentromeric heterochromatin of the X chromosome, while KV123 is near the pericentromeric heterochromatin of chromosome arm 3L (Yan et al. 2002). PEV effects on wm4h are determined by comparing the number of pigmented ommatidia in flies of different genotypes (Figure 7A) or by extracting and quantitating the pigment (Figure 7B). PEV effects on y are determined by comparing the number of darkly pigmented spots on abdomens of flies of different genotypes (Figure 7C). The level of variegation is very sensitive to mutations that directly or indirectly affect chromatin organization.
Figure 7.—
The BEAFAB-KO mutation enhances variegation of wm4h and variegating y transgenes. (A) Males heterozygous for BEAFAB-KO and hemizygous for wm4h show mildly enhanced variegation of wm4h. Males homozygous for BEAFAB-KO and hemizygous for wm4h show a stronger enhancement of wm4h variegation. Eyes of 4- to 5-day-old males are shown. (B) Enhancement of wm4h variegation was quantitated by extracting pigment from male heads of the indicated genotypes and measuring the OD480. (C) Variegation of a y transgene located in the pericentromeric heterochromatin of chromosome arm 3L is enhanced in males heterozygous for BEAFAB-KO and more strongly enhanced in males homozygous for BEAFAB-KO. The y transgene is in the KV123 transposon and is heterozygous. Abdomens of 2- to 3-day-old males are shown. Similar results were obtained with two other variegating y transgenes located on the X chromosome (KV732 and KV600).
The phenotypes of males with one copy of the PEV reporter gene were recorded. In all cases, we found a slight enhancement of PEV in BEAFAB-KO/BEAF males and a stronger enhancement in BEAFAB-KO males (Figure 7 and data not shown). Thus the lack of BEAF allows heterochromatin to spread and to silence the reporter genes in a larger number of cells. This is consistent with our previous results, in which we found that BEAF is a triplo-suppressor of PEV while a dominant-negative form of BEAF is an enhancer of PEV (Gilbert et al. 2006). These PEV assays are consistent with the model that BEAF forms barrier elements, which maintain genes in transcriptionally active states by isolating them from surrounding silent heterochromatin.
DISCUSSION
As a tool for studying BEAF function, we generated the BEAFAB-KO knockout allele by homologous recombination. In the process, we also isolated the BEAFA-KO allele that cannot produce the 32A protein. Flies homozygous for the BEAFA-KO allele are healthy and viable, indicating that the 32B protein is sufficient for normal development. In contrast, flies homozygous for the BEAFAB-KO allele cannot be maintained as a stable line. Maternal BEAF is sufficient to obtain fertile adults, although the resulting BEAFAB-KO flies eclose 1–2 days later than their BEAFAB-KO/CyO siblings and are sickly. Also, although equal numbers of males and females are obtained, the fertility of the BEAFAB-KO females is compromised. Crosses with these females demonstrated that zygotic BEAF is also sufficient to obtain equal numbers of fertile males and females, and fertile males can be obtained even in the absence of BEAF. However, in the absence of maternal BEAF, less than half of the embryos hatch and there is a drastic reduction in the number of adults obtained. The absence of all BEAF results in female lethality by the pharate adult stage or shortly after eclosing. In addition, driving expression of a transgene encoding a dominant-negative form of BEAF by daughterless-GAL4 leads to embryonic lethality (Gilbert et al. 2006). Thus BEAF plays an important role during development, particularly in females, although sickly adults can be obtained that lack BEAF.
The lowered female fertility led us to inspect ovaries from BEAFAB-KO flies. A number of different phenotypes were observed, although ovaries from a given BEAFAB-KO female normally exhibited only one phenotype. We conclude that BEAF plays an important role during oogenesis as well as during development. While the defects in oogenesis could be due to deregulation of genes in the absence of BEAF, it could also be at least partly related to the genetic interaction that we found between BEAF and spindle-E (spn-E) (Roy et al. 2007). The protein encoded by spn-E is a helicase subunit of an RNA interference complex that plays a role in oogenesis (Kennerdell et al. 2002) and heterochromatin formation (Pal-Bhadra et al. 2004). It is of interest to note that a genetic interaction between the RNAi machinery and gypsy insulator function has been reported (Lei and Corces 2006) and that the su(Hw) insulator protein also plays a role in oogenesis (Harrison et al. 1993). In addition, the JIL-1 histone H3 kinase plays a role in modulating chromatin structure and is essential at all stages of development as well as for oogenesis (Zhang et al. 2003).
The scs′ insulator was originally identified because it forms a special chromatin structure that appeared to localize to one end of the heat-shock puff at 87A of polytene chromosomes (Udvardy et al. 1985). It was subsequently shown to function as an insulator in the first transgenic enhancer-blocking and position-independent expression assays to be done (Kellum and Schedl 1991, 1992). This led to the identification of the BEAF proteins as scs′-binding proteins (Zhao et al. 1995; Hart et al. 1997). The importance of the BEAF-binding sites in scs′ for insulator activity has been shown using both cultured cells (Zhao et al. 1995) and transgenic flies (Cuvier et al. 1998), and additional genomic BEAF-binding sites were shown to have insulator activity (Cuvier et al. 1998). However, it is possible that some other protein binds to these sites in vivo to confer insulator activity. It was also shown that a dominant-negative form of BEAF interferes with scs′ insulator activity (Gilbert et al. 2006), although this protein might affect proteins in addition to BEAF. Here we show that BEAF is required for the insulator activity of scs′. Using both a position-independent expression assay and an enhancer-blocking assay, we found that scs′ loses insulator activity in the absence of BEAF protein. In the enhancer-blocking assay, we also tested the scs and gypsy insulators, which lack BEAF-binding sites, and found that these insulators work in the absence of BEAF.
The altered appearance of the X polytene chromosome in BEAFAB-KO male mutant larvae provides dramatic evidence for a role for BEAF in chromatin organization. This is further supported by the PEV assays, which indicate that BEAF helps to limit heterochromatin spreading. Mutations in genes encoding other chromatin proteins have a similar effect on the male X chromosome. This includes ISWI, which is the catalytic subunit of multiple chromatin-remodeling complexes, including the nucleosome remodeling factor (NURF) (Deuring et al. 2000); the NURF301 subunit of NURF (Badenhorst et al. 2002); and the heterochromatin proteins Su(var)3–7 and HP1 (Spierer et al. 2005). This supports models in which insulators function by affecting chromatin structure or dynamics.
It is curious that only the male X chromosome is affected, whereas global structural alterations are observed in all chromosomes of males and females when a dominant-negative form of BEAF is produced in larval salivary glands (Gilbert et al. 2006). It is likely that the chromatin organization of the male X chromosome is especially susceptible to disruption due to some feature associated with dosage compensation. A candidate for such a feature is the hyperacetylation of lysine 16 of histone H4 (Bone et al. 1994), which interferes with formation of 30-nm chromatin fibers (Shogren-Knaak et al. 2006). Evidence that the male X chromosome is more sensitive to disruption is derived from mutations in the histone H3 kinase, JIL-1. When polytene chromosomes were observed using an allelic series of JIL-1 mutations, weak mutations were found to affect mainly the male X chromosome and stronger mutations to affect all chromosomes of both males and females (Wang et al. 2001). Also, BEAFA-KO animals are healthier than BEAFAB-KO animals and we observed a weaker effect on the male X chromosome in BEAFA-KO animals. This suggests that the dominant negative has a stronger effect than the lack of BEAF. This is consistent with the lethal effect of producing the dominant-negative protein in embryos, whereas homozygous BEAFAB-KO adults are obtained. We assume that the dominant negative has a stronger effect because it actively interferes with BEAF activity, while the gradual disappearance of maternal BEAF mitigates the effect of the knockout. Perhaps the dominant-negative protein also interferes with the function of proteins in addition to BEAF. If so, it is likely that these proteins normally interact with BEAF since the phenotypes caused by the dominant negative and by BEAFAB-KO are similar and can be rescued by BEAF transgenes. The future identification of any such proteins should provide insight into how BEAF functions.
We have shown here that the BEAF proteins have insulator activity. BEAF binds to hundreds of sites on polytene chromosomes (Zhao et al. 1995), and other genomic binding sites have insulator activity (Cuvier et al. 1998, 2002). Yet 32A is not essential, adults can be obtained with only maternal BEAF, some embryos hatch with only zygotic BEAF, and a small number of fertile males are obtained in the absence of all BEAF. This is somewhat reminiscent of mutations in the su(Hw) insulator protein, which lead to female sterility but otherwise are not lethal (Harrison et al. 1993). BEAF is normally present at all life stages (for example, see the Western blot of adults in Figure 2F). Using several of our GFBF transgenic fly lines in the BEAFAB-KO background, in which the transgene is insulated and driven by 900 bp of BEAF promoter sequences, we observe green fluorescent BEAF in all nuclei of all tissues at all life stages that we have looked at (data not shown). If BEAF is normally ubiquitous and contributes to gene regulation by forming boundaries between hundreds of domains, why are the effects of a lack of BEAF so limited? The answer is not known at present. One possibility is that the misregulation of genes caused by malfunctioning insulators is minor enough that fitness is reduced without being immediately lethal. Another possibility that we find particularly intriguing is that there could be some type of epigenetic memory mechanism, similar to what has been proposed for Polycomb group proteins (Sarge and Park-Sarge 2005; Bantignies and Cavalli 2006). This epigenetic memory has been shown to be meiotically inheritable (Cavalli and Paro 1998). Loss of this “epigenetic memory” could be stochastic, resulting in deregulation of different genes in different individuals or clonal populations of cells. This could result in the variable timing of death in the absence of BEAF and in the single phenotype observed per ovary but different phenotypes in different ovaries. The knockout mutations described here will be useful tools in future studies aimed at discovering proteins that interact with BEAF and for investigating the role of BEAF in gene regulation and chromatin organization. This will ultimately lead to an understanding of the molecular mechanisms used in insulator function.
Acknowledgments
We thank Nan Jiang for comments on the manuscript; Yian Yee Tan for technical assistance and maintaining fly stocks; and members of the Socolofsky Microscopy Center for assistance with confocal microscopy (David Burk and Matt Brown) and SEM (Cindy Henk and Ying Xiao). This work was supported by grants from the Louisiana Board of Regents [LEQSF(2002-04)-RD-A-06 and LEQSF(2004-05)-RD-A-11] and a Louisiana State University Faculty Research Grant.
References
- Ashburner, M., 1989. Drosophia: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Badenhorst, P., M. Voas, I. Rebay and C. Wu, 2002. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 16 3186–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies, F., and G. Cavalli, 2006. Cellular memory and dynamic regulation of polycomb group proteins. Curr. Opin. Cell Biol. 18 275–283. [DOI] [PubMed] [Google Scholar]
- Bell, A. C., and G. Felsenfeld, 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405 482–485. [DOI] [PubMed] [Google Scholar]
- Bone, J. R., J. Lavender, R. Richman, M. J. Palmer, B. M. Turner et al., 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev. 8 96–104. [DOI] [PubMed] [Google Scholar]
- Cavalli, G., and R. Paro, 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93 505–518. [DOI] [PubMed] [Google Scholar]
- Cuvier, O., C. M. Hart and U. K. Laemmli, 1998. Identification of a class of chromatin boundary elements. Mol. Cell. Biol. 18 7478–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuvier, O., C. M. Hart, E. Kas and U. K. Laemmli, 2002. Identification of a multicopy chromatin boundary element at the borders of silenced chromosomal domains. Chromosoma 110 519–531. [DOI] [PubMed] [Google Scholar]
- Deuring, R., L. Fanti, J. A. Armstrong, M. Sarte, O. Papoulas et al., 2000. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol. Cell 5 355–365. [DOI] [PubMed] [Google Scholar]
- Frevel, M. A., S. J. Sowerby, G. B. Petersen and A. E. Reeve, 1999. Methylation sequencing analysis refines the region of H19 epimutation in Wilms tumor. J. Biol. Chem. 274 29331–29340. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., and G. Felsenfeld, 2006. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7 703–713. [DOI] [PubMed] [Google Scholar]
- Gaszner, M., J. Vazquez and P. Schedl, 1999. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer-promoter interaction. Genes Dev. 13 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, M. K., Y. Y. Tan and C. M. Hart, 2006. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics 173 1365–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada, F. N., P. J. Park, P. R. Gordadze and M. I. Kuroda, 2005. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 19 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse et al., 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405 486–489. [DOI] [PubMed] [Google Scholar]
- Harrison, D. A., P. K. Geyer, C. Spana and V. G. Corces, 1989. The gypsy retrotransposon of Drosophila melanogaster: mechanisms of mutagenesis and interaction with the suppressor of Hairy-wing locus. Dev. Genet. 10 239–248. [DOI] [PubMed] [Google Scholar]
- Harrison, D. A., D. A. Gdula, R. S. Coyne and V. G. Corces, 1993. A leucine zipper domain of the suppressor of Hairy-wing protein mediates its repressive effect on enhancer function. Genes Dev. 7 1966–1978. [DOI] [PubMed] [Google Scholar]
- Hart, C. M., K. Zhao and U. K. Laemmli, 1997. The scs' boundary element: characterization of boundary element-associated factors. Mol. Cell. Biol. 17 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga, I., J. Mihaly, S. Barges and F. Karch, 2001. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 8 1145–1151. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64 941–950. [DOI] [PubMed] [Google Scholar]
- Kellum, R., and P. Schedl, 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12 2424–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J. R., S. Yamaguchi and R. W. Carthew, 2002. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 16 1884–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermekchiev, M., M. Pettersson, P. Matthias and W. Schaffner, 1991. Every enhancer works with every promoter for all the combinations tested: Could new regulatory pathways evolve by enhancer shuffling? Gene Expr. 1 71–81. [PMC free article] [PubMed] [Google Scholar]
- Kuhn, E. J., and P. K. Geyer, 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15 259–265. [DOI] [PubMed] [Google Scholar]
- Kuhn, E. J., C. M. Hart and P. K. Geyer, 2004. Studies of the role of the Drosophila scs and scs' insulators in defining boundaries of a chromosome puff. Mol. Cell. Biol. 24 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador, M., and V. G. Corces, 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111 151–154. [DOI] [PubMed] [Google Scholar]
- Lei, E. P., and V. G. Corces, 2006. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat. Genet. 38 936–941. [DOI] [PubMed] [Google Scholar]
- Lerach, S., W. Zhang, H. Deng, X. Bao, J. Girton et al., 2005. JIL-1 kinase, a member of the male-specific lethal (MSL) complex, is necessary for proper dosage compensation of eye pigmentation in Drosophila. Genesis 43 213–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald, A. P., and M. P. Kambysellis, 1980. Oogenesis, pp. 141–209 in The Genetics and Biology of Drosophila, edited by M. Ashburner and T. R. F. Wright. Academic Press, New York.
- Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics and F. Karch, 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124 1809–1820. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303 669–672. [DOI] [PubMed] [Google Scholar]
- Parnell, T. J., and P. K. Geyer, 2000. Differences in insulator properties revealed by enhancer blocking assays on episomes. EMBO J. 19 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa, F., M. J. Pikaart, B. Burgess-Beusse, A. C. Bell, M. D. Litt et al., 2002. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA 99 6883–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik, W., K. W. Brown, H. Schneid, Y. Le Bouc, W. Bickmore et al., 1995. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2–H19 domain. Hum. Mol. Genet. 4 2379–2385. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2000. Gene targeting by homologous recombination in Drosophila. Science 288 2013–2018. [DOI] [PubMed] [Google Scholar]
- Rong, Y. S., and K. G. Golic, 2001. A targeted gene knockout in Drosophila. Genetics 157 1307–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong, Y. S., S. W. Titen, H. B. Xie, M. M. Golic, M. Bastiani et al., 2002. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 16 1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Y. Y. Tan and C. M. Hart, 2007. A genetic screen supports a broad role for the Drosophila insulator proteins BEAF-32A and BEAF-32B in maintaining patterns of gene expression. Mol. Genet. Genomics 277 273–286. [DOI] [PubMed] [Google Scholar]
- Sarge, K. D., and O. K. Park-Sarge, 2005. Gene bookmarking: keeping the pages open. Trends Biochem. Sci. 30 605–610. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie et al., 2006. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311 844–847. [DOI] [PubMed] [Google Scholar]
- Spierer, A., C. Seum, M. Delattre and P. Spierer, 2005. Loss of the modifiers of variegation Su(var)3–7 or HP1 impacts male X polytene chromosome morphology and dosage compensation. J. Cell Sci. 118 5047–5057. [DOI] [PubMed] [Google Scholar]
- Spradling, A. C., 1986. P-element-mediated transformation, pp. 175–197 in Drosophila: A Practical Approach, edited by D. B. Roberts. IRL Press, Oxford.
- Stuckenholz, C., Y. Kageyama and M. I. Kuroda, 1999. Guilt by association: non-coding RNAs, chromosome-specific proteins and dosage compensation in Drosophila. Trends Genet. 15 454–458. [DOI] [PubMed] [Google Scholar]
- Tartof, K. D., C. Bishop, M. Jones, C. A. Hobbs and J. Locke, 1989. Towards an understanding of position effect variegation. Dev. Genet. 10 162–176. [DOI] [PubMed] [Google Scholar]
- Udvardy, A., E. Maine and P. Schedl, 1985. The 87A7 chromomere: identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J. Mol. Biol. 185 341–358. [DOI] [PubMed] [Google Scholar]
- Wang, Y., W. Zhang, Y. Jin, J. Johansen and K. M. Johansen, 2001. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105 433–443. [DOI] [PubMed] [Google Scholar]
- Yan, C. M., K. W. Dobie, H. D. Le, A. Y. Konev and G. H. Karpen, 2002. Efficient recovery of centric heterochromatin P-element insertions in Drosophila melanogaster. Genetics 161 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Y. Jin, Y. Ji, J. Girton, J. Johansen et al., 2003. Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics 165 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, K., C. M. Hart and U. K. Laemmli, 1995. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell 81 879–889. [DOI] [PubMed] [Google Scholar]