Abstract
Massive accumulation of retrotransposons, comprising >40% of human and mouse genomes, is one of the major events in the evolution of the genome. However, most retrotransposons have lost retrotransposition competency, which makes studying their role in genome evolution elusive. Intracisternal A-particle (IAP) elements are long terminal repeat (LTR)-type mouse retrotransposons consisting of full-length and internally deleted types. Some are retrotransposition competent and their upregulated activity has been reported in mutant mice deficient in genome defense systems, suggesting that IAP elements provide a unique platform for studying the interaction between retrotransposons and mammalian genomes. Using the IAP element as a model case, here we show that mobilization of retrotransposons alters the mouse transcriptome. Retrotransposition assay in cultured cells demonstrated that a subset of internally deleted IAP elements, called IΔ1 type, retrotranspose efficiently when supplied with functional IAP proteins. Furthermore, the IΔ1 type IAP element exhibited substantial transcription-inducing activity in the flanking region. Genomewide transcript analysis of embryonic stem (ES) cells identified IAP-induced transcripts, including fusion transcripts between IAP sequence and endogenous genes. Unexpectedly, nearly half of these IAP elements obtained from ES cells derived from 129 mouse strain were absent in the C57BL/6 genome, suggesting that IAP-driven transcription contributes to the unique trait of the individual mouse strain. On the basis of these data, we propose that retrotransposons are one of the drivers that shape the mammalian transcriptome.
RECENT advances in large-scale genome sequencing and genomewide transcriptome analyses have revealed the vast landscape of coding regions within the genome. The next challenge in the postgenomic era is to elucidate the roles of noncoding regions that occupy most of the genome (Waterston et al. 2002). Retrotransposons are major components of noncoding regions, comprising ∼40% of the genome in human and mouse. Retrotransposons are mobile elements that propagate through an RNA intermediate and reverse transcription. Although retrotransposons have been considered as “junk” elements, recent studies suggest that they contribute to genome evolution (Kazazian 2004; Maksakova et al. 2006). Therefore, studying the interactions between retrotransposons and the genome should provide clues for the role of the noncoding regions. However, most retrotransposons have accumulated mutations and are evolutionarily inactivated. For this reason, evaluating the direct effects of retrotranspositions on the genome has been difficult. Isolation of retrotransposition-competent retrotransposons and the establishment of an in vitro assay system for retrotransposon activity will be helpful in studying the role of retrotransposons on genome evolution.
The intracisternal A-particle (IAP) element is a long terminal repeat (LTR)-type mouse retrotransposon containing gag, pro, and pol genes (Kuff and Lueders 1988). There are ∼1000 copies of IAP elements in the mouse genome. IAP elements are classified into either full-length or internally deleted types that lack some regions of the gag, pro, and pol genes. We consider the IAP element as an appropriate retrotransposon for studying the interaction between retrotransposons and the host genome for the following reasons: First, some populations of IAP elements are retrotransposition competent (Maksakova et al. 2006). A recent in silico search of the mouse genome and subsequent identification of retrotransposition-competent IAP elements (Dewannieux et al. 2004) indicate that the mouse genome may contain hundreds of autonomously active IAP elements. Second, IAP retrotransposition not only inactivates inserted genes but also occasionally activates adjacent genes (Maksakova et al. 2006). Third, IAP expression is upregulated in mutant mice deficient in host genome defense systems such as DNA methylation (Walsh et al. 1998) and RNA interference (Kanellopoulou et al. 2005).
We have previously reported a high frequency of IAP retrotransposition in radiation-induced mouse acute myeloid leukemia (AML) in C3H/He mice (Ishihara and Tanaka 1997) and identified eight IAP elements that retrotransposed during AML formation (Ishihara et al. 2004). We hypothesized that these retrotransposition-competent IAP elements could be useful tools for studying the direct effect of retrotransposition on the genome. In this report, we isolated retrotransposition-competent IAP retrotransposons from the AML cells and used them as a model for analyzing their interactions with the genome. IAP-induced transcription of flanking regions, including fusion transcripts with endogenous genes, was observed genomewide with not only autonomous but also nonautonomous IAP elements. Surprisingly, nearly half of these IAP elements were strain specific, suggesting their contribution to the unique trait of individual strains. These results demonstrate active roles of retrotransposons in shaping the mammalian transcriptome.
MATERIALS AND METHODS
Plasmid construction:
The construction process is illustrated in supplemental Figure S1 at http://www.genetics.org/supplemental/. Oligonucleotide sequences are listed in supplemental Table S1 at http://www.genetics.org/supplemental/. The construction process is as follows:
Cloning of a full-length-type IAP element from the AML genome (supplemental Figure S1A): To construct the full-length-type IAP vectors, the IAP element at the Q14 locus of AML line L8065 (Ishihara et al. 2004) was PCR amplified with primers Q14-U-NotI and Q14-P2-SpeI, using the Expand High-Fidelity PCR amplification system (Roche Applied Biosciences) containing Taq DNA polymerase and proofreading Pwo DNA polymerase. These primers correspond to the 5′- and 3′-flanking genomic regions of the Q14-IAP element. Since the wild-type allele is lost at this locus, only the IAP-inserted allele was amplified. The NotI–SpeI-digested PCR fragment was cloned into the NotI–SpeI site of pBluescriptII KS+ (Stratagene, La Jolla, CA), resulting in pQ14.
Insertion of the neo-indicator into the IAP element (supplemental Figure S1B): To isolate the neo-indicator cassette, the 2.2-kb blunt-ended AccI–ApaLI fragment was excised from pJM101/L1.3 (Moran et al. 1996) and cloned into the EcoRV site of pBluescriptII. From the two orientations of the insert, the one containing SalI and NdeI sites at opposite ends of the neo cassette was selected and named pBSmneo-F. The 2.3-kb SalI (blunt-ended)–NotI neo cassette was isolated from pBSmneo-F and ligated with a 6.7-kb NdeI (blunt-ended)–NotI fragment of pQ14, resulting in pBS-mneo-F-3′-IAP. The 3.9-kb NdeI–NotI fragment was isolated from pQ14 and inserted into the NdeI–NotI site of pBS-mneo-F-3′-IAP, resulting in pQ14mneo.
Replacement of the U3 region of the 5′ LTR with a human cytomegalovirus enhancer (supplemental Figure S1C): To replace the U3 region of the 5′ LTR with a human cytomegalovirus enhancer and chicken β-actin promoter (CAG promoter), the CAG promoter sequence was PCR amplified from pCX-EGFP (Okabe et al. 1997) with primers CA-U1 and CA-L2, using proofreading Pfx DNA polymerase (Invitrogen, San Diego). The R-U5 region was also amplified from the pQ14 LTR region with primers R-U4 and R-L1. Each PCR product was mixed and used as a template for fusion PCR with primers CA-U1 and R-L1. The PCR product was digested with NotI and BspEI and used to replace the NotI–BspEI region of pQ14mneo, resulting in pQ14mneoCAR2.
Introduction of PmeI and PacI sites into plasmid backbone (supplemental Figure S1D): pQ14mneoCAR2 was digested with EcoRV and the 6.3-kb fragment was self-ligated, resulting in pQ14CAR2dRV. The PmeI–PacI linker was generated by annealing PP-U and PP-L and cloned into the blunt-ended SalI site of pQ14CAR2dRV, resulting in pQ14CAR2dRV-PP.
Generation of the GFP-indicator cassette (supplemental Figure S1E): To generate the GFP-indicator cassette containing an intron sequence, human γ-globin intron sequence was PCR amplified from pJM101/L1.3 with primers IVS-SapI-U1 and IVS-SapI-L1, using Pfx polymerase. The PCR product was digested with SapI and cloned into the Bpu10I site of the hrGFP gene (Stratagene) of pCAGhrGFP-PGKneo-dSp [containing the hrGFP gene under the control of the CAG promoter (Niwa et al. 1991) followed by rabbit β-globin polyadenylation signal, our unpublished data]. From the two orientations of the intron insertion, the one containing the intron in the opposite orientation relative to the GFP gene was selected and named pCAGhrGFPint. The 2.2-kb SphI (blunt-ended)–NcoI fragment of the pCAGhrGFPint (containing 5′-deleted-hrGFP, human γ-globin intron, and rabbit β-globin polyadenylation signal) was inserted into the BamHI (blunt-ended)–NcoI site of pEF321-FLAG-EX (expression vector using human EF1α promoter; gift from S. Nagata, Osaka University), resulting in pEF-hrGFPint-DEL. The 96-bp NcoI fragment of pCAGhrGFPint was inserted into the NcoI site of pEF-hrGFPint-DEL to restore the partially deleted region of the hrGFP gene, resulting in pEF-hrGFPint.
Insertion of the GFP indicator into the IAP element (supplemental Figure S1F): pEF-hrGFPint was digested with HindIII and blunt-ended with Klenow, and the 4.6-kb fragment (containing the EF1α promoter, hrGFP-intron cassette, and rabbit β-globin polyadenylation signal) was inserted into the Klenow-treated NdeI site of pQ14 between the pol region and 3′ LTR, resulting in pQ14hrGFPint. The 9.3-kb EcoRV fragment of pQ14hrGFPint (containing the hrGFP-intron cassette and flanking IAP sequence) was inserted into the EcoRV site of pQ14CAR2dRV-PP, resulting in pFL.
Generation of the IAP-protein expression vector (supplemental Figure S1G): To generate the expression vector of full-length IAP proteins, the ScaI–BspEI region of pQ14 containing the wild-type 5′ LTR was replaced by the 2-kb ScaI–BspEI fragment of pFL bearing the CAG promoter instead of the 5′ LTR-U3 region, resulting in pQ14CAG.
Construction of IΔ1-type IAP vectors (supplemental Figure S1H): To construct IΔ1-type IAP vectors using endogenous IAP elements, IAP sequences were PCR amplified from the Q10 and Q11 loci of AML lines L9207 and L8002 (Ishihara et al. 2004) with primers Q10/11-IF2 and Q10-FR1-Pac, and Q10/11-IF2 and Q11-FR2-Pac, respectively. Q10/11-IF2 is an IAP-specific primer; Q10-FR1-Pac and Q11-FR2-Pac correspond to 3′-flanking genomic regions of Q10 and Q11 IAP elements. Each PCR product was digested with BspEI and PshAI and used to replace the BspEI–PshAI region of the pFL containing the gag-pro-pol regions, resulting in pDE1 and pDE2, respectively.
Construction of artificially generated IΔ1-type IAP vector (supplemental Figure S1I): To construct the IΔ1-type IAP vector from full-length-type IAP vector, a 1.9-kb internal deletion was introduced by fusion PCR. The 5′ and 3′ regions of the IΔ1-type IAP element were PCR amplified from pFL with primers Q14seq2 and Q14-Id1-Jnc-R, and Q14-Id1-Jnc-F and Q14rev13, respectively. A mixture of the PCR products was subjected to fusion PCR using Q14seq2 and Q14rev13. The PCR product was digested with Bst1107I and SfiI and used to replace the Bst1107I–SfiI region of the pFL, resulting in pDA.
Construction of IAP vectors flanked by the GFP reporter for Cre-mediated site-specific integration (supplemental Figure S1J): To construct IAP vectors for Cre-mediated site-specific integration, the EGFP fragment was isolated from EcoRI-digested blunt-ended pCX-EGFP (Okabe et al. 1997) and cloned into the blunt-ended XbaI site of the pUHG10-3 containing tetracycline operator and human cytomegalovirus minimal promoter cassette (Gossen and Bujard 1992) upstream of the XbaI site and rabbit β-globin polyadenylation signal downstream of the XbaI site, resulting in pUHG-EGFP. The 9.6-kb SpeI (blunt-ended)–NotI fragment of pQ14 containing the full-length IAP fragment was inserted into the BamHI (blunt-ended)–NotI site of pL1-NotI-1L (our unpublished data), which contains the inverted lox511 sequence at each end of the NotI–BamHI site, resulting in pL1-Q14F-1L. The 3.6-kb BspEI–PshAI fragment of pDA (containing the deleted site of the IΔ1-type IAP element) was used to replace the BspEI–PshAI region of the pL1-Q14F-1L, resulting in pL1-Q14Id1-1L. A NotI linker (Takara, Otsu, Japan) was ligated to each end of the 4.8-kb TfiI (blunt-ended)–StuI fragment of pUHG-EGFP (containing a cassette of minimal promoter, EGFP, and polyadenylation signal) and inserted into the NotI site (blunt-ended) of pL1-Q14F-1L and pL1-Q14Id1-1L, resulting in pL1-Q14F-GR-1L and pL1-Q14Id1-GR-1L, respectively. The same linker-ligated fragment was ligated to the 2.7-kb EagI fragment of pL1-Q14Id1-GR-1L consisting of the plasmid backbone and two lox511 sites, resulting in pL1-GR-1L. pL1-Q14F-GR-1L, pL1-Q14Id1-GR-1L, and pL1-GR-1L were used for Cre-mediated site-specific integration of full-length-type IAP-GFP reporter, IΔ1-type IAP-GFP reporter, and GFP reporter alone, respectively.
In all cloning steps that utilized PCR, the PCR-amplified regions were sequenced and plasmids without mutation were selected.
Retrotransposition assay:
HeLa cells were plated onto 12-well plates at a density of 1 × 105/well and transfected with 0.8 μg of IAP vectors bearing the GFP-intron cassette together with 0.8 μg of pBluescriptII (Stratagene) or the aforementioned pQ14CAG, which expresses functional IAP proteins, using LipofectAMINE2000 (Invitrogen). Seven days after transfection, the percentage of GFP-positive cells was quantified by flow cytometry.
Cre-mediated site-specific integration:
Embryonic stem (ES) cells bearing the fusion gene of hygromycin phosphotransferase and herpes simplex virus thymidine kinase (HYTK) flanked by inverted lox511 sites (Feng et al. 1999) were transfected with 2 μg of plasmids containing the IAP-GFP reporter (pL1-Q14F-GR-1L, pL1-Q14Id1-GR-1L) or the GFP reporter alone (pL1-GR-1L) flanked by inverted lox511 sites together with 0.5 μg of pMC-Cre (Gu et al. 1993) using TransFast (Promega), followed by selection with 3 μm of gancyclovir (GANC). To determine the orientation of the integrated plasmid sequence relative to the target locus, we conducted PCR analysis using primers, one of which is inside the integrated plasmids and the other is in the flanking genomic region. For the introduction of pL1-Q14F-GR-1L and pL1-Q14Id1-GR-1L, we used primers Q14RMCE-1 and C4-5′ for the screening of type I orientation integration and primers Q14RMCE-1 and C4-3′ for type II orientation. For the introduction of pL1-GR-1L, we used primers gloRMCE-2 and C4-5′ for the screening of type I orientation and primers gloRMCE-2 and C4-3′ for type II orientation. Primer sequences are listed in supplemental Table S1 at http://www.genetics.org/supplemental/. The lox511-flanked HYTK gene of the parental ES clone was 3.1 kb, and the lox511-flanked sequences of the three plasmids transfected into the ES cells were 2.1, 7.8, and 9.7 kb, respectively. We did not observe any significant difference in recombination efficiency between the three transfected plasmids (data not shown). Recombinants were further analyzed by Southern blotting using standard procedures.
IAP-induced transcript display, determination of transcription start sites, and detection of fusion transcripts between IAP elements and endogenous genes:
To amplify the IAP-induced transcript by PCR, total RNA from one of the ES cell clones containing the IΔ1 IAP element flanked by the GFP reporter gene was reverse transcribed using SuperScriptII (Stratagene) with random primer (Promega) or oligo(dT) primer (Promega), followed by the second-strand synthesis with IAP–LTR-specific primer TDIAP-F5. The cDNA was digested with a 4-base cutter enzyme (MboI, RsaI, AluI, or HaeIII), ligated with a linker DNA compatible with each restriction site, and amplified by nested PCR. Primers for the first PCR reaction were IAP–LTR-specific primer TDIAP-F2 and linker-specific primer Spl-P1. Primers for the second PCR reaction were IAP-specific primer TDIAP-F3 and linker-specific primers that extend into each restriction site followed by different combinations of dinucleotide at their 3′ end. Sixteen independent linker-specific primers could be designed from the 3′-end dinucleotide combinations. Since specific dinucleotide sequence was used in each PCR, PCR amplification of a mixture of IAP-induced transcripts was divided into 16 groups, thereby improving the resolution of PCR product separation in electrophoresis. The IAP-specific second PCR primer TDIAP-F3 was labeled with either FAM or ROX to visualize PCR products. PCR products were fractionated by nondenaturing polyacrylamide gel electrophoresis (PAGE) and detected using an FMBIOII laser scanner (Hitachi, Tokyo). Bands with discrete signals were excised from the gel and cloned by a TA cloning kit (Promega), and sequences of 49 bands were determined. Genomic location was determined by a BLAT search tool using the University of California at Santa Cruz (UCSC) mouse genome browser version mm5 (Karolchik et al. 2003). Since the UCSC mouse genome database is constructed on the C57BL/6J strain genomic sequence, nonmatching sequences by BLAT searches were presumed to be 129 strain-specific IAP elements. In such cases, we PCR amplified the postulated IAP regions from 129 mouse genomic DNA and confirmed the presence of these IAP elements by sequencing. We also used C57BL/6J genomic DNA as a template and verified that 129 strain-specific IAP elements were not amplified. The sequences of linkers and primers for IAP-induced transcript display and amplification of 129 strain-specific IAP elements are shown in supplemental Tables S2 and S3, respectively, at http://www.genetics.org/supplemental/.
The transcription start sites of the above-mentioned transcripts were determined from total RNA of ES cells by 5′-rapid amplification of cDNA ends (5′RACE), using the FirstChoice RLM-RACE kit (Ambion, Austin, TX) according to the protocol provided by the manufacturer. In brief, total RNA was treated with calf intestinal alkaline phosphatase (CIP) to remove 5′-end phosphate from all RNA except capped mRNA (e.g., rRNA, tRNA, and degraded mRNA). The RNA was further treated with tobacco acid phosphatase (TAP) to remove the cap structure and leave a monophosphate at the 5′ end. Thus, only previously capped full-length RNA could be ligated by an adaptor RNA. Following reverse transcription, the full-length cDNA was amplified by nested PCR and the nucleotide sequence was determined. The nucleotide immediately downstream of the adaptor sequence was presumed to be the transcription start site. PCRs of TAP-untreated samples will in principle demonstrate no amplification and were used as negative controls. Primers were designed on the basis of the sequence determined in the aforementioned PAGE analysis and are listed in supplemental Table S4 at http://www.genetics.org/supplemental/. Due to the presence of repetitive sequences in the flanking region of the IAP elements, primers could not be designed for a subset of the loci.
To detect the fusion transcript between IAP elements and endogenous genes, total RNA of ES cells was reverse transcribed using SuperScriptII with a gene-specific primer and amplified by PCR with IAP- and gene-specific primers shown in supplemental Table S5 at http://www.genetics.org/supplemental/. Northern blot analysis was conducted according to standard procedure using 13 μg of total RNA.
Flow cytometry:
GFP-positive cells were detected using FACScan (Becton Dickinson, San Jose, CA) from a propidium iodine-negative population and analyzed using CellQuest (Becton Dickinson).
RESULTS
Isolation of retrotransposition-competent IAP elements from AML cells and comparison between full-length and IΔ1 types:
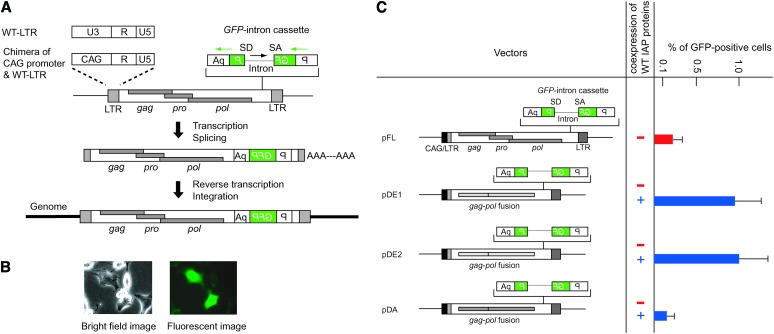
From eight IAP elements retrotransposed in AML cells, one was full-length type, while the remaining were IΔ1-type IAP elements containing a 1.9-kb internal deletion between the gag and the pol region (Ishihara et al. 2004). Although the full-length type constitutes 70% of the entire IAP population in the mouse genome (Kuff and Lueders 1988), mostly IΔ1 type were found as a causative insertion of spontaneous mutant mice (Maksakova et al. 2006), consistent with the predominance of IΔ1 type in our analysis of AML cells. The full-length IAP element, designated as Q14 (Ishihara and Tanaka 1997), retained the open reading frames of gag, pro, and pol genes, suggesting that it is an autonomous IAP element. We therefore isolated the Q14 IAP element from the AML genome and inserted an indicator GFP cassette that contains an intron in the antisense orientation relative to the GFP gene (Ostertag et al. 2000) (Figure 1A). Successful retrotransposition should splice out the intron and generate a functional GFP gene after integration into the genome (Figure 1A). To increase IAP element transcription, we replaced the 5′ LTR-U3 region with a chimeric human cytomegalovirus enhancer and chicken β-actin promoter (CAG promoter) (Figure 1A, pFL in Figure 1C). To avoid any unwanted sequence at the 5′ end of the IAP genomic RNA, the transcription start site of the CAG promoter was joined precisely to the 5′ LTR-R region (supplemental Figure S2A at http://www.genetics.org/supplemental/). Retrotransposition was detected using the CAG/LTR chimeric promoter in HeLa cells that do not contain endogenous IAP elements (Figure 1B), indicating that Q14 is an autonomous IAP element. Replacement of the 5′ LTR-U3 region with the CAG promoter was essential for retrotransposition detection as observed in a separate experiment using an indicator neo-intron cassette (supplemental Figure S2B at http://www.genetics.org/supplemental/). We further cloned two independent IΔ1-type IAP elements [previously reported as Q10 and Q11 (Ishihara et al. 2004)] from AML genomes and inserted their gag–pol region in place of the corresponding full-length Q14-IAP vector region (pDE1 and pDE2, Figure 1C). Retrotransposition was not detected by transfection of IΔ1 IAP elements alone, but coexpression with wild-type IAP proteins resulted in 5-fold higher retrotransposition efficiency compared with the full-length IAP element (Figure 1C). Coexpression of wild-type IAP proteins with the full-length IAP vector was conducted in a separate experiment and GFP-positive cells increased by only 1.7-fold (data not shown), indicating that the expression level of IAP proteins is not the determinant for different retrotransposition activity between the full-length and IΔ1 types. These results are consistent with the high incidence of IΔ1-type retrotransposition in AML cells (Ishihara et al. 2004). Since sequence variation in the gag–pol region may affect the retrotransposition efficiency, we artificially generated an IΔ1-type IAP element from the full-length-type Q14-IAP element by deleting a 1.9-kb sequence between the gag and pol region (pDA, Figure 1C). Retrotransposition was trans-complemented by the expression of functional IAP proteins to a level comparable with that of the full-length-type IAP element (Figure 1C), indicating substantial retrotransposition activity of the IΔ1 type.
Figure 1.—
Retrotransposition of IAP elements isolated from AML cells and comparison between full-length and IΔ1 types. (A) The U3 region of the 5′ LTR was replaced with the CAG promoter consisting of human cytomegalovirus enhancer and chicken β-actin promoter. Details are shown in supplemental Figure S2 at http://www.genetics.org/supplemental/. A GFP cassette containing intron sequence in reverse orientation was used as a reporter for retrotransposition. Orientations of the GFP gene and intron are indicated by the green and black arrows, respectively. SD, splice donor; SA, splice acceptor; P, EF1α promoter; wt, wild type. (B) GFP signal detection following transfection into HeLa cells. (C) Comparison of retrotransposition efficiency between the full-length and IΔ1-type IAP elements. pFL was generated from the Q14 full-length-type IAP element. pDE1 and pDE2 are native IΔ1-type IAP elements isolated from the AML cell genome. pDA was generated from pFL by introducing an internal deletion. The same LTR sequences were used in all vectors. The structure of the IAP protein-expression vector is the same as that of pFL except that the GFP-intron cassette is absent. GFP expression was analyzed by FACS at 7 days posttransfection. The black box at the left end of the IAP sequence indicates CAG promoter. Data are mean ± SEM (n = 3).
Effects of full-length and IΔ1-type IAP elements on transcription of flanking regions:
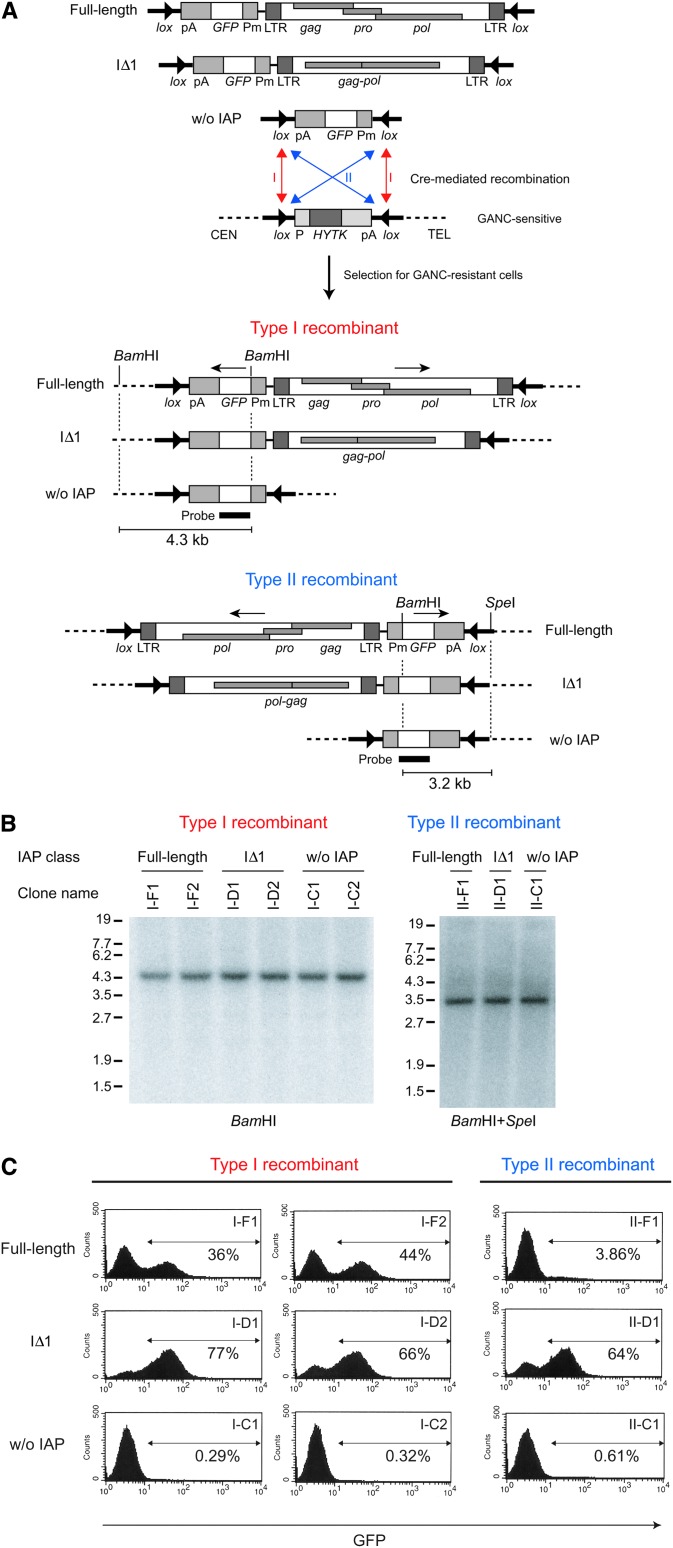
It is widely considered that transposons are suppressed by host-defense systems such as DNA methylation (Yoder et al. 1997). However, previous reports of mutant mouse phenotypes indicated that ectopic expression of cellular genes is occasionally observed with an IAP insertion (Maksakova et al. 2006; Whitelaw and Martin 2001), suggesting that a proportion of IAP insertions escapes the genome-defense system. Interestingly, most IAP elements responsible for ectopic expression were of the IΔ1 type and their orientations were opposite to that of the ectopically expressed gene (Maksakova et al. 2006). These observations suggest that IΔ1 IAP-element insertion in a reverse orientation has substantial activity in inducing flanking genes compared to full-length IAP element. To investigate this possibility, we placed a minimal promoter-GFP reporter cassette upstream of the full-length and artificially generated IΔ1-type elements in the inverted orientation relative to these elements and examined the effect of IAP sequence on GFP expression in ES cells (Figure 2A). The IAP-GFP sequence was flanked by inverted lox511 sites and introduced by Cre recombinase into a specific location of the genome bearing inverted lox511 sites (Feng et al. 1999) (Figure 2A). This eliminates the positional effect and allowed us to compare the effect of different types of IAP elements at the same chromosomal location. Since two lox511 sites outside the replaced sequence were inversely orientated to each other, the vector DNAs were inserted in either orientation (Figure 2, A and B). In either orientation, the percentage of GFP-positive cells was higher with the IΔ1 IAP element compared with the full-length IAP element (Figure 2C), consistent with the notion that the IΔ1 IAP element has substantial activity for inducing the expression of flanking genes. Interestingly, GFP expression was mosaic: Both GFP-positive and -negative cells were observed in each clonal population (Figure 2C). Phenotypes of mutant mice caused by IAP-induced ectopic gene expression were reported to vary between individual mice despite the fact that they were genetically identical (Whitelaw and Martin 2001). This observation led to the proposal that IAP activity is susceptible to epigenetic regulation (Whitelaw and Martin 2001). The mosaic pattern of IAP-induced GFP expression (Figure 2C) may recapitulate such epigenetic regulation.
Figure 2.—
Effects of full-length and IΔ1-type IAP elements on transcription of the flanking region. (A) Cre-mediated introduction of vector DNAs into the same genomic locus. GFP reporter without or with the IAP element (full-length or IΔ1 type) was flanked by inverted lox511 sites and introduced into ES cells containing the HYTK gene also flanked by inverted lox511 sites. Cre-mediated recombinants were selected by GANC and classified into either type I or type II recombinants, according to the orientation of integrated plasmid sequences relative to the target site. Pm, minimal promoter of human cytomegalovirus; CEN, centromere; TEL, telomere. (B) Southern blot analysis of the GANC-resistant clones. GANC-resistant clones were prescreened by PCR as described in materials and methods and classified as type I or type II. This classification was verified by Southern blot analysis as shown. The location of the probe and predicted restriction maps are shown in A. (C) FACS analysis of GFP expression. The regions indicated by arrows are GFP positive and the percentages are presented.
Genomewide screening for IAP-induced transcripts and their strain divergence:
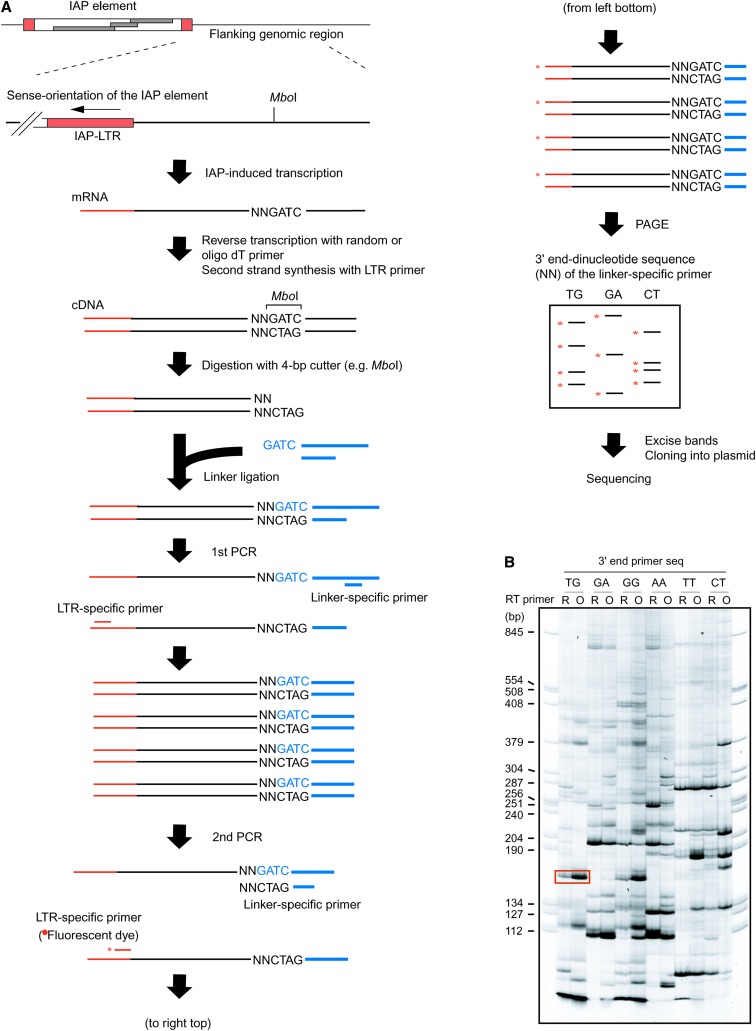
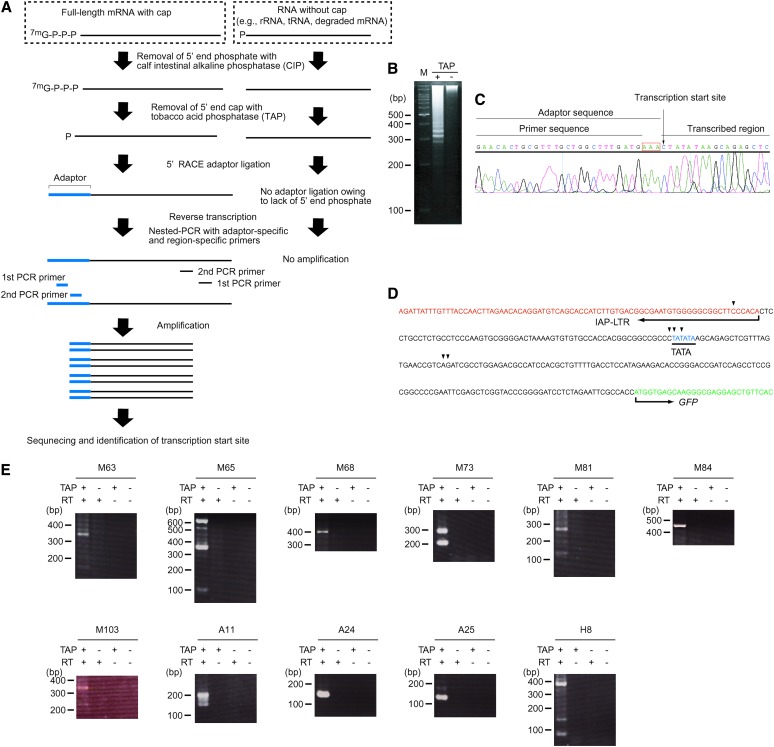
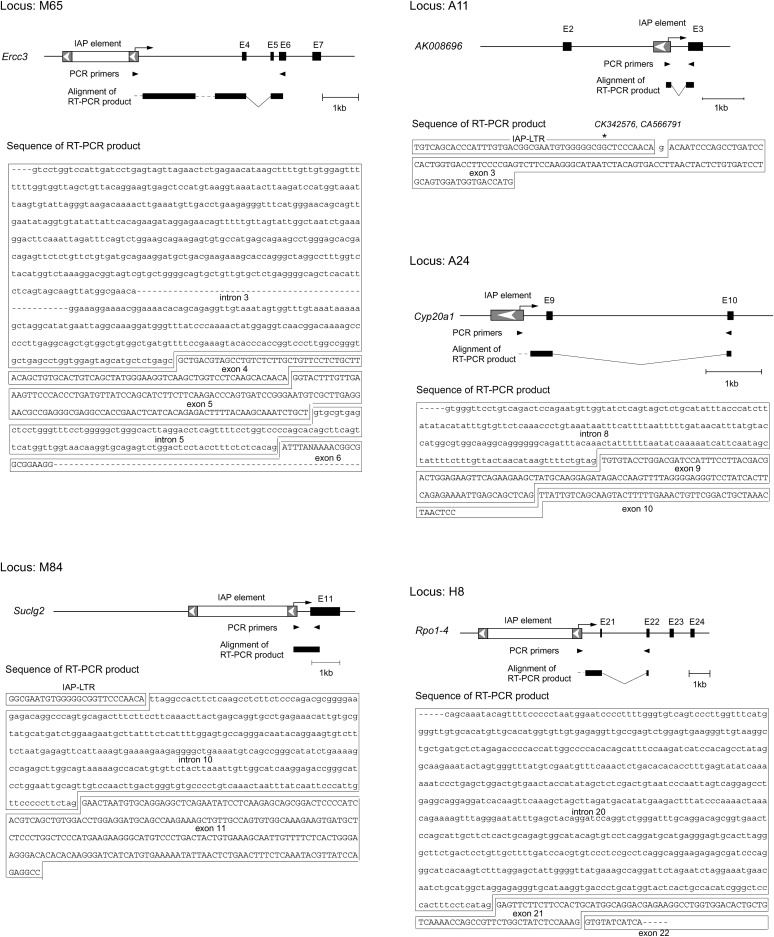
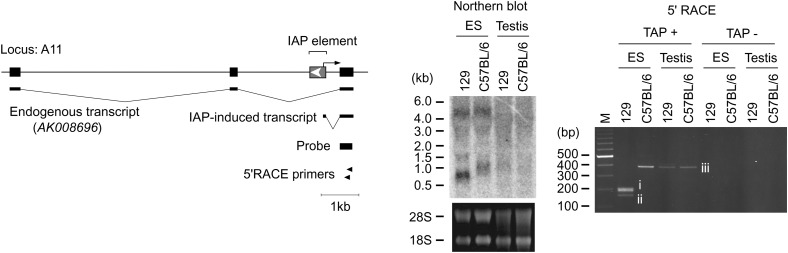
The above results imply that ∼1000 copies of IAP elements residing genomewide could influence the mouse transcriptome by inducing transcription in the flanking region. To address this possibility, we isolated IAP-induced transcripts by PCR using the scheme presented in Figure 3A (see Figure 3 legend and materials and methods for details). One of the ES cell clones expressing GFP (I-D1 in Figure 2) was analyzed because detection of the IAP-induced GFP transcript serves as a positive control. A large number of transcripts were amplified (Figure 3B), including a GFP transcript of predicted size (boxed in red, Figure 3B), and sequences of 49 transcripts were determined (supplemental Table S6 at http://www.genetics.org/supplemental/). While some transcripts could not be mapped due to the repetitive nature or short query size, the rest were mapped to 29 independent loci. It is possible that these transcripts were derived not only from IAP-induced transcripts but also from any readthrough transcripts initiated upstream of the IAP elements such as unspliced cellular transcript containing an IAP element in intronic sequences. For this reason, we conducted 5′RACE for the candidate loci and tried to identify IAP-induced transcripts (Figure 4). This 5′RACE protocol is dependent on the ligation of an RNA adaptor to the 5′ end of capped mRNA (Figure 4A). Therefore, the nucleotide immediately downstream of the RNA adaptor sequence should in principle represent the transcription start site (Maruyama and Sugano 1994; Schaefer 1995). This principle was tested in IAP-induced GFP transcript, which was also used as a positive control in Figure 3B. Multiple bands were amplified (Figure 4B), and transcription start sites were successfully mapped upstream of the GFP gene by sequencing each band (Figure 4, C and D). Reproducibility of the amplification of IAP-induced GFP transcript was confirmed as shown in supplemental Figure S3 at http://www.genetics.org/supplemental/. Next, we tried to determine the transcription start sites for the aforementioned 29 loci, which were identified by the procedure illustrated in Figure 3. Transcription start sites were mapped to the IAP elements in 11 loci (Figure 4E, Table 1). For the remaining loci, transcription starting from endogenous genes could be determined only for one locus, while the rest could not be determined and were not analyzed further. Our screening procedure is likely to isolate only a small portion of the entire population of IAP-induced transcripts, because 5′RACE analysis at the control GFP locus showed predominant transcription downstream of the IAP element (Figure 4D), which theoretically cannot be identified by our screening method (Figure 3A). Most of the IAP elements were located within genes in a reverse orientation, and fusion transcripts between IAP elements and endogenous genes were found in 5 loci (Table 1, Figure 5), suggesting the possibility of encoding truncated proteins. Among the 5 loci, only the A11 locus showed splicing between the IAP element and the downstream endogenous exon (Figure 5). Unexpectedly, 6 of 11 IAP elements were not found in the mouse genome database (UCSC mm5) (Table 1). Since the mouse genome database was constructed from the C57BL/6 mouse strain and 129 mouse strain-derived ES cells were used in the present study, this result suggests that around half of the IAP elements are differentially distributed between these two strains. This observation was further verified by PCR analysis of C57BL/6 and 129 mouse strain-derived genomic DNAs (supplemental Figure S4 at http://www.genetics.org/supplemental/). The significance of this differential IAP distribution was implicated by the higher level of the 129-specific transcript over C57BL/6-derived endogenous gene transcript at the A11 locus (Figure 6). The predicted sizes of the 129-specific IAP-induced transcript and the C57BL/6J-derived endogenous transcript were observed in Northern blot analysis (see the Figure 6 legend), suggesting that the 129-specific IAP-induced transcript really exists at this locus. Considering that only parts of entire IAP-induced transcripts can be identified by our screening parameters (Figure 4D), these results suggest that a large number of strain-specific IAP-induced transcripts exist in the genome and may contribute to different genetic traits among various mouse strains.
Figure 3.—
Screening for IAP-induced transcripts. (A) Procedure for the isolation of IAP-induced transcript. Total RNA from clone I-D1 expressing IAP-induced GFP transcript (Figure 2) was analyzed. As shown in B and Figure 4, detection of IAP-induced GFP transcript serves as a positive control for the experimental procedure. Following reverse transcription and second-strand synthesis, cDNA was digested with a 4-base cutter restriction enzyme (MboI is shown as an example) and ligated with a linker DNA. The cDNA was amplified by nested PCR with LTR-specific primer and linker-specific primer and separated by polyacrylamide gel electrophoresis (PAGE). The linker-specific primer in the second PCR contains the restriction site sequence and selective dinucleotide (shown by “NN”) at the 3′ end. The specific dinucleotide sequence was used in each second PCR and only cDNA fragments that contain the same dinucleotide sequence next to the restriction site were amplified. As a result, only a subset of the cDNA fragment was amplified in each reaction, resulting in improvement of the resolution of PCR products in PAGE analysis. The LTR-specific second PCR primer was fluorescent labeled (asterisk) for visualization. (B) PAGE analysis of the PCR products. A representative PAGE image utilizing six different linker-specific second PCR primers is shown here and the list of linker-specific second PCR primers is presented in supplemental Table S2 at http://www.genetics.org/supplemental/. The band corresponding to the predicted size of IAP-induced GFP transcript is boxed. R, random primer; O, oligo(dT) primer.
Figure 4.—
Start-site determination of IAP-induced transcripts by 5′RACE. (A) The principle of 5′RACE. Note that only capped mRNA is amplified with PCR by treatment with calf intestinal alkaline phosphatase (CIP) and tobacco acid phosphatase (TAP). (B) 5′RACE of the IAP-induced GFP transcript. The 5′RACE product was loaded onto a 3% agarose gel. M, 100-bp marker. TAP treatment is required for RACE-adaptor ligation to capped mRNA as shown in A. Therefore, lack of amplification in the TAP-untreated reaction indicates that the bands in the TAP-treated reaction were derived from capped mRNA. (C) A representative analysis of 5′RACE product. The band shown in B was excised from the gel and sequenced. The presence of “AAA” (boxed in red), which is located at the 3′ end of the adaptor but does not exist in the primer, indicates that the amplification was specific to adaptor-ligated RNA. (D) Transcription start sites of the IAP-induced GFP transcript (arrowheads) obtained by sequencing the bands shown in B. (E) 5′RACE products of the IAP-induced transcripts. The bands were separated in a 2% agarose gel. Sequencing results are summarized in Table 1.
TABLE 1.
Summary of the transcripts initiated from IAP elements
| IAP element |
Gene |
||||||
|---|---|---|---|---|---|---|---|
| Transcript | Sizea (kb) | Type | Presence in C57BL/6 genome | Name | Orientationb | Transcription start sitec | Fusion transcriptd |
| M63 | 7 | Full length | No | D14Ertd500e | Reverse | −56, −251 | ND |
| M65 | 2 | Deleted | No | Ercc3 | Reverse | −20, −264, −558 | + |
| M68 | 5.3 | Deleted | Yes | Kif24 | Reverse | −284 | ND |
| M73 | 0.43 | Solitary LTR | Yes | Acly | Reverse | −8, +28 | ND |
| M81 | 0.34 | Solitary LTR | No | Pcaf | Reverse | −55, −185 | ND |
| M84 | 4 | Deleted | Yes | Suclg2 | Reverse | −363 | + |
| M103 | 5 | Deleted | No | None | NA | −69, −173 | NA |
| A11 | 0.52 | Solitary LTR | No | AK008696 | Reverse | −36, −65 | + |
| A24 | 0.33 | Solitary LTR | Yes | Cyp20a1 | Reverse | −90 | + |
| A25 | 7 | Full length | No | ENSMUSG00000025246 | Reverse | +155 | ND |
| H8 | 5 | Deleted | Yes | Rpo1-4 | Reverse | +10, +73, −13, −223 | + |
Sizes of IAP elements not present in the C57BL/6 genome database were estimated by PCR amplification of IAP elements from the 129 mouse genome.
Orientation of IAP elements relative to the endogenous genes. NA, not applicable because of the absence of genes at the locus under study.
Transcription start sites were determined by sequencing 5′RACE reaction product and the distance from the end of the IAP–LTR is shown in nucleotides. Plus numbers represent the start sites mapped outside the IAP–LTR, and minus numbers represent the start sites mapped inside the IAP–LTR.
ND, not detected by PCR analysis; NA, not analyzed because of the absence of genes at the locus under study.
Figure 5.—
Fusion transcripts between IAP elements and endogenous genes. Initiation of transcription from the IAP element is schematically shown by arrows. Dashed lines indicate the regions of the RT–PCR products that were not sequenced. Splicing was observed in all cases except for M84. In A11, the corresponding transcripts were found in GenBank (accession nos. CK342576 and CA566791), and the 5′ end is indicated by the asterisk. E, exon.
Figure 6.—
129-specific transcript at locus A11 detected in ES cells with Northern blot and 5′RACE. The predicted sizes of the IAP-induced transcript and the AK008696 transcript are ∼0.6 and ∼1.1 kb, respectively, assuming that the GenBank-deposited AK008696 sequence represents full-length mRNA and the size of poly(A) is ∼0.2 kb. Bands i, ii, and iii were sequenced: i and ii were IAP-induced transcripts, whereas iii was a nonspecific product. M, 100-bp marker.
DISCUSSION
The functional study of retrotransposons is often ambiguous because of the multiplicity in copy numbers in the genome and sequence divergence between individual copies. To overcome this difficulty, we isolated retrotransposition-competent IAP elements from AML cells and examined their activity in the same vector backbone or at the same genomic locus. The influence of IAP elements on the mouse transcriptome was further consolidated by genomewide analysis of IAP-induced transcripts.
The main finding of this study is the prevalence of IAP-induced transcripts. Although there are several reports of ectopic expression of cellular genes caused by novel IAP insertions, such as induction of the agouti gene in spontaneous mutant mice (Whitelaw and Martin 2001), it has not been addressed whether IAP elements residing across the genome have a similar activity. A recent study reported the presence of fusion transcripts between retrotransposons and endogenous genes by analyzing expressed sequence tags (ESTs) from full-grown oocytes and two-cell-stage embryos, which are rich in maternal transcripts (Peaston et al. 2004). In contrast, our study was conducted using ES cells derived from the inner cell mass of blastocysts, suggesting that retrotransposon-mediated host gene regulation is also prevalent in later developmental stages. Antisense promoter activity was reported in other retrotransposons such as wheat LTR-type retrotransposon W2-1A (Kashkush et al. 2003) and human non-LTR-type retrotransposon LINE1 (Speek 2001). Thus, transcription induction shown in our study may apply to a wide variety of retrotransposons. Most retrotransposons have accumulated mutations and lost autonomous retrotransposability. However, our data demonstrating substantial transcription-inducing activity of the IΔ1-type IAP element (Figure 2) suggest that the transcription-inducing activity was retained in a large population of nonautonomous retrotransposons. Considering that retrotransposons constitute a large proportion of mammalian genomes, our results suggest significant influence of retrotransposons on the mammalian transcriptome.
Multiple transcription start sites were observed in some of the IAP elements (Table 1). A recent report involving genomewide analysis of promoter regions in mouse and human genomes revealed that multiple transcription start sites were a common feature (Carninci et al. 2006) and that antisense promoter activity of the IAP element may represent one of these cases. Consistent with this, a previous study of antisense transcripts from a cloned IAP–LTR also showed multiple transcription start sites (Christy and Huang 1988). We also observed variations in the location of transcription start sites between IAP elements (Table 1). This variation is due to the sequence divergence between IAP elements or different regulatory effects of flanking genomic sequences at each IAP element. It should be noted that the 5′RACE used to identify the transcription start site has potential problems inherent to PCR-based techniques such as inefficient amplification of particular base composition or overrepresentation of a small amount of template by efficient amplification. Although the result of Northern blot analysis was comparable to the 5′RACE result for the locus A11 (Figure 6), such validation will be necessary for further assessment of the biological significance of each transcript.
Our results also address some of the interesting aspects of IAP elements. Although 70% of IAP elements are full-length type, most of the previous reports of IAP retrotransposition were on the IΔ1 type (Maksakova et al. 2006). Consistent with this, higher retrotransposition efficiency of endogenous IΔ1-type IAP elements over full-length type were observed in our retrotransposition assay (Figure 1C, pDE1 and pDE2 vs. pFL). However, internal deletion per se did not enhance retrotransposition efficiency (Figure 1C, pFL vs. pDA), indicating that the difference in retrotransposition efficiency between endogenous IΔ1 and full-length types was due to the sequence divergence outside the deleted region. The transcription-inducing activity of the IΔ1 type was higher than that of the full-length type (Figure 2), which recapitulates ectopic expression of adjacent cellular genes in spontaneous mutant mice (Maksakova et al. 2006). The region absent in the IΔ1 type may contain a target site for the silencing of IAP activity by the host or the close proximity of two LTRs may have enhanced antisense transcription-inducing activity. It should be noted that our results were obtained by analyzing particular IAP elements that were cloned from ∼1000 copies of endogenous IAP elements. Therefore, further studies are needed to generalize our findings to all IAP elements.
The IAP elements identified in this study and their retrotransposition assay may provide a useful method to study how IAP elements have propagated in the genome. Phylogenetic analysis has been utilized to speculate about the amplification process of repetitive elements. Some of them, such as SINE, LINE, and human endogenous retrovirus family HERV-K, were explained by the “master gene” model in which most copies of each repetitive element family are produced by only one or a few source genes (the master genes) (Deininger et al. 1992; Medstrand and Mager 1998). However, phylogenetic analysis does not necessarily take into account the retrotransposition activity of each copy. Therefore, another functional assay to assess the master gene model will be helpful. With the retrotransposition assay presented here, we can evaluate retrotransposition competency of each copy, which allows us to categorize IAP copies in terms of their functional activity. Such an approach will be complementary to the phylogenetic analysis and facilitate understanding of the evolution of IAP elements. IAP elements identified in our study may also provide a useful model for the understanding of orientation bias of retroelements in cellular genes. It has been reported that retroelements located in gene introns are preferentially oriented antisense to the enclosing gene (Smit 1999; Medstrand et al. 2002; Cutter et al. 2005). van de Lagemaat et al. (2006) recently conducted bioinformatic analysis of human endogenous retroviruses (ERVs) in the genome. Their results indicated that transcriptional disruption of cellular genes by splice sites occurring on either strand of the ERV elements is an important factor for selection against fixation of ERVs in introns. Interestingly, splicing between an ERV and the cellular gene was significantly downregulated when the ERVs were located in antisense orientation, suggesting the possibility that splicing was sterically hindered by annealing of an ERV-derived transcript to a cellular gene transcript. The IAP elements identified in our genomewide screening were located in antisense orientation relative to the cellular gene (Table 1). Therefore, the IAP elements described in our study may serve as a model for further investigation of the finding described by van de Lagemaat et al. (2006).
Although retrotransposons occupy ∼40% of the genome in both human and mouse, retrotransposon sequences are dissimilar between human and mouse, indicating their mobilization after divergence of the two species (Waterston et al. 2002). Therefore, it would be reasonable to consider that most of the retrotransposon-induced transcripts would not be essential for the viability of each species. Rather, we consider that retrotransposon-induced transcripts would confer unique properties to each species. Furthermore, the high frequency of 129-specific IAP-induced transcripts provides insight into genetic variation in mouse strains and suggests the contribution of retrotransposons to the genetic trait of each strain. Mouse ERVs are considered to contribute to phenotypic differences between strains such as tumor susceptibility (Rowe et al. 1972) and predisposition to autoimmune diseases (Wu et al. 1993). Whereas these phenotypes result from activation or inactivation of specific genes, the present study suggests variation of expression in multiple genes due to genomewide differences in retrotransposon distribution. It would be interesting to examine whether such variation of retrotransposon distribution exists in a human population. If it does, it may account for some of the different genetic traits between individuals, such as disease susceptibility and therapeutic efficacy. Although recent advances in genomics revealed significant genetic variations between individuals as exemplified by a large-scale single-nucleotide polymorphism analysis (Altshuler et al. 2005), repetitive sequences have not drawn much attention in such studies. Our hypothesis of the active role of retrotransposons on the shaping of the transcriptome may provide a new point of view for interpretation of genome variations.
Acknowledgments
We thank J. Moran for providing the JM101/L1.3 plasmid, S. Nagata for the pEF321-FLAG-EX plasmid, S. Fiering for 129 ES cells, M. Ikawa for C57BL/6 ES cells, K. Yusa for technical assistance, and D. Mager and C. Kokubu for comments on the manuscript. This work was supported in part by a grant from the New Energy and Industrial Technology Development Organization of Japan and by a Grant-in-Aid for Scientific Research from the ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Altshuler, D., L. D. Brooks, A. Chakravarti, F. S. Collins, M. J. Daly et al., 2005. A haplotype map of the human genome. Nature 437 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci, P., A. Sandelin, B. Lenhard, S. Katayama, K. Shimokawa et al., 2006. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38 626–635. [DOI] [PubMed] [Google Scholar]
- Christy, R. J., and R. C. Huang, 1988. Functional analysis of the long terminal repeats of intracisternal A-particle genes: sequences within the U3 region determine both the efficiency and direction of promoter activity. Mol. Cell. Biol. 8 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter, A. D., J. M. Good, C. T. Pappas, M. A. Saunders, D. M. Starrett et al., 2005. Transposable element orientation bias in the Drosophila melanogaster genome. J. Mol. Evol. 61 733–741. [DOI] [PubMed] [Google Scholar]
- Deininger, P. L., M. A. Batzer, C. A. Hutchison, 3rd and M. H. Edgell, 1992. Master genes in mammalian repetitive DNA amplification. Trends Genet. 8 307–311. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., A. Dupressoir, F. Harper, G. Pierron and T. Heidmann, 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36 534–539. [DOI] [PubMed] [Google Scholar]
- Feng, Y. Q., J. Seibler, R. Alami, A. Eisen, K. A. Westerman et al., 1999. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 292 779–785. [DOI] [PubMed] [Google Scholar]
- Gossen, M., and H. Bujard, 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, H., Y. R. Zou and K. Rajewsky, 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73 1155–1164. [DOI] [PubMed] [Google Scholar]
- Ishihara, H., and I. Tanaka, 1997. Detection and cloning of unique integration sites of retrotransposon, intracisternal A-particle element in the genome of acute myeloid leukemia cells in mice. FEBS Lett. 418 205–209. [DOI] [PubMed] [Google Scholar]
- Ishihara, H., I. Tanaka, H. Wan, K. Nojima and K. Yoshida, 2004. Retrotransposition of limited deletion type of intracisternal A-particle elements in the myeloid leukemia cells of C3H/He mice. J. Radiat. Res. 45 25–32. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin et al., 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19 489–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik, D., R. Baertsch, M. Diekhans, T. S. Furey, A. Hinrichs et al., 2003. The UCSC genome browser database. Nucleic Acids Res. 31 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush, K., M. Feldman and A. A. Levy, 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33 102–106. [DOI] [PubMed] [Google Scholar]
- Kazazian, Jr., H. H., 2004. Mobile elements: drivers of genome evolution. Science 303 1626–1632. [DOI] [PubMed] [Google Scholar]
- Kuff, E. L., and K. K. Lueders, 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51 183–276. [DOI] [PubMed] [Google Scholar]
- Maksakova, I. A., M. T. Romanish, L. Gagnier, C. A. Dunn, L. N. van de Lagemaat et al., 2006. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, K., and S. Sugano, 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138 171–174. [DOI] [PubMed] [Google Scholar]
- Medstrand, P., and D. L. Mager, 1998. Human-specific integrations of the HERV-K endogenous retrovirus family. J. Virol. 72 9782–9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medstrand, P., L. N. van de Lagemaat and D. L. Mager, 2002. Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res. 12 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke et al., 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87 917–927. [DOI] [PubMed] [Google Scholar]
- Niwa, H., K. Yamamura and J. Miyazaki, 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108 193–199. [DOI] [PubMed] [Google Scholar]
- Okabe, M., M. Ikawa, K. Kominami, T. Nakanishi and Y. Nishimune, 1997. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 407 313–319. [DOI] [PubMed] [Google Scholar]
- Ostertag, E. M., E. T. Prak, R. J. DeBerardinis, J. V. Moran and H. H. Kazazian, Jr., 2000. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28 1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaston, A. E., A. V. Evsikov, J. H. Graber, W. N. de Vries, A. E. Holbrook et al., 2004. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev. Cell 7 597–606. [DOI] [PubMed] [Google Scholar]
- Rowe, W. P., J. W. Hartley and T. Bremner, 1972. Genetic mapping of a murine leukemia virus-inducing locus of AKR mice. Science 178 860–862. [DOI] [PubMed] [Google Scholar]
- Schaefer, B. C., 1995. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal. Biochem. 227 255–273. [DOI] [PubMed] [Google Scholar]
- Smit, A. F., 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9 657–663. [DOI] [PubMed] [Google Scholar]
- Speek, M., 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21 1973–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Lagemaat, L. N., P. Medstrand and D. L. Mager, 2006. Multiple effects govern endogenous retrovirus survival patterns in human gene introns. Genome Biol. 7 R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, C. P., J. R. Chaillet and T. H. Bestor, 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20 116–117. [DOI] [PubMed] [Google Scholar]
- Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril et al., 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520–562. [DOI] [PubMed] [Google Scholar]
- Whitelaw, E., and D. I. Martin, 2001. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 27 361–365. [DOI] [PubMed] [Google Scholar]
- Wu, J., T. Zhou, J. He and J. D. Mountz, 1993. Autoimmune disease in mice due to integration of an endogenous retrovirus in an apoptosis gene. J. Exp. Med. 178 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder, J. A., C. P. Walsh and T. H. Bestor, 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13 335–340. [DOI] [PubMed] [Google Scholar]