Abstract
Interactions between specific maize purple plant1 (pl1) alleles result in heritable changes of gene regulation that are manifested as differences in anthocyanin pigmentation. Transcriptionally repressed states of Pl1-Rhoades alleles (termed Pl′) are remarkably stable and invariably facilitate heritable changes of highly expressed states (termed Pl-Rh) in Pl′/Pl-Rh plants. However, Pl′ can revert to Pl-Rh when hemizygous, when heterozygous with pl1 alleles other than Pl1-Rhoades, or in the absence of trans-acting factors required to maintain repressed states. Cis-linked features of Pl1-Rhoades responsible for these trans-sensing behaviors remain unknown. Here, genetic tests of a pl1 allelic series identify two potentially separate cis-linked features: one facilitating repression of Pl-Rh and another stabilizing Pl′ in trans. Neither function is affected in ethyl-methanesulfonate-induced Pl1-Rhoades derivatives that produce truncated PL1 peptides, indicating that PL1 is unlikely to mediate trans interactions. Both functions, however, are impaired in a spontaneous Pl1-Rhoades derivative that fails to produce detectable pl1 RNA. Pl′-like states can also repress expression of a pl1-W22 allele, but this repression is not meiotically heritable. As the Pl′ state is not associated with unique small RNA species representing the pl1-coding region, the available data suggest that interactions between elements required for transcription underlie Pl1-Rhoades epigenetic behaviors.
TRANS-sensing behaviors represent exceptional forms of gene control (Henikoff and Comai 1998), often involving transfer of regulatory information between chromosomes. Paramutation is an example of a trans-sensing allelic interaction resulting in meiotically heritable epigenetic changes in gene regulation. Specifically, epigenetic regulatory states of one allele can facilitate heritable changes of homologous alleles in trans, resulting in a departure from Mendelian inheritance. This type of behavior is well documented for specific alleles of the Zea mays (maize) red color1 (r1) (Brink 1956), booster1 (b1) (Coe 1959), pericarp1 (p1) (Das and Messing 1994), and purple plant1 (pl1) (Hollick et al. 1995) loci. Each of these loci encode tissue-specific transcriptional activators of flavonoid biosynthetic enzyme genes (Dooner et al. 1991). However, paramutation is restricted to neither maize nor regulators of pigment biosynthesis as paramutation-like interactions exist in other eukaryotes (reviewed in Chandler and Stam 2004).
While most documented examples of paramutation represent interhomolog events causing acquisition of repressive states, the term also encompasses reversals from repressed to active states. For certain examples, interactions between distinct alleles of a given locus can result in enhanced pigment phenotypes not expected from simple dominance (Styles and Brink 1969; Hollick and Chandler 1998). Such allelic relationships typify overdominance, in which the heterozygote phenotype exceeds that of a homozygote (East 1936; Shull 1948). Overdominance may contribute to heterosis (East 1936; Shull 1948), also known as hybrid vigor, which has been critical for successful maize-breeding programs (Duvick 2001). While the underlying biology of heterosis is unknown, trans-sensing behaviors (Birchler et al. 2003) or epigenetic complementation similar to that observed at r1 and pl1 (Kermicle and Alleman 1990; Hollick and Chandler 1998) may play a role.
Specific alleles of pl1, such as Pl1-Rhoades, can exhibit epigenetically distinct activity states. All pl1 alleles encode R2R3-type MYB-domain proteins (Cone et al. 1993a) that function in concert with either B or R basic helix-loop-helix proteins to activate anthocyanin biosynthetic genes (Goff et al. 1992). Using pigmentation as a proxy for pl1 action, most pl1 alleles found in the U. S. Corn Belt inbred lines display weak, light-dependent expression and are collectively referred to as sun-red types (Cone et al. 1993b). However, a highly expressed state of Pl1-Rhoades, referred to as Pl-Rh, confers robust, light-independent pigmentation to plant tissues and anthers (Cone et al. 1993b). Pl-Rh is unstable and can spontaneously change to a transcriptionally repressed state, referred to as Pl′, which confers relatively weaker and light-dependent anthocyanin pigmentation (Hollick et al. 1995, 2000). Activity states of Pl1-Rhoades are quantified using a 1–7 graded anther color score (ACS) (Hollick et al. 1995), a visual assay of anther pigmentation patterns that mirrors abundance of pl1 transcripts (Hollick et al. 2000). Pl′ invariably facilitates change of Pl-Rh to Pl′ in a Pl′/Pl-Rh heterozygote, a hallmark of paramutation (Hollick et al. 1995). Both Pl1-Rhoades alleles from a Pl′/Pl-Rh heterozygote are transmitted in a heritable Pl′ state. The ability of Pl′ to facilitate change of Pl-Rh is referred to as paramutagenic activity or paramutagenicity; thus Pl′ (ACS 1–4) is operationally a fully paramutagenic state, while Pl-Rh (ACS 7) is a nonparamutagenic state. Pl1-Rhoades alleles transmitted from plants with intermediate color scores (ACS 5 and 6) do not facilitate changes of Pl-Rh to Pl′ with 100% efficiency, resulting in a broad distribution of ACS phenotypes in testcross progeny sets. Either the ACS 5 and 6 classes represent metastable states with weak paramutagenicity (Hollick et al. 1995) or a significant portion of gametes from these plants carry nonparamutagenic Pl-Rh states. Maintenance of Pl′ is dependent upon trans-acting factors, including those encoded by required to maintain repression1 (rmr1), rmr2 (Hollick and Chandler 2001), rmr6 (Hollick et al. 2005), and mediator of paramutation1 (mop1) (Dorweiler et al. 2000) loci. Identification of MOP1 as the presumptive maize ortholog of the Arabidopsis RNA-dependent RNA polymerase RDR2 (Alleman et al. 2006; Woodhouse et al. 2006b) implicates a role for small RNAs in maintaining Pl′ states.
Alleles displaying paramutation are exceptional; thus most alleles are incapable of acquiring paramutagenicity (reviewed in Chandler and Stam 2004) and are designated “neutral” (Hollick et al. 1995). Previous studies show that Pl′ is unstable when heterozygous with neutral alleles and, as an example of overdominance, can revert to a fully expressed Pl-Rh state (Hollick et al. 1995; Hollick and Chandler 1998). Pl′ is also unstable in hemizygous configuration (Hollick and Chandler 1998; J. B. Hollick, unpublished results), suggesting that reversion of Pl′ to Pl-Rh is a passive process. The stability of Pl′ may be sensitive to either gene dosage or direct homolog interactions. A simple model predicts that neutral alleles lack specific DNA features that stabilize paramutant states in trans (Hollick and Chandler 1998).
An unresolved issue is whether pl1 RNA or PL1 protein mediates the above-mentioned trans-sensing behaviors. To address these issues, we isolated loss-of-function Pl1-Rhoades derivatives and assayed their allelic interactions with Pl′ and Pl-Rh states. Two ethyl methanesulfonate (EMS)-induced derivatives are predicted to encode truncated PL1 proteins, yet they still acquire paramutagenic activity and maintain the Pl′ state in trans, demonstrating that the PL1 protein is unlikely to mediate these trans-sensing behaviors. One spontaneous Pl1-Rhoades derivative fails both to acquire paramutagenicity and to maintain Pl′ states in trans and thus genetically defines a cis-linked feature critical for pl1 allelic interactions. This derivative coincidently does not produce detectable pl1 RNA, suggesting that either transcriptional regulatory elements or the pl1 RNA itself is responsible for these trans-sensing behaviors. We similarly assayed nine additional pl1 alleles from maize inbred lines for their trans-interaction properties. Although no tested pl1 allele acquires paramutagenicity, two functional classes are apparent: one that maintains the Pl′ state in pl1/Pl′ heterozygotes and one that facilitates Pl′ reversion to a nonparamutagenic Pl-Rh state at high frequency. These data show that the two trans-sensing behaviors of paramutagenicity and Pl′ stability are functionally distinct. Consistent with an RNA interference (RNAi)-type role for the pl1 RNA itself, paramutagenic pl1 alleles transiently repress expression of pl1-W22. However, sense-oriented small RNAs homologous to the pl1-coding region are found in plants homozygous for Pl-Rh, Pl′, and neutral pl1 alleles from both functional classes. Absence of an obvious Pl′-specific small RNA species leads us to hypothesize that trans-sensing between pl1 alleles occurs through cis-linked elements affecting transcription.
MATERIALS AND METHODS
Genetic nomenclature:
Following standard conventions (http://www.maizegdb.org/maize_nomenclature.php), maize loci are designated by lowercase italics (i.e., pl1). Specific recessive alleles are designated with a hyphen, followed by a descriptor of the allele, usually the inbred line from which the allele originated (i.e., pl1-B73). Dominant alleles begin with uppercase letters (e.g., Pl1-Rhoades). Loss-of-function derivative alleles of Pl1-Rhoades are noted with a lowercase prefix (e.g., pl1-Rhoades). Translocation breakpoints are indicated with a “T” and paramutagenic states with a prime symbol (′). Plant phenotypes displayed by particular states of Pl1-Rhoades are written in nonitalic text (i.e., Pl′). All diploid genotypes are presented with pistillate (female)-derived alleles preceding staminate (male)-derived alleles.
Germplasm:
All stocks are homozygous for alleles encoding functional enzymes of the anthocyanin biosynthetic pathway and each contains either R-r-like haplotypes conferring kernel aleurone color in combination with dominant colored aleurone1 (c1) alleles or r-r-like haplotypes (nonfunctional for seed color only) unless indicated otherwise. Specific coding regions of both R-r and r-r haplotypes are expressed in somatic tissues and confer pigment to seedling sheaths and anthers in combination with functional PL1 protein. Three Pl-Rh/Pl-Rh-converted inbred lines (W23, A619, and A632) have been previously described (Hollick et al. 2005). The Pl1-Rhoades allele in each line spontaneously changes to Pl′ at distinct frequencies (A619, ∼1/5000; W23, ∼1/1500; A632, ∼1/10). All other Pl1-Rhoades lines have been previously described (Hollick et al. 1995; Hollick and Chandler 1998). Other inbred lines and/or pl1 alleles were obtained from the following sources: the North Central Regional Plant Introduction Station, U. S. Department of Agriculture-Agricultural Research Station (USDA-ARS), Ames, Iowa (A619, B73, Mo17); the Maize Genetics Cooperation Stock Center, USDA-ARS, University of Illinois, Urbana, Illinois (KWF, Pl1-Blotched); Arnel Hallauer, Iowa State University, Ames, Iowa (4Co63); Jerry Kermicle, University of Wisconsin, Madison, Wisconsin (W22 R-r:standard converted); and Inna Golubovskaya, University of California, Berkeley, California (KYS). pl1-CO159 was maintained as described (Cone et al. 1993a; Hollick and Chandler 1998). Lines used for mutagenesis are described in Hollick and Chandler (2001) and where indicated below. The T6-9 (043-1) interchange (referred to here as T Pl′) was used to identify chromosomes carrying Pl1-Rhoades of the Pl′ state as previously described (Hollick et al. 2005). On the basis of recombination frequencies and new linkage relationships, Pl1-Rhoades is 1.5 cM (±1.1 cM) distal to the 6L breakpoint and 5.3 cM from the waxy1 (wx1) locus on chromosome 9S. The breakpoint acts as a dominant semisterility locus in translocation heterozygotes (Patterson 1994). T Pl′/T Pl′ plants used in this study derive from a single T Pl′ homozygote. All other chromosomes are structurally normal unless indicated otherwise and are noted as “+” where appropriate.
Genetic analyses:
Hand pollinations were used for all genetic crosses. For genetic tests of individual pl1 alleles, inbred plants or plants homozygous for pl1-Rhoades or Pl1-Blotched alleles were crossed by T Pl′ homozygotes. In the sole exception to this scheme, a W22 inbred plant was crossed by a pl1-Rhoades(ems9710)/T Pl′ pollen source. F1 heterozygotes (pl1/T Pl′) were crossed with Pl-Rh/Pl-Rh testers (either W23 or A619 converted lines, depending on current availability at time of flowering, were used) and Pl1-Rhoades expression was visually assessed for each progeny individual as described (Hollick et al. 1995). Individuals were subsequently scored for pollen sterility (Hollick et al. 2005) to determine the structural genotype (+/T, ∼50% sterile or +/+, fully fertile). For tests of pl1-B73, pl1-Mo17, and pl1-4Co63, the specific T Pl′/T Pl′ plants used for the assays came from F2 generations derived from prior crosses of T Pl′/T Pl′ plants to the respective inbred lines (B73, Mo17, 4Co63). Given tight linkage of a recessive wx1 mutation to Pl′, selection of kernels with opaque, nonstarchy endosperms assisted in selection of T Pl′/T Pl′ F2 individuals. These F2 plants derived from opaque kernels (T Pl′/T Pl′) were used in the genetic assays for the cognate inbred pl1 allele. All pl1 alleles not directly derived from Pl1-Rhoades, with the possible exception of pl1-W22 (see results), displayed no paramutagenic activity. The pl1 alleles tested, the specific genetic background of the Pl-Rh/Pl-Rh tester used, and the number of fully fertile ACS 7 progeny individuals resulting from pl1/T Pl′ × Pl-Rh/Pl-Rh crosses are as follows: Pl1-Blotched (W23, 18), pl1-KYS (W23, 10), pl1-4Co63 (A619, 18), pl1-B73 (A619, 5), pl1-KWF (A619, 20), pl1-Mo17 (A619, 16), and pl1-CO159 (W23, 54). Test of the pl1-A619 allele (W23 Pl-Rh/Pl-Rh tester) resulted in 15 ACS 7 and 1 ACS 6 fully fertile testcross progeny individuals.
Mutagenesis:
Both Mutator (Mu) lines and EMS pollen mutagenesis were used to generate loss-of-function Pl1-Rhoades derivatives. One derivative, pl1-Rhoades(mum9515), was isolated as follows. Pistillate parents (pl1-CO159/pl1-CO159 ; c1/c1 ; r-r/r-r) were pollinated by plants (Pl1-Rhoades/Pl1-Rhoades ; C1/C1 ; R-r/r-g) containing hypomethylated Mu elements. Colored (r-r/r-r/R-r) kernels were subsequently planted in greenhouse sand benches and seedlings lacking sheath color—putatively containing a loss-of-function Pl1-Rhoades derivative—were rescued and grown to maturity. Plants carrying putative pl1-Rhoades alleles were screened for fully fertile pollen to eliminate maternal haploids and then tested by RFLP analysis (see below) to identify any potential pl1-CO159/pl1-CO159 contaminants. A screen of ∼20,000 seedlings derived from Mu stocks described in Dorweiler et al. (2000) recovered 14 individuals with putative nonfunctional pl1-Rhoades alleles. Eight of these plants survived, but only one new allele, pl1-Rhoades(mum9515), was recovered. The pl1-CO159/pl1-Rhoades(mum9515) M1 individual was backcrossed to a pl1-CO159/pl1-CO159 plant and only colorless progeny seedlings (7 of 7) were found, indicating that the pigment deficiency maps to the pl1 locus. A pl1-CO159/pl1-Rhoades(mum9515) plant was crossed to a Pl′/Pl′ individual and a RFLP genotyped Pl′/pl1-Rhoades(mum9515) F1 was crossed to a Pl-Rh/Pl-Rh tester. All testcross progeny (20 of 20) had colored anthers, indicating that pl1-Rhoades(mum9515) is a recessive allele.
Twelve loss-of-function Pl1-Rhoades derivatives were recovered from EMS treatments. Pollen from Pl-Rh/Pl-Rh ; C1/C1 ; R-r/R-r plants (W23 background; Hollick and Chandler 1998) was treated with EMS as described (Neuffer and Coe 1978) and applied to receptive ears of pl1-CO159/pl1-CO159 ; c1/c1 ; r-r/r-r plants. Approximately 14,000 colored kernels (r-r/r-r/R-r) were planted and screened for colorless seedlings. Colorless seedlings were grown to maturity and absence of anther color was confirmed. Pollen was verified to be fully fertile to avoid selection of maternal haploids. EMS-derived alleles recovered include pl1-Rhoades(ems9703), ems9704, ems9707, ems9710, ems9711, ems9714, ems9715, ems9716, ems9718, ems9725, ems9727, and ems9729. pl1-CO159/pl1-Rhoades(ems) individuals (with the exception of ems9729) were also crossed to Pl-Rh/Pl-Rh (W23) plants. All progeny from this cross have colored anthers, indicating that the colorless phenotypes observed in the genetic screen were not caused by dominant mutations. Plants homozygous for pl1-Rhoades(ems9703), ems9711, and ems9718 were backcrossed to pl1-CO159/pl1-CO159 ; r-r/r-r plants and all progeny were colorless (ems9703, 9/9; ems9711, 34/34; ems9718, 32/32), confirming that the colorless phenotypes are due to recessive loss-of-function lesions at the pl1 locus. Two additional testcrosses using pl1-CO159 similarly showed that the loss-of-function lesion associated with the pl1-Rhoades(ems9710) allele maps to the pl1 locus (see results).
Molecular analyses:
Genomic DNA was extracted from leaves as described (Voelker et al. 1997). A 3′ 1.1-kb XhoI fragment derived from pl1-Tx303 (Cone et al. 1993a) and a 5′ 0.9-kb HindIII fragment (pJH1) (Hollick et al. 2000) were used to distinguish pl1-CO159, Pl1-Rhoades, and their derivatives by RFLP analysis. The pl1-W22, pl1-CO159, and Pl1-Rhoades alleles are distinguished by intron 2 simple sequence length polymorphisms detectable with PCR using primers phi031-F (5′-GCA ACA GGT TAC ATG AGC TGA CGA-3′) and phi031-R (5′-CCA GCG TGC TGT TCC AGT AGT T-3′) (Chin et al. 1996).
For sequencing, DNA was prepared from pl1-Rhoades(ems9703), ems9711, ems9718, and mum9515 homozygotes and the first two pl1 exons were amplified using primers nc009-F (5′-CGA AAG TCG ATC GAG AGA CC-3′) and phi031-R (see above) (Chin et al. 1996) and high-fidelity VentR DNA polymerase (New England Biolabs, Beverly, MA). Reaction conditions were 94° for 30 sec, 61° for 30 sec, and 72° for 1 min. Amplicons of at least two independent PCR reactions for each allele were extracted from 1.5% agarose gels using a QIAquick gel extraction kit (QIAGEN, Valencia, CA) and sequenced from both strands using nc009-F and phi031-R primers to achieve complete 2× coverage. Sequences of pl1-Rhoades alleles were deposited and assigned the following GenBank accession nos.: pl1-Rhoades(ems9703), DQ394071; pl1-Rhoades(ems9711), DQ379502; pl1-Rhoades(ems9718), DQ470841; and pl1-Rhoades(mum9515), DQ379499.
Small RNAs from either husk leaves or seedlings were analyzed by Northern blots. Husk leaves from silking ears of field-grown isogenic Pl-Rh/Pl-Rh and Pl′/Pl′ (A632) plants were harvested at mid-day. The cob, silks, shank, and the two most-exterior husk leaves were removed from each ear and the remaining husk leaves were lightly pulverized in a mortar using liquid nitrogen prior to the extraction procedure. Five 2-week-old seedlings homozygous for pl1-CO159, pl1-B73, or pl1-W22 were pooled and lightly pulverized in a mortar using liquid nitrogen prior to the extraction procedure. Total RNA was extracted from 3 g of husk or seedling tissue using TRIZOL reagent (Invitrogen, Carlsbad, CA). RNA was resuspended in 500 μl 50% formamide, and 150 μl of 50% PEG 8000 was added to precipitate high-molecular-weight RNA. Following incubation on ice and centrifugation at 18,000 × g, supernatant was transferred to a new tube and low-molecular-weight (LMW) RNA was precipitated by adding 3 vol of 100% ethanol. LMW RNA was resuspended in 100 μl 50% formamide and quantified by spectrophotometric analysis at 260 nm. A total of 50 μg LMW RNA was separated at 700 V on 15% polyacrylamide gels with 0.33× TBE and 7 m urea. Gels were stained with ∼1 μg/ml ethidium bromide in 0.5× TBE for 20 min. RNA was transferred to a Magnaprobe nylon membrane (GE Osmonics, Minnetonka, MN) using a Trans-Blot SD electroblotter (Bio-Rad Laboratories, Hercules, CA) at 10 V and 0.5 A for 60 min. Size standards are 25 pmol DNA oligos as follows: antisense 22 nt, phi031-R described above (Chin et al. 1996), and sense 24 nt, 5′-GAA CTA CTG GAA CAG CAC GCT GGG-3′. pl1 RNA probes labeled with [α32P]UTP were synthesized from a linearized plasmid containing 5′ sequences of pl1 cDNA (pJH7) (Hollick et al. 2000) using either T7 or T3 RNA polymerase for sense and antisense probes, respectively. Probes were hydrolyzed in 100 mm sodium carbonate buffer, pH 10.2, for 30 min at 65° prior to hybridization at 40° and washing as described (Hamilton and Baulcombe 1999). Hybridized blots were exposed to Molecular Dynamics phosphor detection screens (GE Healthcare Bio-Sciences, Piscataway, NJ) for 72 hr prior to image acquisition using a Molecular Dynamics PhosphorImager 445SI.
For transcript analysis, RNA was extracted from florets as described (Hollick et al. 2000). cDNAs were generated using Omniscript reverse transcriptase kit (QIAGEN) and PCR amplified with pl1-specific primers, 5′-CAC GGC GAA GGC AAA TGG AG-3′ and 5′-CTG TTG CCG AGG AGC TTG TG-3′ as specified (Cocciolone and Cone 1993). Seedlings were grown in vermiculite and sand for 2 weeks. RNA was extracted from tissues above ground but below the first leaf. RNA from a single individual was harvested using TRIZOL reagent. A total of 10 μg RNA was treated with DNase I (Roche Applied Science, Indianapolis), primed with oligo(dT)15 (Promega, Madison, WI), and reverse transcribed with SuperScript II (Invitrogen). Samples were PCR amplified using pl1 primers (5′-ACC CTG CTG CTA GCT AGC TG-3′ and 5′-CTG TTG CCG AGG AGC TTG TG-3′) (Cone et al. 1993b) and 35 cycles with the following conditions: 95° for 30 sec, 60° for 1 min, and 72° for 1 min. The control alanine aminotransferase (aat) transcript was amplified as described (Woodhouse et al. 2006a) using 35 cycles. PCR amplicons were separated on 3% agarose gels, stained with ethidium bromide, and imaged.
RESULTS
Allele screens identify loss-of-function Pl1-Rhoades derivatives:
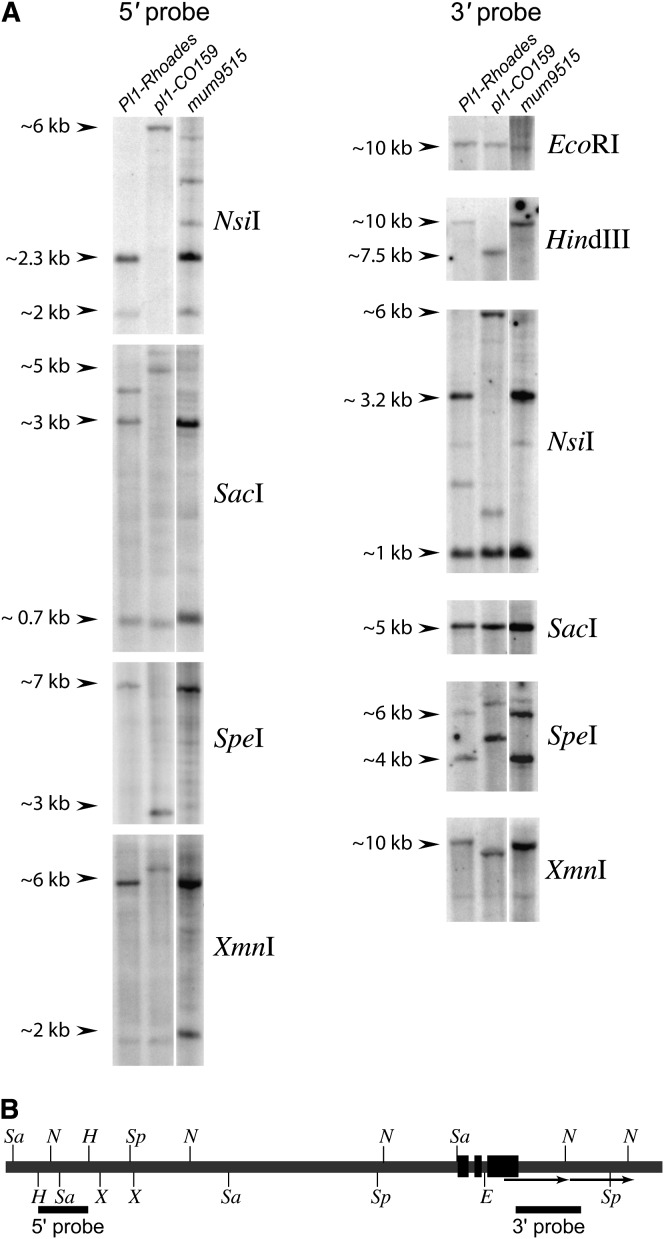
To test whether cis-elements required for the acquisition and maintenance of paramutagenicity are related to Pl1-Rhoades expression, we isolated nonfunctional Pl1-Rhoades derivatives (collectively referred to as pl1-Rhoades alleles) and tested their ability to facilitate heritable change of Pl-Rh and to stabilize Pl′ in trans. The pl1-CO159 allele, which confers no pigment to either seedling sheaths or anthers (Cone et al. 1993a), was used as a reference null. Plants homozygous for pl1-CO159 were crossed by Pl1-Rhoades homozygotes either treated with EMS or carrying Mu transposons. EMS mutagenesis generated 12 putative nonfunctional pl1-Rhoades alleles from ∼14,000 M1 seedlings. Six of these colorless M1 individuals were selfed and M2 progeny were RFLP genotyped to recover homozygotes for the derivative alleles pl1-Rhoades(ems9703), ems9707, ems9710, ems9711, ems9718, and ems9729 (materials and methods). RFLP analysis of the EMS derivative alleles indicates that they are structurally similar, if not identical, to Pl1-Rhoades (data not shown). An allele screen using a pollen parent carrying hypomethylated Mu transposons yielded pl1-Rhoades(mum9515). Genetic crosses show that pl1-Rhoades(mum9515) is recessive (materials and methods). DNA from pl1-Rhoades(mum9515) homozygotes was examined by Southern blot hybridization with 5′ and 3′ pl1 probes. pl1-Rhoades(mum9515) has RFLP patterns distinct from pl1-CO159, but has very similar, if not identical, restriction patterns to Pl1-Rhoades (Figure 1). This finding strongly suggests that no Mu elements have inserted within ∼12 kb 5′ and 10 kb 3′ of the Pl1-Rhoades coding region.
Figure 1.—
pl1-Rhoades derivative mum9515 is structurally identical to Pl1-Rhoades. (A) Southern blots were hybridized with either 5′ or 3′ pl1-specific probes (materials and methods). Approximate sizes of hybridizing fragments are indicated. (B) Restriction map of Pl1-Rhoades and probe locations based on genomic sequence (GenBank accession no. L19494). Arrows below gene structure indicate a 3′ tandem duplication. Only restriction sites generating fragments hybridizing to either the 5′ or 3′ probe are shown. E, EcoRI; H, HindIII; N, NsiI; Sa, SacI; Sp, SpeI; X, XmnI.
Genetic tests of EMS- and Mutator-derived Pl1-Rhoades alleles show that only pl1-Rhoades(mum9515) is impaired in trans-sensing behaviors:
To assay allelic interactions of pl1-Rhoades alleles, each allele was exposed to a reference Pl1-Rhoades allele of Pl′ state carried on a T6-9 (043-1) interchange chromosome (referred to as T Pl′; Hollick et al. 2005) and then evaluated for paramutagenicity following meiotic transmission to stable Pl-Rh/Pl-Rh testers. Because the reference Pl1-Rhoades allele is linked (∼1.5 cM) to the 6L breakpoint, testcross progeny segregate 1:1 for fully fertile (pl1-Rhoades/Pl-Rh) and semisterile (T Pl′/Pl-Rh) plants. Anther pigmentation of testcross progeny measures paramutagenicity of both the tested pl1-Rhoades allele (fully fertile class) and the reference Pl1-Rhoades allele (semisterile class). The ACS of pl1-Rhoades/Pl-Rh progeny indicates the paramutagenicity of each pl1-Rhoades allele. Low ACS values (ACS 1–4) suggest that the pl1-Rhoades allele can still acquire paramutagenicity following exposure to Pl′, while high ACS values (ACS > 4) suggest that the pl1-Rhoades allele is defective in acquiring or transferring paramutagenicity. Using these interpretations, all but one derivative acquires paramutagenicity similar to the progenitor Pl1-Rhoades allele (Table 1). The pl1-Rhoades(mum9515) allele is completely nonparamutagenic as all 23 fully fertile progeny have ACS 7 phenotypes. All other derivatives behave identically to the progenitor in acquiring strong paramutagenicity. From the same testcross, ACS values of semisterile progeny indicate paramutagenicity of the reference Pl1-Rhoades allele. If Pl′ retains paramutagenicity in pl1-Rhoades/T Pl′ plants, all semisterile progeny will be Pl′-like (ACS 1–4), while loss of paramutagenicity will result in higher ACS values (ACS 5–7). The data show that all derivatives except pl1-Rhoades(mum9515) retain the ability to stabilize Pl′ states in trans (Table 1). Among the 24 semisterile progeny derived from pl1-Rhoades(mum9515)/T Pl′ heterozygotes, one was scored ACS 7 and 11 were scored ACS 5 or ACS 6. Hence, pl1-Rhoades(mum9515) behaves like previously described neutral pl1 alleles by facilitating Pl′ reversion (Hollick et al. 1995; Hollick and Chandler 1998).
TABLE 1.
Anther phenotypes of progeny from pl1-Rhoades/T Pl′ × Pl-Rh/Pl-Rh crosses
| pl1-Rhoades allele | Progeny structural genotype | No. of progeny individuals with indicated ACS |
||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| ems9703 | +/+ | 1 | 10 | 8 | 0 | 0 | 0 | 0 |
| ems9707 | +/+ | 4 | 8 | 4 | 0 | 0 | 0 | 0 |
| ems9710 | +/+ | 3 | 9 | 5 | 0 | 0 | 0 | 0 |
| ems9711 | +/+ | 5 | 16 | 11 | 1 | 0 | 0 | 0 |
| ems9718 | +/+ | 4 | 17 | 4 | 0 | 0 | 0 | 0 |
| ems9729 | +/+ | 4 | 13 | 5 | 1 | 0 | 0 | 0 |
| mum9515 | +/+ | 0 | 0 | 0 | 0 | 0 | 0 | 23 |
| ems9703 | T/+ | 1 | 9 | 12 | 1 | 1 | 0 | 0 |
| ems9707 | T/+ | 1 | 8 | 9 | 0 | 0 | 0 | 0 |
| ems9710 | T/+ | 0 | 4 | 15 | 2 | 0 | 0 | 0 |
| ems9711 | T/+ | 1 | 8 | 7 | 0 | 0 | 0 | 0 |
| ems9718 | T/+ | 1 | 6 | 11 | 1 | 0 | 0 | 0 |
| ems9729 | T/+ | 5 | 14 | 4 | 1 | 0 | 0 | 0 |
| mum9515 | T/+ | 0 | 4 | 8 | 0 | 3 | 8 | 1 |
Results for each pl1-Rhoades allele tested are combined from two independent progeny ears derived using the Pl-Rh/Pl-Rh (W23) tester.
Paramutagenic activity of some pl1-Rhoades alleles does not require cis-encoded PL1 protein:
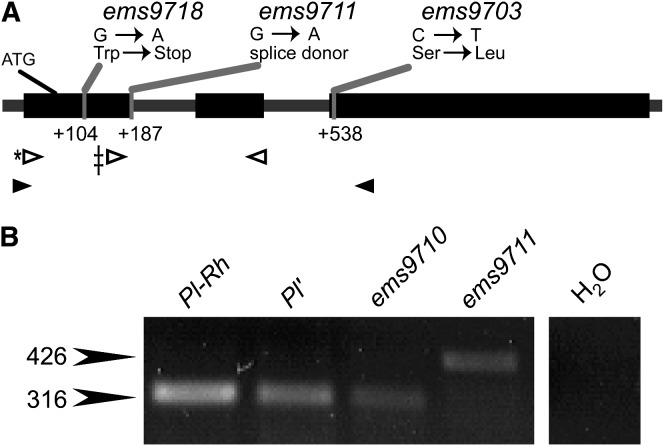
As EMS treatment commonly results in guanine-to-adenine transitions (Kohalmi and Kunz 1988) that can yield dysfunctional proteins, we suspected our paramutagenicity tests indicated that functional PL1 protein was neither required to transfer paramutagenicity nor required to stabilize a Pl′ state in trans. We sequenced N-terminal coding regions of several pl1-Rhoades alleles and discovered that pl1-Rhoades(ems9718) and pl1-Rhoades(ems9711) are not predicted to encode full-length PL1 proteins (Figure 2A). pl1-Rhoades(ems9718) has a guanine-to-adenine transition mutation at position +104, resulting in a nonsense mutation, and should encode a 16-amino-acid peptide. pl1-Rhoades(ems9711) has a guanine-to-adenine transition mutation at the intron 1 splice donor site (position +187), which should prevent the first intron from being properly spliced from the nascent RNA transcript. As predicted, reverse transcription PCR shows that the pl1-Rhoades(ems9711) mRNA is larger by the size of intron 1 (Figure 2B). The resulting protein should be truncated due to an in-frame termination codon within the unspliced intron, and thus pl1-Rhoades(ems9711) should encode a 46-amino-acid polypeptide. pl1-Rhoades(ems9703) contains a cytosine-to-thymine transition mutation within codon 90 (position +538), predicted to result in a nonconservative serine-to-leucine substitution. The S90L alteration occurs within a conserved α-helix of the MYB repeat shown in the PL1 functional paralogue, COLORED ALEURONE1, to be required for RED COLOR1-dependent transcriptional activation (Grotewold et al. 2000), suggesting that this mutation in pl1-Rhoades(ems9703) should disrupt protein function. These results point to cis-linked features, independent of the encoded PL1 transcription factor, responsible for the Pl1-Rhoades trans-sensing behaviors.
Figure 2.—
pl1-Rhoades loss-of-function alleles. (A) Molecular lesions of pl1-Rhoades(ems9718), ems9711, and ems9703. Exons (thick boxes), translation start site, and positions of lesions are relative to +1 as defined by Cone et al. (1993b). Solid arrows indicate positions of sequencing primers; open arrows indicate positions of primers used for RT–PCR reactions (*, the forward primer used for anther RNA RT–PCR; ‡, the forward primer used for seedling RT–PCR; see materials and methods). (B) RT–PCR of Pl1-Rhoades and two derivative alleles from anther RNA. The notations 316 bp and 426 bp are the sizes of properly spliced and unspliced pl1 transcript, respectively. H2O designates the negative control in the PCR reaction.
pl1-Rhoades(mum9515) is defective in RNA expression:
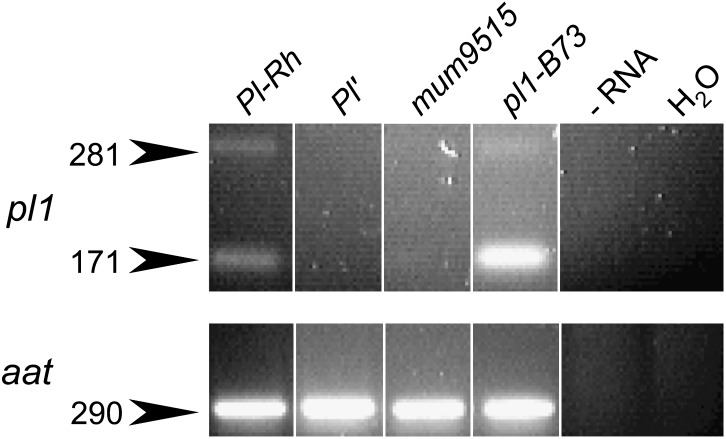
We sequenced the genomic DNA corresponding to the N-terminal coding region of pl1-Rhoades(mum9515) and found that it was identical to Pl1-Rhoades. We therefore wondered if the mutational lesion of pl1-Rhoades(mum9515) affected pl1 mRNA expression. Using RT–PCR analysis, we detected polyadenylated pl1 transcripts from seedlings homozygous for Pl-Rh and pl1-B73, but not for Pl′ or pl1-Rhoades(mum9515) (Figure 3). Additionally, pl1 RNA from pl1-Rhoades(mum9515) homozygotes is undetectable in anther florets using RNAse protection assays (J. B. Hollick, unpublished results). These results are consistent with a regulatory lesion prohibiting RNA expression of pl1-Rhoades(mum9515). Plants homozygous for pl1-Rhoades(mum9515) are always colorless at the seedling stage, yet can sometimes have weakly pigmented anthers, consistent with a mutation affecting developmental regulation or expression levels. The observations that polyadenylated pl1 RNA levels from Pl′ states are found in anther florets but not in seedling tissue (Figure 2B) and that unspliced polyadenylated pl1 RNA from Pl-Rh states are found in seedlings but not in anther florets (Figure 3) are consistent with the idea of tissue-specific regulatory differences affecting pl1 mRNA expression. Thus, while the PL1 protein appears unimportant to either trans-sensing behavior, the lack of pl1 RNA, or some feature required for its production, appears necessary for both paramutagenicity and stabilization of Pl′ states.
Figure 3.—
pl1-Rhoades(mum9515) is an RNA null. Reverse transcriptase–PCR analysis of seedling RNA. pl1: spliced, 171 bp; unspliced, 281 bp. aat: spliced, 290 bp; unspliced, 454 bp (not shown). Negative controls: −RNA, full assay in absence of RNA; H2O, no template in PCR stage only.
Genetic analysis of a naturally occurring allelic series reveals functional diversity for trans-sensing behaviors:
The finding that cis-elements of Pl1-Rhoades mediating pl1 allelic interactions could be disrupted by mutations motivated a broader survey of pl1 allelic variation represented among maize inbred lines. No pl1 allele tested to date, aside from Pl1-Rhoades, can acquire paramutagenicity (Hollick et al. 1995, 2000; Hollick and Chandler 1998) but some neutral pl1 alleles have different abilities to stabilize Pl′ in trans (Hollick and Chandler 1998). We tested nine pl1 alleles (Pl1-Blotched, pl1-KYS, pl1-4Co63, pl1-B73, pl1-KWF, pl1-Mo17, pl1-W22, pl1-A619, and pl1-CO159) in the same genetic assay described above. These alleles are resident in a diversity of inbred lines commonly used in our studies, which represent extreme early (KWF) and late (KYS) flowering lines, Reid Yellow Dent (B73) and Lancaster Sure Crop (Mo17 and A619) varieties, and well-studied anthocyanin lines (4Co63 and W22). Both pl1-CO159 and Pl1-Blotched, an allele structurally identical to Pl1-Rhoades (Cocciolone and Cone 1993), have been evaluated in previous cosegregation assays (Hollick and Chandler 1998; Hollick et al. 2000). If a given pl1 allele can acquire paramutagenicity upon exposure to Pl′, the fully fertile class of testcross progeny (pl1/Pl-Rh) has low ACS values (Pl′-like phenotypes). Conversely, inability to acquire paramutagenicity results in fully fertile progeny with fully colored anthers (ACS 7; Pl-Rh-like phenotypes). With one exception, results show that no fully fertile progeny have Pl′-like phenotypes, indicating that none of the tested pl1 alleles acquire paramutagenicity (materials and methods). This confirms previous results using both Pl1-Blotched and pl1-CO159 (Hollick and Chandler 1998; Hollick et al. 2000). The exceptional ACS 2 individual from the fully fertile pl1-W22/Pl1-Rhoades progeny class likely represents a rare recombinant chromosome in which the reference Pl1-Rhoades allele of the Pl′ state has become unlinked from the T6-9 breakpoint [χ2 = 0.6 (P > 0.05; not significant) for the null hypothesis that the number of observed ACS 7 exceptions is greater than the number expected from recombination between the translocation breakpoint and pl1]. However, two additional fully fertile progeny are ACS 5 and ACS 6, suggesting that pl1-W22 may acquire weak paramutagenicity. The specific Pl-Rh/Pl-Rh line (A619 inbred conversion) used for the pl1-W22 test exhibits a very low level of spontaneous paramutation (materials and methods).
As documented previously (Hollick et al. 1995; Hollick and Chandler 1998), we found that pl1 alleles other than Pl1-Rhoades differ in their ability to maintain Pl′ states in trans. If the tested pl1 allele stabilizes Pl′ in pl1/T Pl′ F1 plants, then semisterile testcross progeny (T Pl′/Pl-Rh) are Pl′-like (ACS 1–4). In contrast, reversion of Pl′ in pl1/T Pl′ plants results in higher ACS values (ACS 5, 6, and 7) among semisterile (T Pl′/Pl-Rh or T Pl-Rh/Pl-Rh) progeny. Results (Table 2) suggest that Pl1-Blotched and pl1 alleles resident in KYS, B73, and W22 inbreds maintain paramutagenicity of Pl′ in pl1/T Pl′ plants as all semisterile testcross progeny individuals (T Pl′/Pl-Rh) have Pl′-like phenotypes (ACS 1–4). In contrast, pl1-4Co63, pl1-KWF, pl1-A619, pl1-CO159, and pl1-Mo17 appear to allow reversion of Pl′ to Pl-Rh at high frequency. Of 16 semisterile testcross progeny from pl1-4Co63/T Pl′ plants, 2 are ACS 7 (12.5%), and an additional 2 individuals are ACS 5. Similarly, 8 of 20 (40%) from pl1-KWF/T Pl′ heterozygotes are ACS 7, and an additional 4 (20%) are ACS 5 or ACS 6. One of 13 (7.6%) from pl1-Mo17/T Pl′ are ACS 7 and an additional one is ACS 5. One of 18 (5.5%) from pl1-A619/T Pl′ are ACS 7, while an additional 7 (38%) are ACS 5 or ACS 6. Consistent with prior results (Hollick and Chandler 1998), 11 of 52 (21%) semisterile individuals from pl1-CO159/T Pl′ parents are ACS 7 and an additional 11 individuals are ACS 5 or ACS 6. These high reversion frequencies stand in sharp contrast to results seen with Pl1-Blotched, pl1-B73, pl1-KYS, and pl1-W22, but they are not a priori due to allelic differences at the pl1 locus. However, previous cosegregation data clearly showed that the chromosome region linked to pl1-CO159 is responsible for facilitating Pl′ reversion (Hollick and Chandler 1998) and that 6L segmental deficiencies, including the pl1 locus, allow similar reversions (Hollick and Chandler 1998; J. B. Hollick, unpublished results). We did not see any phenotypic reversion of Pl′ to Pl-Rh in pl1/T Pl′ plants used for our genetic assays, but we clearly observed that Pl′ had lost meiotically heritable paramutagenicity while heterozygous with some pl1 alleles (Table 2). We grew additional F1 pl1/T Pl′ plants and observed no pigmentation values greater than ACS 3 in the following genotypes: pl1-CO159/T Pl′, 61 plants; pl1-KWF/T Pl′, 39 plants; pl1-A619/T Pl′, 50 plants; pl1-Mo17/T Pl′, 64 plants; and pl1-4Co63/T Pl′, 40 plants. Thus, we found no evidence for unlinked dominant modifiers affecting somatic Pl′ stability. Collectively, with the pl1-Rhoades(mum9515) results, these data suggest that functional diversity exists among naturally occurring pl1 alleles with regards to stabilization of Pl′ states in trans.
TABLE 2.
Anther phenotypes of semisterile progeny from pl1/T Pl′ × Pl-Rh/Pl-Rh crosses
| No. of progeny ears | Progeny structural genotype | No. of progeny individuals with indicated ACS |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| pl1 allele | 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Pl1-Blotched | 1 | +/T | 1 | 11 | 7 | 0 | 0 | 0 | 0 |
| pl1-KYS | 1 | +/T | 2 | 3 | 6 | 0 | 0 | 0 | 0 |
| pl1-4Co63 | 1 | +/T | 1 | 6 | 2 | 3 | 2 | 0 | 2 |
| pl1-B73 | 1 | +/T | 3 | 4 | 3 | 1 | 0 | 0 | 0 |
| pl1-KWF | 2 | +/T | 1 | 3 | 1 | 3 | 2 | 2 | 8 |
| pl1-Mo17 | 1 | +/T | 1 | 6 | 1 | 3 | 1 | 0 | 1 |
| pl1-W22 | 4 | T/+ | 1 | 18 | 8 | 0 | 0 | 0 | 0 |
| pl1-A619 | 2 | T/+ | 0 | 2 | 3 | 5 | 2 | 5 | 1 |
| pl1-CO159 | 6 | T/+ | 1 | 5 | 17 | 7 | 4 | 7 | 11 |
pl1-W22 expression is repressed in trans by pl1 paramutagenic states:
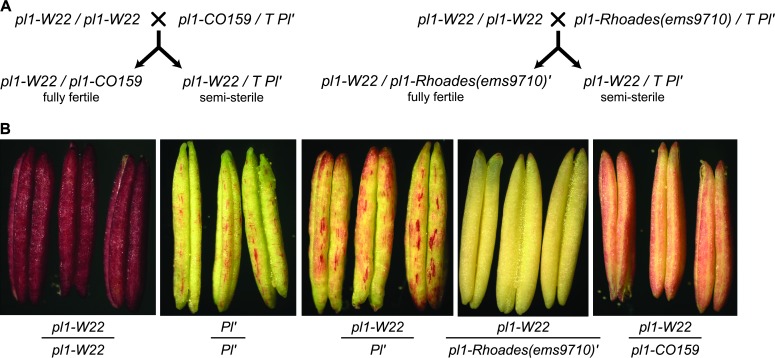
Because several inbred pl1 alleles appear to stabilize Pl′ states, we wondered if their expression might be affected in pl1/Pl′ plants. This is not easily addressed with most neutral pl1 alleles, given their already low levels of expression. The pl1-W22 allele, however, confers strong, uniform pigmentation in the presence of light (Figure 4B). We could therefore ask whether pl1-W22 expression was suppressed in heterozygous combination with paramutagenic states. To control for potential differences in pl1 gene dosage, we compared anther phenotypes of sibling pl1-W22/pl1-CO159 and pl1-W22/T Pl′ genotypes derived from a cross between pl1-W22/pl1-W22 and pl1-CO159/T Pl′ parents (Figure 4A). A dosage effect is seen as pl1-W22/pl1-CO159 plants have pink, rather than brick-red, anthers. However, Pl′ appears to suppress expression of pl1-W22 (Figure 4B, Table 3). Anthers of pl1-W22/T Pl′ individuals have a characteristic Pl′-like phenotype in which a variegated pattern of strongly pigmented epidermal cells exists within a field of near-colorless cells. If pl1-W22 and Pl′ are codominant, then darkly colored cells should be surrounded by pink-colored cells. The observation that Pl′ appears dominant to pl1-W22 is consistent with the idea that Pl′ can also suppress expression of alleles other than Pl1-Rhoades. We reexamined this apparent trans-suppression using a nonfunctional pl1-Rhoades derivative. By using a pl1-Rhoades(ems9710)/T Pl′ pollen parent (Figure 4A) to confer a paramutagenic state to a pigment-defective allele, the phenotype of pl1-W22/pl1-Rhoades(ems9710)′ progeny provides a clearer assessment of pl1-W22 expression. Results show both Pl′ and pl1-Rhoades(ems9710)′ repress pl1-W22 expression as pl1-W22/T Pl′ individuals have Pl′-like anthers and pl1-W22/pl1-Rhoades(ems9710)′ individuals have completely colorless anthers (Table 3, Figure 4B). Both data sets suggest that pl1 paramutagenic states suppress pl1-W22 expression in trans. pl1-W22 does not, however, acquire paramutagenicity following exposure to Pl′, and subsequent crosses of two separate pl1-W22/pl1-Rhoades(ems9710)′ heterozygotes with pl1-CO159/pl1-CO159 testers show that pigmenting action is typically restored in pl1-CO159/pl1-W22 and pl1-W22/pl1-CO159 progeny [15 of 16 plants scored with pink anthers were subsequently genotyped as having pl1-W22, while 17 of 18 plants scored with colorless anthers were genotyped as having pl1-Rhoades(ems9710)′]. These results indicate that the repression of pl1-W22 expression by pl1 paramutant states is not meiotically heritable.
Figure 4.—
pl1-W22 is sensitive to dosage and paramutagenic pl1 alleles. (A) Crosses, progeny, and diagnostic pollen phenotypes used to expose pl1-W22 to both Pl′ and pl1-Rhoades(ems9710)′ states. (B) Representative anthers of the given genotypes collected from single mature florets at the beginning of dehiscence.
TABLE 3.
Progeny anther phenotypes from crosses to pl1-W22/pl1-W22 pistillate parents
| No. of progeny individuals with indicated anther phenotypes |
||||||||
|---|---|---|---|---|---|---|---|---|
| Staminate parent genotype | Progeny structural genotype | Anther color score |
||||||
| Colorless | Pink | 1 | 2 | 3 | 4 | 5 | ||
| pl1-CO159/T Pl′ | +/+ | 0 | 34 | 0 | 0 | 0 | 0 | 0 |
| +/T | 0 | 0 | 10 | 18 | 6 | 1 | 1 | |
| pl1-Rhoades(ems9710)/T Pl′ | +/+ | 30 | 0 | 0 | 0 | 0 | 0 | 0 |
| +/T | 0 | 0 | 0 | 7 | 12 | 3 | 1 | |
Data represent a single progeny ear for the pl1-CO159/T Pl′ cross and two independent progeny ears for the pl1-Rhoades(ems9710)/T Pl′ cross.
All tested pl1 alleles produce sense-oriented small RNAs:
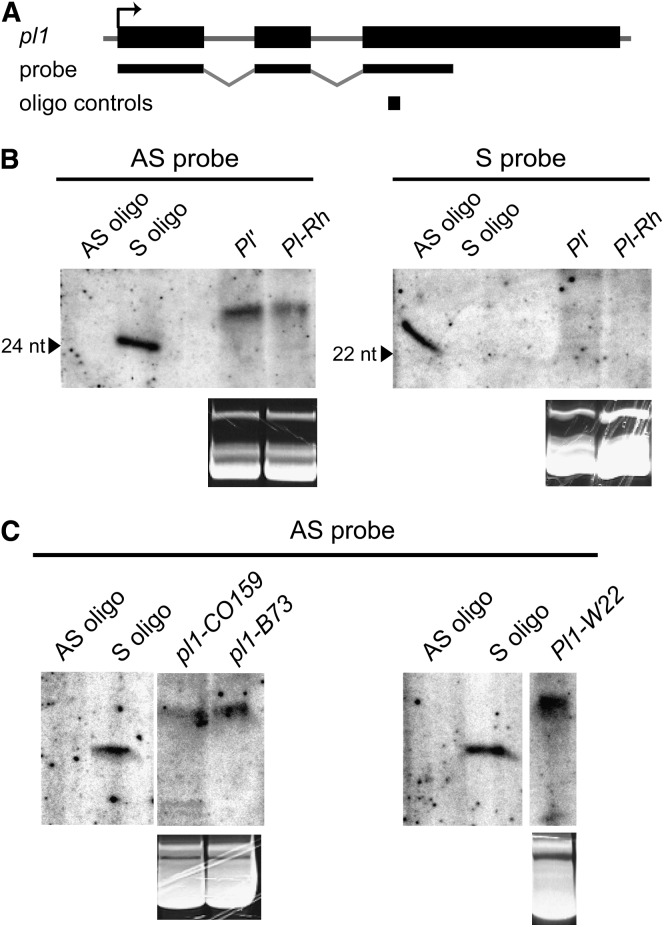
The findings that pl1-Rhoades(mum9515) is defective in pl1 RNA production and that Pl′ states repress pl1-W22 phenotypic expression are consistent with an RNAi-type allelic interaction involving pl1 RNA. Small 21- to 24-nt RNAs, hallmarks of RNAi, can act in trans to both trigger destruction of homologous RNA transcripts and facilitate epigenetic change of homologous DNA sequences within the genome (reviewed in Brodersen and Voinnet 2006). If allelic interactions between pl1 alleles are RNAi mediated, then small RNAs are predicted to correlate with pl1 paramutagenic states. We compared small RNA profiles of the Pl-Rh and Pl′ states in otherwise isogenic Pl1-Rhoades/Pl1-Rhoades plants (materials and methods) using low-molecular-weight RNA isolated from mature husk leaves. Results show that both Pl-Rh and Pl′ are associated with sense-oriented small RNAs >24 nt that hybridize with the pl1-specific probe, yet no antisense pl1 small RNAs were detected (Figure 5A). We also found sense-oriented small RNAs in seedlings homozygous for pl1-CO159, pl1-B73, and pl1-W22 (Figure 5B), indicating that these RNAs are not unique to Pl1-Rhoades. Thus, these detected small RNAs representing the pl1-coding sequence do not appear to be involved in pl1 trans-sensing behaviors.
Figure 5.—
All pl1 alleles produce sense-oriented small RNAs. (A) Map of pl1-transcribed region, partial cDNA probe, and location of oligo controls. (B) Northern blot of husk tissue small RNAs from isogenic Pl-Rh/Pl-Rh and Pl′/Pl′ plants. AS (antisense), S (sense). (C) Northern blot of seedling small RNAs from pl1-CO159, pl1-B73, and pl1-W22 homozygotes. Ethidium-bromide-stained tRNA for the relevant samples is shown below blots in B and C as a loading control. See materials and methods for additional details.
DISCUSSION
The dynamic relationships among Pl1-Rhoades paramutagenic states (Pl′), the nonparamutagenic state (Pl-Rh), and specific pl1 alleles highlight the concept that different allele combinations can lead to different meiotically heritable changes in gene regulation. Interactions between Pl′ and Pl-Rh result in the heritable change of Pl-Rh to Pl′, and Pl′ itself is maintained through interactions with either Pl1-Rhoades or specific pl1 alleles on the alternate homolog. The additional findings that certain pl1 alleles appear to stabilize the Pl′ state suggest a model in which genetically separable elements mediate distinct trans-sensing interactions responsible for paramutation and stabilization of paramutagenic states. Overdominant relationships between Pl′ and other pl1 alleles, documented here and in previous studies (Hollick et al. 1995; Hollick and Chandler 1998), provide further examples of single-locus heterosis, in which heterozygotes confer a phenotype measurably greater than that of either homozygote alone. The pl1-Rhoades(mum9515) mutation, in particular, defines a feature that normally prevents overdominance at the pl1 locus.
One diffusible molecule that could potentially mediate trans-interactions at pl1 is PL1 protein. As a transcription factor, PL1 protein might act as a heritable auto-regulator, establishing and maintaining paramutant states of pl1. However, although pl1-Rhoades(ems9711) and pl1-Rhoades(ems9718) are not predicted to make functional DNA-binding proteins, they still transmit paramutagenicity through meiosis and stabilize Pl′ in trans. Additionally, sequenced pl1 alleles (notably Pl1-Blotched, pl1-W22, and pl1-B73) are predicted to produce proteins identical or similar to Pl1-Rhoades (data not shown), yet they do not acquire paramutagenicity. Collectively, our results strongly argue against PL1 polypeptides acting as a meiotically heritable agent of paramutagenicity or affecting the trans-stabilization of Pl′.
Prior to this study, there were indications that some pl1 alleles were less efficient at stabilizing Pl′ than others (Hollick and Chandler 1998). Our results suggest that two classes of neutral pl1 alleles exist: a stabilizing class that effectively maintains paramutagenic Pl′ states on the homologous chromosome in trans and an amorphic class (Brink 1964) that acts similarly to a deficiency in facilitating overdominance by allowing Pl′ to revert to nonparamutagenic states. Our results with various pl1 alleles highlight this apparent functional diversity even though many inbred lines used in this study derive from the same racial complex of U. S. Corn Belt Dents (Goodman and Brown 1988; Hallauer et al. 1988). While it is still formally possible that specific background modifiers affect Pl′ stability, the fact that we isolated an amorphic pl1-Rhoades derivative demonstrates that such cis-linked diversity can exist. Moreover, we found no evidence of dominant modifiers affecting Pl′ reversion as no pl1/T Pl′ heterozygotes had ACS 7 anthers. This is a difference from previous cosegregation studies (Hollick et al. 1995; Hollick and Chandler 1998) in that certain heterozygous combinations can lead to a meiotically heritable loss of paramutagenicity in the absence of Pl1-Rhoades expression increases in those heterozygotes. This suggests that either the Pl′ state, in the context of the T6-9 interchange, is exceedingly stable or reversion to Pl-Rh occurs very late in somatic development and is therefore not manifest by pigment increases in the F1 heterozygote. In light of our genetic analysis of neutral pl1 alleles, we propose that Pl1-Rhoades has distinct cis-elements for stabilizing Pl′ states in trans and for facilitating heritable paramutational change in trans. If our bipartite hypothesis is correct, the coincident impairment of paramutagenicity and Pl′ trans-stabilizing functions in pl1-Rhoades(mum9515) implies that the lesion disrupts either two, presumably close, distinct elements or a single element with epistatic properties. Because the RFLP patterns of pl1-Rhoades(mum9515) appear identical to those of Pl1-Rhoades, the lesion either is relatively small or exists in more distant 5′ or 3′ flanking regions for which there are no cloned reagents available for more detailed analysis. Pl1-Blotched, which is nearly identical to Pl1-Rhoades and is predicted to produce an identical RNA transcript (Cocciolone and Cone 1993), fails to acquire paramutagenicity yet acts to stabilize Pl′ (this study and Hollick et al. 2000). In terms of our bipartite model, Pl1-Blotched has cis-elements for Pl′ stabilization, but not for paramutagenicity. Given the strong sequence identity between Pl1-Rhoades and Pl1-Blotched, it seems likely that the element(s) responsible for paramutagenicity is far removed from the pl1-coding sequence. This is not without precedence as tandem repeats affecting both expression and paramutagenicity of the B1-Intense (B1-I) allele are nearly 100 kb 5′ of the b1-coding region (Stam et al. 2002a,b).
The finding that mop1 encodes a putative RDR2 ortholog (Alleman et al. 2006; Woodhouse et al. 2006b) implicates RNAi in the maintenance of Pl′ states. The pl1 RNA could provide the target specificity necessary for a trans-acting substance, and the apparent trans-suppression of pl1-W22 by paramutagenic pl1 alleles is consistent with this proposal. Our Northern analysis shows that small sense-oriented pl1 RNA species are produced from Pl-Rh, Pl′, stabilizing alleles (pl1-B73 and pl1-W22), and an amorphic allele (pl1-CO159). As these small RNAs representing the pl1-coding sequence are not specific to either paramutagenic or stabilizing alleles, it appears unlikely that they act as mediators of pl1 allelic interactions. Interestingly, recent studies of Drosophila have discovered Piwi-interacting RNAs (piRNAs) that are typically more abundant in one strand and are associated with silencing of repetitive elements and proper germline cell development (reviewed in Parker and Barford 2006). Whether the sense-oriented pl1 small RNAs are analogous to piRNAs remains to be discovered.
Our inability to detect small RNAs associated with Pl′ states does not preclude RNA-mediated trans interactions in pl1 paramutation. One possibility is that small RNAs are generated from Pl1-Rhoades regulatory regions rather than from the coding region. Provided that pl1 alleles share regulatory sequence homology with Pl1-Rhoades, they could be sensitive to small RNAs generated by paramutagenic states of Pl1-Rhoades. Alignments between Pl1-Rhoades and pl1-B73 genomic sequences show that much of the 5′ proximal sequences in pl1-B73 are present but rearranged in Pl1-Rhoades (not shown). Thus there appear to be significant stretches of sequence similarity between the two alleles upon which homology-dependent mechanisms such as RNAi may operate. The observation that pl1 paramutagenic states suppress pl1-W22 in trans also supports this hypothesis. Alternatively, temporal small RNAs or longer pl1 transcripts, not detected by our analysis of husk and seedling tissues, may be operationally important. The regulatory mutation of pl1-Rhoades(mum9515) either may directly affect a region generating trans-acting RNAs or simply prohibit transcription of the locus, resulting in a failure to generate appropriate trans-acting RNAs from the coding sequence. Functional identification of the lesion disrupting pl1-Rhoades(mum9515) expression should help to distinguish between these possibilities.
Although our observation that paramutagenic states repress pl1-W22 is consistent with an RNAi-type trans-sensing interaction, it is also possible that pl1-W22 cannot compete effectively against Pl′ for a limited pool of transcriptional activators. The apparent dosage sensitivity of pl1-W22 when heterozygous with pl1-CO159 is consistent with such competition models. Whether the apparent trans-interaction between paramutant states and pl1-W22 is mediated by RNAi-type repression or due to interallelic competition between binding sites, the processes are not mutually exclusive. One way to discriminate between these processes will be to examine the trans-repressive interaction between Pl′ and pl1-W22 in mop1 and rmr mutant plants. A requirement for functional Mop1 and Rmr alleles to maintain repression of pl1-W22 by Pl′ would suggest that the trans-repression is related to paramutagenic activity and not a result of promoter binding site competition.
Given that trans-sensing behaviors can lead to overdominant-type variation, it will be important to understand the molecular nature of these interactions and the genomic features responsible. Reversion of repressed epigenetic states is not limited to Pl′. Like Pl′, paramutant R-r′ states become unstable when heterozygous with neutral r1 haplotypes or the r-x1 deficiency (Styles and Brink 1969). Some silenced transgenes can also be reactivated in hemizygous states (de Carvalho et al. 1992; Nap et al. 1997; Sutherland et al. 2000; De Wilde et al. 2001), suggesting that certain forms of repressive epigenetic information are generally maintained by trans-stabilizing interactions. However, not all alleles displaying paramutation require trans-stabilization; paramutant states of B1-I (B′) are exceedingly stable and have not been observed to revert when heterozygous with neutral b1 alleles (Chandler et al. 2000). Insight into these interactions at pl1 will guide an understanding of similar behaviors within the maize genome, which may contribute to general heterosis. We are currently using recombination-based strategies to genetically map these functionally relevant regions at Pl1-Rhoades, and we can now use mutations affecting paramutation (Hollick and Chandler 1998; Dorweiler et al. 2000; Hollick et al. 2005) to identify other genomic targets through differential transcript profiling. Ultimately, a better understanding of these interallelic relationships and the features responsible for them should prove useful to mine epigenetic diversity for plant improvement through standard marker-assisted selection. Our work with pl1 shows that specific allelic combinations can have trans-generational consequences through modifying heritable sources of epigenetic variation.
Acknowledgments
Thanks go to Barbara Rotz and the staff of the College of Natural Resources Oxford Tract Facilities for plant care; Galina Ishkhanova, Ruba Khenaisser, and Bill Pickle for field and lab assistance; Karen Cone (University of Missouri, Columbia) and Ingrid Assmundson for sequence information and clones from Pl1-Rhoades and Pl1-Blotched; R. Keith Slotkin for advice on small RNA analyses; and members of the Hollick laboratory for comments on the manuscript. This work was supported by the National Science Foundation (MCB-0419909) and the National Research Initiative of the U. S. Department of Agriculture Cooperative State Research, Education and Extension Service (99-35301-7753, 2001-35301-10641). The views expressed are solely those of the authors and are not endorsed by the sponsors of this work.
References
- Alleman, M., L. Sidorenko, K. McGinnis, V. Seshadri, J. E. Dorweiler et al., 2006. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442 295–298. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., D. L. Auger and N. C. Riddle, 2003. In search of the molecular basis of heterosis. Plant Cell 15 2236–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., 1956. A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41 872–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink, R. A., 1964. Genetic repression of R action in maize, pp. 183–230 in The Role of Chromosomes in Development, edited by M. Locke. Academic Press, New York.
- Brodersen, P., and O. Voinnet, 2006. The diversity of RNA silencing pathways in plants. Trends Genet. 22 268–280. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., and M. Stam, 2004. Chromatin conversations: mechanisms and implications of paramutation. Nat. Rev. Genet. 5 532–544. [DOI] [PubMed] [Google Scholar]
- Chandler, V. L., W. B. Eggleston and J. E. Dorweiler, 2000. Paramutation in maize. Plant Mol. Biol. 43 121–145. [DOI] [PubMed] [Google Scholar]
- Chin, E. C., M. L. Senior, H. Shu and J. S. Smith, 1996. Maize simple repetitive DNA sequences: abundance and allele variation. Genome 39 866–873. [DOI] [PubMed] [Google Scholar]
- Cocciolone, S. M., and K. C. Cone, 1993. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135 575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E. H., 1959. A regular and continuing conversion-type phenomenon at the B-locus in maize. Proc. Natl. Acad. Sci. USA 45 828–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., S. M. Cocciolone, F. A. Burr and B. Burr, 1993. a Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K. C., S. M. Cocciolone, C. A. Moehlenkamp, T. Weber, B. J. Drummond et al., 1993. b Role of the regulatory gene pl in the photocontrol of maize anthocyanin pigmentation. Plant Cell 5 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, O. P., and J. Messing, 1994. Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics 136 1121–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho, F., G. Gheysen, S. Kushnir, M. Van Montagu, D. Inze et al., 1992. Suppression of beta-1,3-glucanase transgene expression in homozygous plants. EMBO J. 11 2595–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wilde, C., N. Podevin, P. Windels and A. Depicker, 2001. Silencing of antibody genes in plants with single-copy transgene inserts as a result of gene dosage effects. Mol. Genet. Genomics 265 647–653. [DOI] [PubMed] [Google Scholar]
- Dooner, H. K., T. P. Robbins and R. A. Jorgensen, 1991. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25 173–199. [DOI] [PubMed] [Google Scholar]
- Dorweiler, J. E., C. C. Carey, K. M. Kubo, J. B. Hollick, J. L. Kermicle et al., 2000. mediator of paramutation1 is required for establishment and maintenance of paramutation at multiple maize loci. Plant Cell 12 2101–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick, D. N., 2001. Biotechnology in the 1930s: the development of hybrid maize. Nat. Rev. Genet. 2 69–74. [DOI] [PubMed] [Google Scholar]
- East, E. M., 1936. Heterosis. Genetics 21 375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S. A., K. C. Cone and V. L. Chandler, 1992. Functional analysis of the transcriptional activator encoded by the maize B gene: evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6 864–875. [DOI] [PubMed] [Google Scholar]
- Goodman, M. M., and W. L. Brown, 1988. Races of corn, pp. 33–79 in Corn and Corn Improvement, edited by G. F. Sprague and J. W. Dudley. American Society of Agronomy, Madison, WI.
- Grotewold, E., M. B. Sainz, L. Tagliani, J. M. Hernandez, B. Bowen et al., 2000. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc. Natl. Acad. Sci. USA 97 13579–13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallauer, A. R., W. A. Russell and K. R. Lamkey, 1988. Corn breeding, pp. 463–564 in Corn and Corn Improvement, edited by G. F. Sprague and J. W. Dudley. American Society of Agronomy, Madison, WI.
- Hamilton, A. J., and D. C. Baulcombe, 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., and L. Comai, 1998. Trans-sensing: the ups and downs of being together. Cell 93 329–332. [DOI] [PubMed] [Google Scholar]
- Hollick, J. B., and V. L. Chandler, 1998. Epigenetic allelic states of a maize transcriptional regulatory locus exhibit overdominant gene action. Genetics 150 891–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J. B., and V. L. Chandler, 2001. Genetic factors required to maintain repression of a paramutagenic maize pl1 allele. Genetics 157 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J. B., G. I. Patterson, E. H. Coe, Jr., K. C. Cone and V. L. Chandler, 1995. Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141 709–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J. B., G. I. Patterson, I. M. Asmundsson and V. L. Chandler, 2000. Paramutation alters regulatory control of the maize pl locus. Genetics 154 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollick, J. B., J. Kermicle and S. E. Parkinson, 2005. Rmr6 maintains meiotic inheritance of paramutant states in Zea mays. Genetics 171 725–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle, J. L., and M. Alleman, 1990. Gametic imprinting in maize in relation to the angiosperm life cycle. Dev. Suppl.: 9–14. [PubMed]
- Kohalmi, S. E., and B. A. Kunz, 1988. Role of neighbouring bases and assessment of strand specificity in ethylmethanesulphonate and N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis in the SUP4-o gene of Saccharomyces cerevisiae. J. Mol. Biol. 204 561–568. [DOI] [PubMed] [Google Scholar]
- Nap, J. P., A. J. Conner, L. Mlynarova, W. J. Stiekema and R. C. Jansen, 1997. Dissection of a synthesized quantitative trait to characterize transgene interactions. Genetics 147 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuffer, M. G., and E. H. Coe, 1978. Paraffin oil technique for treating mature corn pollen with chemical mutagens. Maydica 23 21–28. [Google Scholar]
- Parker, J. S., and D. Barford, 2006. Argonaute: a scaffold for the function of short regulatory RNAs. Trends Biochem. Sci. 31 622–630. [DOI] [PubMed] [Google Scholar]
- Patterson, E. B., 1994. Translocations as genetic markers, pp. 361–363 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Shull, G. H., 1948. What is heterosis? Genetics 33 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., C. Belele, J. E. Dorweiler and V. L. Chandler, 2002. a Differential chromatin structure within a tandem array 100kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 16 1906–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, M., C. Belele, W. Ramakrishna, J. E. Dorweiler, J. L. Bennetzen et al., 2002. b The regulatory regions required for B′ paramutation and expression are located far upstream of the maize b1 transcribed sequences. Genetics 162 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E. D., and R. A. Brink, 1969. The metastable nature of paramutable R alleles in maize. IV. Parallel enhancement of R action in heterozygotes with r and in hemizygotes. Genetics 61 801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, H. G., M. Kearns, H. D. Morgan, A. P. Headley, C. Morris et al., 2000. Reactivation of heritably silenced gene expression in mice. Mamm. Genome 11 347–355. [DOI] [PubMed] [Google Scholar]
- Voelker, R., J. Mendel-Hartvig and A. Barkan, 1997. Transposon-disruption of a maize nuclear gene, tha1, encoding a chloroplast SecA homologue: in vivo role of cp-SecA in thylakoid protein targeting. Genetics 145 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse, M. R., M. Freeling and D. Lisch, 2006. a The mop1 (mediator of paramutation1) mutant progressively reactivates one of the two genes encoded by the MuDR transposon in maize. Genetics 172 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse, M. R., M. Freeling and D. Lisch, 2006. b Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4 1678–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]