Abstract
Whole-mount fluorescence in situ hybridization (FISH) was applied to Arabidopsis thaliana seedlings to determine the three-dimensional (3D) interphase chromosome territory (CT) arrangement and heterochromatin location within the positional context of entire tissues or in particular cell types of morphologically well-preserved seedlings. The interphase chromosome arrangement was found to be similar between all inspected meristematic and differentiated root and shoot cells, indicating a lack of a gross reorganization during differentiation. The predominantly random CT arrangement (except for a more frequent association of the homologous chromosomes bearing a nucleolus organizer) and the peripheric location of centromeric heterochromatin were as previously observed for flow-sorted nuclei, but centromeres tend to fuse more often in nonendoreduplicating cells and NORs in differentiated cells. After mitosis, sister nuclei revealed a symmetric arrangement of homologous CTs waning with the progress of the cell cycle or in the course of differentiation. Thus, the interphase chromosome arrangement in A. thaliana nuclei seems to be constrained mainly by morphological features such as nuclear shape, presence or absence of a nucleolus organizer on chromosomes, nucleolar volume, and/or endopolyploidy level.
EUKARYOTIC chromosomes undergo condensation toward nuclear division (for review see Belmont 2006), enabling microscopic visualization of individual chromosomes from prometaphase to anaphase. Fluorescence in situ hybridization (FISH) with probes specific for entire chromosomes and advanced imaging technology revealed distinct three-dimensional (3D) chromosome territories (CTs) after chromosome decondensation in interphase nuclei of animals (Cremer et al. 1993, 2001; Habermann et al. 2001; Kozubek et al. 2002; Mahy et al. 2002a,b; Tanabe et al. 2002b) and plants (Abranches et al. 1998; Pecinka et al. 2004; Berr et al. 2006).
In several cell types of vertebrates, a nonrandom radial interphase CT arrangement was found, with chromosomes of high gene density located more centrally than chromosomes with less (active) genes (Croft et al. 1999; Bridger et al. 2000; Boyle et al. 2001). Due to chromosome-size constraints, small chromosomes are often more centrally and large ones more peripherally positioned (Sun et al. 2000; Cremer et al. 2001; Bolzer et al. 2005). The arrangement and internal structure of interphase CTs was proposed to play a role in regulation of gene expression during differentiation (for review see Bártová and Kozubek 2006; Cremer et al. 2006). The features of chromosome arrangement seem to be evolutionarily conserved in many mitotically active cell types of vertebrates (Cremer et al. 2001; Habermann et al. 2001; Kozubek et al. 2002; Mahy et al. 2002a,b; Tanabe et al. 2002a,b, 2005; Mora et al. 2006). In mammalian cell lines, photobleaching of fluorescently labeled histones and painting of particular CTs have shown that the relative chromosome positions can be at best partially transmitted through mitosis and at least transiently maintained as a mirror-symmetrical pattern in sister nuclei (Bickmore and Chubb 2003; Gerlich et al. 2003; Parada et al. 2003; Walter et al. 2003; Williams and Fisher 2003; Thomson et al. 2004; Essers et al. 2005).
While nuclei of many plant species with large genomes and metacentric chromosomes display the so-called Rabl orientation with centromeric regions clustered at one pole and telomeric regions clustered at the opposite pole (reviewed in Dong and Jiang 1998), Arabidopsis thaliana and A. lyrata interphase nuclei do not expose a Rabl orientation. Instead, within their distinct CTs, heterochromatic centromeric regions are randomly positioned at the nuclear periphery and chromosome arms may form loops of varying size, emanating from the heterochromatic chromocenters (Fransz et al. 2002; Pecinka et al. 2004; Berr et al. 2006; Schubert et al. 2006). FISH applied to spread (Fransz et al. 2002) or flow-sorted (Pecinka et al. 2004; Berr et al. 2006; Schubert et al. 2006) Arabidopsis nuclei allowed for study of a large number of nuclei with a good accessibility to the target DNA for the labeled probes. Flow sorting additionally allowed to distinguish nuclei according to their ploidy level (C-value). The disadvantages of both preparation techniques are insufficient 3D information due to flattening of nuclei and loss of the spatial context given within native tissues or in particular cell types. Furthermore, it cannot be excluded that the mainly random arrangement of CTs within large samples of flow-sorted leaf or root nuclei is due to combination of nuclei from different tissues and/or cell types, which, separately investigated, might display distinct features.
To circumvent these shortcomings, for A. thaliana seedlings, we adapted whole-mount in situ hybridization, a technique originally developed to detect transcripts in Drosophila embryos (Tautz and Pfeifle 1989), according to existing protocols (Ludevid et al. 1992; Kwart et al. 1993; Bauwens et al. 1994; Friml et al. 2003). Applying whole-mount FISH and chromosome painting (CP), we traced the 3D arrangement of major heterochromatic blocks and entire or partial CTs in interphase nuclei of diverse cell types in well-preserved differentiated and meristematic tissues. The results revealed a CT arrangement similar to that obtained for flattened flow-sorted or spread A. thaliana and A. lyrata nuclei (Fransz et al. 2002; Pecinka et al. 2004; Berr et al. 2006; Schubert et al. 2006). The largely random CT positioning [except for a more frequent association of the chromosomes bearing a homologous nucleolus organizer region (NOR)] and the arrangement of heterochromatic domains in all differentiated and meristematic cell types studied suggests that cellular differentiation has no severe impact on these parameters of nuclear organization within the studied tissues of A. thaliana. Moreover, observing the dynamics of CT arrangement in sister cells (in pairs of meristematic initial cells or in guard cells of stomata), we found a mirror-image symmetry of homologous CTs immediately after mitosis, which decays with time and is no longer obvious between adjacent related, but non-sister nuclei or between sister nuclei of fully differentiated guard cells. We conclude that the chromosome arrangement in A. thaliana interphase nuclei follows mainly morphological constraints, exerted, e.g., by nuclear shape and nucleolar volume, in a random manner.
MATERIALS AND METHODS
Probes:
Contiguous bacterial artificial chromosomes (BACs) of the Arabidopsis Biological Resource Center (Columbus, OH) selected for negligible amounts of repeats (Lysak et al. 2003) were pooled for painting individual A. thaliana chromosomes or chromosome arms. The list of BACs used for painting will be provided by the authors upon request. BAC DNA isolation and labeling either by nick translation or directly by rolling-circle amplification were performed as described (Pecinka et al. 2004; Berr and Schubert 2006). DNA from the BAC clone T15P10 (AF167571) bearing 45S rRNA genes was used for the localization of NORs. The 5S rDNA and 180-bp centromeric repeat probes were separately generated by PCR with specific primers from genomic DNA (Gottlob-McHugh et al. 1990; Kawabe and Nasuda 2005, respectively). Prior to FISH, labeled probes were precipitated and resuspended in hybridization mix (50% formamide, 10% dextran sulfate, 2× SSC, 50 mm sodium phosphate, pH 7.0).
Whole-mount in situ hybridization:
Seeds of the A. thaliana accession Columbia were sterilized and germinated on the medium of Murashige and Skoog (1962) in a greenhouse under a regimen of 16:8 hr light:dark. Whole seedlings 3–6 days after germination were fixed in 4% formaldehyde in 1× PBS (50 mm NaH2PO4 and 150 mm NaCl, pH 7.4) for 20 min and washed two times for 5 min in 1× PBS. Incubations in MeOH (two times for 5 min), EtOH (two times for 5 min), and rehydratation in 1× PBS (two times for 10 min) followed. Seedlings (∼10) were rinsed in distilled water (two times for 5 min) and citric buffer (10 mm sodium citrate, pH 4.8; 2 times for 5 min) and digested in 1% (w/v) pectolyase, cellulose, and cytohelicase (Sigma, Steinheim, Germany) in citric buffer at 37° for 0.5–3 hr (depending on the target tissue). Seedlings were washed in 1× PBS (two times for 10 min), postfixed in 4% formaldehyde in 1× PBS (20 min), and prehybridized in SF50 (2× SSC and 50% formamide, pH 7) for 1 hr at 50°. Probes (∼100 ng/ml) were denaturated together with the target preparation (approximately four seedlings) in hybridization solution for 4 min at 96°, directly placed on ice for 5 min, and hybridized for 48 hr at 37° in a moist chamber. Posthybridization washes and detection steps were as described (Schubert et al. 2001). Biotin-dUTP was detected by goat-anti-avidin conjugated with Texas Red (1:1000; Vector Laboratories, Burlingame, CA), goat-anti-avidin conjugated with biotin (1:200; Vector Laboratories), and again with avidin conjugated with Texas Red. Cy3-dUTP was detected directly. Fixation, prehybridization, hybridization, and signal detection steps were achieved in 1.5-ml Eppendorf tubes. After detection, seedlings were carefully placed on a slide, counterstained with DAPI (1 μg/ml in Vectashield, Vector Laboratories), and covered with a coverslip. To avoid crushing a specimen between the slide and coverslip, some adhesive tape was applied to create a support for the coverslip.
Microscopy, image processing, and computer simulation:
Fluorescence signals were analyzed using an Axiophot or an Axioplan 2 (Zeiss, Oberkochen, Germany) epifluorescence microscope equipped with a cooled charge coupled device camera (either Sony DXC-950P or Spot 2e Diagnostic Instruments). Images were captured separately for each fluorochrome using appropriate excitation and emission filters. Single-plane images and stacks of optical sections through tissues were acquired with MetaVue (Universal Imaging, West Chester, PA) or with the Digital Optical 3D Microscope system (Schwertner GbR, Germany) and pseudocolored and merged using MetaMorph (Universal Imaging) and/or Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA). The deconvolution of image stacks was performed with the point spread function algorithm. Values predicted by the spherical (1 Mb) chromatin domain (SCD) model (Cremer et al. 2001; Kreth et al. 2004) for the random association of entire chromosomes/chromosome arms in nuclei of the three predominant nuclear shapes (spherical, spindle, and rod shaped) were taken from Pecinka et al. (2004).
RESULTS
The association frequency of homologous chromosome arm territories is random for A. thaliana chromosome AT1 and higher for NOR-bearing chromosome AT2 in all tested differentiated cell types:
According to previous results obtained for flow-sorted spherical-, spindle-, and rod-shaped leaf and root nuclei, the association frequency of homologous and heterologous CTs is random in A. thaliana except for the association frequency of homologous arms of the NOR-bearing chromosomes AT2 and AT4, which was significantly higher (Pecinka et al. 2004).
To elucidate the CT arrangement within the context of a particular tissue or in defined cell types, differently labeled probes for both arms of chromosome AT1 and of the NOR-bearing chromosome AT2 were applied to morphologically well-preserved A. thaliana seedlings (Figure 1). The following situations were analyzed: (i) association of both arms, (ii) of only top arms, (iii) of only bottom arms, or (iv) separation of both arms. The frequencies observed for each of the four situations were compared with the data obtained for flow-sorted nuclei (Pecinka et al. 2004) among nuclei of leaf, stem, or root tissues, in particular among vascular, cortex, epidermal, guard, or root-hair cells, as well as among nuclei of different shapes (spherical, spindle, or rod shaped).
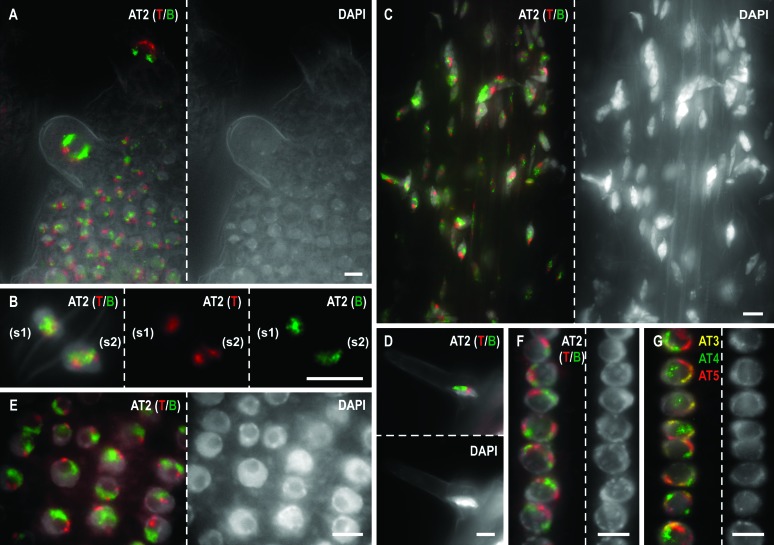
Figure 1.—
Chromosome territory organization in differentiated and meristematic A. thaliana tissues. (A–F) Differently labeled probes for the top (red) and bottom (green) arm of chromosome AT2 were hybridized to A. thaliana seedlings. Possible arrangements of homologous arm territories are shown for part of a differentiated primary leaf (A, with premature trichomes), for a pair of guard cells (B, sister cells s1 and s2), for part of a differentiated primary root (C, front view), for a root hair (D), and for shoot- and root-tip meristematic cells (E and F, respectively). (G) Association of homologous or heterologous chromosome territories for AT3 (yellow), AT4 (green), and AT5 (red) was analyzed in meristematic root-tip nuclei. Bars, 5 μm.
Nuclei of similar shape showed no significant difference (P > 0.05) as to the frequency of the four situations of homologous CT arrangement among the cell types of the three organs (Table 1). Moreover, the observed values did not significantly deviate from those previously reported for flattened flow-sorted nuclei (Pecinka et al. 2004). In all cell types, the NOR-bearing chromosome AT2 revealed significantly more frequent association of homologous arms than the chromosome without NOR (AT1) as reported previously for flow-sorted nuclei of A. thaliana (Pecinka et al. 2004). The same difference between chromosomes with NOR compared to those without NOR was found for flow-sorted nuclei of the closely related species A. lyrata (Berr et al. 2006). For the symmetric chromosome AT1, the frequencies of homologous CT association in the different tissues and cell types were similar (P > 0.05) to the random expectation according to the SCD model considering the different nuclear shapes (Pecinka et al. 2004). For the NOR-bearing chromosome AT2, association of homologs, and particularly of top arms, occurred more often than expected at random (Table 1).
TABLE 1.
Association frequencies of homologous chromosome arm territories in differentiated and meristematic tissues of A. thaliana seedlings
| Whole A. thaliana seedling |
SCD model (n = 103) |
Flow-sorted nuclei: | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Association frequency (%) |
Association frequency (%) |
|||||||||||||
| Homologs | Organ | Cell type | Nuclei shape | n | T+B+ | T+B− | T−B+ | T−B− | T+B+ | T+B− | T−B+ | T−B− | χ2 testa | χ2 testa |
| AT1 | Cotyledon | Vascular | Rod | 100 | 32.0 | 15.0 | 13.0 | 40.0 | 23.6 | 9.1 | 10.3 | 57.0 | * | – |
| Cortical and epidermal | Spindle | 100 | 51.0 | 16.0 | 10.0 | 23.0 | 48.2 | 10.8 | 11.6 | 29.4 | – | – | ||
| Spherical | 100 | 53.0 | 12.0 | 14.0 | 21.0 | 59.9 | 13.1 | 14.9 | 12.1 | – | – | |||
| Guard | Spherical | 100 | 50.0 | 16.0 | 18.0 | 16.0 | 59.9 | 13.1 | 14.9 | 12.1 | – | * | ||
| Shoot | Meristematic | 100 | 38.0 | 16.0 | 21.0 | 25.0 | 43.9 | 11.0 | 12.3 | 32.8 | –b | |||
| Stem | Vascular | Rod | 100 | 36.0 | 10.0 | 11.0 | 43.0 | 23.6 | 9.1 | 10.3 | 57.0 | – | ||
| Cortical and epidermal | Spindle | 100 | 46.0 | 15.0 | 15.0 | 24.0 | 48.2 | 10.8 | 11.6 | 29.4 | – | |||
| Spherical | 100 | 49.0 | 11.0 | 17.0 | 23.0 | 59.9 | 13.1 | 14.9 | 12.1 | – | ||||
| Root | Vascular | Rod | 100 | 29.0 | 16.0 | 10.0 | 45.0 | 23.6 | 9.1 | 10.3 | 57.0 | * | – | |
| Cortical and epidermal | Spindle | 100 | 44.0 | 14.0 | 17.0 | 25.0 | 48.2 | 10.8 | 11.6 | 29.4 | – | – | ||
| Spherical | 100 | 50.0 | 11.0 | 18.0 | 21.0 | 59.9 | 13.1 | 14.9 | 12.1 | * | – | |||
| Root hair | Spindle | 36 | 25.0 | 13.9 | 11.1 | 50.0 | 48.2 | 10.8 | 11.6 | 29.4 | – | – | ||
| Meristematic | 100 | 45.0 | 15.0 | 17.0 | 23.0 | 43.9 | 11.0 | 12.3 | 32.8 | –b | ||||
| AT2 | Cotyledon | Vascular | Rod | 100 | 26.0 | 9.0 | 31.0 | 34.0 | 18.7 | 1.8 | 21.1 | 58.4 | *** | – |
| Cortical and epidermal | Spindle | 100 | 36.0 | 9.0 | 18.0 | 37.0 | 26.3 | 1.8 | 33.2 | 38.7 | *** | – | ||
| Spherical | 100 | 47.0 | 11.0 | 22.0 | 20.0 | 39.1 | 3.3 | 43.6 | 21.2 | *** | – | |||
| Guard | Spherical | 100 | 50.0 | 13.0 | 10.0 | 27.0 | 39.1 | 3.3 | 43.6 | 21.2 | *** | – | ||
| Shoot | Meristematic | 120 | 34.2 | 15.8 | 9.2 | 40.8 | 28.0 | 2.3 | 32.6 | 39.4 | ***b | |||
| Stem | Vascular | Rod | 100 | 26.0 | 9.0 | 24.0 | 41.0 | 18.7 | 1.8 | 21.1 | 58.4 | *** | ||
| Cortical and epidermal | Spindle | 100 | 44.0 | 9.0 | 18.0 | 29.0 | 26.3 | 1.8 | 33.2 | 38.7 | *** | |||
| Spherical | 100 | 41.0 | 12.0 | 20.0 | 27.0 | 39.1 | 3.3 | 43.6 | 21.2 | ** | ||||
| Root | Vascular | Rod | 100 | 26.0 | 10.0 | 22.0 | 42.0 | 18.7 | 1.8 | 21.1 | 58.4 | *** | ||
| Cortical and epidermal | Spindle | 100 | 43.0 | 5.0 | 19.0 | 33.0 | 26.3 | 1.8 | 33.2 | 38.7 | *** | |||
| Spherical | 100 | 46.0 | 7.0 | 25.0 | 22.0 | 39.1 | 3.3 | 43.6 | 21.2 | ** | ||||
| Root hair | Spindle | 29 | 41.4 | 13.8 | 24.1 | 20.7 | 26.3 | 1.8 | 33.2 | 38.7 | *** | |||
| Meristematic | 120 | 31.7 | 11.0 | 19.0 | 38.3 | 28.0 | 2.3 | 32.6 | 39.4 | ***b | ||||
Simulated and flow-sorted leaf or root nuclei classified according to their shapes (Pecinka et al. 2004) in comparison to whole-mount FISH data. T, top arm, B, bottom arm, +, associated, −, separated. For individual columns under “Whole A. thaliana seedling” and “SCD model”: regular type, P > 0.05; italic type, P < 0.05; underlined type, P < 0.01; boldface type, P < 0.001.
Significance level of differences between the entirety of observed association frequencies in whole-mount seedlings vs. the random expectation for the corresponding nuclear shape or vs. corresponding frequencies in flow-sorted nuclei in a column-wise comparison: –, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Association frequencies in meristematic cells were compared to the random association frequency of homologous chromosome arm territories calculated as an average of predicted association values for spherical-, spindle-, and rod-shaped nuclei.
The CT arrangements do not differ between meristematic and differentiated cells:
Whole-mount CP in A. thaliana seedlings was also applied to investigate the degree of similarity of CT arrangement among nuclei of mitotically active cells and differentiated cells (Figure 1; see also supplemental Figures S1 and S2 at http://www.genetics.org/supplemental/). Since nuclear DNA content and nuclear volume are positively correlated in angiosperms (Jovtchev et al. 2006), the volumes of nuclei were measured in shoot- and root-tip meristematic cells and, after subtraction of nucleolar volumes (on average ∼50% of the nuclear volume in root-tip and 12% in shoot-tip meristematic nuclei), used to assess the nuclear ploidy level. Thus, we classified for further evaluation the smallest root meristematic nuclei (30–42 μm3) and shoot meristematic nuclei (26–36 μm3) as G1 nuclei (corresponding with a DNA content of 2C; see supplemental Figure S3 at http://www.genetics.org/supplemental/).
Chromosomes AT3, AT4, and AT5 were painted in different colors within whole A. thaliana seedlings and the association frequencies for all possible homologous and heterologous CT combinations were scored in meristematic root-tip nuclei presumably in G1 phase (Table 2) and compared to those previously reported according to the random SCD simulation (Pecinka et al. 2004). The association frequencies observed for the individual CT combinations were rather high in meristematic nuclei (58.3–98.3%) and not significantly different (P > 0.05) from that of the 103 previously simulated nuclei (68.8–98.4%). Despite the large dimension of nucleoli in meristematic cells, no significant differences (P > 0.05) were observed between the meristematic and previously flow-sorted differentiated nuclei (78.4–96.1%). Thus, in A. thaliana meristematic nuclei, the side-by-side positioning of CTs is random.
TABLE 2.
Comparison of pairwise association frequencies of A. thaliana chromosome territories AT3, AT4, and AT5 observed in root-tip meristems with values from flow-sorted differentiated nuclei and with random simulation values
| Association frequency (%)a |
|||
|---|---|---|---|
| Chromosome combination | Flow-sorted nuclei (n = 51)b | Meristematic 2C nuclei (n = 60) | SCD model (n = 103)b |
| AT3–AT3 | 80.4 | 75.0 | 77.5 |
| AT3–AT4 | 96.1 | 96.6 | 98.4 |
| AT3–AT5 | 98.0 | 95.0 | 98.5 |
| AT4–AT4 | 78.4 | 58.3 | 68.8 |
| AT4–AT5 | 96.1 | 98.3 | 97.5 |
| AT5–AT5 | 88.2 | 70.0 | 78.8 |
All differences were not significant (regular type, P > 0.05; italic type, P < 0.05) in Fisher's exact test.
Data in this column are from Pecinka et al. (2004).
Then the association frequency of homologous chromosome arm territories was investigated in mitotically active cells. Differently labeled probes for both arms of chromosome AT1 and of the NOR-bearing chromosome AT2 were applied to shoot- and root-tip cells. The frequencies of the four situations of homologous CT arrangement were scored as mentioned above for differentiated tissues in at least 100 meristematic nuclei (Table 1). In both shoot- and root-tip meristematic nuclei, the association frequency of homologous arm territories for chromosome AT1 was not significantly different from the random simulation values. The association of top arms and entire homologs for the NOR-bearing chromosome AT2 occurred significantly more often (P < 0.001) and complete separation less often than expected at random. Thus, the CT arrangement in root- and shoot-tip meristematic cells appears to be random, except for NOR-bearing chromosomes, and similar as observed for differentiated cell types.
NORs tend to fuse more frequently in differentiated vs. meristematic cells and centromeres in nonendoreduplicating vs. endoreduplicating cells of A. thaliana:
In flow-sorted or spread interphase nuclei from differentiated cells of A. thaliana, the individual centromeric chromocenters were reported to be localized at the nuclear periphery, while NORs were found to be associated around the nucleolus (Fransz et al. 2002; Berr et al. 2006).
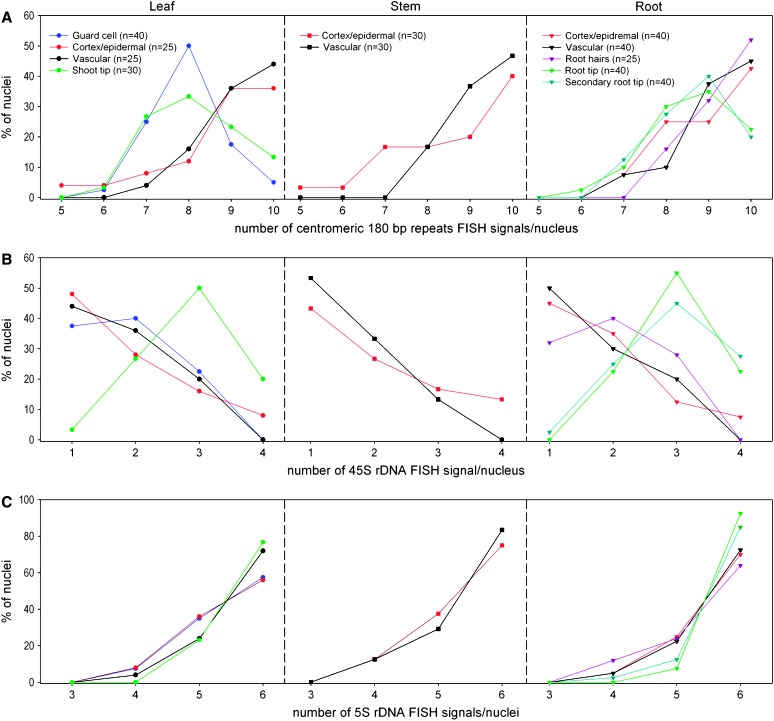
To test whether the number of the major heterochromatic blocks is specific for a particular tissue or cell type, the number and the arrangement of FISH signals obtained with differently labeled centromeric 180-bp satellite, 45S rDNA, and 5S rDNA probes (expected maximum number: 10, 4, and 6 FISH signals, respectively, per 2C nucleus) were scored for vascular cells, cortex and epidermal cells, guard cells, and root-hair and meristematic cells of leaves, stems, and roots, respectively (see Figures 2 and 3 and supplemental Table S1 at http://www.genetics.org/supplemental/). Centromeric and 5S rDNA FISH signals, generally located at the nuclear periphery, were found to be often separated (on average 9.0 and 5.6 signals/nucleus, respectively), while 45S rDNA FISH signals were frequently associated (on average 1.8 signals/nucleus) in all tested endoreduplicating cell types. In cells without endoreduplication (meristematic and guard cells), a tendency toward less centromeric signals per nucleus (on average 8.4 signals/nucleus) was observed, indicating a higher degree of interchromosomal centromere association. In meristematic cells, FISH signals for 45S rDNA (on average 2.7 signals/nucleus) were less often associated and more dispersed than in differentiated cell types. Finally, we observed in the particularly elongated root-hair nuclei extended 5S rDNA FISH signals, sometimes connecting each other and their homing chromocenters (Figure 2E) of AT3, AT4, and AT5 in the accession Columbia (Cloix et al. 2000).
Figure 2.—
Interphase arrangement of the major heterochromatic blocks in differentiated and meristematic A. thaliana tissues. As shown for cortex and epidermal cells (A, spherical- shaped, and B, spindle-shaped nucleus), for guard cells (C), for root meristematic cells (D), and for root hairs (E), 45S rDNA signals (green) are in most nuclei associated with a single nucleolus and in meristematic cells (D) are more dispersed than in differentiated cells. Centromeric 180-bp repeats (red) are preferentially localized at the nuclear periphery (visible in 3D) in all tested cell types. 5S rDNA (yellow) is associated with the flanking centromeres. In root-hair nuclei (E), 5S rDNA FISH signals appear to be particularly elongated (right, with higher magnification as insert). Bars, 5 μm.
Figure 3.—
Numbers of centromeric, 45S rDNA, and 5S rDNA FISH signals in several differentiated and meristematic cell types from leaves, stems, and roots of entire A. thaliana seedlings. (A) Number of centromeric FISH signals. Centromeric 180-bp repeat FISH signals reveal a lower tendency to fuse in endoreduplicated cells (cortex and epidermal cells, vascular cells from leaf, stem, and root as well as root hairs) than in nonendoreduplicating cell types (guard cells, shoot-tip, root-tip, and secondary root-tip meristems). (B) Number of 45S rDNA FISH signals. Mitotically active cells of the shoot tip, root tip, and secondary root tip revealed the largest nucleoli, and 45S rDNA signals were less often associated than in differentiated cells. (C) Number of 5S rDNA FISH signals. Compared to centromeric signals, the number of 5S rDNA signals was more homogenous among the different cell types. 5S rDNA signals show a lower association frequency between heterologous loci.
Chromosome positions are not inherited from mother to daughter cells but are mirror-symmetric between sister nuclei immediately after mitosis:
To find out whether interphase chromosome arrangement can be transmitted to subsequent cell generations, differently labeled probes for the top and the bottom arm of chromosome AT2 were hybridized to whole A. thaliana seedlings.
For the highly differentiated pair of guard cells, forming stomata, and representing sister cells (Zhao and Sack 1999), a similar arrangement of homologous chromosome arm territories in relation to each other was observed in only 14% of guard cells (7 of 50 pairs of sister cells) without clear mirror-image symmetry (Figure 1B).
Within the primary root meristem, initial cells undergo a longitudinal division (along the root axis) followed by a transverse division of the upper daughter cell, yielding the basis cells for cell chains forming the cortex and endodermis (Figure 4, A1–A3; Dolan et al. 1993; Scheres et al. 1994). For a possible symmetric arrangement of homologous chromosome arm territories within meristematic cells, we investigated two positions within the root tip. First, the arrangements of homologous AT2 arm territories between neighboring cells within the longitudinal chains formed by meristematic cells were compared (e.g., Figure 1F; Scheres et al. 1994). Homologous chromosome arm territories were similarly arranged in relation to each other in many cells of the same chain (71.4–81.8% of nuclei from a total of 138 cells of six chains coincided with the most frequent homologous CT arrangement situation) without displaying obvious mirror-image symmetry between any of the adjacent cells descending from the same initial cell. Second, to distinguish between actual mirror-symmetry and possible rotation of nuclei (Chytilova et al. 2000), 10 adjacent BACs from the AT3 top arm were used as a reference point in addition to the AT2 arm-specific FISH probes. A mirror-symmetric positional arrangement of the homologous AT2 arm territories and the 10 adjacent BACs from AT3 was observed in all six investigated pairs of sister cells derived immediately from initial cells (two pairs shown in Figure 4, A and B, sister cells s1, s2, s3, and s4). However, the symmetry was not always perfect regarding the shape of the homologous regions. The differences increased when comparing sister nuclei from the preceding transverse division (one pair shown in Figure 4B).
Figure 4.—
Transient mirror symmetry between sister nuclei in A. thaliana primary root meristem through nuclear division. Differently labeled probes for the top (T, red) and bottom (B, green) arm of chromosome AT2 and 10 adjacent BACs from AT3 top arm (yellow) were hybridized to the A. thaliana root tip. (A1) In the primary root meristem (DAPI staining), initial cells (i) undergo a longitudinal division followed by a transverse division of the upper sister cell, yielding a pair of sister cells (s1 and s2). (A2) CT arrangement of a pair of sister cells (s1 and s2) magnified. Superimposed maximum intensity projections of 24 serial sections (Z-interval = 0.2 μm) reveal a mirror-symmetric positional arrangement of the homologous AT2 arm territories and the 10 adjacent BACs of AT3. (A3) Observing this arrangement from front (left), top (center), and side (right) views of sister cells s1 and s2, the symmetry appears to be imperfect as to the shape and the strict positioning of the homologous regions. (B) Differences increased when the pair s3 and s4 were compared with its closest neighbor pair s5 and s6. (C1–C4) The mirror symmetry between sister nuclei could be related to the symmetry observed between the two sets of chromatids during a different stage of mitotic division: metaphase (C1), early anaphase (C2), late anaphase (C3), and telophase (C4). Bars, 5 μm.
DISCUSSION
We show that chromosome territory arrangement does not significantly differ among nuclei of similar shape (rod, spindle, or sphere) either when isolated from different organs (Pecinka et al. 2004) or within several distinct differentiated and meristematic cell types of intact tissues of A. thaliana. This is true for side-by-side arrangement as well as for association of homologous arms. As previously reported for isolated nuclei of A. thaliana and of A. lyrata, only NOR-bearing chromosomes, attached to a single nucleolus in most cells, and in particular the NOR-bearing arms, associate significantly more often than expected at random (Pecinka et al. 2004; Berr et al. 2006).
The number of the peripherically located centromeric heterochromatin blocks increased in endoreduplicating cells, likely because spatial centromere association is hindered by the increased number of identical chromatids, which in turn might become separated from each other (Schubert et al. 2006). The tendency of NOR signals to fuse more often in differentiated cells might be explained by their attachment to a single nucleolus (in ∼94% of all investigated cells), which is much smaller in differentiated than in meristematic cells. Furthermore, the terminal position of NORs in Arabidopsis chromosomes might favor their association because of less spatial constraints compared to those exerted, e.g., on centromeres that are flanked by chromosome arms on either side.
All together, these results do not reflect an obvious specificity as to the CT arrangement and the organization of the major heterochromatic blocks in interphase nuclei of the tested tissues or cell types. Thus, the nuclear architecture appears to be random in so far as it is not determined by morphologic constraints such as nuclear shape, absence or presence of NORs on chromosomes, nucleolar volume, and/or endopolyploidy level. Remarkably, differences in CT arrangement among nuclei of different shape (Table 1; Pecinka et al. 2004; Berr et al. 2006) are not significantly different from random expectation for these particular shapes according to the SCD model. Nevertheless, we cannot exclude exceptions that might occur in cell types that we did not investigate. Because between A. thaliana and A. lyrata CT and chromatin arrangement in flow-sorted interphase nuclei seems to be evolutionarily conserved (Berr et al. 2006) since their divergence ∼5 MYA (Koch et al. 2000) and because the A. lyrata karyotype is very similar to the proposed ancestral karyotype of the genus Arabidopsis (Lysak et al. 2006), a similar nuclear architecture is expected for other closely related diploid Brassicaceae species.
The maintenance of mirror symmetry between daughter nuclei, at least for a brief period after mitosis, together with the symmetry observed between the two sets of chromatids during anaphase and telophase (Figure 4, C1–C4), supports Boveri's assumption that chromosome arrangement in the metaphase plate leads to rather symmetrically structured daughter nuclei (Boveri 1909) and is in agreement with evidence from mammalian cell lines of the symmetric arrangement of CTs (e.g., Sun and Yokota 1999; Habermann et al. 2001; Gerlich et al. 2003; Walter et al. 2003) and of chromosomal domains (Essers et al. 2005) in sister nuclei. Conversely, metaphase congression and (Brownian) movement of chromatin might be responsible for the lack of mirror symmetry between nuclei of related neighbor cells. Using transgenic A. thaliana lines that express a fluorescently labeled recombinant centromeric histone H3 variant (GFP-AtCENH3), Fang and Spector (2005) observed asymmetry of the 3D centromere distribution between sister cells. The apparently contradictory findings of Fang and Spector's (2005) study with our observations could result from the different behavior of the chromosome domains under investigation. Indeed, large-scale chromatin arrangements were described as highly stable during the cell cycle (Shelby et al. 1996; Abney et al. 1997; Zink and Cremer 1998; Bornfleth et al. 1999; Chubb et al. 2002; Lucas and Cervantes 2002), while considerable movement of chromosomal subregions, such as centromeres, was observed (Martou and De Boni 2000; Cremer et al. 2003). Our observation of a lower level of similarity of the CT arrangement among adjacent nuclei within chains of meristematic cells compared to sister pairs resulting from initial cells, together with the clear lack of symmetry between pairs of sister guard cells, indicates that symmetry is lost through mitotic divisions, as reported for HeLa cells (Walter et al. 2003), and decays during cell differentiation.
Acknowledgments
We thank Martina Kühne, Rita Schubert, and Joachim Bruder for technical assistance and Armin Meister for stimulating discussion. This work was in part supported by a Deutschen Forschungsgemeinschaft grant to I.S. (Schu 951/10-1).
References
- Abney, J. R., B. Cutler, M. L. Fillbach, D. Axelrod and B. A. Scalettar, 1997. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J. Cell Biol. 137 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches, R., A. F. Beven, L. Aragón-Alcaide and P. J. Shaw, 1998. Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bártová, E., and S. Kozubek, 2006. Nuclear architecture in the light of gene expression and cell differentiation studies. Biol. Cell 98 323–336. [DOI] [PubMed] [Google Scholar]
- Bauwens, S., K. Katsanis, M. Van Montagu, P. Van Oostveldt and G. Engler, 1994. Procedure for whole mount fluorescence in situ hybridization of interphase nuclei on Arabidopsis thaliana. Plant J. 6 123–131. [Google Scholar]
- Belmont, A. S., 2006. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 18 632–638. [DOI] [PubMed] [Google Scholar]
- Berr, A., and I. Schubert, 2006. Direct labelling of BAC-DNA by rolling-circle amplification. Plant J. 45 857–862. [DOI] [PubMed] [Google Scholar]
- Berr, A., A. Pecinka, A. Meister, G. Kreth, J. Fuchs et al., 2006. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J. 48 771–783. [DOI] [PubMed] [Google Scholar]
- Bickmore, W. A., and J. R. Chubb, 2003. Chromosome position: Now, where was I? Curr. Biol. 13 R357–R359. [DOI] [PubMed] [Google Scholar]
- Bolzer, A., G. Kreth, I. Solovei, D. Koehler, K. Saracoglu et al., 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3 826–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornfleth, H., P. Edelmann, D. Zink, T. Cremer and C. Cremer, 1999. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys. J. 77 2871–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri, T., 1909. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch. Zellforschung 3 181–268. [Google Scholar]
- Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis et al., 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10 211–219. [DOI] [PubMed] [Google Scholar]
- Bridger, J. M., S. Boyle, I. R. Kill and W. A. Bickmore, 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 10 149–152. [DOI] [PubMed] [Google Scholar]
- Chubb, J. R., S. Boyle, P. Perry and W. A. Bickmore, 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12 439–445. [DOI] [PubMed] [Google Scholar]
- Chytilova, E., J. Macas, E. Sliwinska, S. M. Rafelski, G. M. Lambert et al., 2000. Nuclear dynamics in Arabidopsis thaliana. Mol. Biol. Cell 11 2733–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix, C., S. Turois, O. Mathieu, C. Cuvillier, M. C. Espagnol et al., 2000. Analysis of 5S rDNA arrays in Arabidopsis thaliana: physical mapping and chromosome-specific polymorphisms. Genome Res. 10 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer, M., J. von Hase, T. Volm, A. Brero, G. Kreth et al., 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9 541–567. [DOI] [PubMed] [Google Scholar]
- Cremer, M., I. Solovei, L. Schermelleh and T. Cremer, 2003. Chromosomal arrangement during different phases of the cell cycle, pp. 451–457 in Nature Encylopedia of the Human Genome, edited by D. N. Cooper. Nature Publishing Group, London.
- Cremer, T., A. Kurz, R. Zirbel, S. Dietzel, B. Rinke et al., 1993. Role of chromosome terrritories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp. Quant. Biol. 58 777–792. [DOI] [PubMed] [Google Scholar]
- Cremer, T., M. Cremer, S. Dietzel, S. Müller, I. Solovei et al., 2006. Chromosome territories: a functional nuclear landscape. Curr. Opin. Cell Biol. 18 307–316. [DOI] [PubMed] [Google Scholar]
- Croft, J. A., J. M. Bridger, S. Boyle, P. Perry, P. Teague et al., 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan, L., K. Janmaat, V. Willemsen, P. Linstead, S. Poethig et al., 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119 71–84. [DOI] [PubMed] [Google Scholar]
- Dong, F., and J. Jiang, 1998. Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6 551–558. [DOI] [PubMed] [Google Scholar]
- Essers, J., W. A. van Cappellen, A. F. Theil, E. van Drunen, N. G. J. Jaspers et al., 2005. Dynamics of relative chromosome position during the cell cycle. Mol. Biol. Cell 16 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y., and D. L. Spector, 2005. Centromere positioning and dynamics in living Arabidopsis plants. Mol. Cell. Biol. 16 5710–5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz, P., J. H. de Jong, M. Lysak, M. Ruffini Castiglione and I. Schubert, 2002. Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml, J., A. Vieten, M. Sauer, D. Weijers, H. Schwarz et al., 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426 147–153. [DOI] [PubMed] [Google Scholar]
- Gerlich, D., J. Beaudouin, B. Kalbfuss, N. Daigle, R. Eils et al., 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112 751–764. [DOI] [PubMed] [Google Scholar]
- Gottlob-McHugh, S. G., M. Lévesque, K. MacKenzie, M. Olson, O. Yarosh et al., 1990. Organization of the 5S rRNA genes in the soybean Glycine max (L.) Merrill and conservation of the 5S rDNA repeat structure in higher plants. Genome 33 486–494. [DOI] [PubMed] [Google Scholar]
- Habermann, F. A., M. Cremer, J. Walter, G. Kreth, J. von Hase et al., 2001. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res. 9 569–584. [DOI] [PubMed] [Google Scholar]
- Jovtchev, G., V. Schubert, A. Meister, M. Barow and I. Schubert, 2006. Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114 77–82. [DOI] [PubMed] [Google Scholar]
- Kawabe, A., and S. Nasuda, 2005. Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol. Genet. Genomics 272 593–602. [DOI] [PubMed] [Google Scholar]
- Koch, M. A., B. Haubold and T. Mitchell-Olds, 2000. Comparative evolutionary analysis of the chalocone synthase and alcohol dehydrogenase loci among different lineages of Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kozubek, S., E. Lukásová, P. Jirsová, I. Koutná, M. Kozubek et al., 2002. 3D structure of the human genome: order in randomness. Chromosoma 111 321–331. [DOI] [PubMed] [Google Scholar]
- Kreth, G., J. Finsterle, J. von Hase, M. Cremer and C. Cremer, 2004. Radial arrangement of chromosome territories in human cell nuclei: a computer model approach based on gene density indicates a probabilistic global positioning code. Biophys. J. 86 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwart, M., B. Hirner, S. Hummel and W. B. Frommer, 1993. Differential expression of 2 related amino-acid transporters with differing substrate-specificity in Arabidopsis thaliana. Plant J. 4 993–1002. [DOI] [PubMed] [Google Scholar]
- Lucas, J. N., and E. Cervantes, 2002. Significant large-scale chromosome territory movement occurs as a result of mitosis, but not during interphase. Int. J. Radiat. Biol. 78 449–455. [DOI] [PubMed] [Google Scholar]
- Ludevid, D., H. Höfte, E. Himelblau and M. J. Chrispeels, 1992. The expression pattern of the totoplast intrinsic protein γ-TIP in Arabidopsis thaliana is correlated with cell enlargement. Plant Physiol. 100 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak, M. A., A. Pecinka and I. Schubert, 2003. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 11 195–204. [DOI] [PubMed] [Google Scholar]
- Lysak, M. A., A. Berr, A. Pecinka, R. Schmidt, K. McBreen et al., 2006. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 103 5224–5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy, N. L., P. E. Perry, S. Gilchrist, R. A. Baldock and W. A. Bickmore, 2002. a Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy, N. L., P. E. Perry and W. A. Bickmore, 2002. b Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159 753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martou, G., and U. De Boni, 2000. Nuclear topology of murine, cerebellar Purkinje neurons: changes as a function of development. Exp. Cell Res. 256 131–139. [DOI] [PubMed] [Google Scholar]
- Mora, L., I. Sanchez, M. Garcia and M. Ponsa, 2006. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma 115 367–375. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and F. Skoog, 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plantarum 15 473–497. [Google Scholar]
- Parada, L. A., J. J. Roix and T. Misteli, 2003. An uncertainty principle in chromosome positioning. Trends Cell Biol. 13 393–396. [DOI] [PubMed] [Google Scholar]
- Pecinka, A., V. Schubert, A. Meister, G. Kreth, M. Klatte et al., 2004. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113 258–269. [DOI] [PubMed] [Google Scholar]
- Scheres, B., H. Wolkenfelt, V. Willemsen, M. Terlouw, E. Lawson et al., 1994. Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120 2475–2487. [Google Scholar]
- Schubert, I., P. F. Fransz, J. Fuchs and J. H. de Jong, 2001. Chromosome painting in plants. Methods Cell Sci. 23 467–475. [PubMed] [Google Scholar]
- Schubert, V., M. Klatte, A. Pecinka, A. Meister, Z. Jasencakova et al., 2006. Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby, R. D., K. M. Hahn and K. F. Sullivan, 1996. Dynamic elastic behavior of [alpha]-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H. B., and H. Yokota, 1999. Correlated positioning of homologous chromosomes in daughter fibroblast cells. Chromosome Res. 7 603–610. [DOI] [PubMed] [Google Scholar]
- Sun, H. B., J. Shen and H. Yokota, 2000. Size-dependent positioning of human chromosomes in interphase nuclei. Biophys. J. 79 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, H., F. A. Habermann, I. Solovei, M. Cremer and T. Cremer, 2002. a Non-random radial arrangements of interphase chromosome territories: evolutionary considerations and functional implications. Mutat. Res. 504 37–45. [DOI] [PubMed] [Google Scholar]
- Tanabe, H., S. Müller, M. Neusser, J. von Hase, E. Calcagno et al., 2002. b Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA 99 4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, H., K. Kupper, T. Ishida, M. Neusser and H. Mizusawa, 2005. Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet. Genome Res. 108 255–261. [DOI] [PubMed] [Google Scholar]
- Tautz, D., and C. Pfeifle, 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98 81–85. [DOI] [PubMed] [Google Scholar]
- Thomson, I., S. Gilchrist, W. A. Bickmore and J. R. Chubb, 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14 166–172. [DOI] [PubMed] [Google Scholar]
- Walter, J., L. Schermelleh, M. Cremer, S. Tashiro and T. Cremer, 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 160 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. R. E., and A. G. Fisher, 2003. Chromosomes, positions please! Nat. Cell Biol. 5 388–390. [DOI] [PubMed] [Google Scholar]
- Zhao, L. M., and F. D. Sack, 1999. Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am. J. Bot. 86 929–939. [PubMed] [Google Scholar]
- Zink, D., and T. Cremer, 1998. Chromosome dynamics in nuclei of living cells. Curr. Biol. 8 R321–R324. [DOI] [PubMed] [Google Scholar]