Abstract
Cysteine transport in the yeast Saccharomyces cerevisiae is mediated by at least eight different permeases, none of which are specific for cysteine. We describe a novel, high-affinity, (Km = 55 μm), cysteine-specific transporter encoded by the ORF YLL055w that was initially identified by a combined strategy of data mining, bioinformatics, and genetic analysis. Null mutants of YLL055w, but not of the other genes encoding for transporters that mediate cysteine uptake such as GAP1, GNP1, MUP1, or AGP1 in a met15Δ background, resulted in a growth defect when cysteine, at low concentrations, was provided as the sole sulfur source. Transport experiments further revealed that Yll055wp was the major contributor to cysteine transport under these conditions. The contributions of the other transporters became relevant only at higher concentrations of cysteine or when YLL055w was either deleted or repressed. YLL055w expression was repressed by organic sulfur sources and was mediated by the Met4p-dependent sulfur regulatory network. The results reveal that YLL055w encodes the principal cysteine transporter in S. cerevisiae, which we have named YCT1 (yeast cysteine transporter). Interestingly, Yct1p belongs to the Dal5p family of transporters rather than the amino acid permease family to which all the known amino acid transporters belong.
CYSTEINE, with its free sulphydryl group, is an important amino acid residue for the structural and functional properties of proteins. The free thiol group of cysteine is involved in the formation of disulphide bonds, crucial for the stability of certain proteins, and is also an important catalytic and redox center in various enzymes, cofactors, and regulatory proteins. Cysteine is also the rate-limiting nutrient in glutathione biosynthesis (Alfafara et al. 1992; Wen et al. 2004), the major redox buffer and detoxification molecule in the cell. Studies have revealed that the entire sulfur assimilation pathway leading to cysteine biosynthesis is upregulated during increased cellular demands of glutathione upon exposure to heavy metals or other toxic compounds in yeasts and plants (Vido et al. 2001; Fauchon et al. 2002; Aranda and del Olmo 2004; Mendoza-Cozatl et al. 2005). Despite the importance of cysteine in cellular metabolism, detoxification, and stress response, an increased cysteine level has been shown to be toxic to cells (Ono et al. 1991; Kumar et al. 2006). The intracellular cysteine levels are thus tightly regulated. In addition to the de novo synthesis of cysteine from inorganic sulfur in Saccharomyces cerevisiae, the transport of cysteine from the extracellular medium also contributes to the cellular cysteine homeostasis.
Several studies have been carried out to biochemically characterize cysteine transport and to identify the transporter proteins responsible for uptake of cysteine in S. cerevisiae. However, these studies have been complicated by the different strain backgrounds and growth conditions employed and have led to quite contradictory conclusions. In one of the earliest reports, cysteine uptake by a brewing strain of S. cerevisiae was found to be slow and nonsaturable over a large concentration range of cysteine (Maw 1963). Similarly, During-Olsen et al. (1999) showed that cysteine uptake was nonsaturable in the presence of ammonia as the nitrogen source and that two or more transport systems for cysteine uptake existed. In contrast, Ono and Naito (1991) reported the presence of a specific and a saturable cysteine transport system in a wild-type S288C strain background, which is derepressed only in the absence of sulfur in the growth medium.
Several broad-specificity transporter proteins that can mediate the transport of cysteine in S. cerevisiae have been identified (During-Olsen et al. 1999; Regenberg et al. 1999; Kosugi et al. 2001). During-Olsen et al. (1999) employed an overexpression strategy that identified several members of the amino acid permease (AAP) family that could mediate cysteine uptake under different growth conditions. However, these transporters mediated the uptake of cysteine in a nonspecific manner. They included the general amino acid permease, Gap1p, and the asparagine and glutamine amino acid permease, Agp1p, as the main transporters in proline medium (non-nitrogen repressing); the glutamine high-affinity permease, Gnp1p; the branched-chain amino acid transporters, Bap1p and Bap2p; and the amino acid transporters, Tat1p and Tat2p, which contributed to cysteine transport under ammonia-rich conditions (During-Olsen et al. 1999). In another study, Kosugi et al. (2001) employed a genetic strategy to show the involvement of Mup1p, a high-affinity methionine permease, also belonging to the AAP family, in cysteine uptake. On the basis of these studies, it appears that cysteine is not taken up by a specific permease, but rather by multiple permeases with broad specificity, each active under different sets of growth conditions. However, no specific high-affinity transporter for cysteine uptake has been proposed or characterized in the yeast S. cerevisiae so far.
The absence of a specific trasnporter for this important amino acid has appeared puzzling to us, and we have therefore considered it worthwhile to reexamine the yeast genome for the existence of a cysteine-specific transporter. Starting with an analysis that mined existing genomewide data, we sought out membrane transporters that were being derepressed under conditions of increased cysteine requirements in the cell. This analysis identified a candidate transporter, Yll055wp of unassigned function, belonging to the Dal5p transporter family. Although all the amino acid transporters described so far in S. cerevisiae fall into the amino acid permease family, we nevertheless investigated the possibility that YLL055w might encode a cysteine trasnporter. Detailed genetic, molecular, and biochemical analyses of this protein along with studies on its regulation, which are described in this report, reveal that the YLL055w ORF encodes a high-affinity, cysteine-specific transporter.
MATERIALS AND METHODS
Chemicals and reagents:
All the chemicals used in this study were obtained from commercial sources and were of analytical grade. Media components were purchased from Difco (Detroit). Oligonucleotides were purchased from Biobasic (Markham, ON, Canada). Restriction enzymes, Vent DNA polymerase, and Taq DNA polymerase and other modifying enzymes were obtained from New England Biolabs (Beverly, MA). A DNA sequencing kit (ABI PRISM 310 with dye termination cycle-sequencing ready-reaction kit) was obtained from Perkin-Elmer (Norwalk, CT). Gel-extraction kits and plasmid miniprep columns were obtained from QIAGEN (Valencia, CA) or Sigma (St. Louis). [35S]Cysteine (specific activity 37MBq mmol−1) was obtained from Bhabha Atomic Research Centre, Mumbai, India.
Strains, media, and growth conditions:
The Escherichia coli strain DH5α was used as a cloning host. Yeast strains used in the study are described in Table 1. The yeast was regularly maintained on nonselective yeast extract, peptone and dextrose medium. For yeast transformation and induction experiments, synthetically defined minimal medium containing yeast nitrogen base and dextrose supplemented with adenine, histidine, leucine, lysine, tryptophan, and uracil (when not used as auxotrophic marker) at 50 mg/liter was used with either 5 g/liter ammonium sulfate as the nitrogen source (minimal ammonium medium) or 2 g/liter proline as nitrogen source (minimal proline medium; During-Olsen et al. 1999). Growth, handling of bacteria and yeast, and all the molecular techniques used in the study were according to the standard protocols (Sambrook et al. 1989; Guthrie and Fink 1991).
TABLE 1.
Strains used in this study
|
Strain |
Genotype |
Source |
|---|---|---|
| ABC 734 (BY4742) | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | J. Boeke |
| ABC 733 (BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | J. Boeke |
| ABC 1080 (CC718-1A) | MATa his 3 leu2 ura3 ade2 trp1 cbf1∷TRP1 | Y. Surdin-Kerjan |
| ABC 1081 (CC950-2A) | MATa his 3 leu2 ura3 ade2 trp1 met 4∷TRP1 | Y. Surdin-Kerjan |
| ABC 1082 (CD130-7D) | MATa his 3 leu2 ura3 ade2 trp1 met 28∷LEU2 | Y. Surdin-Kerjan |
| ABC 1097 | MATa his3Δ1 leu 2Δ0 lys2Δ 0 ura3Δ 0 met31Δ∷KanMX4 | EUROSCARF |
| ABC 1098 | MATa his3Δ1 leu 2Δ 0 lys2Δ 0 ura3Δ 0 met32Δ∷KanMX4 | EUROSCARF |
| ABC 1381 (W303) | MATa his 3 leu2 ura3 ade2 trp1 | Y. Surdin-Kerjan |
| ABC 1580 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yll055wΔ∷KanMX4 | EUROSCARF |
| ABC 1814 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLL055w∷GFP-HIS3MX | Invitrogen (San Diego) |
| ABC 1827 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gnp1Δ∷KanMX4 | EUROSCARF |
| ABC 1839 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mup1Δ∷KanMX4 | EUROSCARF |
| ABC 1842 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gap1Δ∷KanMX4 | EUROSCARF |
| ABC 1846 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 agp1Δ∷KanMX4 | EUROSCARF |
| ABC 1902 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gnp1Δ∷KanMX4 yll055w∷HIS4 | This study |
| ABC 1903 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mup1Δ∷KanMX4 yll055w∷HIS4 | This study |
| ABC 1904 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yll055w∷HIS4 | This study |
| ABC 1905 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 agp1Δ∷KanMX4 yll055w∷HIS4 | This study |
| ABC 2090 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gnp1Δ∷KanMX4 mup1Δ∷LEU2 | This study |
| ABC 2091 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gnp1Δ∷KanMX4 mup1Δ∷LEU2 yll055w∷HIS4 | This study |
| ABC 2067 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yal067cΔ∷KanMX4 | EUROSCARF |
| ABC 2072 |
MATa
his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 yil166cΔ∷KanMX4 |
EUROSCARF |
Cloning of YLL055w and construction of the yll055w∷HIS3 disruption plasmid:
The ORF YLL055w was PCR amplified from yeast genomic DNA using the primer pair YLL055W-F and YLL055W-R (Table 2). The 1.6-kb PCR product obtained was digested with XbaI and XhoI and cloned downstream of the TEF promoter in the single copy, URA3-based expression vector (p416TEF; Mumberg et al. 1995).
TABLE 2.
Oligonucleotides and their sequences in this study
|
Oligomer name |
Sequence (5′–3′) |
|---|---|
| YLL055W-F | GCT AGC TCT AGA AAT GTC AAA AGT TGA CGT AAA AATTG |
| YLL055W-R | CGA GTC TCG AGT GTC AGA TAG AAC ATT TAC ACA AC |
| MUP1dLEU2-F | TCA ACA AGG AGA ACT ATC AAT TTT CTT CTT CTA CTACAA ATC GAC TAC GTC GTA AGG CCG |
| MUP1dLEU2-R | GGC AAT TTT GAC TCT CCA GAA CCC ATC TTC ACC AAG CAC AAA ATG GAA TCC CAA CAA TTA C |
| Xho1YLL055wP600F | AAC ACT CGA GTT CAG CAT CCG AGC |
| XhoYLLwP-387F | ATT GCT CGA GCA AAA ATG TGT GGC TTC TG |
| XhoYLLwP-372F | TGT GCT CGA GTC TGA AAA AAA AAA TAG GCA CCC C |
| BamH1YLL055wP-R |
AAT CGG ATC CCA TTT CTT TTT TGT TAT ATT TTC |
The yll055w∷HIS3 disruption plasmid was prepared by cloning the HIS3 disruption cassette at the BamHI site in the ORF YLL055w, leading to insertional disruption of YLL055w. The HIS3 disruption cassette was released by BamHI digestion of the plasmid pCEN3-HIS3 (Futcher and Carbon 1986).
Construction of strains:
The YLL055w ORF was disrupted in the met15Δ, met15Δ agp1Δ, met15Δ gap1Δ, met15Δ gnp1Δ, and met15Δ mup1Δ strain backgrounds using the yll055w∷HIS3 disruption plasmid. The pyll055w∷HIS3 disruption plasmid was digested with XbaI and EcoRI and the resulting 2.9-kb fragment was transformed into the strains. The transformants were selected on minimal ammonia medium without histidine and confirmed for the disruption by diagnostic PCR using the primer pair YLL055W-F and YLL055W-R.
The MUP1 gene was disrupted in met15Δ gnp1Δ and met15Δ gnp1Δ yll055wΔ strains using one-step PCR-mediated gene disruption (Baudin et al. 1993). The mup1Δ∷LEU2 disruption cassette was generated using the plasmid pair MUP1dLEU2-F and MUP1dLEU2-R and the plasmid pSP1 as a template (Cottarel et al. 1993). The 2.2-kb PCR product obtained was transformed into the strains and the resulting transformants were selected on minimal ammonia medium without leucine. The transformants were confirmed by their growth defect on a low concentration of methionine (2 μm) as a sole source of sulfur.
Construction of the YLL055w promoter–LacZ fusion constructs for the β-galactosidase reporter assay:
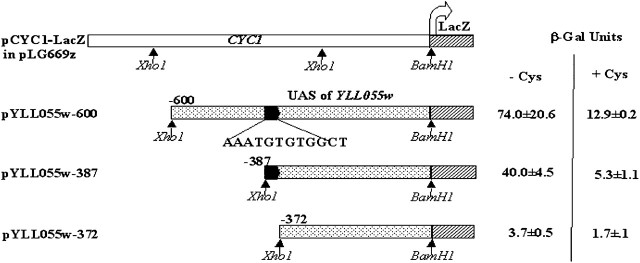
The YLL055w upstream activating sequences were PCR amplified using different primer pairs, using oligonucleotides listed in Table 2. Briefly, for pYLL055w-600, pYLL055w-387, and pYLL055w-372 reporter constructs, the 600, 387, and 372 bp upstream of YLL055w were amplified from yeast genomic DNA, using the forward primers XhoIYLL055wP-600F, XhoYLLwP-387F, and XhoYLLwP-372F, respectively, and the reverse primer BamH1YLL055wP-R. The PCR products were purified, digested with XhoI and BamHI, and cloned into pLG669z (Guarente and Ptashne 1981) (Figure 8).
Figure 8.—
β-Galactosidase reporter assay of different deletion constructs of the YLL055w promoter to identify the functionally important cis motifs in it. Multicopy plasmids bearing different upstream sequences fused to the LacZ ORF were transformed into the met15Δ strain and β-galactosidase enzyme activity was determined after growing cells in a poor sulfur source (2 μm methionine) and in a rich sulfur source (250 μm cysteine).
For construction of an integrative YLL055w promoter–LacZ fusion construct, the 600-bp BamHI–XhoI YLL055w promoter fragment cloned above was excised out and cloned into BamHI–XhoI sites of the integrative vector pLacZi (CLONTECH, Palo Alto, CA). This integrative plasmid pYLL055w-600i was linearized by digestion with SphI, which cuts in the YLL055w promoter. The linearized plasmid was transformed into the met15Δ (ABC 733) strain and the transformants were selected on minimal media plates without uracil. The β-galactosidase reporter assay was done with eight independent transformants.
Growth assay by dilution spotting:
For growth assay, the different strains were grown overnight in minimal ammonia medium without uracil and reinoculated in fresh medium to an OD600 of 0.1 and grown for 6 hr. The exponential-phase cells were harvested, washed with water, and resuspended in water to an OD600 of 0.2. These were serially diluted to 1:10, 1:100, and 1:1000. Of these cell resuspensions, 10 μl were spotted on minimal medium containing cysteine, dl-homocysteine, or methionine as sole sulfur source. The plates were incubated at 30° for 3 days and photographs were taken.
Induction conditions and β-galactosidase assay:
Fresh yeast transformants were used in all β-galactosidase experiments. The transformants were picked, grown overnight in minimal ammonia medium without uracil, and reinoculated in fresh minimal ammonia medium (without any organic sulfur source or with 0.25 mm cysteine, 0.25 mm methionine, or 0.25 mm glutathione) to an initial OD600 of 0.1. They were grown for an additional 6–7 hr to induce β-galactosidase. Since the met15Δ, met31Δ, met32Δ, met28Δ, cbf1Δ, and met4Δ strains used in this study were methionine/organic sulfur auxotrophs, the transformants were initially grown in medium containing methionine and were washed during induction and resuspended in medium containing a very low concentration of methionine (0.02 mm, a concentration that did not affect induction). The cells were then harvested and washed twice in Lac-Z buffer (60 mm Na2HPO4·7H2O, 40 mm NaH2PO4·H2O, 10 mm KCl, 1 mm MgSO4·7H2O, 0.27% 2-mercaptoethanol, pH 7) and ∼5 × 107 cells were taken for the reporter gene assay. β-Galactosidase activity was assayed in permeabilized yeast cells essentially as described previously (Guarente and Ptashne 1981). Briefly, the cells were resuspended in Lac-Z buffer, permeabilized by the addition of 50 μl chloroform and 20 μl SDS (0.1%). They were then vigorously vortexed for 20 sec. These samples were equilibrated for 10 min at 30° and then o-nitrophenyl β-galactopyranoside was added sequentially to the reaction samples. β-Galactosidase units are given as OD420 × 1000 min−1 ml−1/OD600 at 30°. The experiments were repeated with a minimum of three independent colonies.
Cysteine uptake measurement:
Yeast transformants were grown overnight in minimal ammonia medium without uracil and reinoculated to an OD600 of 0.1 in fresh medium containing 2 μm methionine (nonrepressive sulfur conditions) or 200 μm methionine (repressive sulfur conditions). Cultures were incubated at 30° for 6 hr and exponentially growing cells were harvested and washed with a large volume of ice-cold MES buffer (20 mm MES/KOH, 0.5 mm CaCl2, 0.25 mm MgCl2, pH 5.5). Cells were finally resuspended in the MES buffer containing 2% glucose (resuspension buffer) at 2 OD600/ml, aliquoted in 100-μl samples, and kept on ice. After a 5-min preincubation of cells at 30°, cysteine uptake was initiated by addition of 100 μl of assay medium. The assay medium contained radiolabeled cysteine ([35S]cysteine, specific activity 37MBq mmol−1) in MES resuspension buffer at a concentration of 100 μm, such that the final concentration of cysteine was 50 μm in the reaction vial. At selected times, the uptake was stopped by diluting the medium with a 20-fold volume of ice-cold water and cells were collected on the glass fiber prefilter (Advanced Microdevices, Ambala, India) by vacuum filtration. The harvested cells were washed twice with the same volume of ice-cold water. The filters were immersed in 3 ml of scintillation fluid (Sigma-Fluor Universal LSC cocktail, Sigma) and radioactivity was measured using a liquid scintillation counter (Wallac Microbeta, 1450 LSC and luminescence counter, Perkin-Elmer Life Sciences, Groningen, The Netherlands). The results were expressed as nanomoles of cysteine · mg · protein−1 min−1.
For saturation kinetics, the initial rate of cysteine uptake was measured at a range of cysteine concentrations from 10 to 100 μm, with specific activity being kept constant at each concentration. The initial rate of cysteine uptake was determined by measuring the radioactive cysteine accumulated in the cells at 30- and 90-sec time points.
For inhibition studies with different sulfur compounds and amino acids, the competing ligand was added at 20-fold excess along with assay medium and initial rate of cysteine uptake was measured. The metabolic inhibitors were preincubated with the cells for 15 min at 30°, before measuring the initial rate of cysteine uptake.
As we observed that the cysteine transport activity rapidly fell when the cells were incubated in resuspension buffer on ice, the uptake experiments were performed within half an hour of the incubation of cells on ice.
Phylogenetic footprinting:
The identification of conserved sequences in the multiple sequence alignment of YLL055w ortholog promoters of the Saccharomyces species was carried out using the webserver (http://203.90.127.21/anand/sacch_prom_pat.html; Kohli et al. 2004).
RESULTS
Identification of Yll055wp, a member of the Dal5p transporter family, as a possible cysteine transporter:
To identify putative transporters in the yeast genome that were induced under conditions leading to insufficiency for cysteine in the cell, we examined the published literature for different genomewide expression profiling studies carried out in S. cerevisiae. This analysis revealed an uncharacterized ORF, YLL055w, encoding a putative transporter that was specifically upregulated under sulfur-limiting conditions under both aerobiosis and anaerobiosis, similarly to other sulfur assimilatory enzymes (Boer et al. 2003; Tai et al. 2005). In addition, this ORF was also found to be induced in a gsh1Δ strain background (Wheeler et al. 2003) (which has reduced intracellular levels of glutathione) and upon exposure to cadmium (Vido et al. 2001; Fauchon et al. 2002) or acetaldehyde (Aranda and del Olmo 2004), conditions under which most of the genes in the sulfur assimilation pathway were upregulated.
The induction profiles of the YLL055w ORF in these different studies were consistent with a role in sulfur assimilation, and therefore indicated a candidate cysteine transporter. However, this ORF did not belong to the amino acid permease family, to which all the known amino acid transporters have been found to belong. Yll055wp, which is predicted to encode a polypeptide of 531 amino acids, shows homology to Dal5p, an allantoate permease, in S. cerevisiae and is a member of the DAL5 gene family. The DAL5 gene family is a subfamily of the anion:cation symporter family, which in turn belongs to the major facilitator superfamily (Nelissen et al. 1997; Saier et al. 1999). The members encode putative weak acid permeases, which share between 20 and 30% amino acid identity. Some of the eight Dal5p family members have been functionally characterized and shown to transport substrates such as allantoate, biotin, pantothenate, and nicotinic acid in S. cerevisiae (Table 3). Although none have been shown to transport amino acids, two—YLL055w and YIL166c with unknown function—have been shown to be under sulfur regulation (Boer et al. 2003) while a third, YAL067c, has been picked in a mutagenic screen based on a toxic analog of methionine, the same screen in which Mup1p was identified as the high-affinity methionine permease (Isnard et al. 1996). These observations raised the possibility that one or more of these uncharacterized ORFs could probably encode for the transporters involved in uptake of sulfate or sulfur-containing molecules, such as cysteine.
TABLE 3.
DAL5 gene family members: sulphur amino acid composition and regulation pattern
|
ORF |
Gene name |
Reported substrate |
Cys + Met residues (% Cys + Met residues) |
Regulation |
Reference |
|---|---|---|---|---|---|
| YJR152w | DAL5 | Allantoate ureidosuccinate | 8 + 18 (4.9) | Nitrogen catabolite repression | Rai et al. (1987, 1988) |
| YLR004c | THI73 | Unknown | 14 + 15 (5.6) | Thiamine | Llorente and Dujon (2000) |
| YLL055w | — | Unknown | 4 + 12 (3.0) | Sulphur source | Boer et al. (2003) |
| YGR260w | TNA1 | Nicotinic acid | 13 + 18 (5.8) | Nicotinic acid | Llorente and Dujon (2000) |
| YIL166c | — | Unknown | 13 + 18 (3.7) | Sulphur source | Boer et al. (2003) |
| YCR028c | FEN2 | Pantothenate | 9 + 20 (5.7) | Pantothenate | Stolz and Sauer (1999) |
| YAL067c | SEO1 | Unknown | 8 + 11 (3.2) | — | Isnard et al. (1996) |
|
YGR065c |
VHT1 |
Biotin |
13 + 11 (4.0) |
Biotin |
Stolz
et al. (1999) |
A characteristic feature of the sulfur assimilatory enzymes and proteins of yeast is that they have a lower sulfur amino acid content (Baudouin-Cornu et al. 2001). We therefore analyzed the sulfur amino acid content of all the members of the Dal5p family to see if further possibilities for these ORFs being a part of the sulfur assimilatory pathway might be suggested. Among the members of the Dal5p family, Yll055wp showed the lowest sulfur content, further suggesting that the protein indeed might play a role in sulfur assimilation (Table 3).
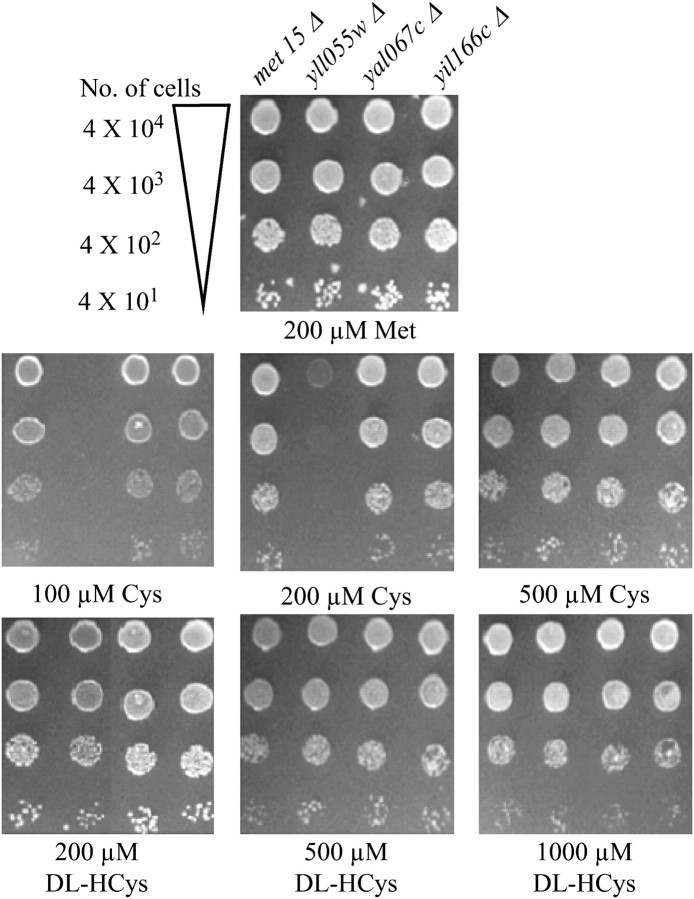
On the basis of these analyses, a preliminary genetic investigation into the role of these uncharacterized ORFs of the Dal5p family was carried out to determine their role in cysteine or homocysteine uptake. A met15Δ strain is unable to assimilate inorganic sulfur and is an organic sulfur auxotroph (Thomas and Surdin-Kerjan 1997). It can grow on organic sulfur sources such as methionine, glutathione, cysteine, and homocysteine. The deletion strains yll055wΔ, yil166cΔ, and yal067cΔ, each in a met15Δ background, were procured from the EUROSCARF (European S. cerevisiae Archive for Functional Analysis) deletion collection and checked for their ability to utilize cysteine or homocysteine as a sole sulfur source (Figure 1). The disruptant strains along with the wild type (met15Δ) were serially diluted and spotted on minimal ammonia medium with cysteine or homocysteine as the sole source of organic sulfur over a range of concentrations. The wild type and the different disruptants were able to use methionine as an organic sulfur source efficiently. However, a clear growth defect was observed for the yll055wΔ mutant at low concentrations of cysteine (100 and 200 μm), as compared to the wild type (Figure 1). At higher cysteine concentrations, both strains grew equally well. As multiple permeases have been shown to be capable of transporting cysteine (During-Olsen et al. 1999; Regenberg et al. 1999; Kosugi et al. 2001), it is likely that the phenotype of yll055w disruption was being masked by the activity of these permeases at the higher cysteine concentrations. The disruptions of the other two ORFs, YIL166c and YAL067c, did not result in any growth defect at any of the cysteine concentrations examined. Furthermore, no difference was observed in the utilization of homocysteine between the wild type and the three disruptants. Taken together, these results strongly suggested the possibility that Yll055wp might in fact be a cysteine transporter.
Figure 1.—
YLL055w disruption in the met15Δ background results in a growth defect at low cysteine concentrations. The met15Δ, met15Δ yll055wΔ, met15Δ yil166cΔ, and met15Δ yal067cΔ strains were grown to exponential phase in minimal ammonia medium containing methionine, harvested, washed, and resuspended in water and serially diluted to give 0.2, 0.02, 0.002, and 0.0002 OD600 of cells. A total of 10 μl of these dilutions was spotted on minimal ammonia medium containing different concentrations of cysteine (Cys) and dl-homocysteine (dl-Hcys). The photographs were taken after 3 days of incubation at 30°.
YLL055w encodes the major cysteine transporter of S. cerevisiae:
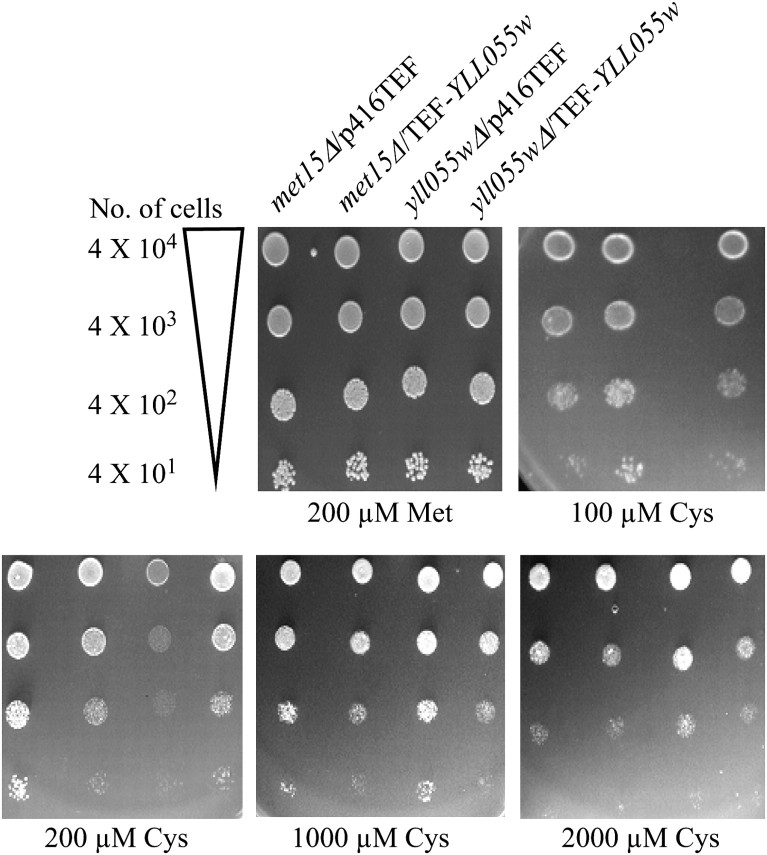
To evaluate the role of YLL055w in cysteine uptake, the YLL055w ORF was cloned downstream of the TEF promoter in a centromeric expression vector (p416TEF). Upon introduction into a yll055wΔ mutant, the gene complemented the growth defect at low cysteine concentrations (Figure 2), confirming the role of the YLL055w ORF in the utilization of cysteine in this strain. At high concentrations of cysteine, transformation with the YLL055w gene expressed from the strong TEF promoter also showed cessation of growth of both the met15Δ yll055wΔ and the met15Δ strain. This is likely to be a consequence of the increased cysteine uptake and cysteine toxicity, a phenomenon similar, although less acute, than what was observed with the yeast glutathione transporter (Srikanth et al. 2005). Complementation of the growth defect of the met15Δ yll055wΔ strain and the cysteine-mediated toxicity upon overexpression of YLL055w gave further support to the notion that Yll055wp might indeed be a cysteine transporter.
Figure 2.—
The YLL055w ORF expressed from TEF promoter complements the growth defect observed in met15Δ yll055wΔ at low cysteine concentrations. Plasmid-bearing YLL055w under the TEF and the corresponding vector were transformed into met15Δ and met15Δ yll055wΔ (yll055wΔ). The dilution spotting was done as described in materials and methods and in the legend to Figure 1.
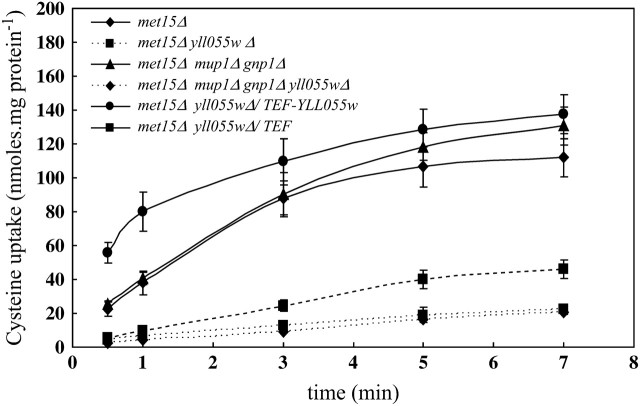
To gain more direct information on the role of YLL055w-encoded protein in cysteine uptake, we next measured the accumulation of radiolabeled cysteine ([35S]cysteine) in the met15Δ cells and met15Δ yll055wΔ cells. Cells were grown in a limiting organic sulfur source (20 μm methionine) and cysteine uptake was measured at different time intervals as described in materials and methods. In agreement with the growth assays done on the plates, the disruption of YLL055w in the met15Δ background led to a significant decrease in the uptake of radiolabeled cysteine (50 μm cysteine) as compared to the wild-type strain (Figure 3). The initial rate of cysteine uptake in met15Δ cells was 31.4 nmol · mg protein−1 min−1 compared to 5.3 nmol · mg protein−1 min−1 in the met15Δ yll055wΔ cells. We attributed the residual uptake observed in the deletant strain to the presence of the other nonspecific permeases and nonspecific binding of the radiolabeled ligand to the cell. However, the amount of cysteine taken up by the other permeases appeared to be insufficient in meeting the sulfur demands of the cells needed for growth when cysteine is present at low concentrations. Complementation of the yll055wΔ strain with YLL055w expressed from the TEF promoter led to a two- to threefold higher accumulation of [35S]cysteine in cells as compared to the wild-type cells (Figure 3). These observations confirm that YLL055w encodes a major yeast cysteine transporter, and we refer to the gene as YCT1 in the remaining sections of this article.
Figure 3.—
YLL055w disruption in met15Δ and met15Δ gnp1Δ mup1Δ results in decreased uptake of [35S]cysteine, which is restored by the YLL055w ORF expressed from TEF promoter. Strains—met15Δ, met15Δ yll055wΔ, met15Δ gnp1Δ mup1Δ, met15Δ gnp1Δ mup1Δ yll055wΔ, and met15Δ yll055wΔ complemented with YLL055w under the TEF promoter and the corresponding vector—were grown in minimal ammonia medium containing 2 μm methionine and used for the transport assay as described in materials and methods. Log-phase cells were incubated with 50 μm [35S]cysteine for different time intervals and counts were taken to determine the intracellular cysteine accumulation with time. Data are shown as mean ± SD (n = 3).
The previously reported cysteine permeases such as Gap1p, Gnp1p, Mup1p, and Agp1p play a negligible role in cysteine uptake, as compared to Yct1p, in different nitrogen sources:
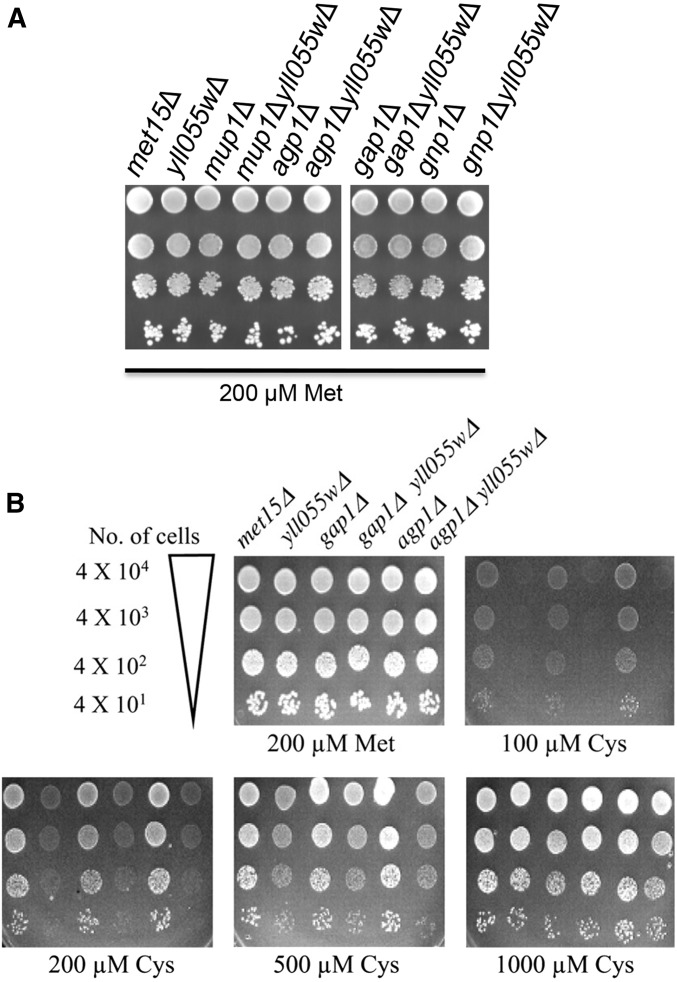
Earlier studies have revealed Gap1p, the general amino acid permease, as the major cysteine transporter (Greasham and Moat 1973; During-Olsen et al. 1999). However, Gap1p is inactive in yeast growing on a preferred nitrogen source, e.g., ammonia (Courchesne and Magasanik 1983; Jauniaux and Grenson 1990). Gap1p, along with Agp1p, mediates cysteine uptake in media containing nonrepressive nitrogen sources (e.g., proline medium), with Agp1p being induced in the presence of leucine in the medium (During-Olsen et al. 1999; Regenberg et al. 1999). In ammonia-based medium devoid of amino acids, Agp1p, Gnp1, and Mup1p have been shown to be the major participants in cysteine uptake (During-Olsen et al. 1999; Regenberg et al. 1999; Kosugi et al. 2001). Hence, to evaluate the contribution of Yct1p in cysteine uptake in comparison to the other permeases mediating cysteine transport, we decided to disrupt the major cysteine transporters reported in the literature and check the growth on cysteine as the sulfur source. Accordingly, the deletions of AGP1, GAP1, GNP1, and MUP1 in the met15Δ background were procured from the EUROSCARF deletion collection, and YLL055w (YCT1) deletions were also created in these backgrounds as described in materials and methods. As the HIS3 marker was used to disrupt the YLL055w gene in these strains, we used the same disruption cassette to make a met15Δ yll055w∷HIS3 strain. The met15Δ, met15Δ agp1Δ, met15Δ gap1Δ, met15Δ gnp1Δ, and met15Δ mup1Δ, as well as the met15Δ yll055wΔ, met15Δ agp1Δ yll055wΔ, met15Δ gap1Δ yll055wΔ, met15Δ gnp1Δ yll055wΔ, and met15Δ mup1Δ yll055wΔ disruptants, were analyzed for their ability to utilize cysteine as an organic sulfur source in the presence of different nitrogen sources.
As shown in Figure 4A, the disruption alone of AGP1, GAP1, GNP1, or MUP1 in the met15Δ background did not have any effect on growth on cysteine as a sole source of organic sulfur in minimal ammonia medium. However, disruption of YCT1 in the respective deletion strains led to a complete growth defect at low cysteine concentrations, similar to what was observed upon disruption of YCT1 in the met15Δ strain alone (Figure 1). Interestingly, the disruption of MUP1 and GNP1 led to better growth at the high-cysteine concentrations (1 and 2 mm; Figure 4A) as compared to the met15Δ cells, which we take to be due to decreased cysteine-associated toxicity in these strains. This observation implies that Mup1p and Gnp1p have some contribution in cysteine uptake at high concentrations, as previously reported. However, the role of Agp1p in cysteine uptake is not evident, despite the fact that leucine, which is known to induce AGP1 expression, has been included in the growth medium to meet the auxotrophic demands of the strains.
Figure 4.—
Effect of YLL055w disruption on utilization of cysteine as the sole sulfur source in different strain backgrounds deleted in the major cysteine permeases in the presence of different nitrogen sources. (A) Strains—met15Δ, met15Δ yll055wΔ, met15Δ mup1Δ, met15Δ mup1Δ yll055wΔ, met15Δ agp1Δ, met15Δ agp1Δ yll055wΔ, met15Δ gap1Δ, met15Δ gap1Δ yll055wΔ, met15Δ gnp1Δ, and met15Δ gnp1Δ yll055wΔ—were used for the dilution spotting on ammonia as the nitrogen source in minimal media. (B) Strains—met15Δ, met15Δ yll055wΔ, met15Δ gap1Δ, met15Δ gap1Δ yll055wΔ, met15Δ agp1Δ, and met15Δ agp1Δ yll055wΔ were analyzed for growth on minimal proline medium by dilution spotting.
As Mup1p and Gnp1p appeared to contribute to cysteine uptake in the plate assay at higher cysteine concentrations (as seen on the basis of decreased toxicity), MUP1 was disrupted in met15Δ gnp1Δ and met15Δ gnp1Δ yll055wΔ strains to give met15Δ gnp1Δ mup1Δ and met15Δ gnp1Δ mup1Δ yll055wΔ strains. The radiolabeled cysteine (50 μm cysteine) uptake was determined in these strains and compared to uptake in met15Δ (Figure 3). No difference was observed in cysteine accumulation with time between the met15Δ cells and the met15Δ gnp1Δ mup1Δ cells. Moreover, the rate of cysteine uptake in the latter strain was 30.8 nmol · mg protein−1 min−1, which was similar to the rate of cysteine uptake in the met15Δ strain (31.4 nmol · mg protein−1 min−1). The disruption of YCT1 in met15Δ gnp1Δ mup1Δ decreased the rate of cysteine uptake to 3.8 nmol · mg protein−1 min−1, underlining the predominant role played by Yct1p in cysteine uptake at low concentrations.
As only Gap1p and Agp1p have been implicated in cysteine uptake in media with a nonpreferred nitrogen source (During-Olsen et al. 1999), independent gap1 and agp1 disruptants in the met15Δ yll055wΔ background were checked for their ability to grow on cysteine as a sulfur source in the presence of proline as the sole nitrogen source. Despite the presence of Gap1p in met15Δ agp1Δ yll055wΔ and vice versa, yct1 disruption prevents growth at low concentrations and results in a marginal growth defect even at a higher cysteine concentration (500 μm cysteine) (Figure 4B). met15Δ agp1Δ and met15Δ gap1Δ by themselves did not show any obvious growth defect on cysteine and grew as well as met15Δ alone. From these results, it can be concluded that Yct1p is the major contributor to cysteine uptake in both nitrogen-rich and nitrogen-poor medium, especially at low-cysteine concentrations, at which the different cysteine permeases reported so far have a very limited role to play.
Biochemical characterization of Yct1p reveals that it is a high-affinity, cysteine-specific transporter:
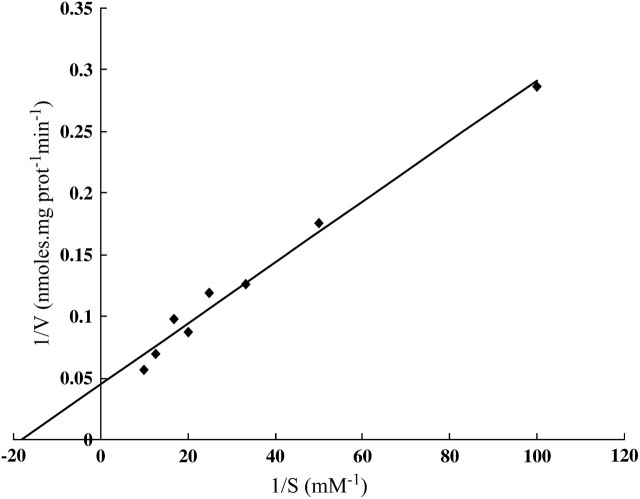
From our genetic data it is evident that at high-cysteine concentrations (>500 μm), multiple permeases are involved in cysteine uptake; this probably explains the observed nonsaturation in cysteine uptake by During-Olsen et al. (1999). However, at low-cysteine concentrations (<200 μm), only Yct1p mediates cysteine uptake. Hence, for kinetic studies of Yct1p, we chose a narrow range of substrate concentrations, 10–100 μm, and measured the initial rate of cysteine uptake in the met15Δ and the met15Δ yll055wΔ background. The Lineweaver–Burke plot, obtained after subtracting the values of the initial rate of cysteine uptake in the met15Δ yll055wΔ double-delete strain from the values for the met15Δ strain, was linear, implying that a single uptake system is involved in uptake in this narrow range of concentrations (Figure 5). A Km of 55.4 ± 6.1 μm and a Vmax of 22.4 ± 6.3 nmol of cysteine · mg · protein−1 min−1 were calculated from this curve.
Figure 5.—
Lineweaver–Burke plot for cysteine uptake in the met15Δ strain to determine kinetic parameters of Yct1p. The initial rate of cysteine uptake was measured at cysteine concentrations ranging from 0.01 to 0.1 mm by harvesting cells incubated with radiolabeled cysteine at 30- and 90-sec time intervals. Data are representative of three experiments (done in duplicates).
To define the substrate specificity of the transporter, competitive transport studies were undertaken in which the initial rate of cysteine uptake was measured in the presence of 20-fold excess of unlabeled competing ligand. Taking into account the participation of other amino acid permeases in cysteine uptake, we undertook these studies in the met15Δ gnp1Δ mup1Δ triple-delete strain to decrease the interference from these permeases in the competition assay. Moreover, these studies were done at low substrate concentration (40 μm) so that we could preferentially measure the cysteine uptake activity of the Yct1p.
The effect of the different sulfur amino acids homocysteine and methionine and the nonsulfur amino acids such as glycine, proline, valine, leucine, phenylalanine, serine, glutamate, glutamine, and lysine was evaluated on the cysteine uptake by Yct1p. Among the various amino acids used, only homocysteine (and, to a lesser extent, methionine) inhibited the cysteine uptake (Table 4). However, the other amino acids checked had little or no effect on the uptake of cysteine. Moreover, sulfur compounds with modified carboxy group (cysteamine), amino group (cysteic acid), or higher molecular weight (glutathione, cystathionine, cystine) did not have any effect on cysteine uptake in the met15Δ gnp1Δ mup1Δ triple-delete strain. Hence, unlike the other known amino acid permeases that transport cysteine, Yct1p is a highly cysteine-specific transporter.
TABLE 4.
Effect of various compounds on cysteine uptake in the met15Δ gnp1Δ mup1Δ triple-delete strain
|
Compound |
% activity |
|---|---|
| No inhibitor | 100 |
| Amino acidsa | |
| l-cysteine | 45 ± 2 |
| dl-homocysteine | 58 ± 2 |
| l-methionine | 76 ± 2 |
| Glycine | 90 ± 3 |
| l-proline | 100 ± 1 |
| l-valine | 85 ± 2 |
| l-leucine | 97 ± 1 |
| l-phenylalanine | 89 ± 1 |
| l-serine | 100 ± 1 |
| l-glutamine | 99 ± 4 |
| l-gluatamic acid | 98 ± 3 |
| l-lysine | 89 ± 1 |
| Sulphur compoundsa | |
| l-cysteamine HCl | 92 ± 10 |
| dl-cysteic acid | 111 ± 15 |
| l-glutathione | 98 ± 2 |
| l-cystineb | 102 ± 3 |
| l-cystathionineb | 104 ± 4 |
| Buffer (50 mm KCl–HCl pH 1)b | 109 ± 2 |
| Metabolic inhibitorsc | |
| Sodium azide (40 μm) | 64 ± 5 |
| Sodium azide (100 μm) | 42 ± 4 |
| CCCP (40 μm)d | 62 ± 5 |
| CCCP (100 μm)d | 48 ± 1 |
| DMSOd |
101 ± 5 |
The rate of uptake of cysteine (40 μm) was measured in met15Δ gnp1Δ mup1Δ in the presence of the different compounds listed in the table. The cells were harvested at 30- and 90-sec intervals. The results were normalized to the rate of uptake measured in the absence of any other compound (no inhibitor). Data are shown as mean ± SD (n = 4).
All competitors were added to a final concentration of 800 μm in a 10-μl volume, i.e., 20-fold excess over the labeled substrate, and were added simultaneously with uptake medium.
l-cystine and l-cystathionine were dissolved in buffer (50 mm KCl–HCl, pH 1); hence the buffer was analyzed for its effect on cysteine uptake by the cells.
Cells were preincubated with the indicated concentrations of metabolic inhibitors for 15 min prior to the addition of the uptake medium.
CCCP was dissolved in DMSO and hence DMSO was analyzed for its effect on cysteine uptake by the cells.
The metabolic inhibitors, such as sodium azide and CCCP, which deplete the cellular ATP level and collapse the transmembrane proton gradient, were also examined for any inhibitory effect on cysteine uptake by Yct1p. A 15-min preincubation of the inhibitor with the cells resulted in a considerable loss in the cysteine uptake. This implies that Yct1p-mediated cysteine uptake is an energy-dependent process (Table 4).
The competitive transport studies done in the met15Δ gnp1Δ mup1Δ triple-delete strain retained ∼40% activity with both cysteine (the strongest inhibitor) and metabolic inhibitors (even at sodium azide concentrations of 1 mm; data not shown) and is a consequence of background activity in the strain due to contribution from the other cysteine transporters as well as nonspecific binding of the radiolabeled ligand. (The cysteine uptake activity in the met15Δ gnp1Δ mup1Δ yll055wΔ tetra-delete strain was found to be nearly 15–25% of the cysteine uptake observed in the met15Δ gnp1Δ mup1Δ triple-delete strain, hence contributing significantly to background cysteine observed in the triple-delete strain.)
YCT1 is under sulfur regulation and is the major cysteine transporter under sulfur-derepressed conditions:
As mentioned earlier, genomewide expression profiling studies have revealed that YCT1 is expressed under conditions of sulfur limitation, cadmium toxicity, low-glutathione levels, and acetaldehyde exposure, conditions under which the entire sulfur assimilation pathway has been shown to be upregulated (Vido et al. 2001; Fauchon et al. 2002; Boer et al. 2003; Wheeler et al. 2003; Aranda and del Olmo 2004; Tai et al. 2005). To gain further insights into the physiological role of Yct1p, we decided to undertake a more detailed study on the expression pattern of YCT1 in response to different sulfur sources.
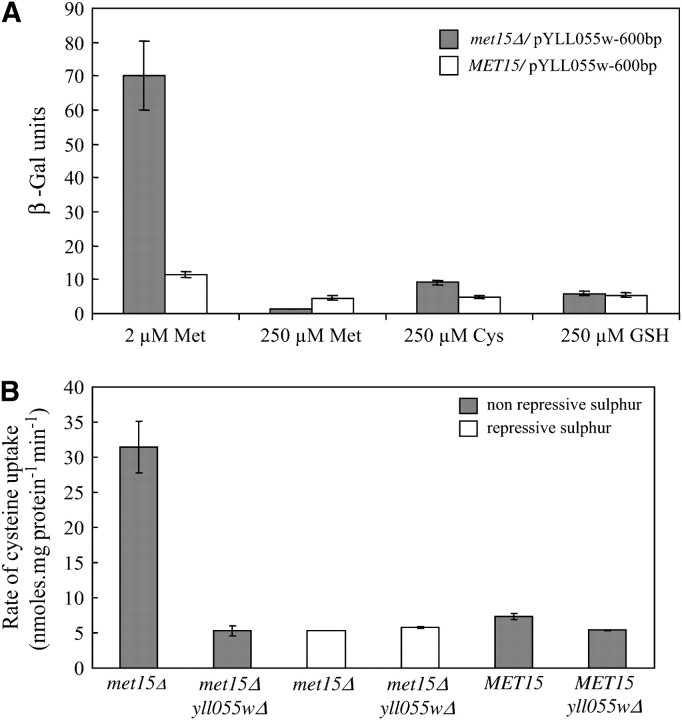
A YCT1 promoter–β-galactosidase reporter fusion was constructed in which the 600-bp upstream sequences of YCT1 were fused upstream of the β-galactosidase gene in the multicopy plasmid pLG699Z. The transformants were grown in different sulfur sources and examined for β-galactosidase activity in both a met15Δ and a MET15 background. As shown in Figure 6A, expression of YCT1 was maximum in nonrepressing sulfur conditions in both the met15Δ and the MET15 background and considerably repressed in the presence of organic sulfur in the medium. Among the organic sulfur sources, methionine strongly repressed the expression of YCT1, whereas cysteine had a milder repressing effect in the met15Δ background. However, this differential repression pattern was not observed in the wild-type (MET15) background, in which methionine, cysteine, and glutathione resulted in mild repression of YCT1. Moreover, the fold induction in the YCT1 expression between the poor methionine and the rich methionine medium was ∼60-fold in the met15Δ background, whereas it was just 2-fold in the MET15 background. This could be the result of sulfur starvation that is being experienced by cells in the met15Δ background due to the impairment of the sulfur assimilation pathway in this strain, implying that YCT1 is under tight sulfur regulation.
Figure 6.—
Expression of YLL055w is under strong sulfur regulation in both met15Δ (ABC 733) and MET15 (ABC 734) strains. (A) β-Galactosidase reporter assay–plasmid pYLL055w-600, bearing 600-bp upstream sequences of YLL055w fused upstream of the LacZ ORF, was transformed into met15Δ and MET15 strains and the transformants were grown in the presence of different sulfur sources and assayed for β-galactosidase enzyme activity as described in materials and methods. Data are shown as mean ± SD (n = 5). (B) Cysteine uptake. Strains met15Δ and met15Δ yll055wΔ were grown in minimal ammonia medium containing 2 μm methionine (nonrepressive sulfur) and 200 μm methionine (repressive sulfur) and the initial rate of [35S]cysteine uptake was calculated by measuring the radiolabeled cysteine accumulated in the cells at 30- and 90-sec time intervals. Wild-type (MET15) and yll055wΔ strains were also grown in minimal ammonia medium and the initial rate of cysteine uptake was determined. Data are shown as mean ± SD (n = 3).
To confirm if the sulfur repression seen with 2μ plasmids was also observed in a single-copy integrated promoter–LacZ fusion at the YLL055w site, we also constructed a 600-bp YLL055w promoter–β-galactosidase reporter fusion in an integrating plasmid and integrated this construct in the YLL055w promoter in a met15Δ strain (materials and methods). We observed >13-fold repression in the YCT1 expression when the integrative transformants were grown in methionine-rich medium (data not shown). However, as the sensitivity obtained with the integrated reporter construct was very low, we worked with 2μ-based constructs for the subsequent studies on YCT1 expression.
The repression of YCT1 by organic sulfur sources was also observed in actual transport measurements. Cysteine uptake was measured in the met15Δ strain after growing it in rich methionine medium (Figure 6B). No difference was observed in cysteine uptake between the met15Δ strain and the met15Δ yll055wΔ when these strains were grown in 200 μm methionine containing minimal ammonia medium. Interestingly, although Mup1p, the methionine transporter that has also been shown to transport cysteine (Kosugi et al. 2001), is also derepressed in sulfur-limiting conditions (Boer et al. 2003), it did not appear to contribute to cysteine transport to any significant extent under these conditions. The tight sulfur regulation of the YCT1 gene was also reflected by the low rate of cysteine uptake in the MET15 strain background, which was found to be just marginally more than the rate of cysteine uptake in the yll055wΔ background (Figure 6B). These results imply that the major role of Yct1p in cysteine transport is under conditions of sulfur limitations in the cell.
The YCT1 gene is under regulation by the Met4p-based sulfur regulatory network and involves cis-elements corresponding to the known cis-regulatory elements:
Studies on the regulation of the different structural genes in the sulfur assimilation pathway have led to identification of three cis-acting elements: TCACGTG, AAANTGTGG, and TGACTC, which are present in different combinations in the upstream regions of the sulfur assimilatory pathway genes (Thomas and Surdin-Kerjan 1997). Of these, the “TCACGTG” sequence is the binding site for the heteromeric transcriptional activation complex, Cbf1p–Met4p–Met28p (Kuras et al. 1996). Met4p is a leucine zipper transcriptional activator and is absolutely essential for activation of the sulfur assimilation genes in nonrepressing conditions (Thomas et al. 1992). The second motif, “AAANTGTGG,” is recognized by the Met4p–Met28p–Met31p/Met32p complex, although the role of this complex in gene regulation has been shown to be gene dependent (Blaiseau et al. 1997). The role of the third motif and the trans-factors bound to it are yet unknown (Thomas and Surdin-Kerjan 1997). In addition, a recent study in our lab on regulation of HGT1, a high-affinity glutathione transporter, has led to the identification of a novel cis-element, CGCCACA, present in two copies, and it was shown that the trans-factors Met4p, Met28p, and Met31p control the regulation of this gene (Srikanth et al. 2005). Considering the strong derepression of YCT1 in sulfur-limiting conditions, we decided to examine the cis-elements and trans-factors involved in the transcriptional activation of YCT1 to check whether YCT1 expression is regulated by the same mechanism as the other genes in the sulfur assimilation pathway.
A general sequence inspection of the 1-kb upstream sequence region of YCT1 did not reveal the presence of any of the above cis-motifs reported in the sulfur regulation. Hence we resorted to the phylogenetic footprinting approach to identify any of the conserved motifs in upstream sequences of YCT1. Phylogenetic footprinting, a comparative genomics approach, has been successfully employed to identify regulatory motifs and other functional elements in the noncoding regions of the genomes on the basis of their high degree of conservation across the closely related species during the course of evolution due to selection pressure (Cliften et al. 2003; Kellis et al. 2003; Lenhard et al. 2003).
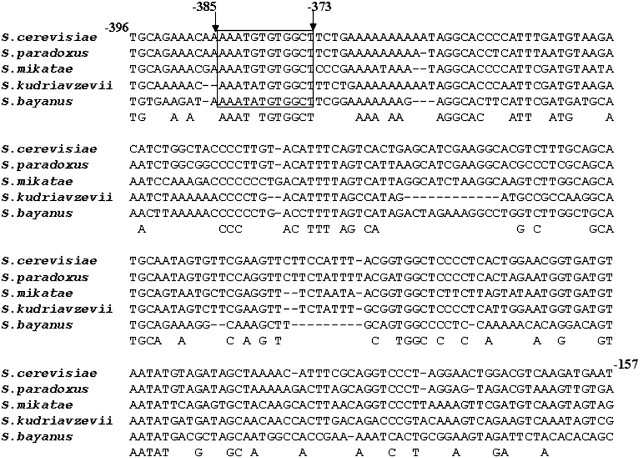
A web-based tool (Kohli et al. 2004) was employed to perform the multiple sequence alignment of the 600-bp upstream sequences of YCT1 with the orthologs of other closely related sensu stricto Saccharomyces species (S. paradoxus, S. mikatae, S. bayanus) and the more distant sensu stricto species S. kudriavzevii to identify any conserved sequence stretches (Figure 7). The analysis identified a conserved sequence stretch, AAATNTGTGGCT, at −385 bp upstream from the translation start site, which closely matched the sulfur regulatory motif II (AAANTGTGG). The similarity of this conserved sequence stretch in the YCT1 orthologous promoters from the Saccharomyces spp. and the repression seen with both methionine and cysteine suggested that these conserved sequence stretches might act as the cis regulatory elements for the expression of YCT1. To test this hypothesis, constructs of YCT1 promoter–β-galactosidase fusions were created only to include and exclude the conserved sequence “AAATNTGTGGCT,” positioned at −385, thereby generating pYLL055w-387 and pYLL055w-372 reporter constructs (Figure 8). These constructs were transformed into the met15Δ strain and the β-galactosidase activity was measured after growing cells in the sulfur-limiting and sulfur-rich medium (Figure 8). We observed that the deletion of the conserved sequence AAATNTGTGGCT in the pYLL055w-372 construct resulted in the complete loss of activation in the sulfur-limiting medium and also decreased the expression in cysteine medium compared to the control, the pYLL055w-600 construct. In contrast, the other deletion construct showed β-galactosidase activity comparable to the control in the sulfur-limiting conditions. This indicates that the motif AAATGTGTGG in the YCT1 upstream region is absolutely essential for the activation of the gene in nonrepressing sulfur conditions, although the full expression of the gene in the sulfur-limiting and sulfur-rich medium requires the presence of some additional upstream elements in the YCT1 promoter.
Figure 7.—
Multiple sequence alignment of the YLL055w upstream sequences of different Saccharomyces species to identify conserved sequence stretches. The YLL055w upstream activating sequences of S. cerevisiae, S. bayanus, S. mikatae, S. paradoxus, and S. kudriavzevii were extracted and multiple sequence alignment was carried out using the webserver (http://203.90.127.21/anand/sacch_prom_pat.html; Kohli et al. 2004). A sequence alignment stretch corresponding to the −396 to −157 upstream region of YLL055w from S. cerevisiae is shown. The conserved sequence stretch at −385, resembling the sulfur regulatory motif II-AAANTGTGG, is boxed.
Since the cis-regulatory sequence that we identified was slightly different from the canonical cis-regulatory sequences involved in regulation of the sulfur assimilatory enzymes, we sought to examine if the trans-factors involved in the sulfur assimilatory pathway play a role in the regulation of YCT1. To examine this, the pYLL055w-600 construct was transformed into different genetic backgrounds bearing mutations in the different sulfur regulatory trans-factors (met4Δ, met28Δ, cbf1Δ, met31Δ, met32Δ) and YCT1 expression was examined (Table 5). Consistent with the absolute requirement of the Met4p for the activation of sulfur assimilation genes in the sulfur-limiting conditions (Thomas and Surdin-Kerjan 1997), significantly reduced enzyme activity was detected in the met4Δ strain. A decreased activation of YCT1 was also observed in the met32Δ strain, whereas expression was significantly increased in the met28Δ strain under the same growth conditions. However, this high induction in enzyme activity in the met28Δ background was not observed in a sulfur-rich medium. Thus it appears that the sulfur regulatory factors Met4p and Met32p are absolutely essential for the expression of YCT1 under nonrepressive sulfur conditions whereas Met28p seems to “restrict” the activity in these inducing conditions.
TABLE 5.
Regulation of YLL055w by different sulphur regulatory trans-factors
|
Strain background |
− Methionine |
+ Methionine |
|---|---|---|
| Wild type (ABC 734) | 8.4 ± 0.4 | 3.1 ± 0.4 |
| Wild type (ABC 1381) | 17.8 ± 1.1 | 5.6 ± 1.6 |
| met4Δ (ABC1081) | 3.7 ± 1.1 | 5.6 ± 1.5 |
| met28Δ (ABC 1080) | 212.5 ± 7.5 | 6.2 ± .08 |
| cbf1Δ (ABC 1082) | 15.0 ± 0.6 | 4.6 ± 2.6 |
| met31Δ (ABC 1031) | 15.0 ± 0.7 | 4.3 ± 0.25 |
|
met32Δ (ABC 1032) |
2.9 ± 0.2 |
1.6 ± 0.2 |
The pYLL055w-600 multicopy plasmid bearing a 600-bp upstream region of YLL055w fused upstream of the β-galactosidase gene was transformed into strains deleted for the different sulphur regulatory trans-factors and their respective wild-type strains. The transformants were grown in the presence and absence of methionine and the β-galactosidase enzyme activity was measured. Data are shown as mean ± SD (n = 3).
DISCUSSION
This study has revealed the existence of a high-affinity cysteine-specific transporter in S. cerevisiae that is also the major transporter of cysteine in the yeast. This transporter (Yct1p, Yll055wp) was initially picked up from data mining and bioinformatics analysis and has been demonstrated to function as a high-affinity, cysteine-specific transporter through both genetic and biochemical studies.
The plate-based loss-of-growth phenotype and the decreased cysteine uptake in the yct1Δ strain, which were complemented by a plasmid bearing YCT1, provided direct evidence for the role of the YCT1-encoded transporter in cysteine uptake. Although cysteine uptake was mediated by the other members of the AAP family at high concentration, the complete loss in growth of the yct1Δ strain at low concentrations of cysteine, coupled with the low Km of the Yct1p for cysteine (55.4 μm), clearly makes it a high-affinity, major cysteine transporter in the yeast. The fact that the deduced Km of Agp1p, the only cysteine transporter in yeast that shows saturation with respect to cysteine uptake, is 200 μm, further signifies the high affinity of Yct1p for cysteine uptake (During-Olsen et al. 1999). Apart from cysteine, significant inhibition in cysteine uptake by Yct1p was exhibited only by homocysteine, a close structural homolog of cysteine (and, to a lesser extent, by methionine). The growth experiments with the yll055wΔ strain, however, suggest that homocysteine is not transported principally by Yct1p (Figure 1).
The transport experiments, which were carried out with whole cells, suggest plasma membrane localization for Yct1p. The possibility that Yct1p might also be localized to other organelles, however, has not been examined. A genomewide analysis of protein localization in S. cerevisiae, using green fluorescent protein (GFP)-tagged proteins expressed from their native promoter in the chromosome itself, has predicted that Yct1p is restricted to the endoplasmic reticulum (Huh et al. 2003). However, functional analyses of the yeast strain bearing the chromosomal copy of the GFP-tagged YLL055w in the met15Δ background gave a yll055w null mutant phenotype for growth on cysteine (data not shown). Hence, it appears that the GFP-tagged Yll055wp is nonfunctional, and hence no conclusion can be drawn regarding the localization of the protein from such a construct.
The fact that this major cysteine transporter in S. cerevisiae has eluded identification so far is quite surprising since several attempts have been made to identify cysteine transporter genes, and several genetic studies have targeted the sulfur assimilation pathway, of which this forms a part (Cherest and Surdin-Kerjan et al. 1992; During-Olsen et al. 1999; Kosugi et al. 2001). One possibility could be that the YLL055w gene under its own promoter is toxic in E. coli (J. Kaur and A. K. Bachhawat, unpublished observations). As a result, E. coli colonies harboring plasmids bearing this gene with its native promoter form pin-point-sized colonies on plates, and hence the gene is likely to be absent from genomic libraries used to isolate this gene. A second aspect is that the phenotypic defect on cysteine plates is seen only at low concentrations of cysteine over a narrow concentration range. Finally, the fact that this gene is under tight regulation by the sulfur regulatory network makes significant contribution in cysteine uptake discernible only in sulfur-limiting conditions such as in the met15Δ background.
The observation that the major cysteine transporter of S. cerevisiae does not fall into the AAP family, to which all the other known amino acid transporters belong, is intriguing and shows that not all the transporters of the 20 common l-α-amino acids in S. cerevisiae need to belong to the AAP family (Regenberg et al. 1999). The Dal5p transporter family is unique to fungi. Its members, in addition to transporting cysteine as described in this report, have been reported to transport different metabolites like nicotinic acid, allantoin, pantothenic acid, and biotin (Table 3). The possibility of Yct1p homologs being cysteine transporters in other yeast and fungi was therefore evaluated by examining possible orthologs in other organisms. BLAST analysis and reverse Best Hit analysis suggests that orthologs of the cysteine transporter probably do exist in other yeasts and fungi (data not shown), although this needs to be experimentally established.
The discovery of this transporter has helped resolve much of the confusion that abounds in the literature about the kinetic aspects of cysteine transport. Kinetic studies of transporters are always complicated when multiple transporters contribute to the transport of the substrate. Yct1p plays the predominant role at lower concentrations of cysteine, as seen from growth experiments and as reflected in the Km of the transporter for cysteine. At higher concentrations, other transporters contribute in a much more significant manner, masking the contributions of Yct1p to a significant extent. The use of higher ranges of substrate concentrations has thus masked the kinetic behavior of cysteine transport (During-Olsen et al. 1999). Hence we have restricted ourselves to a much lower range of concentrations in determining Km, a concentration range that was also employed by one earlier study that was able to observe saturating kinetics with respect to cysteine uptake and reported a Km of 83.3 μm, which is reasonably close to what we observed in this study (Ono and Naito 1991).
A recent study has presented a rational basis for the presence of multiple permeases for methionine in the cell (Menant et al. 2006). Methionine transport is mediated by seven different transporters that include Mup1p, Mup3p, Agp3p, Agp1p, Bap2p, Bap3p, and Gnp1p. Three of them, Mup1p, Mup3p and Agp3p, have been shown to be under methionine repression, while the remaining Agp1p, Bap2p, Bap3p, and Gnp1p, which are nonspecific transporters, are induced severalfold in the presence of methionine (Menant et al. 2006). Hence, by manipulating the activity and regulation of these permeases in response to different extracellular and intracellular conditions, the cell is able to adjust itself to the prevailing environmental conditions and yet maintain the homeostasis of the amino acid inside. The presence of nine transporters mediating cysteine uptake in S. cerevisiae, which include Yct1p that is under tight regulation by sulfur, might suggest the need for a similar mechanism for maintaining cysteine homeostasis (Regenberg et al. 1999).
Studies on regulation of the sulfur assimilation pathway have revealed that between the two common organic sulfur sources, methionine and cysteine, methionine represses the gene transcription more efficiently, although the effector molecule for the transcriptional regulation of these genes is proposed to be cysteine or a derivative thereof (Thomas and Surdin-Kerjan 1997; Hansen and Johannesen 2000). This has been attributed to the slow rate of uptake of cysteine from the extracellular medium, due to the low affinity of the cysteine transporters in the plasma membrane (Ono et al. 1991; During-Olsen et al. 1999). However, the identification of Yct1p as a high-affinity, cysteine-specific transporter questions this assumption. It is also observed that a met15Δ strain grows better on glutathione as an organic sulfur source compared to cysteine, when both are present at the same concentration (Hansen and Johannesen 2000; Ganguli et al. 2006). As glutathione utilization proceeds through cysteine (Ganguli et al. 2006), this difference becomes difficult to explain considering that Hgt1p, the high-affinity glutathione transporter in yeast (Bourbouloux et al. 2000), and Yct1p, the high-affinity cysteine transporter in yeast, have similar Km values. Whether this decreased ability to utilize cysteine as an efficient organic sulfur source arises due to some post-transcriptional regulation of Yct1p, and hence to decreased cysteine uptake or some other step in the cysteine utilization, could be an interesting subject for future studies.
Acknowledgments
This work was supported by a Grant-in-Aid project to A.K.B. from the Department of Biotechnology, Government of India. J.K. was the recipient of a Research Fellowship from Council of Scientific and Industrial Research, Government of India.
Footnotes
Communicating editor: S. Sandmeyer
References
- Alfafara, C. G., A. Kanda, T. Shio, H. Shimizu, S. Shioya et al, 1992. Effect of amino acids on glutathione production by Saccharomyces cerevisiae Appl. Microbiol. Biotechnol. 36: 538–540. [Google Scholar]
- Aranda, A., and M. L. del Olmo, 2004. Exposure of Saccharomyces cerevisiae to acetaldehyde induces sulfur amino acid metabolism and polyamine transporter genes, which depend on Met4p and Haa1p transcription factors, respectively. Appl. Environ. Microbiol. 70: 1913–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute and C. Cullin, 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae Nucleic Acids Res. 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudouin-Cornu, P., Y. Surdin-Kerjan, P. Marliere and D. Thomas, 2001. Molecular evolution of protein atomic composition. Science 293: 297–300. [DOI] [PubMed] [Google Scholar]
- Blaiseau, P., A. Isnard, Y. Surdin-Kerjan and D. Thomas, 1997. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell. Biol. 17: 3640–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer, V. M., J. H. De Winde, J. T. Pronk and M. D. Piper, 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278: 3265–3274. [DOI] [PubMed] [Google Scholar]
- Bourbouloux, A., P. Shahi, A. Chakladar, S. Delrot and A.K. Bachhawat, 2000. Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae J. Biol. Chem. 275: 13259–13265. [DOI] [PubMed] [Google Scholar]
- Cherest, H., and Y. Surdin-Kerjan, 1992. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P. F., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton et al, 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301: 71–76. [DOI] [PubMed] [Google Scholar]
- Cottarel, G., D. Beach and U. Deuschle, 1993. Two new multi-purpose multicopy Schizosaccharomyces pombe shuttle vectors, pSP1 and pSP2. Curr. Genet. 23: 547–548. [DOI] [PubMed] [Google Scholar]
- Courchesne, W. E., and B. Magasanik, 1983. Ammonia regulation of amino acid permeases in Saccharomyces cerevisiae Mol. Cell. Biol. 3: 672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- During-Olsen, L., B. Regenberg, C. Gjermansen, M. C. Kielland-Brandt and J. Hansen, 1999. Cysteine uptake by Saccharomyces cerevisiae is accomplished by multiple permeases. Curr. Genet. 35: 609–617. [DOI] [PubMed] [Google Scholar]
- Fauchon, M., G. Lagniel, J. C. Aude, L. Lombardia, P. Soularue et al, 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9: 713–723. [DOI] [PubMed] [Google Scholar]
- Futcher, B., and J. Carbon, 1986. Toxic effects of excess cloned centromeres. Mol. Cell. Biol. 6: 2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli, D., C. Kumar and A. K. Bachhawat, 2006. The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae Genetics 175: 1137.–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greasham, R. L., and A. G. Moat, 1973. Amino acid transport in a polyaromatic amino acid auxotroph of Saccharomyces cerevisiae J. Bacteriol. 115: 975–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente, L., and M. Ptashne, 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae Proc. Natl. Acad. Sci. USA 78: 2199–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194: 3–37. [PubMed] [Google Scholar]
- Hansen, J., and P. F. Johannesen, 2000. Cysteine is essential for transcriptional regulation of the sulfur assimilation genes in Saccharomyces cerevisiae Mol. Gen. Genet. 263: 535–542. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al, 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Isnard, A.-D., D. Thomas and Y. Surdin-Kerjan, 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 262: 473–484. [DOI] [PubMed] [Google Scholar]
- Jauniaux, J-C., and M. Grenson, 1990. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae Nucleotide sequence, protein similarity with the other bakers yeast amino acid permeases, and nitrogen catabolite repression. Eur. J. Biochem. 190: 39–44. [DOI] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren and E. S. Lander, 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254. [DOI] [PubMed] [Google Scholar]
- Kohli, D. K., C. V. Srikanth and A. K. Bachhawat, 2004. A search tool for identification and analysis of conserved sequence patterns in Saccharomyces spp. orthologous promoter. In Silico Biol. 4: 411–415. [PubMed] [Google Scholar]
- Kosugi, A., Y. Koizumi, F. Yanagida and S. Udaka, 2001. MUP1, high affinity methionine permease, is involved in cysteine uptake by Saccharomyces cerevisiae Biosci. Biotechnol. Biochem. 65: 728–731. [DOI] [PubMed] [Google Scholar]
- Kumar, A., L. John, M. M. Alam, A. Gupta, G. Sharma et al, 2006. Homocysteine- and cysteine-mediated growth defect is not associated with induction of oxidative stress response genes in yeast. Biochem. J. 396: 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., H. Cherest, Y. Surdin Kerjan and D. Thomas, 1996. A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15: 2519–2529. [PMC free article] [PubMed] [Google Scholar]
- Lenhard, B., A. Sandelin, L. Mendoza, P. Engstrom, N. Jareborg et al, 2003. Identification of conserved regulatory elements by comparative genome analysis. J. Biol. 2: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente, B., and B. Dujon, 2000. Transcriptional regulation of the Saccharomyces cerevisiae DAL5 gene family and identification of the high affinity nicotinic acid permease TNA1 (YGR260w). FEBS Lett. 23: 237–241. [DOI] [PubMed] [Google Scholar]
- Maw, G. A., 1963. The uptake of some sulphur-containing amino acids by a brewer's yeast. J. Gen. Microbiol. 31: 247–259. [DOI] [PubMed] [Google Scholar]
- Menant, A., R. Barbey and D. Thomas, 2006. Substrate-mediated remodeling of methionine transport by multiple ubiquitin-dependent mechanisms in yeast cells. EMBO J. 25: 4436–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cozatl, D., H. Loza-Tavera, A. Hernandez-Navarro and R. Moreno-Sanchez, 2005. Sulfur assimilation and glutathione metabolism under cadmium stress in yeast, protists and plants. FEMS Microbiol Rev. 29: 653–671. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., R. Muller and M. Funk, 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. [DOI] [PubMed] [Google Scholar]
- Nelissen, B., R. De Wachter and A. Goffeau, 1997. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae FEMS Microbiol. Rev. 21: 113–134. [DOI] [PubMed] [Google Scholar]
- Ono, B., and K. Naito, 1991. The cysteine transport system of Saccharomyces cerevisiae Yeast 7: 849–855. [DOI] [PubMed] [Google Scholar]
- Ono, B., K. Naito, Y. Shirahige and M. Yamamoto, 1991. Regulation of cystathionine gamma-lyase in Saccharomyces cerevisiae Yeast 7: 843–848. [DOI] [PubMed] [Google Scholar]
- Rai, R., F. S. Genbauffe, H. Z. Lea and T. G. Cooper, 1987. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae J. Bacteriol. 169: 3521–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, R., F. S. Genbauffe and T. G. Cooper, 1988. Structure and transcription of the allantoate permease gene (DAL5) from Saccharomyces cerevisiae J. Bacteriol. 170: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg, B., L. During-Olsen, M. C. Kielland-Brandt and S. Holmberg, 1999. Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae Curr. Genet. 36: 317–328. [DOI] [PubMed] [Google Scholar]
- Saier, M. H., Jr., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne et al 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1: 257–279. [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual, Ed. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Srikanth, C. V., P. Vats, A. Bourbouloux, S. Delrot and A. K. Bachhawat, 2005. Multiple cis-regulatory elements and the yeast sulphur regulatory network are required for the regulation of the yeast glutathione transporter, Hgt1p. Curr. Genet. 47: 345–358. [DOI] [PubMed] [Google Scholar]
- Stolz, J., and N. Sauer, 1999. The fenpropimorph resistance gene FEN2 from Saccharomyces cerevisiae encodes a plasma membrane H+-pantothenate symporter. J. Biol. Chem. 274: 18747–18752. [DOI] [PubMed] [Google Scholar]
- Stolz, J., U. Hoja, S. Meier, N. Sauer and E. Schweizer, 1999. Identification of the plasma membrane H+-biotin symporter of Saccharomyces cerevisiae by rescue of a fatty acid-auxotrophic mutant. J. Biol. Chem. 274: 18741–18746. [DOI] [PubMed] [Google Scholar]
- Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde et al, 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae J. Biol. Chem. 280: 437–447. [DOI] [PubMed] [Google Scholar]
- Thomas, D., and Y. Surdin-Kerjan, 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae Microbiol. Mol. Biol. Rev. 61: 503–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D., I. Jacquemin and Y. Surdin Kerjan, 1992. MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae Mol. Cell. Biol. 12: 1719–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vido, K., D. Spector, G. Lagniel, S. Lopez, M. B. Toledano et al, 2001. A proteome analysis of the cadmium response in Saccharomyces cerevisiae J. Biol. Chem. 276: 8469–8474. [DOI] [PubMed] [Google Scholar]
- Wen, S., T. Zhang and T. Tan, 2004. Utilization of amino acids to enhance glutathione production in Saccharomyces cerevisiae Enzyme Microb. Technol. 35: 501–507. [Google Scholar]
- Wheeler, G. L., E. W. Trotter, I. W. Dawes and C. M. Grant, 2003. Coupling of the transcriptional regulation of glutathione biosynthesis to the availability of glutathione and methionine via the Met4 and Yap1 transcription factors. J. Biol. Chem. 278: 49920–49928. [DOI] [PubMed] [Google Scholar]