Abstract
We have defined the histone acetylation pattern of the endogenous murine β-globin domain, which contains the erythroidspecific β-globin genes. The β-globin locus control region (LCR) and transcriptionally active promoters were enriched in acetylated histones in fetal liver relative to fetal brain, whereas the inactive promoters were hypoacetylated. In contrast, the LCR and both active and inactive promoters were hyperacetylated in yolk sac. Hypersensitive site two of the LCR was also hyperacetylated in murine embryonic stem cells, whereas β-globin promoters were hypoacetylated. Thus, the acetylation pattern varied at different developmental stages. Histone deacetylase inhibition selectively increased acetylation at a hypoacetylated promoter in fetal liver, suggesting that active deacetylation contributes to silencing of promoters. We propose that dynamic histone acetylation and deacetylation play an important role in the developmental control of β-globin gene expression.
Histone acetylation and deacetylation play important roles in transcriptional regulation (1–3). Allis and colleagues (4) proposed a model to explain how histone acetylation can regulate gene-specific transcription despite the ubiquitous distribution of nucleosomes in the genome. In this model, sequence-specific DNA binding proteins physically recruit histone acetylases (HATs) to chromosomal sites, which selectively target promoters for chromatin remodeling. The consequences of targeted HAT recruitment are evident from biochemical studies showing that histone acetylation increases the accessibility of nucleosomal DNA to trans-acting factors (5, 6). Thus, increased histone acetylation at a promoter may enhance the binding of factors that stimulate preinitiation complex assembly or may directly promote binding of the transcriptional machinery. Studies on the role of acetylation in transcription have been facilitated by the development of a chromatin immunoprecipitation (ChIP) assay (7), which allows one to measure the histone acetylation state of specific chromosomal sites in living cells. Analysis of histone acetylation by ChIP has shown that histone hyperacetylation at promoters correlates with transcriptional activity (8–12). Beyond the impact of local histone acetylation on promoter function, little is known about the importance of histone acetylation for long-range transcriptional control. Given that acetylation impairs higher-order chromatin folding (13), which can modulate the accessibility of cis-acting elements, histone acetylation could also control long-range activation. In addition, HATs recruited by enhancers and locus control regions (LCRs) may modify histones surrounding these elements, which could influence the function of the respective nucleoprotein complexes.
An increasing number of genes have been shown to reside within chromosomal domains controlled by LCRs (14). The best example of a locus regulated by a LCR is the β-globin locus containing the embryonic, fetal, and adult β-globin genes. High-level transcription of the β-globin genes is conferred by the β-globin LCR (15–17), which consists of four DNaseI hypersensitive sites (HSs) at the 5′ end of the β-globin locus (18, 19). However, the importance of the LCR in establishing the developmental expression pattern is unresolved.
Studies with mouse transgenes lacking the β-globin LCR (20) have shown that the LCR is not required for the switch from fetal to adult β-globin expression. Thus, the promoters may contain the intrinsic information to confer stage-specific expression. Models for the control of β-globin gene switching have invoked stage-specific factors acting at the β-globin promoters, rendering a promoter susceptible or resistant to LCR-mediated activation (21). We proposed that the β-globin LCR recruits chromatin remodeling enzymes required for transactivation of all β-globin genes, and then developmental control is established via stage-specific factors acting at promoters (22). In this regard, we showed that E1A, an inhibitor of the HATs CREB binding protein (CBP/p300) and p300/CBP-associated factor (P/CAF), abolished LCR-mediated transactivation in transfection assays and strongly decreased endogenous γ-globin expression (23). Further support for a role of HATs in long-range transactivation comes from work implicating the β-globin (24) and growth hormone (25) LCRs in modulating acetylation over long distances on a chromosome.
To understand how HATs recruited by the β-globin LCR regulate the β-globin genes, it is necessary to define the histone acetylation state of the endogenous β-globin locus. We reasoned that the pattern might provide unique insights into the mechanism of LCR function and β-globin gene switching. We measured the levels of histone acetylation throughout the endogenous murine β-globin locus and describe the implications of the histone acetylation pattern vis-à-vis a new model for β-globin gene switching and the regulation of other complex multigene loci.
Materials and Methods
Cell Culture.
Mouse erythroleukemia (MEL) cells (26) were maintained as described (27) and incubated for 4 days in the presence or absence of 1.5% DMSO (Sigma). Murine embryonic stem (ES) cells (strain 129/Sv) were maintained in DMEM media (Biofluids, Rockville, MD) containing 15% FBS (GIBCO/BRL), 1% antibiotic/antimycotic (GIBCO/BRL), 0.1 mM β-mercaptoethanol, and 1000 units/ml murine leukemia inhibitory factor. Approximately 106 ES cells were plated in 100 mm cell culture dishes and harvested 3 days later for ChIP analysis.
Isolation of Fetal Livers and Brains.
Livers and brains from 14.5 days postcoitum (dpc) mouse embryos were prepared as described (27). Pooled livers and brains were passed separately through a 21-gauge needle in 20 ml of DMEM-based medium (28), and cross-linking was performed immediately as described below. Yolk sacs from 11.5 dpc embryos were prepared similarly. For the experiment of Fig. 6, disaggregated cells were incubated in DMEM-based dissection media containing 20% FBS with or without 20 mM sodium butyrate (Sigma) or 165 nM trichostatin A (Wako Chemicals, Osaka) for 4 h before ChIP analysis.
Figure 6.
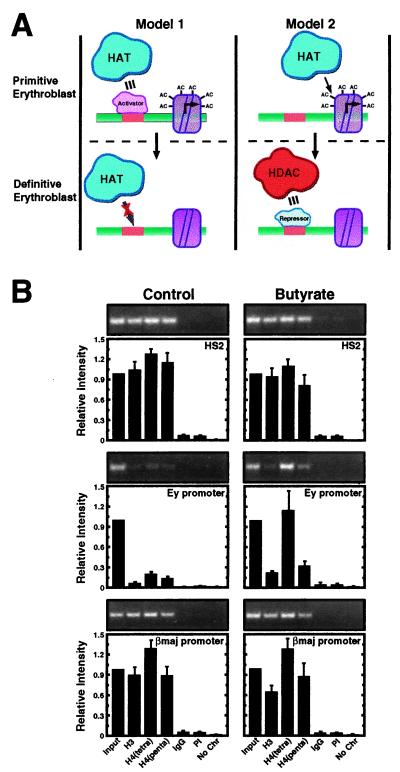
HDAC inhibition selectively induces acetylation at the developmentally silenced Ey promoter. (A) Models to explain the hypoacetylated state of the embryonic β-globin promoters in 14.5 dpc fetal liver. Model 1 assumes that defective HAT recruitment in the definitive erythroblast is responsible for hypoacetylation. The defect would result from limiting amounts of an activator required to recruit HATs to the promoters. Model 2 assumes that a stage-specific repressor recruits HDACs to the embryonic β-globin promoters in definitive erythroblasts and therefore establishes the hypoacetylated state. HATs recruited by the LCR would be sufficient to generate a hyperacetylated state in primitive erythroblasts lacking the repressor. (B) Fetal liver cells (14.5 dpc) were incubated with or without butyrate for 4 h, and then the acetylation state of the endogenous murine β-globin locus was measured by ChIP analysis. Representative ethidium bromide-stained gels are shown. The graphs show the relative intensity of the PCR products from three independent experiments (mean ± SEM). Note that butyrate treatment increased the acetylation state only of the Ey promoter.
Chromatin Immunoprecipitation Assay.
ChIPs were performed as described previously (27) with minor modifications. Protein-DNA cross-linking was performed by incubating MEL cells (2 × 107 per condition), ES cells (6–8 × 106 per condition), or suspensions of fetal liver, brain, or yolk sac cells (approximately 2 livers or brains or 20 yolk sacs per condition) with formaldehyde at a final concentration of 0.4% for 10 min at room temperature with gentle agitation. Glycine (0.125 M) was added to quench the reaction. Cells were then collected by centrifugation at 240 × g for 8 min and washed in PBS. Nuclei were isolated by incubation in cell lysis buffer (10 mM Tris/10 mM NaCl/0.2% Nonidet P-40/10 mM sodium butyrate/1 μg/ml leupeptin/50 μg/ml PMSF, pH 8.0) for 10 min on ice followed by centrifugation at 600 × g for 5 min. Nuclei were lysed in nuclei lysis buffer (50 mM Tris/10 mM EDTA/1% SDS/10 mM sodium butyrate/1 μg/ml leupeptin/50 μg/ml PMSF, pH 8.1) for 10 min on ice. The lysate was sonicated with 8 pulses of 40 seconds each at 50–60% of maximum power with a Heat Wave Systems W185F sonicator (Ultrasonics, Farmingdale, NY) equipped with a microtip to reduce the chromatin fragments to an average size of less than 500 bp. Soluble chromatin was precleared by addition of 50 μl preimmune serum followed by 100 μl Protein A-Sepharose. An aliquot of precleared chromatin was removed (input) and used in the subsequent PCR analysis. The remainder of the chromatin was diluted with IP dilution buffer (20 mM Tris/150 mM NaCl/2 mM EDTA/0.01% SDS/1% Triton X-100/1 μg/ml leupeptin/50 μg/ml PMSF/10 mM sodium butyrate, pH 8.1) and incubated with or without 5 μl antibody, rabbit IgG (Sigma), or rabbit preimmune serum in a final volume of 600 μl for 3 h at 4°C. Immune complexes were collected by incubation with 30 μl Protein A-Sepharose for 2 h at 4°C. A control sample was prepared in all experiments in which IP wash buffer 1 (20 mM Tris/50 mM NaCl/2 mM EDTA/0.1% SDS/1% Triton X-100, pH 8.1) was added instead of chromatin. Protein A-Sepharose pellets were washed twice with 500-μl aliquots of IP wash buffer 1, once with IP wash buffer 2 (10 mM Tris/0.25 M LiCl/1 mM EDTA/1% Nonidet P-40/1% deoxycholate, pH 8.1), and twice with TE (10 mM Tris/1 mM EDTA, pH 8.0). Immune complexes were eluted twice with 150 μl of IP elution buffer (0.1 M NaHCO3/1% SDS). RNaseA (0.03 mg/ml) and NaCl (0.3 M) were added, and cross-links were reversed by incubation for 4–5 h at 65°C. Samples were digested with Proteinase K (0.24 mg/ml) for 2 h at 45°C. DNA was purified by two extractions with phenol:chloroform followed by ethanol precipitation. Purified DNA was resuspended in 30 μl water. Aliquots of 2 μl (livers and brains) or 4 μl (MEL cells) were analyzed by PCR with the appropriate primer pairs. PCR products were resolved on 1.6% agarose gels containing ethidium bromide and quantitated by using NIH IMAGE version 1.61.1. Band intensities are expressed relative to the signal obtained from 0.06% (fetal livers, brains, yolk sacs, or ES cells) or 0.12% (MEL cells) input. Importantly, the signals were proportional to the amount of DNA input in PCR reactions (Fig. 1).
Figure 1.
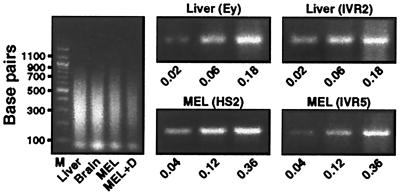
PCR assay of solubilized chromatin from murine fetal liver and MEL cells. (Left) A representative ethidium bromide-stained agarose gel of input DNA isolated from solubilized chromatin. Note that the average size of the DNA fragments is less than 500 bp. (Center and Right) PCR products obtained with the indicated primers and increasing amounts of input DNA from murine fetal liver and MEL cells, respectively. The numbers indicate the percentage of input DNA used in the corresponding PCR reactions. MEL + D, DMSO-treated MEL cells.
Antibodies.
Anti-acetylated histone H3 (06-599) and anti-tetraacetylated histone H4 (06-866) antibodies were obtained from Upstate Biotechnology (Lake Placid, NY). The anti-pentaacetylated histone H4 was a gift from Dr. David Allis. Rabbit IgG and preimmune serum served as controls for the anti-acetylated histone H3 antibody and the two anti-acetylated histone H4 antisera, respectively.
PCR Primers.
The primer pairs used were designed based on Hbbd haplotype sequences (GenBank accession numbers Z13985, X14061, AF128269, and AF133300). For primer sequences see supplementary Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org. Each primer pair amplified a single fragment of the expected size.
Results and Discussion
A Chromosomal Region Encompassing the Developmentally Repressed Embryonic β-globin Promoters Is Deficient in Acetylated Histones H3 and H4.
To assess the consequences of HAT recruitment by the β-globin LCR and intralocus regulatory elements, we used a ChIP assay to determine the histone acetylation pattern of the endogenous β-globin locus in murine fetal liver, fetal brain, and MEL cells. The chromatin used for immunoprecipitation averaged less than 500 bp, with undetectable amounts of DNA larger than 1000 bp (Fig. 1). The ratio of signals obtained from PCR analysis of immunoprecipitated chromatin and the respective input chromatin reflected the relative level of histone acetylation. Importantly, the amount of PCR product quantitated via densitometric analysis was proportional to the amount of input DNA (Fig. 1).
We postulated that LCR-mediated recruitment of HATs induces histone hyperacetylation locally near the site of recruitment. Thus, we asked whether chromatin at the HS2 subregion of the LCR was enriched in acetylated histones. ChIP analysis was done with disaggregated cells from 14.5 dpc fetal liver (Fig. 2B) and brain (Fig. 2C). Primers specific for HS2 revealed a strong enrichment of both acetylated histones H3 and H4 in fetal liver relative to fetal brain. Immunoprecipitations with anti-tetraacetylated and anti-pentaacetylated H4 antibodies yielded similar results. Only very weak H3 and H4 acetylation was apparent in fetal liver using primers for the promoter of a brain-specific protein, necdin (29). In contrast, strong necdin promoter acetylation was detected in fetal brain, where necdin is transcriptionally active. Thus, the chromatin at HS2 is enriched in both acetylated H3 and H4 in an erythroid cell-specific manner.
Figure 2.
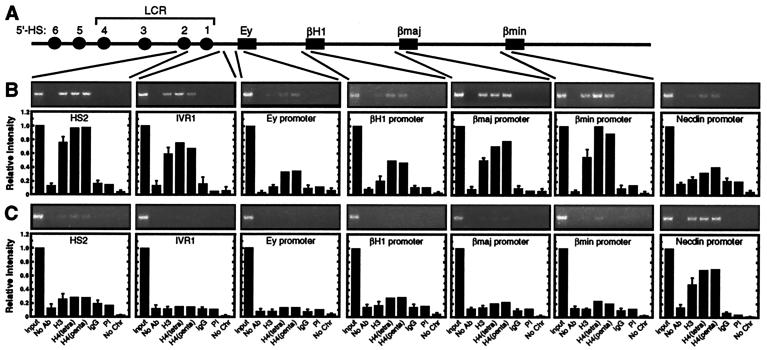
Hypoacetylated chromatin at developmentally silenced embryonic/fetal globin promoters in fetal liver. (A) Structure of the murine β-globin locus. The β-globin genes are depicted as boxes. HSs are depicted as spheres. (B) Histone acetylation pattern of the endogenous murine β-globin locus in 14.5 dpc fetal liver by ChIP analysis. Ethidium bromide-stained gels from one experiment are shown. The graphs show the relative intensity of the PCR products from two to four independent experiments (mean ± SEM). The brain-specific necdin promoter was used as a negative control. (C) Histone acetylation pattern of the endogenous murine β-globin locus in 14.5 dpc fetal brain. The brain-specific necdin promoter was used as a positive control.
We extended the ChIP analysis to determine whether acetylated histones are uniformly distributed throughout the locus or are restricted to specific functional regions. Chromatin immunoprecipitated from fetal liver (Fig. 2B) or brain (Fig. 2C) was analyzed with primers specific for the intervening region between the LCR and the Ey promoter (IVR1) and the Ey, βH1, βmajor, and βminor promoters. The Ey and βH1 genes are embryonic β-globin genes expressed in primitive erythroblasts of the yolk sac before 12 dpc and in circulating primitive erythroblasts. The adult βmajor and βminor genes are highly transcribed in definitive erythroblasts of the fetal liver after 12 dpc (30). No significant acetylation of the β-globin locus was detected in fetal brain. In contrast, histone H3 and H4 hyperacetylation was detected at IVR1 and the βmajor and βminor promoters in fetal liver. Only very weak H3 and H4 acetylation was detected at the Ey and βH1 promoters in fetal liver relative to other sites of the locus. The reduction in acetylated H4 at these sites was less robust than the reduction in acetylated H3. Thus, the embryonic β-globin promoters, which have been developmentally silenced, were strongly depleted of acetylated histone H3 and had reduced levels of acetylated histone H4 in fetal liver. The weak acetylation is consistent with low but detectable levels of Ey- and βH1-globin mRNA detected by reverse transcription (RT)-PCR in fetal liver at this developmental stage (data not shown). However, histone H3 and H4 hyperacetylation did not simply correlate with transcriptional activity at promoters, because strong acetylation was also detected at IVR1 and HS2. Because intergenic transcripts have been detected throughout the β-globin locus (31, 32), it cannot be ruled out that hyperacetylation at IVR1 and HS2 requires transcription through these elements.
The Histone Acetylation Pattern of the β-Globin Locus in Mouse Erythroleukemia Cells Recapitulates That of Fetal Liver.
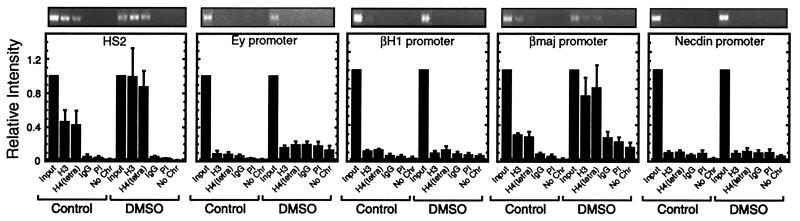
Because 14.5 dpc fetal liver contains a small number of primitive erythroblasts, we wanted to determine whether the acetylation pattern of the β-globin locus in 14.5 dpc fetal liver would be different from a system in which the embryonic β-globin genes are completely repressed. Thus, we used MEL cells, which are erythroleukemia cells expressing βmajor and βminor globin genes but not the embryonic Ey and βH1 genes (26). These cells require induction of erythroid maturation with DMSO or other agents to acquire high-level β-globin gene expression. The inducibility might involve the synthesis of factors required to strongly activate the β-globin genes, the establishment of the appropriate chromatin configuration of the β-globin locus, or a combination of the two mechanisms. ChIP analysis revealed strong acetylation of histones H3 and H4 at HS2 and the βmajor promoter in uninduced MEL cells (Fig. 3). No acetylation was detected at the repressed Ey, βH1, and necdin promoters. DMSO treatment resulted in an ≈2-fold increase in H3 and H4 acetylation at HS2 and the βmajor promoter. By contrast, DMSO did not induce acetylation at the inactive Ey, βH1, or necdin promoters. Similar results were obtained with an anti-pentaacetylated histone H4 antibody (data not shown). Thus, the histone acetylation pattern of the β-globin locus in MEL cells recapitulates that of fetal liver and is only mildly influenced by DMSO treatment. The concordance of the acetylation data from MEL cells and fetal liver makes the MEL cell system an attractive one for defining factors that establish and reconfigure the pattern of histone acetylation.
Figure 3.
The histone acetylation pattern of the β-globin locus in MEL cells recapitulates that of fetal liver. ChIP analysis was used to define the histone acetylation pattern of the endogenous β-globin locus in MEL cells. The brain-specific necdin promoter was used as a negative control. Representative ethidium bromide-stained gels are shown. The graphs show the relative intensity of the PCR products (mean ± SEM). The number of independent determinations for HS2, Ey, βH1, βmajor, and necdin were 5, 4, 5, 7, and 6, respectively.
Identification of a Hypoacetylated Subdomain Within the β-Globin Domain.
The results of Figs. 2 and 3 raise the question of whether hypoacetylated chromatin is restricted to the silenced promoters. In this scenario, deacetylation of histones in one or two promoter-associated nucleosomes could explain the results. To address this, additional ChIP assays were done to obtain a higher-resolution map of the acetylation state of the β-globin locus in DMSO-induced MEL cells (Fig. 4). Similar to the results with HS2, acetylated H3 and H4 were enriched at HSs 1, 3, and 4, as well as the intervening regions between the HSs. To define the limits of the hyperacetylated domain, we extended the analysis to include the 5′ and 3′ ends of the locus. Transitions from hyperacetylated to hypoacetylated chromatin were detected at both the 5′ and 3′ ends. Surprisingly, hypoacetylated sites within the hyperacetylated domain were not restricted to the silenced promoters, but rather an ≈30-kb hypoacetylated subdomain was apparent. Such a broad island of hypoacetylated chromatin within an active domain has not been described in any system. In addition, a site between the active βmajor and βminor genes was hypoacetylated. Thus, hypoacetylated chromatin was not localized solely to the silenced promoters.
Figure 4.
Histone acetylation pattern of the entire β-globin locus in MEL cells. (A) Histone H3 (Upper) and H4 (Lower) acetylation pattern of the β-globin locus in DMSO-induced MEL cells. The relative levels of H3 and H4 acetylation were determined by ChIP analysis and plotted as a function of the position within the locus (mean ± SEM). The PCR primers used are indicated by vertical lines. (B) Representative ethidium bromide-stained gels are shown.
The Histone Acetylation Pattern of the β-Globin Locus Is Developmentally Regulated.
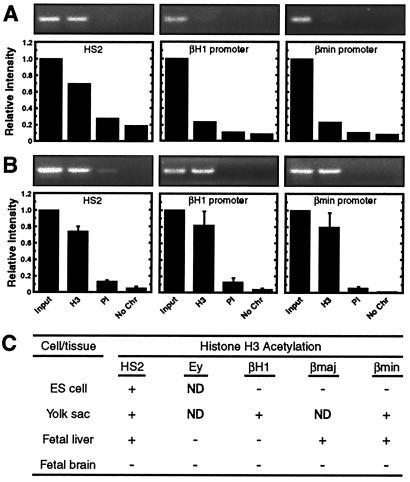
The deficiency of hyperacetylated chromatin at the inactive promoters in fetal liver and MEL cells led us to hypothesize that histone acetylation may be involved in the developmental control of β-globin gene expression. To address whether the acetylation pattern is developmentally dynamic, we analyzed H3 acetylation at functionally distinct sites [HS2, an embryonic promoter (βH1), and an adult promoter (βminor)] of the β-globin locus in undifferentiated embryonic stem (ES) cells and in 11.5 dpc yolk sacs. At this developmental stage, the yolk sac is enriched in primitive erythroid cells. HS2 was enriched in acetylated histone H3 in both ES cells and yolk sac (Fig. 5). No acetylation was detected at either the βH1 or βminor promoters in ES cells, where these promoters are inactive. In contrast, the active βH1 promoter was hyperacetylated in yolk sac. Interestingly, the inactive βminor promoter was also hyperacetylated in yolk sac. Thus, the pattern of histone H3 acetylation of the β-globin locus is unique in ES cells, yolk sac, and fetal liver, supporting a model in which developmentally dynamic changes in acetylation have functional consequences for β-globin gene regulation. There is not a strict correlation between acetylation and active transcription because the transcriptionally inactive βminor promoter was hyperacetylated in yolk sac.
Figure 5.
Developmentally distinct patterns of histone acetylation within the β-globin locus. Histone H3 acetylation of representative β-globin sequences in murine ES cells (A) and 11.5 dpc fetal yolk sac (B). Representative ethidium bromide-stained gels are shown. The graphs show the relative intensity of the PCR products (mean ± SEM). The number of independent determinations for each sequence was two and three for ES cells and yolk sac, respectively. (C) Summary of the developmental and tissue specificity of histone H3 acetylation of the murine β-globin locus.
Does the Deficiency of Acetylated Histones at Developmentally Repressed β-Globin Promoters Result from Active Deacetylation or Defective HAT Recruitment?
The deficiency of acetylated histones at the Ey and βH1 promoters could result from the lack of HAT recruitment (Fig. 6A, Model 1), active deacetylation (Fig. 6A, Model 2), or a combination of the two mechanisms. It is attractive to propose a model in which LCR-mediated long-range activation requires the concerted actions of HATs and HDACs to establish the developmentally dynamic pattern of histone acetylation within the β-globin domain. We reasoned that the LCR-mediated recruitment of HATs might result in acetylation throughout the locus, limited only by insulating sequences at the extremities of the locus (33, 34), which would oppose the spread of acetylated chromatin. If HATs recruited by the LCR acetylate histones throughout the locus, factors bound to cis-acting elements within the locus might establish a specific pattern of acetylation by enhancing or reducing acetylation at specific sites. Such intralocus regulatory elements can be important for LCR-mediated regulation of the β-globin genes (35). How would the respective binding factors modulate acetylation established by the LCR? Stage-specific factors bound to a promoter destined for repression might recruit HDACs that would dominantly erase acetylation at the promoter, establishing an island of repressive chromatin within the active chromosomal domain (Fig. 6A, Model 2).
A prediction of model 2 is that inhibition of HDACs would induce acetylation at the hypoacetylated subdomain. Acetylation at other sites, e.g., HS2 and the active βmajor and βminor promoters, should not be influenced by HDAC inhibition. On the other hand, if hypoacetylation results from defective HAT recruitment, HDAC inhibition should have no effect on acetylation at these sites, unless there is a generalized enhancement at all sites. To distinguish between these models, we isolated fetal liver cells from 14.5 dpc mouse embryos and treated the cells with butyrate (36), an effective inhibitor of multiple HDACs (37). Solubilized chromatin was subjected to ChIP analysis as described above. Similar to the results of Fig. 2B, robust histone H3 and H4 acetylation was detected at HS2 and the βmajor promoter, whereas very weak acetylation was detected at the Ey promoter (Fig. 6B). Butyrate treatment increased both H3 and H4 acetylation at the Ey promoter (3.7-, 5.3-, and 2.9-fold with anti-acetylated H3, anti-tetraacetylated H4, and anti-pentaacetylated H4, respectively), without increasing acetylation at HS2 and the βmajor promoter (Fig. 6B). A similar increase in acetylation at the Ey promoter was measured on treatment of fetal liver cells with the HDAC inhibitor trichostatin A (TSA) (2.5-, 2.6, and 2.9-fold with anti-acetylated H3, anti-tetraacetylated H4, and anti-pentaacetylated H4, respectively); TSA did not influence acetylation at HS2 and the βmajor promoter (data not shown). Although attempts to directly cross-link HDACs 1 or 2 to the Ey promoter and other sites of the β-globin locus failed (data not shown), the selective sensitivity of the Ey promoter to HDAC inhibition supports model 2, in which broad acetylation throughout the locus is subjected to HDAC-mediated deacetylation, generating the hypoacetylated island within the active domain.
HDAC-mediated erasure of acetylation at developmentally repressed promoters might be necessary or sufficient to reactivate embryonic β-globin gene expression. Hyperacetylation of the embryonic β-globin promoters would not suffice to activate the promoters if additional factors present in the yolk sac are limiting in fetal liver. To distinguish between these possibilities, we treated fetal liver cells with butyrate and measured the expression of Ey-globin by RT-PCR. Butyrate did not induce Ey-globin mRNA under conditions in which H3 and H4 acetylation on the Ey promoter increased (data not shown). Thus, reactivation of the Ey-globin gene requires additional factors or specific chromatin modifications.
Implications of the Results Vis-à-Vis the Mechanism of β-Globin Gene Switching.
The acetylation analysis described herein has led to a model in which LCR-mediated recruitment of HATs and HDACs results in developmentally specific and tissue-specific acetylation of histones within the β-globin locus. We propose that hyperacetylation near promoters establishes transcriptional competence, but additional factors would be necessary for transcriptional activation. HDACs would generate a hypoacetylated subdomain encompassing the Ey- and βH1-globin genes as part of the silencing mechanism. However, because butyrate-mediated induction of acetylation at the Ey promoter does not reactivate transcription, clearly this must only be one step in the silencing mechanism. Although we have not examined the role of the LCR in establishing the acetylation pattern, our previous work implicated a HAT, CBP/p300, in LCR-mediated long-range transactivation of the human β-globin genes (23). Recently, Schubeler et al. (24) addressed whether the LCR regulates the acetylation state of the β-globin locus on human chromosome 11 transferred into MEL cells. Chromosome 11 lacking HS2 to HS5 had low levels of acetylated histone H3 at the β-globin promoter and the β-globin gene. It was concluded that H3 acetylation correlated with transcription. In contrast, H4 acetylation was unaffected by the HS2-HS5 deletion, and it was concluded that H4 acetylation correlated with the general DNaseI sensitivity of the locus.
In our analysis of the fetal liver and MEL cell β-globin locus, we measured a reduction of both acetylated H3 and H4 on the repressed promoters and the surrounding region. Thus, in contrast to the study in hybrid MEL cells, fetal liver and MEL cells have a hypoacetylated chromatin island within the DNaseI sensitive β-globin domain. The chromatin within this island may occlude transcription factor binding sites on the promoters and therefore inhibit preinitiation complex assembly. According to this mechanism, stage-specific repressive factors would be required to target HDACs to these sites to generate the repressive chromatin and therefore would be important determinants of the stage-specificity of embryonic globin gene expression.
How does our model relate to what is known about trans-acting factors that control Ey- and βH1-globin expression? Filipe et al. (38) described the importance of a DR-1 element for the developmental silencing of human ɛ-globin in definitive erythroblasts of transgenic mice. This element was found on multiple mouse and human embryonic and fetal globin promoters. Protein–DNA interaction studies revealed that chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII), an orphan nuclear receptor, was a major component interacting with this site. COUP-TFII is one of a number of related COUP-TF factors that can interact with corepressors containing HDAC subunits (39). Thus, COUP-TFII can function as a repressor and may mediate HDAC recruitment at Ey and βH1 promoters, leading to localized deacetylation and developmental silencing. Because an NF-κB complex was implicated in the developmental silencing of the human ζ-globin gene in transgenic mice (40), NF-κB family members may also mediate silencing of embryonic globin genes in definitive erythroblasts. It will be important to identify the repressors that establish the hypoacetylated subdomain of the β-globin locus and to determine whether other complex multigene loci have internal hypoacetylated subdomains.
Supplementary Material
Acknowledgments
We thank Kirby Johnson for a critical review of the manuscript. We thank David Allis for providing the anti-pentaacetylated histone H4 antibody. We acknowledge support from the Milwaukee Foundation, the Leukemia and Lymphoma Society of America, and the National Institutes of Health (Grants DK50107 and DK55700 to E.H.B. and Grant HD36847 to K.M.D.). E.H.B. is a Leukemia and Lymphoma Society of America Scholar and a Shaw Scientist. E.C.F. is a predoctoral fellow of the American Heart Association.
Abbreviations
- ChIP
chromatin immunoprecipitation
- ES cell
embryonic stem cell
- HAT
histone acetylase
- HDAC
histone deacetylase
- HS
hypersensitive site
- LCR
locus control region
- MEL
mouse erythroleukemia
- dpc
days postcoitum
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 2.Howe L, Brown C E, Lechner T, Workman J L. Crit Rev Eukaryot Gene Expr. 1999;9:231–243. doi: 10.1615/critreveukargeneexpr.v9.i3-4.80. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe A P, Hayes J J. Nucleic Acids Res. 1999;27:711–720. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 5.Lee D Y, Hayes J J, Pruss D, Wolffe A P. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 6.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Nature (London) 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 7.Kuo M H, Allis C D. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 8.Alberts A S, Geneste O, Treisman R. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Lin R J, Xie W, Wilpitz D, Evans R M. Cell. 1999;98:675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 10.Krebs J E, Kuo M H, Allis C D, Peterson C L. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parekh B S, Maniatis T. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 13.Tse C, Sera T, Wolffe A P, Hansen J C. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Harju S, Peterson K R. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 15.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 16.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 17.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 18.Forrester W C, Thompson C, Elder J T, Groudine M. Proc Natl Acad Sci USA. 1986;83:1359–1363. doi: 10.1073/pnas.83.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuan D, London I M. Proc Natl Acad Sci. USA. 1984;81:2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starck J, Sarkar R, Romana M, Bhargava A, Scarpa A L, Tanaka M, Chamberlain J W, Weissman S M, Forget B G. Blood. 1994;84:1656–1665. [PubMed] [Google Scholar]
- 21.Fraser P, Gribnau J, Trimborn T. Curr Opin Hematol. 1998;5:139–144. doi: 10.1097/00062752-199803000-00009. [DOI] [PubMed] [Google Scholar]
- 22.Bresnick E H, Tze L. Proc Natl Acad Sci USA. 1997;94:4566–4571. doi: 10.1073/pnas.94.9.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forsberg E C, Johnson K, Zaboikina T N, Mosser E A, Bresnick E H. J Biol Chem. 1999;274:26850–26859. doi: 10.1074/jbc.274.38.26850. [DOI] [PubMed] [Google Scholar]
- 24.Schubeler D, Francastel C, Cimbora D M, Reik A, Martin D I, Groudine M. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 25.Elefant F, Cooke N E, Liebhaber S A. J Biol Chem. 2000;275:13827–13834. doi: 10.1074/jbc.275.18.13827. [DOI] [PubMed] [Google Scholar]
- 26.Marks P A, Sheffrey M, Rifkind R A. Prog Clin Biol Res. 1985;191:185–203. [PubMed] [Google Scholar]
- 27.Forsberg E C, Downs K M, Bresnick E H. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- 28.Lawson K A, Meneses J, Pederson R A. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 29.Uetsuki T, Takagi K, Sugiura H, Yoshikawa K. J Biol Chem. 1996;271:918–924. doi: 10.1074/jbc.271.2.918. [DOI] [PubMed] [Google Scholar]
- 30.Orkin S H, Zon L I. Annu Rev Genet. 1997;31:33–60. doi: 10.1146/annurev.genet.31.1.33. [DOI] [PubMed] [Google Scholar]
- 31.Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong S, Bohl D, Li C, Tuan D. Mol Cell Biol. 1997;17:3955–3965. doi: 10.1128/mcb.17.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prioleau M N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitoh N, Bell A C, Recillas-Targa F, West A G, Simpson M, Pikaart M, Felsenfeld G. EMBO J. 2000;19:2315–2322. doi: 10.1093/emboj/19.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Bungert J, Engel J D. Proc Natl Acad Sci. USA. 1997;94:169–174. doi: 10.1073/pnas.94.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boffa L C, Vidali G, Mann R S, Allfrey V G. J Biol Chem. 1978;253:3364–3366. [PubMed] [Google Scholar]
- 37.Ayer D E. Trends Cell Biol. 1999;9:193–198. doi: 10.1016/s0962-8924(99)01536-6. [DOI] [PubMed] [Google Scholar]
- 38.Filipe A, Li Q, Deveaux S, Godin I, Romeo P-H, Stamatoypannopoulos G, Mignotte V. EMBO J. 1999;18:687–697. doi: 10.1093/emboj/18.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata H, Nawaz Z, Tsai S Y, O'Malley B W, Tsai M J. Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Liebhaber S A. EMBO J. 1999;18:2218–2228. doi: 10.1093/emboj/18.8.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.