Abstract
The proapoptotic protein Bim is expressed de novo following withdrawal of serum survival factors. Here, we show that Bim−/− fibroblasts and epithelial cells exhibit reduced cell death following serum withdrawal in comparison with their wild-type counterparts. In viable cells, Bax associates with Bcl-2, Bcl-xL and Mcl-1. Upon serum withdrawal, newly expressed BimEL associates with Bcl-xL and Mcl-1, coinciding with the dissociation of Bax from these proteins. Survival factors can prevent association of Bim with pro-survival proteins by preventing Bim expression. However, we now show that even preformed BimEL/Mcl-1 and BimEL/Bcl-xL complexes can be rapidly dissociated following activation of ERK1/2 by survival factors. The dissociation of Bim from Mcl-1 is specific for BimEL and requires ERK1/2-dependent phosphorylation of BimEL at Ser65. Finally, ERK1/2-dependent dissociation of BimEL from Mcl-1 and Bcl-xL may play a role in regulating BimEL degradation, since mutations in the BimEL BH3 domain that disrupt binding to Mcl-1 cause increased turnover of BimEL. These results provide new insights into the role of Bim in cell death and its regulation by the ERK1/2 survival pathway.
Keywords: apoptosis, Bcl-xL, Bim, ERK1/2, Mcl-1
Introduction
The pro-survival Bcl-2 proteins, such as Bcl-2 and BclxL, typically contain four Bcl-2 homology (BH) domains, while the pro-death proteins include those with the BH1, 2 and 3 domains (Bax and Bak) and the ‘BH3-only proteins' (BOPs), including Bim, Hrk/DP5, Bmf, Puma, Bid and Bad (Cory and Adams, 2002). Bax and Bak proteins promote the release of apoptogenic factors from the mitochondria, but in viable cells are restrained by their binding to pro-survival Bcl-2 proteins. A variety of studies suggest that the BOPs bind to the pro-survival Bcl-2 proteins and neutralise them, thereby allowing Bax and/or Bak to initiate cell death (Puthalakath and Strasser, 2002). Bim and Puma are the most effective BOPs for cell killing, probably because they can engage with all the pro-survival Bcl-2 proteins (Chen et al, 2005). The different BOPs respond to different forms of cellular stress and are subject to regulation at both the transcriptional and post-translational level (Puthalakath and Strasser, 2002). For example, Puma is a transcriptional target of p53 (Nakano and Vousden, 2001), while Bad is phosphorylated and sequestered by 14-3-3 proteins so that it cannot bind to Bcl-2 proteins (Zha et al, 1996; Scheid et al, 1999; Datta et al, 2000).
Bim is expressed de novo following withdrawal of survival factors (Dijkers et al, 2000; Whitfield et al, 2001; Reginato et al, 2003; Weston et al, 2003; Wang et al, 2004). Bim mRNA increases due to inactivation of the protein kinase B (PKB) (Dijkers et al, 2000; Gilley et al, 2003) or ERK1/2 pathways (Weston et al, 2003), or due to activation of JNK (c-Jun N-terminal kinase) (Whitfield et al, 2001). Alternative splicing of the Bim gene gives rise to multiple isoforms that differ in proapoptotic potency (O'Connor et al, 1998) due to differences in their post-translational regulation (Ley et al, 2005b). Of the short, long and extra-long Bim proteins (BimS, BimL and BimEL), BimS is the most effective killer and consists of the pro-death BH3 domain and a C-terminal membrane-tethering domain. BimL and BimEL contain a dynein light chain 1 (DLC1) interacting domain, allowing their sequestration at microtubules in viable cells (Puthalakath et al, 1999; Lei and Davis, 2003). BimEL, the least effective cell killer, contains a unique exon that encodes an ERK1/2 docking domain (Ley et al, 2005a) and ERK1/2 phosphorylation sites that control the proteasomal turnover of BimEL (Ley et al, 2003; Luciano et al, 2003; Ley et al, 2004; Marani et al, 2004; Ley et al, 2005a).
Here, we define a new role for ERK1/2-dependent phosphorylation of BimEL. We find that Bim is required for optimal cell death upon serum withdrawal and associates with Mcl-1 and Bcl-xL; survival factors prevent this association by preventing Bim expression. However, we now show that survival factors can also promote the rapid dissociation of preformed BimEL/Mcl-1 and BimEL/Bcl-xL complexes by ERK1/2-dependent phosphorylation of BimEL. Dissociation is not a consequence of BimEL degradation, since proteasome inhibitors and mutations that prevent BimEL ubiquitylation do not impair dissociation of BimEL from pro-survival proteins; indeed, dissociation may contribute to BimEL degradation. Our results provide new insights into the regulation of BimEL and cell survival by the ERK1/2 pathway.
Results
Bim expression determines the rate and magnitude of cell death following serum withdrawal
CCl39 fibroblasts die by apoptosis following withdrawal of serum survival factors (Chalmers et al, 2003; Weston et al, 2003). Bim has been implicated in cell death in neurons (Whitfield et al, 2001), epithelial cells (Reginato et al, 2003; Wang et al, 2004) and haematopoietic cells deprived of trophic support (Bouillet et al, 1999). Indeed, following serum starvation of CCl39 cells we observed a rapid increase in BimEL, followed by a slower increase in expression of BimL, whereas the expression of Bmf and Bax did not change (Figure 1A). Of the pro-survival proteins, Bcl-2 and Bcl-xL levels remained unchanged, but there was a gradual decrease in the expression of Mcl-1 (Figure 1A), although the kinetics and magnitude of this varied between experiments.
Figure 1.
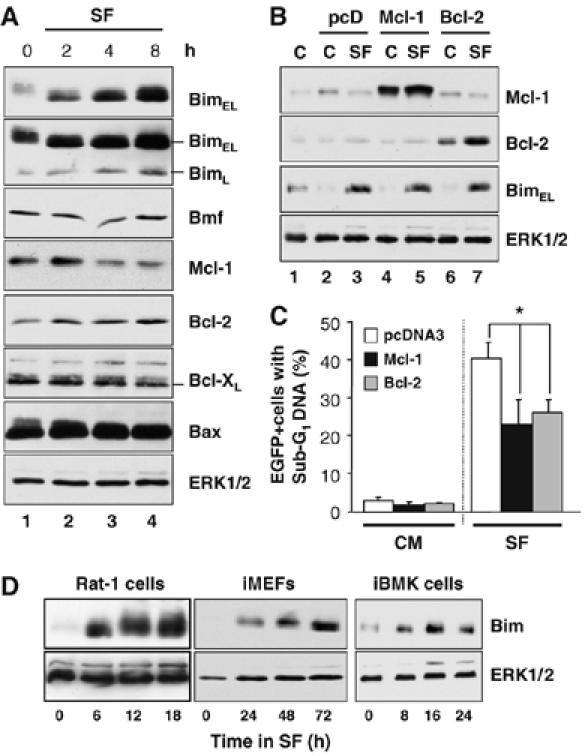
Expression of Bim is an early event during serum withdrawal-induced death. (A) CCl39 cells in complete medium (0 h) were serum starved for 2, 4 or 8 h. Cell extracts were fractionated by SDS–PAGE and immunoblotted with the indicated antibodies. The blot for BimEL (top blot) was overexposed (second blot) to reveal the expression of BimL. (B, C) Cycling CCl39 cells were transfected with EGFP-spectrin and pcDNA3, pcDNA3-Mcl-1 or pcDNA3-Bcl-2; 24 h later cells were switched to serum-free medium (SF) or fresh complete medium (C). (B) Expression of Bcl-2, Mcl-1 and BimEL was confirmed by Western blotting. (C) The percentage of EGFP-positive cells with sub-G1 DNA was determined by flow cytometry from triplicate cell samples (mean±s.d.). The asterisk indicates expression of Bcl-2 or Mcl-1 afforded significant protection against serum withdrawal, by the Student's t-test (P<0.05). (D) Rat-1 cells, iMEFs or iBMK cells were serum starved as indicated and whole-cell extracts were analysed or expression of Bim and total ERK1/2 as a loading control.
Death following withdrawal of cytokines is thought to proceed through the Bcl-2-inhibitable, cell intrinsic pathway. To test this, we transfected CCl39 cells with expression plasmids for Bcl-2 or Mcl-1 (or empty vector, pcD) together with a plasmid encoding EGFP-spectrin to identify transfected cells. Overexpression of Bcl-2 or Mcl-1, confirmed by Western blotting (Figure 1B), caused a reduction in cell death (Figure 1C). Thus, serum withdrawal-induced apoptosis in CCl39 cells proceeds through the cell intrinsic pathway since it is inhibited by Bcl-2 or Mcl-1. It was notable that expression of Bcl-2 or Mcl-1 did not block the increase in Bim expression under these conditions (Figure 1B). Increased Bim expression following serum withdrawal was highly reproducible and was also seen in Rat-1 cells, immortalised mouse embryo fibroblasts (iMEFs) and immortalised baby mouse kidney epithelial (iBMK) cells (Figure 1D), albeit with distinct kinetics in each cell line.
To address directly if Bim was involved in promoting cell death following serum withdrawal, we examined iMEFs derived from wild-type (WT) and Bim−/− mice (Figure 2A). Bim−/− iMEFs exhibited an approximate 90% reduction in peak caspase activation in comparison to their WT counterparts (Figure 2B). The Bim−/− iMEFs also exhibited a delay and reduction in the accumulation of dead cells with sub-G1 DNA (Figure 2C), but this was not persistent, and Bim−/− iMEFs clearly started to die at later time points (72–96 h). Furthermore, there was no difference between WT and Bim−/− iMEFs in long-term (7 day) clonogenic survival assays (K Ewings, K Balmanno and S Cook, unpublished observations). Similar results were obtained by comparing WT and Bim−/− BMK cells (Figure 2D); the Bim−/− cells again exhibited a reduction, but not a complete inhibition, of cell death (Figure 2E). As a comparison, we confirmed that loss of Bim protected iBMK cells for up to 48 h of paclitaxel exposure (Supplementary Figure 1; Tan et al, 2005). In contrast, loss of Bim provided some resistance to serum withdrawal at early time points, but by 48 h there was no difference in viability between WT and Bim−/− BMK cells. These results, in two different cell systems, suggest that there is a major role for Bim in the initiation of caspase activation and cell death following serum withdrawal, but other redundant BOPs or alternative death pathways can substitute for Bim in promoting cell death at later time points.
Figure 2.
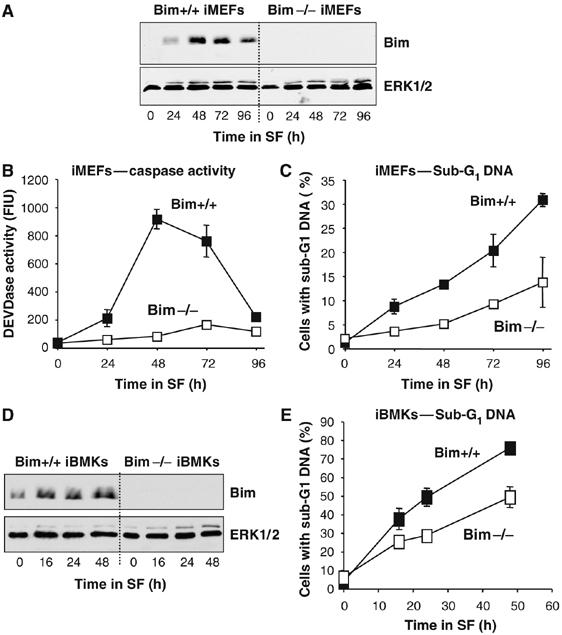
Bim contributes to cell death following serum withdrawal in iMEFs and iBMK epithelial cells. (A–C) WT or Bim−/− iMEFs were serum starved as indicated. (A) Cell extracts were immunoblotted with antibodies to Bim and ERK1/2. (B) Cell extracts were assayed for caspase activity using a DEVDase assay. (C) The percentage of cells with sub-G1 DNA was determined by flow cytometry. (D, E) WT or Bim−/− iBMK cells were serum starved for the times indicated. (D) Whole-cell extracts were immunoblotted with antibodies to Bim and ERK1/2. (E) Cells were fixed, stained with PI and the percentage of cells with sub-G1 DNA was determined by flow cytometry. In panels B, C and E each data point represents the mean±s.d. of triplicate dishes of cells from a single experiment representative of three.
Bim expressed following serum withdrawal associates with Bcl-xL and Mcl-1 but not Bax
Bim promotes apoptosis by binding to pro-survival Bcl-2 proteins and neutralising their protective effect or, in the case of BimS, perhaps by binding directly to Bax (Marani et al, 2002). We examined which Bcl-2 proteins Bim interacted with by immunoprecipitation of Bcl-xL or Mcl-1 from CCl39 whole cell extracts (WCE) (Figure 3A and B). Serum starvation increased BimEL expression and the newly expressed BimEL associated with Mcl-1 and Bcl-xL (Figure 3A and B). While Bim also binds to Bcl-2 (O'Connor et al, 1998), we failed to identify an antibody that could immunoprecipitate Bcl-2 complexes from CCl39 cells.
Figure 3.
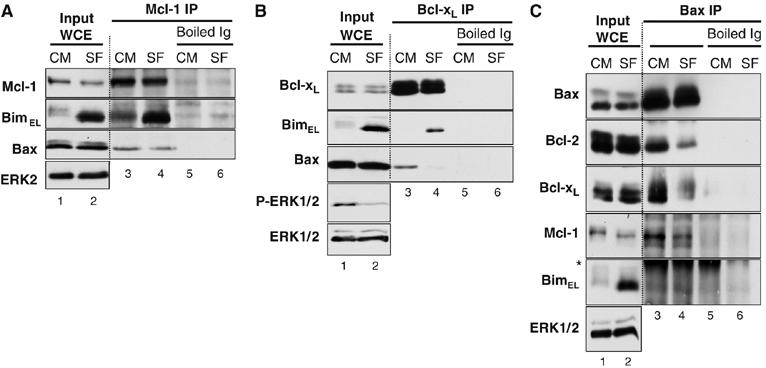
Newly expressed BimEL associates with the pro-survival proteins Bcl-xL and Mcl-1 following serum withdrawal. CCl39 cells in complete medium (CM) were serum starved (SF) for 6 h. Cell extracts (input) were used for immunoprecipitation with antibodies to (A) Mcl-1 or (B) Bcl-xL or (C) Bax (N-20). Antibodies denatured before immunoprecipitation (boiled Ig) served as a control. Input and IP samples were immunoblotted with the indicated antibodies. The asterisk in panel (C) indicates cross-reactivity with the antibody light chain used in the IP. Results are taken from a single experiment representative of three giving similar results.
During these studies we found that Bax associated with Bcl-xL and Mcl-1 in viable cells (Figure 3A and B). Serum withdrawal (SF) caused a small decrease in the amount of Bax bound to Mcl-1 (Figure 3A), but a very pronounced dissociation of Bax from Bcl-xL complexes, commensurate with the binding of BimEL (Figure 3B). To investigate this further, we immunoprecipitated Bax using the N-20 N-terminal antibody. The N-20 epitope is exposed by a conformational change in response to apoptotic insults, but the presence of Triton X-100 in the lysis buffer ensured that all the Bax had undergone this conformational change (Marani et al, 2002). Bax immunoprecipitates (IPs) from viable cells contained Bcl-2, Bcl-xL and Mcl-1 but no BimEL (Figure 3C). Upon serum starvation, a treatment that results in Bim binding to pro-survival proteins (Figure 3A and B), the amount of Bcl-2, Bcl-xL and Mcl-1 bound to Bax decreased (Figure 3C). In addition, while serum starvation increased Bim expression, we failed to detect BimL or BimEL in Bax IPs (Figure 3C); we have been unable to reproducibly detect the low levels of BimS in CCl39 cells. Interestingly, these results indicate that changes in the N-terminal conformation of Bax (monitored by the N-20 antibody) are not sufficient to cause its dissociation from pro-survival Bcl-2 proteins. These results demonstrate that following serum starvation, newly expressed Bim binds to pro-survival proteins but fails to associate with Bax; indeed, the binding of Bim to Bcl-xL and Mcl-1 coincides with a reduction in the binding of Bax to these proteins.
Survival factors promote the rapid dissociation of BimEL from Mcl-1
Thrombin is a survival factor for CCl39 cells (Chalmers et al, 2003; Figure 4A). When added to cells at the time of serum withdrawal, thrombin inhibited Bim expression (Figure 4B, input WCE) and so prevented its recruitment into Mcl-1 complexes (Figure 4B, IP Mcl-1); addition of fresh FBS was equally effective. Quantification of these blots (Figure 4C) confirmed that thrombin blocked Bim expression and so prevented its binding to Mcl-1.
Figure 4.
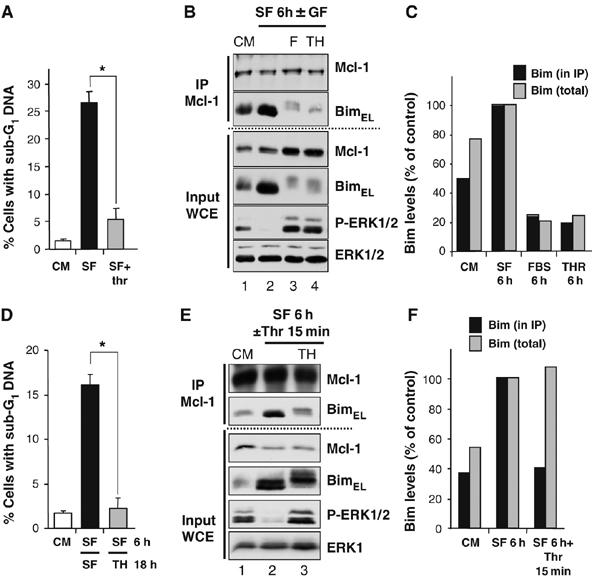
Survival factors block Bim expression but can also promote the dissociation of preformed Bim/Mcl-1 complexes. (A–C) CCl39 cells were subjected to the following condition: serum starvation±10 nM thrombin, which was added at the time of serum withdrawal. (A) Thrombin protected against serum withdrawal-induced cell death (*P<0.01), as judged by cells with sub-G1 DNA after 24 h. (B) Cell extracts (input) were prepared after 6 h and used for immunoprecipitation of Mcl-1 and samples were immunoblotted with antibodies to Mcl-1, Bim, P-ERK1/2 and ERK1/2. (C) The amount of Bim in the input (total) and the Mcl-1 IP was quantified by densitometry. (D–F) CCl39 cells were serum starved for 6 h to allow the expression of Bim and the assembly of BimEL/Mcl-1 complexes. (D) Cells were re-stimulated with 10 nM thrombin for a further 18 h and cell death was assayed by the accumulation of cells with sub-G1 DNA. (E) Cells were re-stimulated with 10 nM thrombin for 15 min. Whole-cell extracts (input) were used for immunoprecipitation of Mcl-1 and samples were immunoblotted with the indicated antibodies. (F) The amount of Bim in the input WCE (total) and the Mcl-1 IP was quantified by densitometry and revealed that a 15 min treatment with thrombin promoted the dissociation of BimEL/Mcl-1 complexes without reducing total BimEL levels.
In the course of these studies, we noted that addition of thrombin to cells that had been serum starved for 6 h was still able to protect cells from cell death (Figure 4D). Since assembly of BimEL/Mcl-1 complexes was already apparent after 6 h of serum withdrawal (Figure 4B and E), we examined the effect of acute thrombin stimulation on the preassembled BimEL/Mcl-1 complex. CCl39 cells were serum starved for 6 h to induce the assembly of a preformed BimEL/Mcl-1 complex. Stimulation with thrombin for 15 min had no impact on the turnover of BimEL (Ley et al, 2003) or the total amount of BimEL in whole-cell extracts (Figure 4E, input WCE, quantified in Figure 4F), but promoted the phosphorylation of BimEL (Figure 4E, input WCE) and caused a clear reduction in the amount of BimEL in Mcl-1 IPs (Figure 4E, IP Mcl-1, quantified in Figure 4F). Stimulation with FBS for 15 min was also effective at promoting the phosphorylation and rapid dissociation of BimEL from Mcl-1 (Supplementary Figure 2A). Furthermore, dissociation of BimEL from Bcl-xL complexes was also seen in starved WT iMEFs re-stimulated with FBS, confirming that this was not unique to CCl39 cells (Supplementary Figure 2B).
Activation of ERK1/2 is necessary and sufficient to promote the dissociation of BimEL from Mcl-1 and Bcl-xL
The ERK1/2 and PKB pathways promote cell survival and regulate Bim (Dijkers et al, 2000; Gilley et al, 2003; Weston et al, 2003), so we used selective inhibitors to determine which pathway was responsible for the dissociation of BimEL from Mcl-1. Stimulation of serum-starved CCl39 cells with thrombin caused dissociation of BimEL from Mcl-1 (Figure 5A, IP Mcl-1) and U0126, an inhibitor of MEK1/2, blocked ERK1/2 phosphorylation and BimEL phosphorylation (Figure 5A, input WCE) and prevented the thrombin-stimulated dissociation of BimEL from Mcl-1 (Figure 5A, IP Mcl-1). In contrast, LY294002, a PI3′-kinase inhibitor, prevented PKB phosphorylation, but did not prevent BimEL dissociation from Mcl-1 (Figure 5A).
Figure 5.
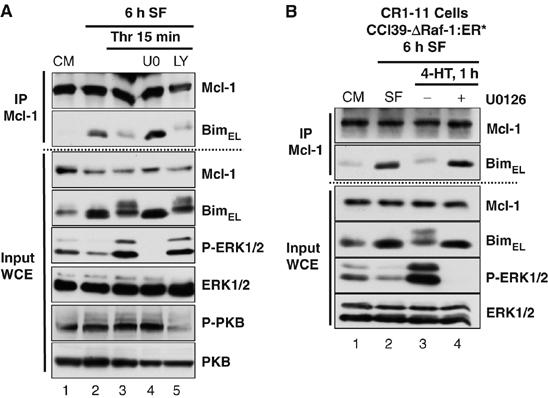
Activation of ERK1/2 pathway is necessary and sufficient to promote the release of BimEL from Mcl-1. (A) Cycling CCl39 cells (CM) were serum starved (SF) for 6 h to induce formation of BimEL/Mcl-1 complexes. Cells were then stimulated for 15 min with 10 nM thrombin, with or without 20 μM U0126 or 30 μM LY294002. Whole-cell extracts (input) were used for immunoprecipitation of Mcl-1 and samples were immunoblotted with the indicated antibodies. (B) CR1-11 cells (CCl39 cells expressing ΔRaf-1:ER*) in complete medium (CM) were serum starved (SF) for 6 h to induce formation of BimEL/Mcl-1 complexes. Cells were then stimulated for 1 h with 100 nM 4-HT±20 μM U0126. Whole-cell extracts (input) were used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with the indicated antibodies. Results are taken from a single experiment representative of three.
To determine if the ERK1/2 pathway was sufficient to dissociate BimEL, we used CR1-11 cells (Weston et al, 2003), a clone of CCl39 cells expressing the conditional protein kinase ΔRaf-1:ER*, which selectively activates ERK1/2 in response to treatment with 4-hydroxytamoxifen (4-HT). A 1 h stimulation with 4-HT promoted ERK1/2, phosphorylation of BimEL (Figure 5B, input WCE) and resulted in dissociation of BimEL from preformed BimEL/Mcl-1 complexes (Figure 5B, IP Mcl-1), although the total amount of Bim in whole-cell extracts was not affected (Figure 5B, input WCE). All these effects were reversed by treatment with U0126. ΔRaf-1:ER* also promoted the ERK1/2-dependent dissociation of preformed BimEL/Bcl-xL complexes (Supplementary Figure 2C). Thus, activation of the ERK1/2 pathway is necessary and sufficient to promote dissociation of endogenous BimEL from endogenous Mcl-1 or Bcl-xL, two major pro-survival proteins.
ERK1/2-dependent dissociation of BimEL/Mcl-1 complexes is not a consequence of BimEL degradation
Phosphorylation of BimEL by ERK1/2 promotes its proteasome-dependent destruction (Ley et al, 2003), so it was important to establish the relationship between dissociation and destruction. Dissociation of BimEL from Mcl-1 was readily apparent within 15 min of FBS stimulation (Supplementary Figure 2A), whereas emetine chase experiments revealed that significant turnover of endogenous BimEL only occurred between 20 min and 1 h (Supplementary Figure 3A and B). However, to be sure, we devised a series of experiments to determine whether the dissociation of BimEL from Mcl-1 was simply a consequence of ERK1/2-dependent BimEL destruction. First, CR1-11 cells were serum starved for 6 h to assemble the BimEL/Mcl-1 complex and then re-stimulated with 4-HT in the presence of the proteasome inhibitor MG132. Following activation of ΔRaf-1:ER*, phosphorylated forms of BimEL accumulated due to the presence of MG132, and were detected in the whole-cell extracts (Figure 6A, input WCE). This phosphorylation of BimEL caused a 60–70% reduction in the amount of BimEL associated with Mcl-1 (Figure 6A and B), similar to that observed in response to thrombin (Figure 4F); dissociation of BimEL was apparent within 30 min, maximal by 1 h and persisted for as long as ΔRaf-1:ER* remained active. These results, using a pharmacological inhibitor of the proteasome, show that dissociation of BimEL takes place even when total BimEL levels are constant and so is not a consequence of it being degraded by the proteasome. In addition, we observed that a mutant version of BimEL, in which both lysines were mutated to arginine (KK/RR), dissociated from Mcl-1 to the same degree as WT BimEL following activation of ΔRaf-1:ER* in HR1 cells (HEK293 cells expressing ΔRaf-1:ER*; Boughan et al, 2006) (Figure 6C). Since this mutant cannot be ubiquitylated (Akiyama et al, 2003), this provides further molecular genetic evidence that ERK1/2-dependent dissociation of BimEL was not a consequence of its ubiquitylation or proteasomal degradation. Finally, growth factor-dependent dissociation of BimEL from Mcl-1 proceeded in the presence of cycloheximide and MG132, indicating that ERK1/2 was promoting BimEL dissociation from pre-existing complexes rather than reducing the stability of newly expressed BimEL before it has a chance to bind (Supplementary Figure 3C). Together, these results indicate that dissociation of BimEL from pro-survival proteins does not result from its destruction.
Figure 6.
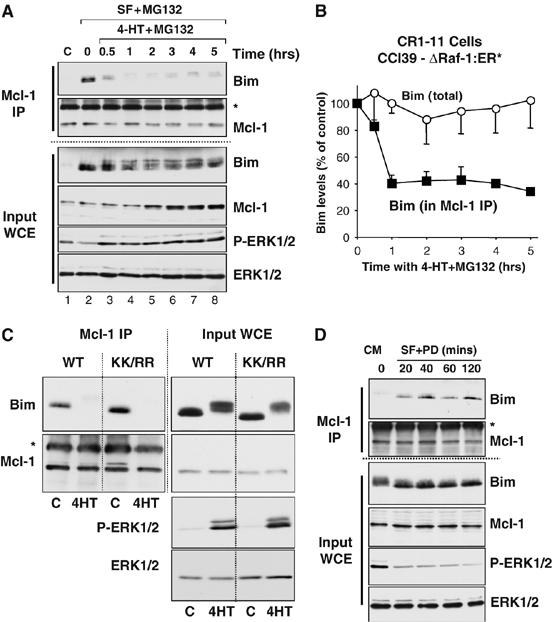
ERK1/2-dependent dissociation of BimEL/Mcl-1 complexes is not a consequence of BimEL degradation. (A, B) CR1-11 cells were serum starved to induce the assembly of BimEL/Mcl-1 complexes. Cells were then re-stimulated with 100 nM 4-HT in the presence of 10 μM MG132, as indicated. Whole-cell extracts (input) were prepared, used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with the indicated antibodies. (A) Results from a single experiment representative of three yielding identical results. (B) Total and Mcl-1-associated BimEL were quantified from live ECL images; data represent the mean±s.d. from three independent experiments. (C) HR1 cells (HEK293 cells expressing ΔRaf-1:ER*) were transfected with wild-type HA-BimEL (WT) or a mutant in which both lysines had been mutated to arginine (KK/RR). Cells were stimulated with 4-HT for 1 h. Whole-cell extracts (input) were used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with the indicated antibodies. (D) CCl39 cells maintained in 10% FBS were serum starved in the presence of 5 μM PD184352 for the times indicated. Whole-cell extracts (input) were used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with the indicated antibodies. The asterisk in panels (A, C, D) indicates cross-reactivity with the antibody light chain used in the IP.
We also performed the reciprocal experiment by examining the kinetics of BimEL/Mcl-1 association following inhibition of the ERK1/2 pathway. CCl39 cells growing in complete medium (10% FBS) were serum starved in the presence of the ERK1/2 pathway-selective inhibitor PD184352 for between 20 and 120 min. This treatment led to the immediate inactivation of ERK1/2, dephosphorylation of BimEL and an increase in the amount of BimEL bound to Mcl-1 (Figure 6D). Significantly, while serum withdrawal (with or without ERK1/2 inhibition) can increase BimEL expression, this is not apparent until approximately 2 h (Ley et al, 2003; Weston et al, 2003), whereas the increase in association of BimEL with Mcl-1 was apparent within 20 min, a time point at which total BimEL and Mcl-1 levels were unchanged (Figure 6D). These results indicate that inhibition of ERK1/2 causes the rapid dephosphorylation of BimEL and its association with its pro-survival target proteins.
BimEL dissociation is reversible upon inhibition of the ERK1/2 pathway
When degradation of BimEL was blocked, we found that the dissociation of BimEL from Mcl-1 was readily reversible. For example, when serum-starved CR1-11 cells were stimulated with 4-HT for 2 or 4 h in the presence of MG132 to prevent BimEL degradation, we observed a 70% reduction in the amount of BimEL bound to Mcl-1, even though total BimEL levels did not change (Supplementary Figure 4A and B). In contrast, if cells were treated with 4-HT for 2 h to phosphorylate and dissociate BimEL and then treated with U0126 for a further 2 h to inhibit ERK1/2, BimEL was dephosphorylated and a significant proportion was found reassociated with Mcl-1 (Supplementary Figure 4A and B). A potential explanation for this came when we subjected these cell extracts to a crude subcellular fractionation. Regardless of its phosphorylation status, BimEL remained in the same COX-IV containing heavy membrane fraction as Mcl-1 rather than being released into the cytosol (Supplementary Figure 4C). These results suggest that when the proteasome is inhibited, both BimEL and Mcl-1 are retained on the mitochondria following their dissociation and so can readily reassociate when BimEL is dephosphorylated following ERK1/2 inhibition. This may explain why we have been unable to visualise the dissociation of BimEL from Mcl-1 using immunofluorescence microscopy. It also demonstrates the dynamic and reversible nature of ERK1/2-dependent BimEL dissociation.
ERK1/2-dependent phosphorylation of BimEL is required for dissociation of the Bim/Mcl-1 and Bim/Bcl-xL complex
The dissociation of BimEL from Mcl-1 could be due to ERK1/2-dependent phosphorylation of BimEL (Luciano et al, 2003; Ley et al, 2004; Marani et al, 2004), Mcl-1 (Domina et al, 2004) or both. To address this, we expressed FLAG-tagged Puma (Nakano and Vousden, 2001) in HR1 cells and examined its binding to Mcl-1. Puma bound to endogenous Mcl-1 but was not phosphorylated and did not dissociate following activation of ΔRaf-1:ER* (Supplementary Figure 5). Furthermore, while BimS, BimL and BimEL (Figure 7A) were all able to bind to Mcl-1 in HR1 cells (Figure 7B), only HA-BimEL dissociated from Mcl-1 following activation of ΔRaf-1:ER* (Figure 7B, IP Mcl-1). Thus, ERK1/2 can promote the dissociation of BimEL/Mcl-1 and BimEL/Bcl-xL complexes but not Puma/Mcl-1 complexes; this is a property unique to BimEL of the three canonical Bim splice variants and is observed in HEK293 cells, CCl39 cells and iMEFs. We have also seen similar effects in human colorectal cancer cells (J Wickenden and S Cook, unpublished observations), so this is a widespread mode of regulation.
Figure 7.
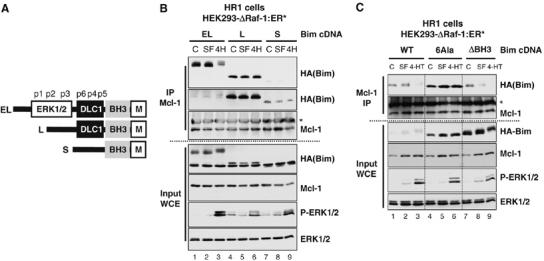
ERK1/2-dependent regulation of BimEL/Mcl-1 interactions is unique to the BimEL isoform. (A) Diagram of the structure of BimS, BimL and BimEL indicating the six proline-directed phosphorylation sites (p1–p6), the ERK1/2 docking domain and phosphorylation sites, the DLC1 binding site, BH3 domain and membrane binding motif. (B) HR1 cells were transfected with HA-BimEL (EL), HA-BimL (L) or HA-BimS (S). After 24 h, cells were serum starved (SF) for 6 h. Finally, one set of cells in serum-free medium was stimulated with 100 nM 4-HT for 15 min to activate ERK1/2 (4H). Cell extracts (input) were used to immunoprecipitate Mcl-1 and samples were subjected to immunoblotting. The anti-HA blot showing the binding of BimEL and BimL to Mcl-1 (top panel) has also been overexposed to reveal the binding of BimS (second panel from top). (C) HR1 cells were transfected with HA-BimEL (WT), HA-BimEL6Ala (6Ala) or HA-BimELΔBH3 (ΔBH3) in complete medium (CM). After 24 h, cells were serum starved (SF) for 6 h. Finally, one set of cells was stimulated with 100 nM 4-HT for 15 min to activate ERK1/2. Whole-cell extracts (input WCE) were used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with antibodies to Mcl-1, HA(Bim), P-ERK1/2 and ERK1/2. The asterisk in panels (B, C) indicates cross-reactivity with the antibody light chain used in the IP.
Phosphorylation of BimEL at Ser65 is required for dissociation from Mcl-1
BimEL, but not BimS or BimL, is a substrate for ERK1/2, a proline-directed protein kinase. BimEL possesses six Ser–Pro or Thr–Pro motifs (p1–p6 in Figure 7A), one of which (Ser65, p2) is an in vitro and in vivo ERK1/2 phosphorylation site (Luciano et al, 2003; Ley et al, 2004; Marani et al, 2004). To determine the role of proline-directed phosphorylation of BimEL, we analysed a mutant (HA-BimEL6Ala) in which all six Ser–Pro or Thr–Pro motifs had been mutated to non-phosphorylatable residues. As a control, we observed that BimEL with three mutations in the BH3 domain (HA-BimELΔBH3) exhibited greatly reduced binding to Mcl-1 in HR1 cells, despite expressing at the highest level of all three constructs (Figure 7C). HA-BimEL6Ala expressed at much higher levels than HA-BimEL (Figure 7C, input WCE), but was not phosphorylated and did not dissociate in response to ERK1/2 activation, whereas HA-BimEL did (Figure 7C, Mcl-1 IP), demonstrating that phosphorylation of proline-directed sites on BimEL is required for its ERK1/2-dependent dissociation from Mcl-1.
Since phosphorylation-dependent dissociation from Mcl-1 was only observed for BimEL, we focused on the three sites (p1–p3) in exon 3 by analysing the effect of individual point mutations in the p1 (Ser55Ala), p2 (Ser65Ala) or p3 (Ser73Ala) sites. All mutants associated with Mcl-1 normally in HR1 cells and the HA-BimEL protein was phosphorylated and dissociated from Mcl-1 following activation of the ERK1/2 pathway (Figure 8A). Mutation of the p1 (Ser55) or p3 (Ser73) sites had a partial effect on the ERK1/2-dependent phosphorylation and dissociation of BimEL. In contrast, mutation of the p2 site (Ser65) strongly inhibited phosphorylation of BimEL and completely prevented its ERK1/2-dependent dissociation from Mcl-1. Mutation of the proline-directed sites (p4–p6) had no effect on the dissociation of BimEL from Mcl-1 (K Ewings and S Cook, unpublished observations), consistent with the fact that BimL did not dissociate upon ERK1/2 activation (Figure 7B). These results demonstrate that phosphorylation of BimEL at Ser65 is absolutely required for ERK1/2-dependent dissociation of BimEL from Mcl-1.
Figure 8.
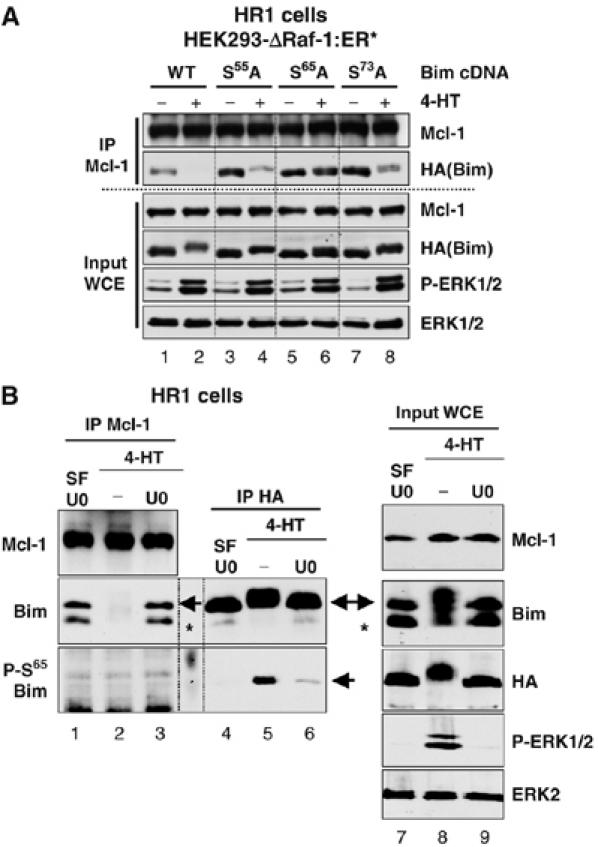
ERK1/2-dependent phosphorylation of BimEL at Ser65 is required for dissociation of BimEL from Mcl-1. (A) HR1 cells were transfected with plasmids encoding HA-BimEL (WT), or HA-BimEL with individual point mutations at Ser55Ala, Ser65Ala or Ser73Ala. After 24 h, cells were serum starved for 6 h before treating for a further 15 min with 100 nM 4-HT to activate ERK1/2. Whole-cell extracts (input WCE) were used for immunoprecipitation of Mcl-1 and samples were then immunoblotted with antibodies to Mcl-1, HA(Bim), P-ERK1/2 and ERK1/2. (B) HR1 cells were transfected with HA-BimEL. Twenty-four hours later, cells were serum starved for 1 h in the presence of 20 μM U0126 to inactivate ERK1/2. Cells were then washed thoroughly and stimulated with 100 nM 4-HT±20 μM U0126 for 1 h. Whole-cell extracts (input WCE) were used to immunoprecipitate Mcl-1 or HA-BimEL. Samples were then immunoblotted with antibodies to Mcl-1, HA(Bim), Bim, Phospho-Ser65-Bim, P-ERK1/2 and ERK1/2. The arrowheads indicate the position of the HA-BimEL, while the asterisk indicates the position of the endogenous BimEL. Note that ΔRaf-1:ER* induces phosphorylation of BimEL at Ser65, but phospho-Ser65-BimEL is not detected in Mcl-1 IPs.
A logical prediction of these experiments is that phospho-mimetic forms of BimEL, in which the p2 or p1–p3 sites are mutated to acidic residues (Asp or Glu), should bind poorly to Mcl-1. However, we found that the binding of such mutant forms of BimEL to Mcl-1 was highly variable so that it was not possible to draw any reliable conclusions. It would appear that acidic substitutions at Ser65, either alone or in combination with mutation of Ser55 and Ser73, cannot faithfully mimic the phosphorylation events in vivo. As an alternative, we examined the distribution of BimEL phosphorylated at Ser65 using a phospho-Ser65-specific antibody. HR1 cells were transfected with HA-BimEL and then serum starved before stimulating with 4-HT in the presence or absence of U0126. Extracts were divided and either Mcl-1 or HA-BimEL was immunoprecipitated. Both the HA-BimEL and the endogenous BimEL underwent an ERK1/2-dependent phosphorylation (Figure 8B, input WCE) and dissociated from Mcl-1 (Figure 8B, Mcl-1 IP). We were unable to detect phospho-Ser65 BimEL in whole-cell extracts, but the antibody worked well when we immunoprecipitated HA-BimEL from these cell extracts; this allowed us to demonstrate 4-HT-dependent increases in phospho-Ser65 BimEL, which were inhibited by U0126 treatment, confirming that Ser65 is a target for ERK1/2 in vivo (Figure 8B, HA IP). Significantly, while we could readily detect 4-HT-stimulated, ERK1/2-dependent phosphorylation of HA-BimEL at Ser65, we always failed to detect any phospho-Ser65 BimEL in Mcl-1 IPs; the appearance of phospho-Ser65 BimEL always correlated with the dissociation of BimEL from Mcl-1 (Figure 8B, HA IP versus Mcl-1 IP). This result alone does not prove that Ser65 phosphorylation is sufficient for dissociation of BimEL from Mcl-1; however, together with Figure 8A in which Ser65 phosphorylation is shown to be required for dissociation from Mcl-1, our results suggest that phosphorylation of BimEL at Ser65 is not compatible with binding to Mcl-1.
BimEL mutants that fail to bind to pro-survival proteins exhibit accelerated turnover
Finally, we investigated whether the dissociation of BimEL was linked to its proteasomal degradation. For example, although phosphorylation might be the direct signal for degradation of BimEL, an alternative possibility was that BimEL that was not bound to a pro-survival Bcl-2 protein was the primary target for degradation, with phosphorylation simply serving to ‘liberate' BimEL from Mcl-1 or Bcl-xL. This possibility was further supported by our observation that phosphorylation of Ser65, a site required for ERK1/2-dependent degradation of BimEL (Ley et al, 2004; Luciano et al, 2003), was also required for its dissociation from Mcl-1 (Figure 8). A logical prediction from this model is that a BimEL mutant defective for binding to pro-survival Bcl-2 proteins should exhibit a shorter half-life than the WT protein, so we compared the half-life of WT HA-BimEL with that of the HA-BimELΔBH3 mutant, which exhibits greatly reduced binding to pro-survival Bcl-2 proteins including Mcl-1 (Figure 7C). These constructs were expressed in HEK293 cells, which were then serum starved and subjected to an emetine chase protocol to examine their turnover. Immunoblotting for total ERK1/2, a relatively stable protein under these conditions, confirmed equal loading. Under serum-free conditions, HA-BimEL was stable in the presence of emetine for up to 12 h, whereas the HA-BimELΔBH3 mutant clearly turned over in the presence of emetine (Figure 9). A trivial explanation for the enhanced turnover of the HA-BimELΔBH3 protein was that mutations in the BH3 domain of BimEL compromised protein folding, so that the mutant protein did not express well. However, we reproducibly observed that HA-BimELΔBH3 actually expressed at considerably higher levels than the WT HA-BimEL (Figures 7C and 9). Furthermore, HA-BimELΔBH3 was still able to undergo ERK1/2-dependent phosphorylation (Figure 7C), suggesting that the mutant protein was folding normally. These results indicate that dissociation of BimEL from pro-survival proteins is sufficient to promote its degradation.
Figure 9.
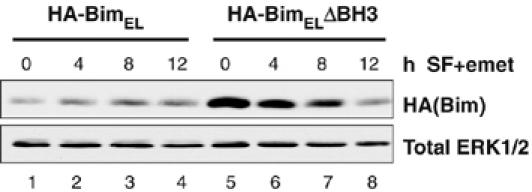
Dissociation from pro-survival proteins is sufficient to promote turnover of BimEL. HEK293 cells were transfected with HA-BimEL or HA-BimELΔBH3. After 24 h, cells were serum starved in the presence of 10 μM emetine for 4, 8 or 12 h. Whole-cell extracts were subjected to SDS–PAGE and immunoblotted with antibodies to HA(Bim) and total ERK1/2.
Discussion
Bim is involved in promoting cell death following serum withdrawal
The initiation of CCl39 cell death following serum withdrawal is a caspase-dependent process that requires new gene expression (Chalmers et al, 2003; Weston et al, 2003) and proceeds through the mitochondrial death pathway (Figure 1). The increase in expression of Bim in CCl39 cells, Rat-1 cells, iMEFs and iBMKs following serum withdrawal implied a potential role for Bim in promoting cell death. By comparing WT and Bim−/− iMEFs and iBMK epithelial cells we have now demonstrated that Bim expression is rate-determining for cell death following serum withdrawal; indeed, Bim appears to play a major role in initiating caspase activation in iMEFs following serum withdrawal. Taken together with other recent studies, it appears that the de novo expression of Bim is involved in initiating apoptosis following withdrawal of trophic support in a variety of cell types (Bouillet et al, 1999; Dijkers et al, 2000; Whitfield et al, 2001; Reginato et al, 2003; Wang et al, 2004). However, in iMEFs and especially iBMKs, the protection afforded by loss of Bim was transient; this is consistent with the role of Bim in promoting apoptosis being dependent on the death stimulus, tissue type and the presence of other redundant BOPs. A likely alternative to Bim in these studies is Bad, which is normally negatively regulated by RSK- and PKB-dependent phosphorylation (Datta et al, 1997, 2000; Bonni et al, 1999; Lizcano et al, 2000). While Bad−/− MEFs are not protected against serum withdrawal-induced death (Ranger et al, 2003), presumably reflecting the prominent role for Bim in MEFs (Figure 2), Bad−/− mammary epithelial cells are protected against withdrawal of EGF (Ranger et al, 2003). Similarly, loss of Bim only affords partial protection against NGF withdrawal in neurons (Whitfield et al, 2001) and this may reflect a role for Hrk/DP5 (Harris and Johnson, 2001). Alternatively, while Bim may regulate a classical caspase-dependent apoptosis at early times following serum withdrawal, other caspase-independent pathways (Chipuk and Green, 2005) may ensure cell death in the long term.
Newly expressed Bim associates with pro-survival proteins following serum withdrawal
In viable CCl39 cells maintained in complete medium, Bax associated with Bcl-2, Bcl-xL and Mcl-1. Following serum withdrawal, newly expressed BimEL associated with Bcl-xL and Mcl-1, and this was accompanied by the dissociation of Bax from the pro-survival proteins, especially Bcl-xL. These results fit well with the features of the ‘displacement' model (Willis and Adams, 2005), since we failed to observe an association between Bax and BimEL (or BimL). However, it is important to point out that only BimS has been proposed to bind directly to Bax (Marani et al, 2002), and we struggled to detect the very low levels of BimS in CCl39 cells. Furthermore, we cannot rule out the possibility that the Bax N-20 antibody disrupts interactions with BimEL, although this seems unlikely since Bax binding to Bcl-xL or Mcl-1 was readily detectable in N-20 immune complexes.
Exposure of an occluded N-terminal region in Bax, including the N-20 epitope, occurs in response to stress but may not be sufficient for commitment to cell death (Makin et al, 2001). Subsequent changes towards the C-terminus occur commensurate with dissociation of Bax from pro-survival Bcl-2 proteins and may be more relevant to the initiation of cell death. A similar multistep process seems to operate for Bak activation (Griffiths et al, 2001). We previously showed that serum withdrawal caused the initial N-terminal change in Bax (Weston et al, 2003). Since this change is reversible (Makin et al, 2001), we studied changes in the repertoire of Bax complexes associated with the later conformational change(s) by using the N-20 antibody to immunoprecipitate Bax from Triton X-100 cell lysates, thereby focusing on the Bax that had already undergone the initial N-terminal conformational change. We could readily detect complexes between N-20-reactive Bax and all three pro-survival proteins (Bcl-2, Bcl-xL and Mcl-1) in viable cells and similar results have been reported in viable cells lysed in the presence of CHAPS (Gomez-Bougie et al, 2005). Thus, while changes at the N-terminus of Bax may be stress responsive (Makin et al, 2001; Marani et al, 2002; Weston et al, 2003), they are not sufficient to release Bax from pro-survival proteins. Rather, even when the N-terminus is exposed, serum starvation provides an additional signal to promote release of Bax; our results are consistent with Bim providing this additional signal to displace Bax from pro-survival proteins following serum withdrawal. The mechanism by which serum starvation induces early changes at the N-terminus and their precise role remains to be defined.
ERK1/2-dependent phosphorylation of BimEL at Ser65 is required for dissociation of BimEL from pro-survival Bcl-2 proteins
Survival factors such as thrombin can prevent the assembly of BimEL/Bcl-xL or BimEL/Mcl-1 complexes by preventing Bim expression (Figure 4A–C). However, we have now shown that the association between BimEL/Bcl-xL or BimEL/Mcl-1 is actually subject to dynamic, post-translational regulation by survival factors. Specifically, even an acute, 15 min stimulation with thrombin or FBS promoted the dissociation of preassembled BimEL/Mcl-1 complexes (Figure 4E and F). Since the total expression level of BimEL or Mcl-1 did not change over these short time points, the dissociation of the BimEL/Mcl-1 complex could not be explained by turnover of BimEL. Indeed, dissociation of BimEL/Mcl-1 proceeded in the presence of the proteasome inhibitor MG132 (Figure 6A and B) and a BimEL mutant that could not be ubiquitylated (Akiyama et al, 2003) underwent ERK1/2-dependent dissociation normally (Figure 6C); thus, dissociation of BimEL is not a consequence of its proteasome-dependent degradation. We also observed that inhibition of ERK1/2 for as little as 20 min was sufficient to promote the binding of BimEL to Mcl-1 without changing total BimEL or Mcl-1 expression (Figure 6D). Since Bim must interact with pro-survival proteins to promote cell death (O'Connor et al, 1998; Willis et al, 2007), these results describe a completely novel mechanism by which ERK1/2 can prevent BimEL-dependent cell death; namely, by reducing the binding of BimEL to its target pro-survival proteins. While it has previously been shown that preassembled Bad/Bcl-xL complexes can be dissociated by phosphorylation of Bad (Scheid et al, 1999), this is the first such demonstration for Bim, or other BOPs, and may explain reports that ERK1/2 can inhibit the activity of BimEL, without apparent changes in its abundance (Wang et al, 2004; Reginato et al, 2005).
ERK1/2-dependent dissociation was unique to BimEL, consistent with exon 3 (unique to BimEL) harbouring the ERK1/2 docking domain and phosphorylation sites (Ley et al, 2005b). Furthermore, two lines of evidence strongly suggest that Ser65 (Ser69 in human BimEL) is the major site controlling dissociation. First, a Ser65Ala BimEL mutant failed to dissociate, indicating that phosphorylation at Ser65 is absolutely required; mutation of the other ERK1/2 sites had only a moderate effect. Second, while we could detect ERK1/2-dependent phosphorylation of BimEL at Ser65 using a phospho-specific antibody, we could not detect phospho-Ser65 BimEL bound to Mcl-1, suggesting that phosphorylation at this site is not compatible with binding to Mcl-1. It is unclear if phosphorylation at Ser65 is sufficient to promote dissociation and so-called phospho-mimetic acidic substitutions at this site were not informative. Ser65 is outside the BH3 domain, so phosphorylation may not interfere with binding to pro-survival proteins by simple charge repulsion; indeed, access to the Bim BH3 domain might be expected to be limited in a preassembled BimEL/Mcl-1 complex. Alternatively, phosphorylation may promote binding of accessory proteins that facilitate dissociation of BimEL, as is seen with phosphorylation-dependent dissociation of Bad/Bcl-xL complexes, which requires 14-3-3 (Datta et al, 1997, 2000; Bonni et al, 1999; Lizcano et al, 2000); in this case, simple acidic substitutions may fail to faithfully mimic phosphorylation-dependent interactions with such proteins. Dissociation of BimEL from Mcl-1 may require multiple phosphorylation events. In addition to Ser65 (so-called p2 site), two additional sites are phosphorylated in an ERK1/2-dependent manner in vivo, and one of these requires prior phosphorylation at Ser65 (Ley et al, 2004). Ser55 (p1) and Ser73 (p3) are likely sites, since their mutation reduces the ERK1/2-dependent mobility shift of BimEL and partially reverses its dissociation from Mcl-1 (Figure 8A).
Phosphorylation-dependent dissociation of BimEL from Bcl-xL and Mcl-1 may be linked to its degradation. For example, while WT BimEL could bind to Mcl-1 and Bcl-xL, and was stable under serum-free conditions, mutation of the BimEL BH3 domain greatly reduced interactions with pro-survival proteins and promoted BimEL degradation under the same conditions. These results are consistent with a model in which in serum-starved cells, BimEL is dephosphorylated, associates with pro-survival proteins, is stabilised and promotes cell death, whereas following activation of ERK1/2, BimEL is phosphorylated, dissociates from pro-survival proteins and in this unbound state is targeted for degradation. A recent study has provided some support for such a model by showing that BimEL belongs to the class of intrinsically unstructured proteins (Hinds et al, 2006); such proteins are extremely sensitive to proteolysis, are more efficiently processed by the proteasome and may be stabilised by interactions with binding partners (Dyson and Wright, 2005). Thus, it may not be ERK1/2-dependent phosphorylation per se that is the signal for degradation but rather the release of BimEL from pro-survival proteins that enhances its turnover, with ERK1/2 serving simply to promote this dissociation. One potential advantage of linking BimEL dissociation to its degradation is that it may prevent BimEL from reassociating with pro-survival proteins to promote apoptosis. Indeed, when degradation of BimEL was blocked by MG132, we found that inhibition of ERK1/2 resulted in dephosphorylation of BimEL and its reassociation with Mcl-1. The reason for this may be that both BimEL and Mcl-1 remain in the mitochondrial membrane fraction after dissociation, probably due to their own autonomous membrane targeting motifs, and so may remain in close proximity to facilitate binding and reassociation following BimEL dephosphorylation. This reversibility further underlines the dynamic nature of the ERK1/2-dependent dissociation of BimEL from pro-survival proteins.
In summary, we have shown that expression of Bim contributes to fibroblast and epithelial cell death following withdrawal of serum survival factors. Furthermore, by demonstrating that ERK1/2-dependent phosphorylation of BimEL causes the rapid dissociation of BimEL/Mcl-1 and BimEL/Bcl-xL complexes, we have defined a completely new mechanism by which survival factors can antagonise BimEL function, providing a further explanation for the reduced efficacy of BimEL relative to BimS and BimL (O'Connor et al, 1998). Mutation of Ser65Ala has been shown to enhance the toxicity of BimEL and it was assumed that this was due to stabilisation of the protein (Luciano et al, 2003; Ley et al, 2004). While this may well be the case, our results suggest that an additional effect of this mutation is to prevent growth factor-dependent dissociation of BimEL from its target pro-survival proteins. This may prove important in a variety of physiological and pathophysiological settings in which survival signals are limiting, including the developing CNS, immune system and in tumour cells.
Materials and methods
Antibodies
The following antibodies were used for Western blotting: Bax N-20, Santa Cruz (sc-493); Bcl-2, Santa Cruz (sc-7382); Bcl-xL, Pharmingen (66491A); Bim, Chemicon (AB17003); phospho-Ser65 Bim, Upstate Biotech; Bmf, Alexis Biochemicals (210-831-R100); ERK1/2 (9102) and ERK1/2 phosphospecific (9106), Cell Signalling Technology; HA, Babraham Institute Mab Facility; Mcl-1, Santa Cruz (sc-819); PKB (9272) and PKB S473 phosphospecific (9271), Cell Signalling Technology.
Cells and cell culture
Culture of CCl39 and CR1-11 (Weston et al, 2003), HEK293 (Ley et al, 2003), HR1 (Boughan et al, 2006) and WT and Bim−/− E1A-immortalised iBMK cells (Tan et al, 2005) have been described previously. WT and Bim−/− iMEFs were provided by David Huang, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia. MEFs were generated from E13-14.5 embryos and immortalised (at passages 2–4) with SV40 large T antigen. The mice used had been backcrossed (>10 generations) to the C57BL/6 genetic background and their genotype determined as described previously (Bouillet et al, 1999).
Assay of cell death
Assay of caspase activity by DEVDase assay and cell death by propidium iodide staining were described previously (Weston et al, 2003; Austin and Cook, 2005).
Cell extracts and immunoprecipitation
Following stimulation, cells were lysed in TG lysis buffer, as described previously (Weston et al, 2003). TG lysates were used for immunoprecipitations with the following antibodies: Bax N-20, Santa Cruz (sc-493) with protein G sepharose beads; Bcl-xL, Cell Signalling Technology (2762) and Mcl-1, Santa Cruz (sc-819) with protein A sepharose beads.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends
Supplementary Information
Acknowledgments
We thank members of the Cook Group for their support and discussions. We are grateful to David Huang for providing the WT and Bim−/− immortalised MEFs and for advice and Karen Vousden (Puma), Martin McMahon (ΔRaf-1:ER*) and Paul Coffer (rat Bim) for provision of plasmids. We also thank Caroline Dive and Guy Makin for discussions about Bax and Bak conformational changes, Martin McMahon and Robert Cartlidge for advice on the use of the phospho-Ser65 Bim antibody and Andrew Gilmore for advice on imaging of Bcl-2 family proteins. This work was supported by grants from Cancer Research UK (SP2458/0201), the Association of International Cancer Research and a competitive strategic grant from the BBSRC to the Babraham Institute. KE was supported by a BBSRC BCB Special Committee Studentship and SJC was supported by a Senior Cancer Research Fellowship from Cancer Research UK (2000–2006) and the Babraham Institute.
References
- Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Hikita A, Seto H, Okada T, Inaba T, Sanjay A, Baron R, Kawaguchi H, Oda H, Nakamura K, Strasser A, Tanaka S (2003) Regulation of osteoclast apoptosis by ubiquitylation of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J 24: 6653–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin M, Cook SJ (2005) Increased expression of Mcl-1 is required for protection against serum starvation in PTEN null mouse embryonic fibroblasts, but repression of Bim is favored in human glioblastomas. J Biol Chem 280: 33280–33288 [DOI] [PubMed] [Google Scholar]
- Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME (1999) Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286: 1358–1362 [DOI] [PubMed] [Google Scholar]
- Boughan PK, Argent RH, Body-Malapel M, Park J-H, Ewings KE, Bowie AG, Ong SJ, Cook SJ, Sorensen OE, Manzo BA, Klein NJ, Nunez G, Atherton JC, Bajaj-Elliott M (2006) Nucleotide-binding oligomerisation domain-1 (NOD-1) and epidermal growth factor receptor (EGFR): critical regulators of β-defensins during H. pylori infection. J Biol Chem 281: 11637–11648 [DOI] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738 [DOI] [PubMed] [Google Scholar]
- Chalmers CJ, Balmanno K, Hadfield K, Ley R, Cook SJ (2003) Thrombin inhibits Bim expression and prevents serum-withdrawal-induced apoptosis via protease-activated receptor 1. Biochem J 375: 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC (2005) Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17: 393–403 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Green DR (2005) Do inducers of apoptosis trigger caspase-independent cell death? Nat Rev Mo Cell Biol 6: 268–275 [DOI] [PubMed] [Google Scholar]
- Cory S, Adams JM (2002) The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer 2: 647–656 [DOI] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 [DOI] [PubMed] [Google Scholar]
- Datta SR, Katsov A, Hu L, Petros A, Fesik SW, Yaffe MB, Greenberg ME (2000) 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell 6: 41–51 [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ (2000) Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 10: 1201–1204 [DOI] [PubMed] [Google Scholar]
- Domina AM, Vrana JA, Gregory MA, Hann SR, Craig RW (2004) MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 23: 5301–5315 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE (2005) Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Gilley J, Coffer PJ, Ham J (2003) FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol 162: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Bougie P, Bataille R, Amiot M (2005) Endogenous association of Bim BH3-only protein with Mcl-1, Bcl-xL and Bcl-2 on mitochondria in human B cells. Eur J Immunol 35: 971–976 [DOI] [PubMed] [Google Scholar]
- Griffiths GJ, Corfe BM, Savory P, Leech S, Esposti MD, Hickman JA, Dive C (2001) Cellular damage signals promote sequential changes at the N-terminus and BH-1 domain of the pro-apoptotic protein Bak. Oncogene 20: 7668–7676 [DOI] [PubMed] [Google Scholar]
- Harris CA, Johnson EM Jr (2001) BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem 276: 37754–37760 [DOI] [PubMed] [Google Scholar]
- Hinds MG, Smits C, Fredericks-Short R, Risk JM, Bailey M, Huang DC, Day CL (2006) Bim, Bad and Bmf: intrinsically unstructured BH3-only proteins that undergo a localized conformational change upon binding to prosurvival Bcl-2 targets. Cell Death Differ 14: 128–136 [DOI] [PubMed] [Google Scholar]
- Lei K, Davis RJ (2003) JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci USA 100: 2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ (2003) Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 278: 18811–18816 [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Cook SJ (2005b) Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ 12: 1008–1014 [DOI] [PubMed] [Google Scholar]
- Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ (2004) Extracellular signal-regulated kinases 1/2 are serum-stimulated ‘Bim(EL) kinases' that bind to the BH3-only protein BimEL causing its phosphorylation and turnover. J Biol Chem 279: 8837–8847 [DOI] [PubMed] [Google Scholar]
- Ley R, Hadfield K, Howes E, Cook SJ (2005a) Identification of a DEF-type docking domain for extracellular signal-regulated kinases 1/2 that directs phosphorylation and turnover of the BH3-only protein BimEL. J Biol Chem 280: 17657–17663 [DOI] [PubMed] [Google Scholar]
- Lizcano JM, Morrice N, Cohen P (2000) Regulation of BAD by cAMP-dependent protein kinase is mediated via phosphorylation of a novel site, Ser155. Biochem J 349: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, Auberger P (2003) Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 22: 6785–6793 [DOI] [PubMed] [Google Scholar]
- Makin GW, Corfe BM, Griffiths GJ, Thistlethwaite A, Hickman JA, Dive C (2001) Damage-induced Bax N-terminal change, translocation to mitochondria and formation of Bax dimers/complexes occur regardless of cell fate. EMBO J 20: 6306–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marani M, Hancock D, Lopes R, Tenev T, Downward J, Lemoine NR (2004) Role of Bim in the survival pathway induced by Raf in epithelial cells. Oncogene 23: 2431–2441 [DOI] [PubMed] [Google Scholar]
- Marani M, Tenev T, Hancock D, Downward J, Lemoine NR (2002) Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol Cell Biol 22: 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH (2001) PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- O'Connor L, Strasser A, O'Reilly LA, Hausmann G, Adams JM, Cory S, Huang DC (1998) Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 17: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3: 287–296 [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Strasser A (2002) Keeping killers on a tight leash: transcriptional and post-translational control of the pro-apoptotic activity of BH3-only proteins. Cell Death Differ 9: 505–512 [DOI] [PubMed] [Google Scholar]
- Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, Kutok JL, Le Beau MM, Greenberg ME, Korsmeyer SJ (2003) Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acd Sci USA 100: 9324–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, Brugge JS (2005) Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol 25: 4591–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J, Muthuswamy SK, Brugge JS (2003) Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol 5: 733–740 [DOI] [PubMed] [Google Scholar]
- Scheid MP, Schubert KM, Duronio V (1999) Regulation of Bad phosphorylation and association with Bcl-x(L) by the MAPK/Erk kinase. J Biol Chem 274: 31108–31113 [DOI] [PubMed] [Google Scholar]
- Tan TT, Degenhardt K, Nelson DA, Beaudoin B, Nieves-Neira W, Bouillet P, Villunger A, Adams JM, White E (2005) Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell 7: 227–238 [DOI] [PubMed] [Google Scholar]
- Wang P, Gilmore AP, Streuli CH (2004) Bim is an apoptosis sensor that responds to loss of survival signals delivered by epidermal growth factor but not those provided by integrins. J Biol Chem 279: 41280–41285 [DOI] [PubMed] [Google Scholar]
- Weston CR, Balmanno K, Chalmers C, Hadfield K, Molton SA, Ley R, Wagner EF, Cook SJ (2003) Activation of ERK1/2 by ΔRaf-1:ER* represses Bim expression independently of the JNK or PI3K pathways. Oncogene 22: 1281–1293 [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J (2001) Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron 29: 629–643 [DOI] [PubMed] [Google Scholar]
- Willis SN, Adams JM (2005) Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol 17: 617–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315: 856–859 [DOI] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87: 619–628 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure Legends
Supplementary Information


