Abstract
YscU is an essential component of the export apparatus of the Yersinia injectisome. It consists of an N-terminal transmembrane domain and a long cytoplasmic C-terminal domain, which undergoes auto-cleavage at a NPTH site. Substitutions N263A and P264A prevented cleavage of YscU and abolished export of LcrV, YopB and YopD but not of Yop effectors. As a consequence, yscUN263A mutant bacteria made needles without the LcrV tip complex and they could not form translocation pores. The graft of the export signal of the effector YopE, at the N-terminus of LcrV, restored LcrV export and assembly of the tip complex. Thus, YscU cleavage is required to acquire the conformation allowing recognition of translocators, which represent an individual category of substrates in the hierarchy of export. In addition, yscUN263A mutant bacteria exported reduced amounts of the YscP ruler and made longer needles. Increasing YscP export resulted in needles with normal size, depending on the length of the ruler. Hence, the effect of the yscUN263A mutation on needle length was the consequence of a reduced YscP export.
Keywords: effectors, targeting, translocation, type III secretion, Yops
Introduction
The injectisome allows pathogenic or symbiotic bacteria to inject effector proteins across eukaryotic cell membranes, a process called type III secretion (T3S). It is evolutionary related to the bacterial flagellum (reviewed by Macnab (2003)). Both structures share a similar basal body consisting of several rings embedded in the two bacterial membranes connected by a central rod (Blocker et al, 1999, 2001; Marlovits et al, 2004; Morita-Ishihara et al, 2006). Depending on the family of injectisomes, a hollow stiff needle (Kubori et al, 1998, 2000; Blocker et al, 1999, 2001; Hoiczyk and Blobel, 2001), a filament (Knutton et al, 1998; Daniell et al, 2001; Crepin et al, 2005) or a pilus (Van Gijsegem et al, 2000) terminate the structure. The length of the needle is genetically determined (Magdalena et al, 2002; Tamano et al, 2002; Journet et al, 2003).
The proximal rings are thought to contain the T3S export apparatus, made of a number of integral membrane proteins and soluble components (reviewed by Tampakaki et al (2004); Cornelis (2006); Galan and Wolf-Watz (2006)). In the Yersinia Ysc injectisome, a ca 60-nm long stiff hollow needle, assembled from the 9-kDa protein YscF, projects from the basal body into the exterior milieu (Hoiczyk and Blobel, 2001). The needle terminates in a tip structure (the tip complex) made of the protein LcrV (Mueller et al, 2005). Upon contact with a eukaryotic cell membrane, the injectisome translocates a set of proteins called Yops (reviewed by Mota and Cornelis (2005)). These include the translocators (YopB and YopD) that form a pore in the target membrane and the effectors that traffic through this pore. The tip complex is thought to act as a scaffold for the folding and assembly of YopB and YopD into a functional pore (Goure et al, 2005).
During morphogenesis, the components of the transmembrane rings are handled by the Sec machinery, while those of the rod and the needle are sequentially exported by the T3S apparatus itself (Sukhan et al, 2001), traveling through the growing structure and polymerizing at its distal end (Li et al, 2002). There is no clear hierarchy in the synthesis of the components. Thus, the export apparatus is expected to switch its substrate specificity over time from early to late substrates, so that needle subunits (early substrates) are exported before LcrV and before the Yops (late substrates). This substrate specificity switch presumably leads to the arrest of needle growth, which determines the needle length. Hence, it is only triggered when the needle has reached its genetically defined length. The trigger is provided by YscP, another early substrate of the export apparatus. The N-terminal sequence of YscP is thought to act as a molecular ruler (Journet et al, 2003), while the C-terminal domain triggers the substrate specificity switch (Agrain et al, 2005a).
Mutations affecting FliK, the flagellar YscP homolog, lead to extra-long hooks (called polyhooks) but no filament and hence bacteria are not motile (Hirano et al, 1994). Motile revertants appear as consequence of extragenic suppressive mutations in the integral membrane protein FlhB, suggesting that FlhB is involved in specificity switching (Kutsukake et al, 1994; Williams et al, 1996). FlhB has a long C-terminal cytosolic domain, which undergoes an autoproteolytic cleavage between N269 and P270; however, the resulting subdomains remain tightly associated with each other (Minamino and Macnab, 2000; Fraser et al, 2003; Ferris et al, 2005). This cleavage is abolished by the mutation N269A and cells producing FlhBN269A assemble polyhook structures lacking filaments, leading to the proposal that cleavage and interaction of the two fragments generates conformational changes important for the specificity switching process (reviewed by Ferris and Minamino (2006)).
The FlhB homolog in the Yersinia injectisome is YscU, a 354-residue polypeptide with four transmembrane helices and a long cytoplasmic tail (Allaoui et al, 1994) (Figure 1A). Like FlhB, YscU undergoes autoproteolytic cleavage before P264, generating a 10-kDa C-terminal fragment (Lavander et al, 2002). YscUN263A is not cleaved at this site, but nevertheless retains the capacity to secrete Yops (Lavander et al, 2002).
Figure 1.
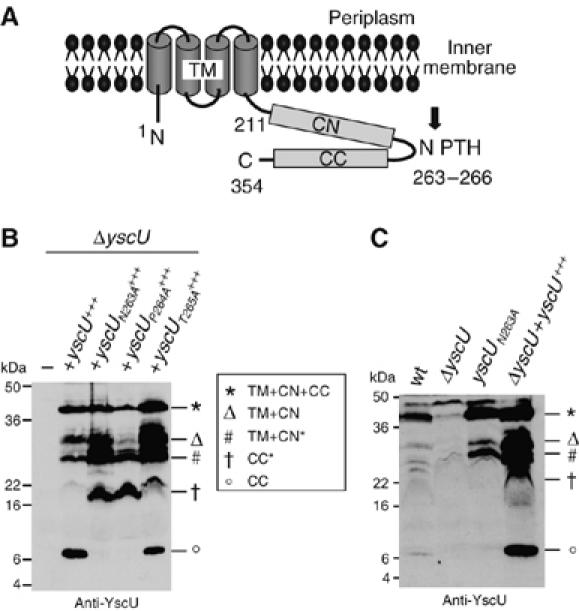
(A) Schematic representation of YscU based on studies by Allaoui et al (1994) and Lavander et al (2002). Letters indicate N-terminus (N), C-terminus (C), conserved NPTH motif (NPTH), transmembrane domain (TM, residues 1–210), N-terminal half of the cytoplasmic domain (CN, residues 211–263) and the C-terminal half of the cytoplasmic domain (CC, residues 264–354). Numbers indicate amino-acid positions in YscU from Y. enterocolitica W22703 (NCBI NC_002120). The black arrow represents the putative cleavage site at the NPTH motif. (B) Total membrane proteins of Y. enterocolitica E40 ΔyscU mutant bacteria, complemented in trans with wt or mutated alleles under the arabinose inducible pBAD promoter, were purified after 4 h of induction of the yop regulon and analyzed by immunoblot with anti-YscU antibodies. The different forms of YscU are indicated as follows: YscU (star), YscUTM+CN (triangle), YscUCC (circle), YscUTM+CN* (#) and YscUCC* (cross). The latter two result from cleavage at the alternative site. (C) Total membrane proteins of the indicated Y. enterocolitica strains. Strains and plasmids used: wt (pYV40); ΔyscU (pLY4001); yscUN263A, mutation inserted at the yscU locus (pISO4007); yscU+++ (pLY7); yscUN263A+++ (pSTW7); yscUP264A+++ (pSTW8); yscUT265A+++ (pSTW9).
The position of LcrV, at the tip of the needle, implies that LcrV is exported immediately after YscF but before the Yops. Thus, the hierarchy must consist of at least three categories of substrates and not two as in the flagellum; however, to date there are no genetic data to support this assumption. In this paper, we show that YscUN263A is impaired in the export of the translocators LcrV, YopB and YopD but not in the export of the effector Yops. Accordingly, cells with YscUN263A produce needles without tip complexes and this effect can be assigned to a lack of LcrV recognition. Thus, YscU is specifically involved in substrate recognition and has to be cleaved to acquire the conformation required for translocator recognition. This is the first genetic evidence that translocators represent an individual category of substrates in the hierarchy of export. Accordingly, the translocators, and not the Yop effectors, occupy the same rank as flagellin in the assembly of the flagellum.
Results
Mutation of residue N263 or P264 changes the autocleavage properties of YscU
To analyze the phenotype of a non-cleavable YscU mutant, we generated different point mutations in the NPTH motif of YscU and overexpressed the resulting yscUN263A, yscUP264A and yscUT265A genes in trans in Y. enterocolitica ΔyscU mutant bacteria. The yscUwt gene and the mutated alleles were overexpressed from the pBAD promoter only at the end of the logarithmic growth phase when synthesis of the T3S system was induced by shifting to 37°C. In those conditions, we did not observe any clear toxicity of either the YscUwt or the mutated YscU, as reported by others (Lavander et al, 2002). This discrepancy presumably results from differences in the experimental conditions used in the two reports. It is indeed likely that early overexpression of an integral membrane protein is toxic.
We then analyzed YscU in membranes purified from the different Y. enterocolitica strains incubated in secretion permissive conditions by immunoblotting with anti-YscU antibodies (Figure 1B). In strains expressing yscU or yscUT265A, an YscU fragment of about 10 kDa was clearly detectable. This could be assigned to the C-terminal part (CC) after cleavage at or around the NPTH motif. In contrast no 10-kDa fragment was present in bacteria expressing yscUN263A or yscUP264A. Here, a protein fragment of about 16 kDa (indicated as CC*), probably resulting from cleavage at an alternative site, was observed, as already shown for an yscUΔNPTH mutant from Y. pseudotuberculosis (Lavander et al, 2002). This 16-kDa fragment was also present in lower amounts in membranes of strains expressing yscUwt or yscUT265A. The 24-kDa N-terminal part (TM+CN*) remaining after cleavage at this alternative site was detected in all YscU expressing strains. The cleavage at or around the NPTH motif, which results in the C-terminal 10-kDa (CC) fragment described above, would leave a 30-kDa N-terminal fragment (TM+CN). This 30-kDa fragment was not only found in strains overexpressing yscUwt or yscUT265A that contained the small 10-kDa fragment, but also in samples from bacteria overexpressing yscUN263A or yscUP264A, where we did not observe the 10-kDa fragment. Furthermore, uncleaved YscUwt (TM+CN+CC) could be detected in all bacterial samples expressing yscU, demonstrating that cleavage was never complete. The data presented in Figure 1 are representative of a number of highly reproducible experiments.
Up to now, due to detection problems the cleavage of YscU and its flagellar homolog FlhB has only been demonstrated after overexpression of the protein (above and Minamino and Macnab, 2000; Lavander et al, 2002). We also analyzed the cleavage of wild-type (wt) amounts of YscUwt. As shown in Figure 1C, the 10-kDa (CC) and the 30-kDa (TM+CN) fragments of YscUwt could be detected in purified membranes. In addition, we observed cleavage at the alternative site, resulting in the 24-kDa (TM+CN*) and the 16-kDa (CC*) fragments, as shown for overproduced YscUwt (Figure 1B, lane 2 and Figure 1C, lane 4). We then analyzed a mutant expressing yscUN263A from its native promoter on the virulence plasmid of Y. enterocolitica. In agreement with the data obtained after overproduction of YscUN263A, a 30-kDa protein was present in the membranes from yscUN263A mutant bacteria, suggesting cleavage at or around the NPTH motif, but again no 10-kDa fragment co-purified. In addition, the 24-kDa signal corresponding to the N-terminal part after cleavage at the alternative site was present. Importantly, the uncleaved YscU protein was always detected at 40 kDa (TM+CN+CC), demonstrating that even when expressed at natural levels, YscU cleavage was not complete. These data thus confirm the data obtained with the overexpressed protein presented above. Together, they suggest that YscU is cleaved at least at two different sites. Although the 10-kDa fragment only co-purifies with the remaining N-terminal part in strains producing YscUwt or YscUT265A, it does not formally rule out that the first cleavage occurs in YscUN263A or YscUP264A, because the sequence alteration could prevent the binding of the cleaved 10-kDa fragment. In contrast, the C-terminal 16-kDa fragment produced by the cleavage at the alternative site co-purifies with its N-terminal half, even in strains producing YscUN263A or YscUP264A. Although the data are not quantitative, they show that a preferred point of cleavage exists, which depends on the protein examined.
yscUN263A and yscUP264A mutants export effector Yops but no translocators
Next we tested whether export of Yop proteins was affected in the different yscU mutants. To do this, the yop regulon was induced in the different Y. enterocolitica strains and the culture supernatants were analyzed by SDS–PAGE and Coomassie staining. In agreement with previously reported data, no Yops were secreted by ΔyscP and ΔyscU mutant bacteria (Allaoui et al, 1994; Stainier et al, 2000), and the yscUT265A mutant showed the same pattern of exported proteins as the wt (Figure 2A). In contrast, the yscUN263A and yscUP264A mutant bacteria did not export translocators (Figure 2A). The defect in translocator export was not due to overexpression of yscUN263A, since expression of yscUN263A from its native promoter had the same effect (Figure 2A). Analysis of YopE and LcrV in culture supernatants by immunoblotting confirmed this result (Figure 2B). The intra-bacterial levels of YopE and LcrV were comparable in wt and yscUN263A mutant bacteria. Whereas the export of YopE in yscUN263A mutant bacteria was only slightly decreased compared to the wt, no exported LcrV could be detected (Figure 2B). To check whether overexpression of lcrV in an yscUN263A mutant background could overcome the lack of export, as previously shown for export compromised yscP mutants (Agrain et al, 2005b), lcrV was overexpressed from the pBAD promoter by adding varying concentrations of L-arabinose (Figure 2C). Only on maximal lcrV overexpression with 0.5% L-arabinose a faint LcrV band could be observed in the supernatant fraction of yscUN263A mutant bacteria (Figure 2C). Overexpression of lcrV in wt Yersinia did not change the export level, possibly because the export capacity of the wt was already saturated.
Figure 2.
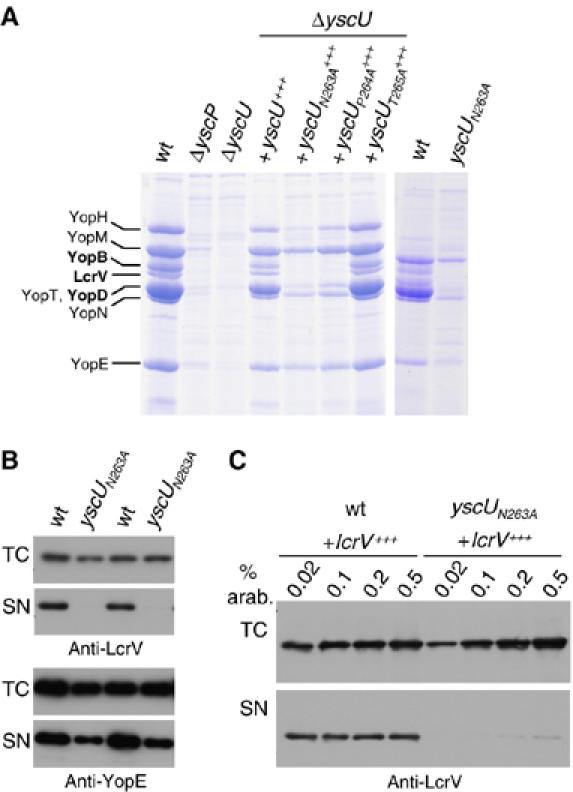
Analysis of the Yop proteins secreted by different yscU mutants. (A) Coomassie-stained 12% SDS–PAGE of Yops secreted by wt, ΔyscP, ΔyscU mutant bacteria, ΔyscU mutant bacteria overexpressing mutated yscU alleles in trans from the pBAD promoter, or bacteria carrying the yscUN263A allele at the yscU locus (natural yscU promoter). (B) Expression and export of LcrV and YopE in Y. enterocolitica E40 wt and yscUN263A mutant bacteria. Total cell (TC) and supernatant fractions (SN) were analyzed by immunoblotting using anti-LcrV or anti-YopE antibodies. Shown are samples from two independent experiments. (C) Expression and export of LcrV after overexpression of lcrV in trans from the pBAD promoter in Y. enterocolitica wt and yscUN263A mutant bacteria. L-arabinose concentrations ranging from 0.02 to 0.5% were used to induce LcrV synthesis when bacteria were shifted to 37°C and again 2 h later. TC and SN were analyzed by immunoblotting using anti-LcrV antibodies. Strains and plasmids used: wt (pYV40); ΔyscP (pLJ4036); ΔyscU (pLY4001); yscU+++ (pLY7); yscUN263A+++ (pSTW7); yscUP264A+++ (pSTW8); yscUT265A+++ (pSTW9); yscUN263A (pISO4007); lcrV+++ (pPB42).
The yscUN263A mutant does not assemble a tip complex and fails to induce lysis in red blood cells
The fact that the yscUN263A mutant did not export the translocators LcrV, YopB and YopD suggests that this mutant may not be able to form the translocation pore. Formation of the translocation pore can be studied by infection of red blood cells (RBCs), which undergo hemolysis (Hakansson et al, 1996). Since Y. enterocolitica bacteria do not adhere to RBCs, a ΔyopN mutant that secretes Yops in a contact-independent manner has been used previously (Marenne et al, 2003).
Therefore, a ΔyopNyscUN263A mutant was generated. As expected from the ΔyopN deletion, secretion of Yops by the ΔyopNyscUN263A mutant was independent of the presence of calcium, whereas secretion of Yops by the yscUN263A mutant and wt bacteria was calcium dependent (data not shown). The pattern of proteins secreted by the ΔyopNyscUN263A mutant was identical to that of proteins secreted by the yscUN263A mutant, except that YopN was missing (Figure 3B). We analyzed the capacity of the ΔyopNyscUN263A mutant to induce lysis in RBCs. In agreement with the loss of translocator export, lytic activity was neither observed for the ΔyopNyscUN263A mutant nor for the negative control ΔHOPEMNV mutant lacking LcrV (Figure 3C). Like for wt, and ΔyopN mutant bacteria, needles could be observed on the surface of ΔyopNyscUN263A mutant bacteria (Figure 3A). The needles from yscUN263A mutant bacteria were purified and analyzed by scanning transmission electron microscopy (STEM) (Figure 3D). Out of 51 needles analyzed, none had a tip and 16 had a clear pointed end (Figure 3D), as previously reported for the lcrV mutant (Mueller et al, 2005). These data confirm the defect in the export of LcrV.
Figure 3.
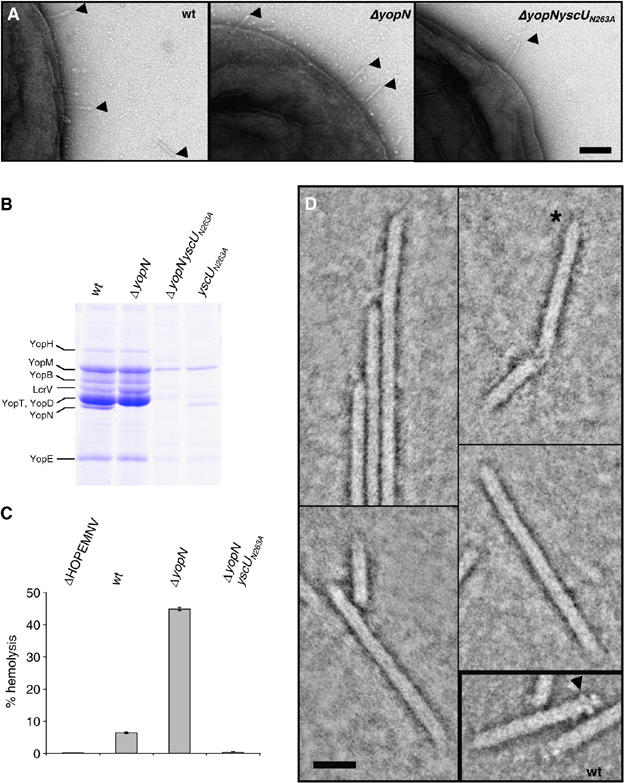
The yscUN263A mutant does not induce hemolysis in RBC due to a missing tip complex. (A) Transmission electron micrographs of Y. enterocolitica E40 WT, ΔyopN and ΔyopNyscUN263A bacteria negatively stained with 2% uranyl acetate. Scale bar, 100 nm. Needles are indicated by arrowheads. (B) Yops secreted by Y. enterocolitica wt, ΔyopN, yscUN263A and ΔyopNyscUN263A bacteria. Coomassie-stained 12% SDS–PAGE. The position of YopN is indicated on the left. (C) Percentage lysis of RBCs after 1 h of contact with the indicated Y. enterocolitica strains. (D) STEM dark-field image of Y. enterocolitica yscUN263A needles. Protein is displayed in bright shades. Inset: wt needles similarly imaged. The yscUN263A mutant needles had rather pointed ends (asterisk); the tip complexes so characteristic of wt needles (arrowhead) were not detected. Scale bar, 20 nm. Strains: wt (pYV40); ΔyopN (pIM41); ΔyopNyscUN263A (pISO4010); yscUN263A (pISO4007); ΔHOPEMNV (pMN4002).
The impaired export of LcrV is due to a lack of recognition by the YscUN263A protein
There might be at least two reasons why LcrV is not exported in the yscUN263A mutant; either LcrV is not recognized by the YscUN263A protein or the YscUN263A protein has an altered conformation, which blocks the export channel for some substrates. To investigate if the channel is still permissive for LcrV, we fused the export signal of the effector YopE to the N-terminus of LcrV and analyzed the export of the hybrid protein in the yscUN263A mutant. As shown in Figure 4A, a protein of the expected size was detected by Coomassie-stained SDS–PAGE of the supernatant fraction. The identity of the protein was confirmed by immunoblot using anti-LcrV antibodies (Figure 4B). In contrast to the YopE-hybrid protein, LcrV alone was not exported in the yscUN263A mutant. These data suggest that the loss of LcrV export in the yscUN263A mutant is not due to changes of the channel properties, but rather due to a defect in recognition of LcrV as export substrate. We then wondered whether the YopE1−15LcrV hybrid protein would be functional in the yscUN263A mutant, that is, whether this protein would be able to form a functional tip complex. We first investigated the functionality of the hybrid protein in an lcrV mutant (here ΔHOPEMNV), by testing its ability to induce lysis of RBCs. The multi-effector mutant ΔHOPEMN was used as positive control and induced 46% hemolysis, whereas the lcrV mutant ΔHOPEMNV only led to 1% hemolysis (Figure 4C). Overexpression of either lcrV or yopE1−15lcrV in lcrV mutant bacteria could restore hemolysis up to wt level, indicating that the hybrid protein was functional (Figure 4C). As expected, expression of yopE1−15lcrV in the ΔyopNyscUN263A mutant could not restore the lytic activity, because not only LcrV but also the two other translocators YopB and YopD were not exported by this mutant (Figure 4C). When purified needles of an yscUN263A mutant expressing yopE1−15lcrV were analyzed using STEM, tip complexes were detected on 43 out of 76 analyzed needles (Figure 4D). This shows that LcrV export by yscUN263A mutant bacteria could be restored by using the export signal of YopE. Furthermore, the yopE1−15lcrV fusion resulted in a functional protein that was able to form a tip complex. From these data we cannot say that not all the needles have a tip, because we can never exclude that some needles analyzed broke during purification.
Figure 4.
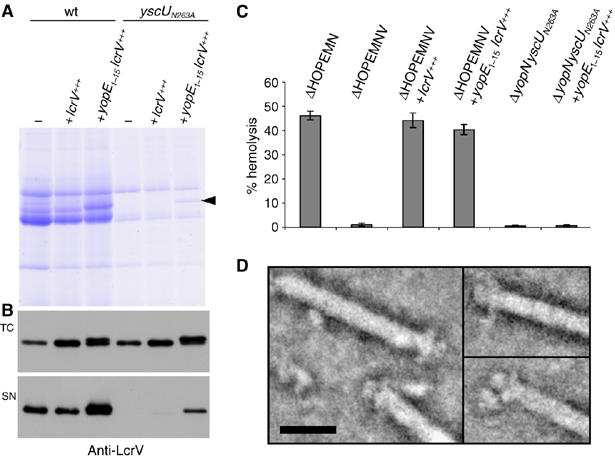
YopE1−15LcrV is exported by Y. enterocolitica yscUN263A mutant bacteria and forms a tip complex. (A) Yops secreted by Y. enterocolitica E40 wt (pYV40) and Y. enterocolitica E40 yscUN263A (pISO4007) mutant bacteria overexpressing lcrV (pPB42) or yopE1−15lcrV (pISOA132) in trans from the pBAD promoter. The position of the YopE1−15LcrV protein is indicated by the arrowhead. (B) Total cell (TC) and supernatant fractions (SN) of the same cultures as in (A) were analyzed by immunoblot with anti-LcrV antibodies. (C) Lysis of RBCs after 1 h of contact with the indicated Y. enterocolitica E40 strains. (D) STEM images of needles isolated from Yersinia yscUN263A expressing YopE1−15LcrV from the pBAD promoter. Protein is displayed in bright shades. Scale bar, 20 nm. Strains and plasmids: wt (pYV40); yscUN263A (pISO4007); ΔHOPEMN (pIM417); ΔHOPEMNV (pMN4002); ΔyopNyscUN263A (pISO4010); lcrV+++ (pPB42); yopE1−15lcrV+++ (pISOA132).
Needles of Yersinia yscUN263A and yscUP264A mutants are longer and less regulated
Next, we were interested in whether the different yscU mutations described above would affect the length of the injectisome needle. Therefore, needles from Y. enterocolitica ΔyscU bacteria overexpressing yscU, yscUN263A, yscUP264A or yscUT265A were measured. Needles of yscUT265A mutant bacteria had wt length (yscUwt: median 61±15 nm; yscUT265A: median 63±15 nm), whereas needles from bacteria overexpressing yscUN263A or yscUP264A were longer and less controlled than the wt (yscUN263A: median 107±51 nm; yscUP264A: median 119±53 nm) (Supplementary Figure S1). These data reflect the differences that we reported above for the cleavage of YscU and the export of the translocators.
Needle length in a yscUN263A mutant is still controlled by YscP
To analyze whether YscP still exerts its ruler function in a yscUN263A mutant, we combined yscP alleles of different sizes with the yscUN263A allele. As shown before (Journet et al, 2003; Mota et al, 2005), yscUwt bacteria producing the 388-amino acid ruler YscP388 made shorter needles (49±7 nm), whereas bacteria producing the 680-amino acid ruler yscP680 made longer needles (149±45 nm) than yscUwt bacteria endowed with the wt ruler yscP515 (67±11 nm) (Figure 5A, histograms I–III).
Figure 5.
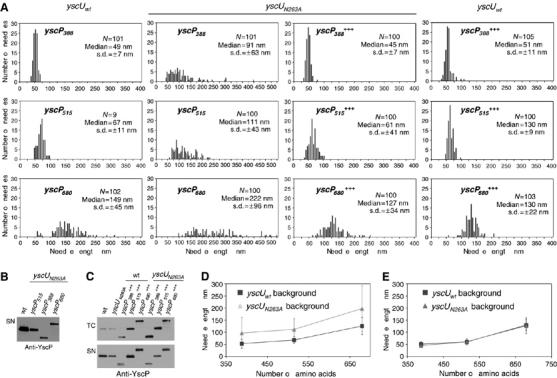
(A) Histograms showing the length of needles from Y. enterocolitica E40 yscUwt and E40 yscUN263A mutant bacteria expressing three different YscP proteins either from their native promoter (histograms I–VI) or additionally in trans from the pBAD promoter (histograms VII–XII). Overexpression of yscP in trans from the pBAD promoter was induced with 0.1% L-arabinose. N, number of needles measured; s.d., standard deviation. (B) The supernatant (SN) of culture of Y. enterocolitica E40 strains expressing the three indicated yscP alleles from their native promoter in an yscUN263A mutant background were analyzed by immunoblot with anti-YscP antibodies. (C) Total cell (TC) and supernatant fractions (SN) of Y. enterocolitica yscUwt and yscUN263A mutant bacteria expressing the indicated yscP alleles from their native promoter and additionally in trans from the pBAD promoter. Overexpression of yscP alleles from the pBAD promoter was induced with 0.1% L-arabinose. Samples were analyzed by immunoblotting using anti-YscP antibodies. (D) Plot of the needle length versus the number of residues in YscP for Y. enterocolitica strains expressing different yscP alleles from their native promoter in either yscUwt or yscUN263A mutant bacteria. The medians and s.d.s of histograms I–VI that are given in (A) are shown. (E) Plot of the needle length versus the number of residues in YscP for Y. enterocolitica yscUwt and yscUN263A mutant bacteria expressing indicated yscP alleles from their native promoter and additionally in trans from the pBAD promoter, which was induced with 0.1% L-arabinose. The medians and s.d.s of histograms VII–XII that are given in (A) are shown. Strains and plasmids used: wt (pYV40); yscP388 (pLJ4022); yscP515 (pYV40); yscP680 (pLJM4001); yscUN263AyscP388 (pISO4011); yscUN263AyscP515 (pISO4007); yscUN263AyscP680 (pISO4012); yscUN263A and yscP388+++ (pISO4011+pCA20); yscUN263A and yscP515+++ (pISO4007+pLJ6); yscUN263A and yscP680+++ (pISO4012+pLJ19); yscUwt and yscP388+++ (pLJ4022+pCA20); yscUwt and yscP515+++ (pYV40+pLJ6); yscUwt and yscP680+++ (pLJM4001+pLJ19).
yscUN263A mutant bacteria producing the different YscP proteins made longer needles than yscUwt bacteria. Length still correlated with the number of residues of YscP, but the control was not as tight, as shown by the larger standard deviations (yscUN263AyscP388: 91 nm±63 nm; yscUN263AyscP515: 111±43 nm; yscUN263 yscP680: 222±96 nm) (Figure 5A, histograms IV–VI). By plotting the medians of needle length against the number of amino acids in YscP, we obtained parallel slopes for the yscUwt and the yscUN263A mutant (Figure 5D). In addition to these changes in needle length, yscUN263A mutant bacteria exported less YscP than wt bacteria (Figure 5B and C). The increase in needle length as well as the decrease in length control could be due to this decrease in YscP export, as shown previously (Agrain et al, 2005b). We therefore tested whether an increase in YscP production would compensate the defects in length control. Therefore, yscP388, yscP515 and yscP680 were overexpressed from the pBAD promoter in yscUN263A mutant bacteria harboring the corresponding yscP allele on the virulence plasmid. Overproduction of the different YscP proteins increased their export to wt level, as shown by immunoblot analysis of the total cell and supernatant fractions using anti-YscP antibodies (Figure 5C). Increased production of YscP388, YscP515 and YscP680 in yscUN263A mutant bacteria (Figure 5A, histograms VII–IX) led to needles that were comparable to those of the corresponding yscUwt strains (Figure 5A, histograms X–XII). Although yscUN263A mutant bacteria overexpressing yscP had needles of the expected length (Figure 5A, histogram VIII), they still did not export translocators (Supplementary Figure S2).
The plot in Figure 5E shows that overexpression of YscP can indeed restore needle length control in the yscUN263A mutant to wt level. This showed that the defect in needle length control of the yscUN263A mutant was rather due to a failure in YscP export than to a failure of the substrate specificity switch.
Discussion
The fact that the injectisome exports its own distal components, before it exports the effectors, implies that the T3S apparatus can recognize and sequentially export different categories of substrates. To achieve this, it is believed to switch its substrate specificity when assembly is completed.
To better understand this sequential export process, we introduced mutations into the NPTH cleavage site of YscU and analyzed the phenotypes. At variance with previous reports on FlhB (Minamino and Macnab, 2000) and YscU (Lavander et al, 2002), we not only analyzed the phenotype after overexpression of cloned yscU alleles, but also after replacement of the yscU alleles on the pYV plasmid, ensuring physiological expression levels. To detect minute protein quantities, we probed the proteins with an anti-YscU antiserum rather than an antibody directed against the C-terminus or a C-terminal His-tag. Our results confirmed that the 40-kDa YscU is naturally cleaved into a TM+CN 30-kDa fragment and a 10-kDa CC fragment, as shown previously for FlhB (Minamino and Macnab, 2000) and for YscU from Y. pseudotuberculosis (Lavander et al, 2002). In good agreement with the estimated stoichiometry of two FlhB molecules per flagellum (Zhu et al, 2002), unless overexpressed, the CC product from YscU could only be detected after enrichment by purification of the membrane fraction from Y. enterocolitica. Surprisingly, the cleavage of YscU was never complete, even at physiological expression levels, although bacteria were harvested at a stage where all the needles analyzed had their normal length and tip complex. No CC fragment was detected for the mutant proteins YscUN269A and YscUP264A, as shown before by Lavander et al (2003) using C-terminal tagged YscU. As pointed out by these authors, this suggests that cleavage did not occur. However, since a band corresponding to the size of the TM+CN fragment was still observed, we cannot formally rule out that cleavage was prevented. We observed an alternative cleavage site in the YscUN269A and YscUP264A mutant proteins, yielding a ca 24-kDa (TM+CN*) and a ca 16-kDa (CC*) fragment. This alternative cleavage of YscU is reminiscent of the alternative cleavage observed in FlhBN269A and FlhBP270A, but was not reported for YscU from Y. pseudotuberculosis. Surprisingly, we detected the TM+CN* fragment in extracts from bacteria producing YscUwt, suggesting that even YscUwt could be partially cleaved at this alternative cleavage site.
The yscUN263A mutant bacteria assembled injectisome needles, but their length seemed to be poorly controlled and the median length was 111 nm rather than the 67 nm found for wt bacteria. When this yscUN263A allele was combined with a longer (680 codons) and a shorter (388 codons) allele of yscP, the median needle lengths were 222 nm and 91 nm, respectively, indicating that needle length was still dependent on the length of the YscP ruler, although the standard deviation was much larger. In addition to this loose length control, the yscUN263A bacteria released less YscP into the culture supernatant than wt bacteria do, although the intra-bacterial amount of protein was unchanged. We tried to overcome the poor export efficiency of YscP by overexpressing the gene downstream from the pBAD promoter. As expected from the previous work of Agrain et al (2005b), overexpression of the three different yscP alleles indeed led to the export of more YscP proteins, and also to a better control of needle length. The latter was in fact as good as in yscUwt bacteria. Hence, we conclude that the yscUN263A mutation reduces the efficacy of the T3S system to export YscP, which, indirectly, leads to a less stringent control of needle length. We also conclude that the yscUN263A mutation does not affect the capacity of the T3S export apparatus to switch off the export of the YscF needle subunits when the needle reaches its genetically defined length.
These data demonstrate that the cleavage at the NPTH motif is not required to switch off needle subunit export. Even more, they confirm that the same variants of YscU and YscP can give rise to two different needle lengths, depending on the amount of YscP exported. To us, this rules out the hypothesis that YscU could play a role as a timer to determine needle length, as was proposed for the flagellar hook length control (Moriya et al, 2006).
yscUN263A mutant bacteria also released slightly less effector Yops than wt bacteria, but the export of effectors was still significant. This observation shows that the export machine is still capable of switching on the export of Yops. Hence, the yscUN263A mutation does not affect the substrate specificity switching from early (YscF, YscP) to late (Yops) substrates. This interpretation agrees with the observations reported by Lavander et al (2002) that mutation yscUN263A does not affect Yop secretion in Y. pseudotuberculosis. However, it may seem at odds with the observation that mutation flhBN269A prevents export of flagellin, and with the interpretation that it inhibits the substrate specificity switch.
The most intriguing phenotype of the yscUN263A mutation was the deficiency in export of the translocators LcrV, YopB and YopD. In good agreement with the fact that LcrV was not exported, needles produced by the mutant bacteria had no tip complex.
A deficiency in translocator export is not completely unprecedented. The invE mutant of Salmonella enterica showed reduced secretion of the translocators SipB, SipC and SipD, while the export of other T3S effectors was increased (Kubori and Galan, 2002). Beside these observations and the fact that InvE is not required for the assembly of the needle complex, nothing is known about the actual function of InvE and its putative homologs. Mutants sepL and sepD, mutants of enteropathogenic and enterohemorrhagic Escherichia coli, as well as of Citrobacter rodentium, have a similar phenotype; translocator secretion is completely abolished, while the export of effector proteins is increased (Deng et al, 2005). It was suggested that SepL and SepD are not only necessary for efficient translocator secretion but also control a switch from translocator to effector secretion by sensing certain environmental signals such as low calcium (Deng et al, 2005). A yscW (called earlier virG) mutant of Y. enterocolitica also shows a decrease in the amounts of secreted proteins, especially of the translocators YopB, YopD and LcrV (Allaoui et al, 1995). YscW is the pilot protein of the secretin YscC (Burghout et al, 2004) and hence its absence probably modifies the channel properties.
Here, the failure to export LcrV and to assemble the needle tip could be circumvented by the N-terminal addition of the YopE export signal to LcrV. This indicates that the failure to export LcrV was not due to a change in the channel conformation, making it unsuitable for protein export, but rather to a failure in substrate recognition. This implies that the translocators have a specific type of export signal and a status distinct from the effectors regarding export. This makes sense, since they need to be exported before the effectors (Cornelis and Wolf-Watz, 1997). We already know that LcrV is exported before the Yops, since it forms the tip complex, even in the absence of Yop secretion (Mueller et al, 2005). However, no mutation specifically affecting export of the effectors has been described before. Surprisingly, while considerable effort was made to unravel the export signal of YopE (Michiels and Cornelis, 1991; Sory et al, 1995; Anderson and Schneewind, 1997), YopH (Michiels and Cornelis, 1991; Sory et al, 1995), YopN (Anderson and Schneewind, 1997) and YopQ (Michiels and Cornelis, 1991; Anderson and Schneewind, 1999) in Yersinia, little has been done to decipher the signal of translocators. It is known that LcrV can be exported even with an N-terminal His-tag, and that both the N- and C-termini of LcrV are required for its export (Fields et al, 1999). This can be taken as a hint that the signals are different for effectors and translocators, although more work is needed to characterize the export signals of the translocators and hence to understand the hierarchy of assembly. The export signal of YscP, an early substrate, was recently characterized and turned out to be totally different from the known export signal of effectors (Agrain et al, 2005b).
As mentioned above, the phenotype of the yscUN263A mutation may appear, at first sight, to be different from the phenotype of the flhBN269A mutation, in the sense that YscUN263A allows Yop (late substrate) export, while FlhBN269A does not allow flagellin export. However, the two observations can easily be reconciled, given the additional observation that YscUN263A prevents export of the translocators. Keeping in mind that there are two hierarchy classes in the assembly of the flagellum (hook/rod and filament) and three hierarchy classes in the operation of the injectisome (needle, translocators, effectors), both mutants are simply deficient in the export of the second hierarchy class.
Finally, our results confirm that YscU, and especially the structure of the CC fragment, plays a critical role in substrate recognition. They also show that the translocators are specifically recognized by YscU and thus, the injectisome has at least three classes of substrates. However, they do not provide any evidence that cleavage of the CC fragment is involved in the substrate specificity switch.
Materials and methods
Bacterial strains, plasmids and genetic constructions are listed in Supplementary Table 1.
Alleles to be inserted in the pYV plasmids were subcloned into the pKNG101 suicide vector and the allelic exchange was selected by plating diploid bacteria on sucrose (Kaniga et al, 1991).
E. coli Top10 was used for plasmid purification and cloning. E. coli BL21 Rosetta was used for protein expression. Bacteria were routinely grown on Luria–Bertani agar plates and in liquid Luria–Bertani medium. Ampicillin was used at a concentration of 200 μg/ml to select for expression vectors.
Plasmids were generated using either Pfu turbo polymerase (Stratagene) or Vent DNA polymerase (New England Biolabs). The oligonucleotides used for genetic constructions are listed in Supplementary Table 2. All constructs were confirmed by sequencing using a 3100-Avant genetic analyzer (ABI Prism).
Yop secretion
Induction of the yop regulon was described by Cornelis et al (1987). Expression of the different genes cloned downstream from the pBAD promoter was routinely induced by adding 0.2% L-arabinose to the culture just before the shift to 37°C, and again 2 h later. The carbon source was glycerol (4 mg/ml) when expressing genes from the pBAD promoter, and glucose (4 mg/ml) in the other case. Total cell and supernatant fractions were separated by centrifugation at 20 800 g for 10 min at 4°C. The cell pellet was taken as total cell fraction. Proteins in the supernatant were precipitated with trichloroacetic acid 10% (w/v) final for 1 h at 4°C.
Secreted proteins were analyzed by Coomassie-stained 12% SDS–PAGE; in each case, proteins secreted by 3 × 108 bacteria were loaded per lane. For detection of YscP in total cells, 1.6 × 108 bacteria were loaded per lane. For YscP detection in supernatants, the supernatants from 2.5 × 107 bacteria were loaded per lane. For analysis of LcrV and YopE, 2.5 × 107 bacteria and the supernatants from 2 × 107 bacteria were loaded per lane on a 12% SDS–PAGE. Immunoblotting was carried out using rabbit polyclonal antibodies against LcrV (MIPA220; 1:2000) and YscP (MIPA57; 1:3000), or rat polyclonal antibodies against YopE (MIPA94; 1:10000). Detection was performed with the respective secondary antibodies conjugated to horseradish peroxidase (1:5000; Dako), before development with supersignal chemiluminescent substrate (Pierce).
Electron microscopy
Needles at the cell surface of bacteria were visualized by transmission electron microscopy, as described by Hoiczyk and Blobel (2001) and Agrain et al (2005b). After 4 h of induction of the yop regulon at 37°C, bacteria were harvested at 2000 g and resuspended gently in 20 mM Tris–HCl, pH 7.5. Droplets were applied for 1 min to freshly glow-discharged, formvar-carbon-coated grids, and negatively stained with 2% (w/v) uranyl acetate. Bacteria were visualized in a Philips CM100 electron microscope at a nominal magnification of × 20 000 and an acceleration voltage of 80 kV. Sizes were measured with the ‘Soft imaging system' software (Hamburg, Germany).
Needle purification
Needles were purified from Y. enterocolitica cultures incubated under secretion permissive conditions. Bacteria from 300 ml culture were harvested by centrifugation (10 min at 5700 g) and washed once with 20 mM Tris–HCl, pH 7.5 (1/30 of initial culture volume). The washing supernatant was passed through a 0.45 μm mesh filter (cellulose acetate membrane) and then centrifuged for 30 min at 20 000 g. The resulting pellet was resuspended in 20 mM Tris–HCl, pH 7.5 (1/3000 of initial culture volume) and analyzed by electron microscopy (Mueller et al, 2005).
STEM
The purified needles were diluted with buffer (20 mM Tris–HCl, pH 7.5), as required, adsorbed to thin carbon film, washed with four droplets of quartz double-distilled water and stained with 2% (w/v) sodium phosphotungstate. Digital dark-field images were generated using a Vacuum Generators HB5 STEM interfaced to a modular computer system (Tietz Video and Image Processing Systems GmbH, D-8035 Gauting). The microscope was operated at 100 kV and a nominal magnification of × 500 000, using doses that ranged between 4400 and 13 500 electrons/nm2.
Hemolysis
Hemolytic assays were carried out as described by Goure et al (2005).
Purification of total membrane proteins
To purify total cell membranes, Yersinia bacteria were cultivated in secretion permissive conditions (BHI-Ox), as described before. Bacteria from 200 ml culture were harvested by centrifugation (20 min/5000 g/4°C) and washed once with phosphate-buffered saline (PBS). After resuspending the cells in 5 ml buffer I (50 mM Hepes pH 7.6, 500 mM potassium acetate, 5 mM magnesium acetate) containing the protease inhibitor cocktail complete Mini (Roche), 0.7 mg/ml lysozyme was added, followed by a 30-min incubation at 4°C. Then cells were lysed by sonication on ice. After removal of unbroken cells by low-speed centrifugation (30 min/6000 g/4°C), the supernatant was passed through a 0.45 μm mesh filter (cellulose acetate membrane) and centrifuged at high speed (2 h/150 000 g/4°C). The pellet containing the total cell membranes was resuspended in 400 μl buffer I. Lipids were extracted with 400 μl n-hexan for 30 min/4°C on a rotating wheel. After isolation of the lower hydrophilic phase, proteins were precipitated by addition of four volumes acetone (1 h/4°C), centrifuged (10 min/20−800 g/4°C) and resuspended in 160 μl buffer II (7 M urea, 2 M thiourea, 2% CHAPS), and supplemented with 40 μl 5 × SDS loading buffer (5 × SDS loading buffer: 225 mM Tris–HCl, pH 6.8, 5% SDS, 50% glycerol, 50 mM DTT, bromophenol blue). Samples were separated on a 15% SDS–PAGE, transferred onto nitrocellulose membrane and analyzed by immunoblotting with anti-YscU antibodies.
YscU antibodies
To produce polyclonal anti-YscU antibodies (MIPA 221), YscU211−354 was expressed from the pBAD promoter with a C-terminal His-tag, using plasmid pLY1. A soluble His-protein was produced in E. coli Top10 and purified on chelating sepharose beads (Amersham Biosciences). A rabbit was immunized by four injections with a total of 1 mg of YscU211−354 (CER, Marloie, Belgium). For immunoblot analysis, anti-YscU antibodies were used at a dilution of 1:1000.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Table 1
Supplementary Table 2
Acknowledgments
We thank Laure Journet for plasmid pLJ14, Gianni Morson for assistance with the TEM and Philippe Ringler for the STEM microscopy. This work was supported by the Swiss National Science Foundation (grant 32-65393.01 to GC and grant 3100A-108299 to AE) and the Maurice E Müller Foundation of Switzerland).
References
- Agrain C, Callebaut I, Journet L, Sorg I, Paroz C, Mota LJ, Cornelis GR (2005a) Characterization of a type III secretion substrate specificity switch (T3S4) domain in YscP from Yersinia enterocolitica. Mol Microbiol 56: 54–67 [DOI] [PubMed] [Google Scholar]
- Agrain C, Sorg I, Paroz C, Cornelis GR (2005b) Secretion of YscP from Yersinia enterocolitica is essential to control the length of the injectisome needle but not to change the type III secretion substrate specificity. Mol Microbiol 57: 1415–1427 [DOI] [PubMed] [Google Scholar]
- Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis GR (1995) VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to exsB of Pseudomonas aeruginosa. J Bacteriol 177: 4230–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaoui A, Woestyn S, Sluiters C, Cornelis GR (1994) YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol 176: 4534–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Schneewind O (1997) A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278: 1140–1143 [DOI] [PubMed] [Google Scholar]
- Anderson DM, Schneewind O (1999) Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol Microbiol 31: 1139–1148 [DOI] [PubMed] [Google Scholar]
- Blocker A, Gounon P, Larquet E, Niebuhr K, Cabiaux V, Parsot C, Sansonetti P (1999) The tripartite type III secreton of Shigella flexneri inserts IpaB and IpaC into host membranes. J Cell Biol 147: 683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A (2001) Structure and composition of the Shigella flexneri ‘needle complex', a part of its type III secreton. Mol Microbiol 39: 652–663 [DOI] [PubMed] [Google Scholar]
- Burghout P, Beckers F, de Wit E, van Boxtel R, Cornelis GR, Tommassen J, Koster M (2004) Role of the pilot protein YscW in the biogenesis of the YscC secretin in Yersinia enterocolitica. J Bacteriol 186: 5366–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis G, Vanootegem JC, Sluiters C (1987) Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb Pathog 2: 367–379 [DOI] [PubMed] [Google Scholar]
- Cornelis GR (2006) The type III secretion injectisome. Nat Rev Microbiol 4: 811–825 [DOI] [PubMed] [Google Scholar]
- Cornelis GR, Wolf-Watz H (1997) The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol 23: 861–867 [DOI] [PubMed] [Google Scholar]
- Crepin VF, Shaw R, Abe CM, Knutton S, Frankel G (2005) Polarity of enteropathogenic Escherichia coli EspA filament assembly and protein secretion. J Bacteriol 187: 2881–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell SJ, Takahashi N, Wilson R, Friedberg D, Rosenshine I, Booy FP, Shaw RK, Knutton S, Frankel G, Aizawa S (2001) The filamentous type III secretion translocon of enteropathogenic Escherichia coli. Cell Microbiol 3: 865–871 [DOI] [PubMed] [Google Scholar]
- Deng W, Li Y, Hardwidge PR, Frey EA, Pfuetzner RA, Lee S, Gruenheid S, Strynakda NC, Puente JL, Finlay BB (2005) Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 73: 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM (2005) FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280: 41236–41242 [DOI] [PubMed] [Google Scholar]
- Ferris HU, Minamino T (2006) Flipping the switch: bringing order to flagellar assembly. Trends Microbiol 14: 519–526 [DOI] [PubMed] [Google Scholar]
- Fields KA, Nilles ML, Cowan C, Straley SC (1999) Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect Immun 67: 5395–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Gonzalez-Pedrajo B, Tame JR, Macnab RM (2003) Interactions of FliJ with the Salmonella type III flagellar export apparatus. J Bacteriol 185: 5546–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Wolf-Watz H (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- Goure J, Broz P, Attree O, Cornelis GR, Attree I (2005) Protective anti-v antibodies inhibit Pseudomonas and Yersinia translocon assembly within host membranes. J Infect Dis 192: 218–225 [DOI] [PubMed] [Google Scholar]
- Hakansson S, Schesser K, Persson C, Galyov EE, Rosqvist R, Homble F, Wolf-Watz H (1996) The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J 15: 5812–5823 [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa S (1994) Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol 176: 5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiczyk E, Blobel G (2001) Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc Natl Acad Sci USA 98: 4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet L, Agrain C, Broz P, Cornelis GR (2003) The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Kaniga K, Delor I, Cornelis GR (1991) A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109: 137–141 [DOI] [PubMed] [Google Scholar]
- Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G (1998) A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J 17: 2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Galan JE (2002) Salmonella type III secretion-associated protein InvE controls translocation of effector proteins into host cells. J Bacteriol 184: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galan JE, Aizawa SI (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280: 602–605 [DOI] [PubMed] [Google Scholar]
- Kubori T, Sukhan A, Aizawa SI, Galan JE (2000) Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA 97: 10225–10230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, Minamino T, Yokoseki T (1994) Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol 176: 7625–7629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A (2002) Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J Bacteriol 184: 4500–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavander M, Sundberg L, Edqvist PJ, Lloyd SA, Wolf-Watz H, Forsberg A (2003) Characterisation of the type III secretion protein YscU in Yersinia pseudotuberculosis. YscU cleavage—dispensable for TTSS but essential for survival. Adv Exp Med Biol 529: 109–112 [DOI] [PubMed] [Google Scholar]
- Li CM, Brown I, Mansfield J, Stevens C, Boureau T, Romantschuk M, Taira S (2002) The Hrp pilus of Pseudomonas syringae elongates from its tip and acts as a conduit for translocation of the effector protein HrpZ. EMBO J 21: 1909–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM (2003) How bacteria assemble flagella. Annu Rev Microbiol 57: 77–100 [DOI] [PubMed] [Google Scholar]
- Magdalena J, Hachani A, Chamekh M, Jouihri N, Gounon P, Blocker A, Allaoui A (2002) Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J Bacteriol 184: 3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenne MN, Journet L, Mota LJ, Cornelis GR (2003) Genetic analysis of the formation of the Ysc–Yop translocation pore in macrophages by Yersinia enterocolitica: role of LcrV, yscF and YopN. Microb Pathogen 35: 243–258 [DOI] [PubMed] [Google Scholar]
- Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM (2004) Structural insights into the assembly of the type iii secretion needle complex. Science 306: 1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T, Cornelis GR (1991) Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol 173: 1677–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM (2000) Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate specificity switching. J Bacteriol 182: 4906–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita-Ishihara T, Ogawa M, Sagara H, Yoshida M, Katayama E, Sasakawa C (2006) Shigella Spa33 is an essential C-ring component of type III secretion machinery. J Biol Chem 281: 599–607 [DOI] [PubMed] [Google Scholar]
- Moriya N, Minamino T, Hughes KT, Macnab RM, Namba K (2006) The type III flagellar export specificity switch is dependent on FliK ruler and a molecular clock. J Mol Biol 359: 466–477 [DOI] [PubMed] [Google Scholar]
- Mota LJ, Cornelis GR (2005) The bacterial injection kit: type III secretion systems. Ann Med 37: 234–249 [DOI] [PubMed] [Google Scholar]
- Mota LJ, Journet L, Sorg I, Agrain C, Cornelis GR (2005) Bacterial injectisomes: needle length does matter. Science 307: 1278. [DOI] [PubMed] [Google Scholar]
- Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR (2005) The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science 310: 674–676 [DOI] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR (1995) Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA 92: 11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier I, Bleves S, Josenhans C, Karmani L, Kerbourch C, Lambermont I, Totemeyer S, Boyd A, Cornelis GR (2000) YscP, a Yersinia protein required for Yop secretion that is surface exposed, and released in low Ca2+. Mol Microbiol 37: 1005–1018 [DOI] [PubMed] [Google Scholar]
- Sukhan A, Kubori T, Wilson J, Galan JE (2001) Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol 183: 1159–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamano K, Katayama E, Toyotome T, Sasakawa C (2002) Shigella Spa32 is an essential secretory protein for functional type III secretion machinery and uniformity of its needle length. J Bacteriol 184: 1244–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampakaki AP, Fadouloglou VE, Gazi AD, Panopoulos NJ, Kokkinidis M (2004) Conserved features of type III secretion. Cell Microbiol 6: 805–816 [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F, Vasse J, Camus JC, Marenda M, Boucher C (2000) Ralstonia solanacearum produces hrp-dependent pili that are required for PopA secretion but not for attachment of bacteria to plant cells. Mol Microbiol 36: 249–260 [DOI] [PubMed] [Google Scholar]
- Williams AW, Yamaguchi S, Togashi F, Aizawa SI, Kawagishi I, Macnab RM (1996) Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol 178: 2960–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Gonzalez-Pedrajo B, Macnab RM (2002) Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry 41: 9516–9524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Table 1
Supplementary Table 2


