Abstract
Excessive accumulation of sodium in plants causes toxicity. No mutation that greatly diminishes sodium (Na+) influx into plant roots has been isolated. The OsHKT2;1 (previously named OsHKT1) transporter from rice functions as a relatively Na+-selective transporter in heterologous expression systems, but the in vivo function of OsHKT2;1 remains unknown. Here, we analyzed transposon-insertion rice lines disrupted in OsHKT2;1. Interestingly, three independent oshkt2;1-null alleles exhibited significantly reduced growth compared with wild-type plants under low Na+ and K+ starvation conditions. The mutant alleles accumulated less Na+, but not less K+, in roots and shoots. OsHKT2;1 was mainly expressed in the cortex and endodermis of roots. 22Na+ tracer influx experiments revealed that Na+ influx into oshkt2;1-null roots was dramatically reduced compared with wild-type plants. A rapid repression of OsHKT2;1-mediated Na+ influx and mRNA reduction were found when wild-type plants were exposed to 30 mM NaCl. These analyses demonstrate that Na+ can enhance growth of rice under K+ starvation conditions, and that OsHKT2;1 is the central transporter for nutritional Na+ uptake into K+-starved rice roots.
Keywords: HKT, Na+ uptake, salt stress
Introduction
Sodium (Na+) is an alkali cation, which is not accumulated at large concentrations in most plant cells, in contrast to the alkali cation potassium (K+), which is an essential macronutrient for plant growth. High concentrations of external Na+ inhibit K+ absorption (Rains and Epstein, 1965, 1967b) and elevated Na+ concentrations in plant cells disturb functions of vital enzymes (Murguía et al, 1995) and photosynthesis (Tsugane et al, 1999), leading to Na+ toxicity and cell death. On the other hand, classical plant physiological studies have reported a positive effect of low Na+ concentrations on the growth of many plant species, but the underlying Na+ uptake mechanisms remain unknown (Flowers and Läuchli, 1983).
Several genes that encode Na+ permeable transporters/channels have been identified in plants. Biochemical analyses of vacuoles revealed a mechanism for the sequestration of Na+ into vacuoles under salt stress via Na+/H+ antiporters (Blumwald and Poole, 1985, 1987). Genome sequencing of Arabidopsis thaliana led to the identification of the corresponding plant Na+/H+ antiporter genes, AtNHX1 to 6 (Apse et al, 1999; Gaxiola et al, 1999; Yokoi et al, 2002; Aharon et al, 2003). An essential salt tolerance gene of Arabidopsis, salt overly sensitive 1 (SOS1), also belongs to the same H+ exchanger family of transporters (Shi et al, 2000). The SOS1 transporter has been reported to function in long-distance Na+ transport from roots to shoots in xylem parenchyma cells during salt stress (Shi et al, 2002).
Electrophysiological analyses using root cortex cells and suspension cultured cells of monocot plants suggest that toxic Na+ influx into roots under high external Na+ concentrations is mediated by voltage-independent channels (VIC) or non-selective cation channels (NSC) (Amtmann et al, 1997; Roberts and Tester, 1997; Tyerman et al, 1997; Buschmann et al, 2000; Davenport and Tester, 2000). VIC/NSC currents have also been reported in Arabidopsis roots and were shown to be downregulated by addition of cAMP and cGMP (Maathuis and Sanders, 2001). Recently, the cyclic nucleotide-gated channel 3 (CNGC3) gene was proposed to mediate non-selective cation uptake in the root of Arabidopsis plants (Gobert et al, 2006). The detailed molecular identities of VIC/NSC, however, remain unknown and no genetic mutations have been isolated in any plant species, to date, that show a strong reduction in Na+ influx into roots.
HKT-type transporters have been characterized in several plant and bacterial species (Schachtman and Schroeder, 1994; Rubio et al, 1995; Fairbairn et al, 2000; Uozumi et al, 2000; Horie et al, 2001; Golldack et al, 2002; Garciadeblás et al, 2003; Su et al, 2003; Ren et al, 2005; Tholema et al, 2005). TaHKT2;1 (previously named TaHKT1) (Platten et al, 2006), from wheat, has been shown to mediate at least two transport modes in heterologous expression systems, K+–Na+ co-uptake and Na+ influx at high Na+ concentrations (Rubio et al, 1995; Gassmann et al, 1996). Analyses of the ion transport specificity of AtHKT1;1, previously named AtHKT1, in Arabidopsis and OsHKT2;1 (OsHKT1) in rice revealed that these HKT1 transporters/channels showed a relatively Na+-selective transport activity relative to K+ transport activity in yeast and Xenopus oocytes (Uozumi et al, 2000; Horie et al, 2001; Garciadeblás et al, 2003). Null mutations in AtHKT1;1 (Mäser et al, 2002a; Gong et al, 2004) and a reduced-function mutant allele of AtHKT1;1 (Berthomieu et al, 2003) cause Na+ over-accumulation in shoots, resulting in leaf chlorosis. Detailed 22Na+ influx studies showed that AtHKT1;1 does not mediate Na+ influx into Arabidopsis roots (Berthomieu et al, 2003; Essah et al, 2003). Recent studies have shown that AtHKT1;1 in Arabidopsis and its closest homologue, SKC1 or OsHKT1;5 in rice, function by removing Na+ from the xylem sap, thus reducing Na+ accumulation in leaves (Ren et al, 2005; Sunarpi et al, 2005; Horie et al, 2006). Unlike AtHKT1;1, which is a single-copy gene in Arabidopsis thaliana, seven full-length OsHKT genes were identified in the japonica rice genome based on the completed genome sequence (Garciadeblás et al, 2003).
Molecular genetic identification of transporters that mediate Na+ influx into plant roots is crucial for developing models of Na+ uptake mechanisms into plant roots. In this study, we have isolated and characterized OsHKT2;1-disrupted rice plants from a Tos17-tagged population (Hirochika, 1997, 2001; Yamazaki et al, 2001), to determine the physiological role of the OsHKT2;1 transporter in rice. Here, we demonstrate that oshkt2;1 loss-of-function mutant alleles cause a dramatic reduction in Na+ influx into K+-starved rice roots. We further demonstrate that OsHKT2;1 plays an exclusive role in nutritional Na+ uptake into K+-starved rice roots, with an apparent Na+ affinity (Km) of ≈280 to 330 μM. Interestingly, OsHKT2;1-mediated Na+ influx does not cause Na+ toxicity, owing to a rapid downregulation of the OsHKT2;1 transporter upon Na+ stress.
Results
Isolation of oshkt2;1 mutants and corresponding TosWT plants
Ion selectivity analyses of OsHKT2;1 expressed in yeast and Xenopus oocytes showed that OsHKT2;1 functions as a Na+ transporter in these heterologous expression systems (Horie et al, 2001; Mäser et al, 2002b; Garciadeblás et al, 2003). But the in vivo function of OsHKT2;1 in rice remains unknown. We searched the rice Tos17 insertion mutant database (Hirochika, 1997, 2001; Miyao et al, 2003) for a putative OsHKT2;1 gene-disrupted line to be used for uncovering the physiological role of OsHKT2;1 in rice plants. The retrotransposon Tos17 undergoes local transposition events only during tissue culture, but is stable and non-motile in rice plants. We identified five putative OsHKT2;1 insertion alleles. Among them, three oshkt2;1 alleles, named oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 (Figure 1A), were chosen for further characterization, based on the fertility of plants and the germination rate of seeds of the next generation. In addition, we pursued isolation of related OsHKT2;1 wild-type (WT) control plants, named ‘TosWT'. Each Tos17 mutant line comprises an average of 8–10 insertions, which demands additional WT controls for characterization of mutants. TosWT control lines were isolated from the same seed populations as oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 by screening for individuals that show no insertion in the OsHKT2;1 gene. Southern hybridization showed polymorphisms in autoradiographs, confirming the insertion in OsHKT2;1 in each line (data not shown). RT–PCR analyses using the primer set shown in Figure 1A showed that mature OsHKT2;1 mRNA is missing in oshkt2;1-1, oshkt2;1-2 plants (Figure 1B) and oshkt2;1-3 (Supplementary Figure 1A). These data show the disruption of the OsHKT2;1 gene in the oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 mutant lines.
Figure 1.
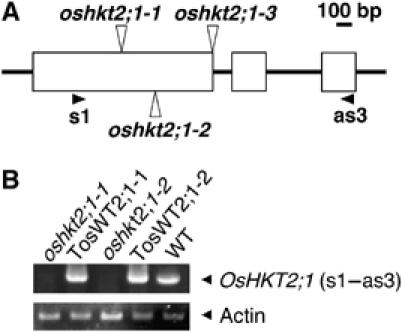
Isolation of homozygous Tos17 insertion mutants in the OsHKT2;1 gene. (A) A schematic diagram of oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 alleles. White boxes represent exons. oshkt2;1-3 has an insertion at the exon–intron boundary. s1 and as3 are primers used for RT–PCR. (B) RT–PCR analyses using cDNA derived from whole plant tissues of oshkt2;1 mutant and WT plants (10-day old plants), which were grown under K+ starvation conditions.
oshkt2;1 mutant plants exhibit reduced growth under low Na+ and K+-starved conditions
oshkt2;1 and TosWT plants showed normal growth and were indistinguishable from WT cv. Nipponbare plants, either on soil or on ordinary nutrient media (Figure 2A and data not shown). A low-affinity Na+ uptake mode of OsHKT2;1 has been hypothesized based on functional analyses, using heterologous expression systems (Horie et al, 2001). We thus first imposed NaCl stress (50–200 mM) on both soil grown and hydroponically grown plants. However, no notable difference in the visual phenotypes was observed between oshkt2;1 and WT plants (data not shown).
Figure 2.
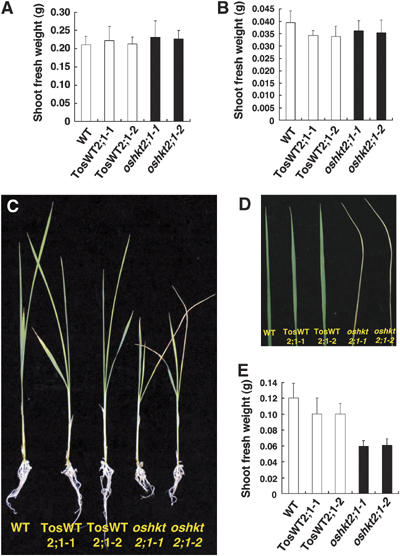
oshkt2;1 mutant plants show reduced growth under low Na+ and K+-starved growth conditions. (A) Fresh weights of 25-day-old WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 plants, which were grown under 1.25 mM K+ conditions for the last 15 days (n=12; ±s.d.). (B) Fresh weights of WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 plants, which were germinated and grown in 1 mM CaSO4 solution for 10 days (n=18; ±s.d.). (C, D) Twenty-five-day-old plants, which were grown under hydroponic conditions for the last 15 days in the presence of 0.5 mM Na+ in a K+-free medium. oshkt2;1 mutant plants showed reduced growth (C) and chlorotic withering of the oldest leaves (D). Representative photographs of plants are shown and were observed in three independent experiments with over 18 total plants of each line. (E) Fresh weights of 25 day-old WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 plants, which were grown under the same 0.5 mM Na+ condition for the last 15 days as in (C) and (D) (n=12; ±s.d.).
The accumulation of OsHKT2;1 mRNA was reported to dramatically increase in response to K+ starvation (Horie et al, 2001; Garciadeblás et al, 2003). Ten-day old oshkt2;1 plants grown in a 1 mM CaSO4 (K+ deprived) solution showed no visible growth defects and no differences in the fresh weight compared with TosWT plants (Figure 2B, P>0.36 for TosWT2;1-1 versus oshkt2;1-1; P>0.08 for TosWT2;1-2 versus oshkt2;1-2; Supplementary Figure 2A, P>0.22 for TosWT2;1-3 versus oshkt2;1-3). The plants were subsequently grown for 15 additional days in hydroponic culture medium containing 0.5 mM NaNO3 (see Materials and methods). Fifteen days later, oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 mutant plants showed remarkably reduced growth, accompanied with a withering of the oldest leaf compared with TosWT2;1-1, TosWT2;1-2, TosWT2;1-3 and WT plants (Figure 2C and D and Supplementary Figure 2B and C). Fresh weights of oshkt2;1 mutant plants were reduced approximately by 30–40% compared with TosWT plants (Figure 2E, P<1.6 × 10−5 for TosWT2;1-1 versus oshkt2;1-1; P<7.1 × 10−7 for TosWT2;1-2 versus oshkt2;1-2; Supplementary Figure 2D, P<5.7 × 10−6 for TosWT2;1-3 versus oshkt2;1-3). These data show that the OsHKT2;1 transporter allows improved growth of WT plants under low nutrient K+-starved conditions when 0.5 mM NaNO3 was added to the growth solution.
oshkt2;1 mutant plants accumulate less Na+ in both shoots and roots
We determined Na+ contents of oshkt2;1 mutant, TosWT and WT plants. Ten-day-old plants grown in 1 mM CaSO4 solution were transferred onto minimal medium containing 0.5 mM Na+ and grown for nine additional days. Interestingly, oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 mutant plants were found to accumulate considerably less Na+ in both roots and shoots compared with TosWT and WT plants (Figure 3 and Supplementary Figure 3). These results show that OsHKT2;1 functions in Na+ accumulation in roots and also in shoots.
Figure 3.
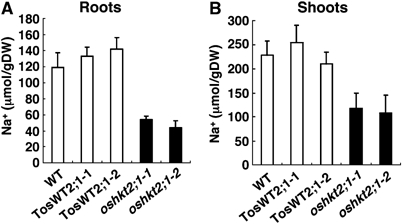
oshkt2;1 mutant plants accumulate less Na+ in roots and shoots. Nineteen-day-old plants, which were hydroponically cultured under 0.5 mM Na+- and K+-free conditions for the last 9 days, were used. Na+ contents of roots (A) and shoots (B) were measured by ICP-OES. Error bars represent standard deviations (n=6).
OsHKT2;1 expression in cortical and endodermal cells in roots and in vascular bundle regions in leaves
Transgenic rice plants carrying the 1.6 kb OsHKT2;1 promoter-β-glucuronidase (GUS) or green fluorescence protein (GFP) gene constructs were produced in order to determine the expression pattern of OsHKT2;1. Strong GUS signals were found in the main root but not in the root tip region (Figure 4A and B). Sections of GUS-stained roots further showed that cortex cells and endodermis cells were strongly stained (Figure 4C, see red stain). Sections of GUS-stained leaves showed the expression of OsHKT2;1 in vascular bundle regions (Figure 4D and E).
Figure 4.

OsHKT2;1 gene expression in the cortex and endodermis of K+-starved roots and in leaf vascular bundles. Transgenic rice plants expressing GUS or GFP reporter genes under the control of a 1.6 kb OsHKT2;1 promoter were grown in 1 mM CaSO4 solution in the presence of hygromycin. (A, B) Strong GUS staining was detected in main roots (A) but not in root tip regions (B). (C) A dark-field microscopic image of sections derived from GUS-stained main roots. Sections of GUS-stained (red staining) main roots showed strong signals in cortical and endodermal cells. (D, E) Dark-field microscopic images of sections derived from GUS-stained leaves. The sections showed strong GUS signals (red stain) in vascular bundle regions. In (E) is an enlarged image of a section of the region shown in (D). Note that the reddish staining in (C–E) shows GUS signals, as these were obtained using dark-field microscopy. (F) A 3D reconstruction image of GFP fluorescence derived from K+-starved roots of OsHKT2;1 promoter-GFP plants, showing strong GFP fluorescence in root cortex cells. (G) A 3D reconstruction image of propidium iodide fluorescence derived from K+-starved roots of the same plant shown in (F). (H) Combined images of GFP and propidium iodide fluorescence shown in (F) and (G). (I) An enlarged image of K+-starved roots shown in (H). (J, K) Combined images of GFP and propidium iodide fluorescence derived from 10-day-old OsHKT2;1 promoter-GFP plants grown in 1 mM CaSO4 solution supplemented with either 30 mM K+ (J) or 30 mM Na+ (K). (L) GFP fluorescence from three different conditions were quantified and normalized relative to propidium iodide fluorescence. Fluorescence intensities of GFP and propidium iodide in (J) were measured using three independent plants for each ionic condition (±s.d.).
Tissue expression of OsHKT2;1 was further analyzed using transgenic plants expressing GFP driven by the 1.6 kb OsHKT2;1 promoter. Roots of hygromycin B-resistant plants were briefly stained with propidium iodide (Swarup et al, 2004). Fluorescence was analyzed by confocal microscopy. A strong GFP fluorescence was found in root cortex cells of K+-starved main roots (Figure 4 F, H and I). The tissue specificity of OsHKT2:1 expression in K+-starved roots of japonica rice presented in this study overlaps with, but also slightly differs from the expression pattern in indica rice varieties (Golldack et al, 2002). The reason for this discrepancy could be the difference in the regulation of OsHKT2;1 expression between the two rice varieties and experimental conditions.
To analyze the transcriptional regulation of OsHKT2;1 in response to external K+ and Na+ concentrations, OsHKT2;1 promoter-GFP plants were grown in 1 mM CaSO4 solution supplemented with either 30 mM K+ or 30 mM Na+. Addition of 30 mM K+ or 30 mM Na+ led to substantial reductions in GFP fluorescence of root cells (Figure 4J and K) compared with those of K+-starved roots (Figure 4H). GFP and propidium iodide fluorescence intensities were quantified based on 3D reconstruction images obtained by z-series stacking and GFP fluorescence intensities and normalized to propidium iodide fluorescence intensities. The average GFP fluorescence intensity of K+-starved roots was approximately five times higher than that of 30 mM K+- or 30 mM Na+-treated roots (Figure 4L). Taken together, these results demonstrate that the OsHKT2;1 gene is mainly expressed at the cortex cell layer and the endodermal cell layer of K+-starved japonica rice roots, and that OsHKT2;1 gene expression is controlled by external K+ and Na+ concentrations at the transcriptional level.
oshkt2;1 mutant plants accumulate less Na+ in the xylem sap
We next determined Na+ contents of the xylem sap. Ten-day-old oshkt2;1 mutant and TosWT plants grown in 1 mM Ca2+ solution were transferred to hydroponic nutrient medium containing 0.5 mM Na+ and cultured for 2 days. Xylem sap was collected and Na+ concentrations were determined by inductively coupled plasma-optic emission spectroscopy (ICP-OES). The results clearly showed that oshkt2;1-1 and oshkt2;1-2 mutant plants contained much less Na+ compared with TosWT plants (Figure 5). Na+ levels in the xylem sap of oshkt2;1-1 were reduced by approximately 81.0% compared with TosWT2;1-1 (Figure 5A, P<0.0008 for oshkt2;1-1 versus TosWT2;1-1) and those of oshkt2;1-2 were reduced by approximately 89% compared with TosWT2;1-2 (Figure 5B, P<0.001 for oshkt2;1-2 versus TosWT2;1-2).
Figure 5.
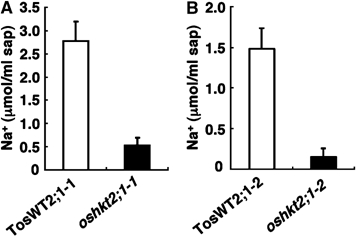
oshkt2;1 mutant plants accumulate less Na+ in the xylem sap. Ten-day-old seedlings grown in 1 mM CaSO4 solution were subsequently grown in 0.5 mM Na+- and K+-free conditions two more days. Xylem sap was collected from WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2. Na+ contents were measured by ICP-OES. (A) TosWT2;1-1 versus oshkt2;1--1. (B) TosWT2;1-1 versus oshkt2;1-1. Xylem sap extractions for (A) were performed at a different time of year than (B). Error bars represent standard error (n=8).
OsHKT2;1 localization at the plasma membrane
To gain insight into the subcellular localization of the OsHKT2;1 protein in plant cells, a chimeric EGFP-OsHKT2;1 cDNA was constructed and placed downstream of the CaMV 35S promoter. Transient expression analyses were performed using tobacco leaf mesophyll cells. A bright fluorescence bordering the cell was found in mesophyll cells expressing EGFP-OsHKT2;1 (Figure 6A). Cells were subsequently stained with the plasma membrane marker FM4-64 (Bolte et al, 2004). As shown in Figure 6B, FM4-64-derived red fluorescence was detected at the border of cells. Combined images showed an overlap of the two different fluorescence emissions at the border of cells (Figure 6C and E), indicating that the EGFP-OsHKT2;1 protein localizes at the plasma membrane. Fluorescence images derived from EGFP and FM4-64 were analyzed in control experiments. In controls, green fluorescence was more broadly observed in the cell, including in the nucleus (Figure 6D), compared with EGFP-OsHKT2;1-derived fluorescence (Figure 6A). Native red chloroplast fluorescence showed the localization of chloroplasts, in addition to the plasma membrane staining with FM4-64 (Figure 6D). Combined images showed that EGFP-derived fluorescence and FM4-64-derived fluorescence did not overlap at the plasma membrane (Figure 6D and F), indicating that the EGFP control protein does not localize at the plasma membrane.
Figure 6.
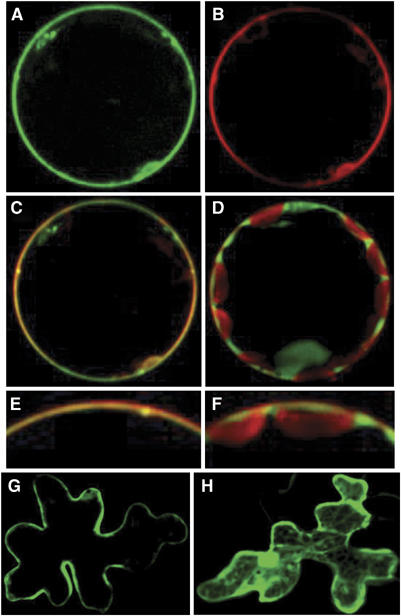
OsHKT2;1 localizes at the plasma membrane of plant cells. EGFP-OsHKT2;1 cDNA and EGFP were transiently expressed in protoplasts of tobacco mesophyll cells (A–F) and Arabidopsis epidermal cells (G, H) under the control of the 35S promoter. Fluorescence was analyzed by confocal microscopy. (A) GFP fluorescence of tobacco mesophyll cell protoplast expressing EGFP-OsHKT2;1. (B) FM4-64 plasma membrane marker fluorescence emitted from the same protoplast shown in (A). (C) Combined images of GFP and FM4-64 fluorescence shown in (A) and (B). (D) Combined images of GFP control and FM4-64 fluorescence, derived from cells harboring a control construct. Note that native red chloroplast autofluorescence was observed in addition to red FM4-64 fluorescence at the plasma membrane (D, F). (E) An enlarged image of (C). (F) An enlarged image of (D). (G) GFP fluorescence of Arabidopsis epidermal cells expressing EGFP-OsHKT2;1. (H) GFP fluorescence of Arabidopsis epidermal cells harboring a GFP control construct.
Subcellular localization of EGFP-OsHKT2;1 was also tested using Arabidopsis leaf epidermal cells. An EGFP-OsHKT2;1 and a control construct were introduced into epidermal cells by particle bombardment. Epidermal cells expressing the EGFP-OsHKT2;1 construct showed fluorescence at the border of cells (Figure 6G). In contrast, cells harboring a control construct showed ubiquitous fluorescence in the cytoplasm and in the nucleus (Figure 6H). Taken together, these transient expression analyses provide evidence that the OsHKT2;1 transporter localizes at the plasma membrane of plant cells.
Severe disruption of Na+ influx in oshkt2;1 mutant roots
Less Na+ accumulation in shoots and roots of oshkt2;1 mutants (Figure 3) and localization analyses of OsHKT2;1 (Figures 4 and 6) indicated that OsHKT2;1 may mediate Na+ influx into rice roots. In order to test this hypothesis, we performed short-term tracer influx experiments using 22Na+. Time-dependent Na+ influx into roots of intact rice plants was monitored at 0.1 mM external Na+ using 10-day-old oshkt2;1-1, oshkt2;1-2, oshkt2;1-3, TosWT2;1-1, TosWT2;1-2, TosWT2;1-3 and WT plants, which were grown in 1 mM CaSO4 solution. Interestingly, short-term Na+ influx into rice roots at 0.1 mM external Na+ was almost completely abolished in oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 plants, while TosWT2;1-1, TosWT2;1-2, TosWT2;1-3 and WT plants exhibited similar Na+ influx time courses (Figure 7A and Supplementary Figure 4A).
Figure 7.
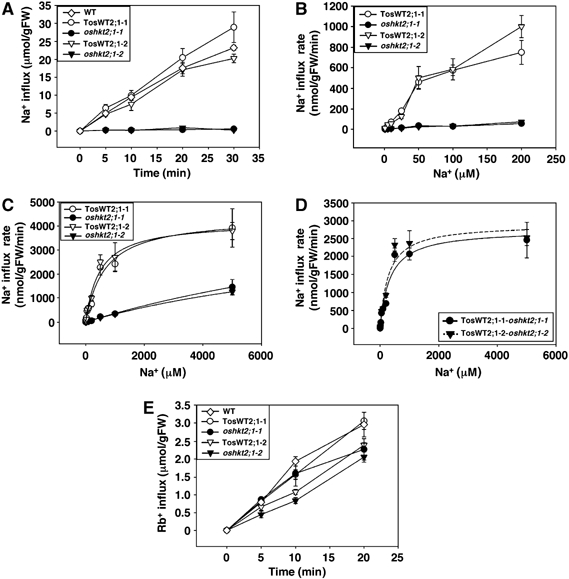
Short-term 22Na+ influx analyses show that OsHKT2;1 functions as a major Na+ uptake pathway in K+-starved rice roots. Plants were grown in 1 mM CaSO4 solution for 10 days. Time-dependent and concentration-dependent 22Na+ influx experiments were performed using 22NaCl as a tracer. (A) Time-dependent Na+ influx into roots of WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 at 0.1 mM external Na+ (n=3; ±s.d.). (B) Concentration-dependent Na+ influx into roots of TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 at 1, 5, 10, 25, 50, 100 and 200 μM external Na+ (n=3; ±s.d.). (C) Concentration-dependent Na+ influx into roots of TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 at 1–5000 μM external Na+ (n=3; ±s.d.). Curves in (C) are plotted separately for each WT and oshkt2;1 mutant allele derived from Michaelis–Menten analyses of the data (Table Ia). (D) OsHKT2;1-dependent Na+ influx rates of rice roots. Differences in Na+ influx rates between TosWT and oshkt2;1 mutant plants are shown (±s.d.). Curves in (D) were derived from Michaelis–Menten analyses (Table Ib). (E) Time-dependent Rb+ (K+) influx into roots of WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2 at 0.1 mM external Rb+ (n=6; ±s.d.).
Detailed concentration-dependent Na+ influx studies were performed in the ‘high-affinity' Na+ uptake range, at 1, 5, 10, 25, 50, 100 and 200 μM external Na+, using oshkt2;1 mutant plants and TosWT plants. oshkt2;1-1 and oshkt2;1-2 plants showed severe Na+ influx reductions compared with TosWT2;1-1 and TosWT2;1-2 plants (Figure 7B; 92.7% reduction at 200 μM). We further characterized 22Na+ influx kinetics of oshkt2;1 mutants and TosWT plants at higher external Na+ concentrations of up to 5000 μM. Sodium influx into roots of oshkt2;1-1 and oshkt2;1-2 plants within this range was severely reduced compared with that of TosWT2;1-1 and TosWT2;1-2 (Figure 7C). Sodium influx in oshkt2;1 mutant lines showed a reduction of 22Na+ influx by approximately 88–90% of WT controls at 500 μM Na+, 86–88% at 1000 μM Na+ and 63–67% at 5000 μM Na+, respectively.
Michaelis–Menten analyses were performed using concentration-dependent Na+ influx data obtained at 1–5000 μM external Na+. A single-uptake phase formalism described the kinetics of Na+ influx better in rice roots than a two-uptake phase analysis (Table Ia). The fitting results are plotted in Figure 7C for each WT and mutant allele. These results demonstrated that the disruption of the OsHKT2;1 gene dramatically affected the affinity of rice roots for Na+ and caused a severe Na+ uptake reduction (average Km shift from 550 μM to 13.6 mM) (Table Ia). To approximate OsHKT2;1-dependent Na+ influx into rice roots, Na+ influx rates of oshkt2;1 mutants were subtracted from the corresponding rates of TosWT plants. Michaelis–Menten analysis predicted an apparent affinity for the OsHKT2;1-dependent Na+ influx into roots of ≈280 to 330 μM Na+ and a Vmax of ≈2750 to 2900 nmol/gFW/min (Figure 7D; Table Ib). These flux analyses demonstrate that OsHKT2;1 mediates Na+ influx into roots and composes a major component for Na+ uptake into K+-starved rice roots in a broad concentration range.
Table 1.
Michaelis–Menten curve-fitting results for 22Na+ influx kinetics in intact rice roots of K+-starved oshkt2;1 mutant and TosWT2;l control plants (Figure 7C), and for OsHKT2; 1-dependent Na+ uptake rates in rice roots (Figure 7D)
| R2 | KM (μM) | Vmax (nmol/gFW/min) | ||
|---|---|---|---|---|
| a | TosWT2;1-l | 0.981 | 655.52±114.17 | 4390.10±272.26 |
| TosWT2;1-2 | 0.985 | 477.82±74.45 | 4176.83±219.04 | |
| oshkt2;1-1 | 0.998 | 15 764.47±3639.95 | 6051.83±1082.91 | |
| oshkt2;1-2 | 0.999 | 11 499.50±579.23 | 4165.29±149.14 | |
| b | TosWT2;1-1-oshkt2;1-1 | 0.958 | 280.4±74,8 | 2901.1±236.6 |
| TosWT2;1-2-oshkt2;1-2 | 0.964 | 330.9±81.6 | 2744.5±213.2 | |
| Curve fitting was performed with SigmaPlot 8.0 (SPSS, Chicago) software. Results from a one-term Michaelis–Menten equation. Fitting results to the data derived from TosWT and oshkt1 mutant plants are shown in (a) (see results and Figure 7C). (b) Fitting results to the values, which represent the difference in Na+ influx rates between oshkt2;1 mutant and TosWT2:l plants. | ||||
Intact K+ (Rb+) uptake in oshkt2;1 mutant plants
To determine whether OsHKT2;1 mutations have an influence on K+ uptake in intact rice roots, tracer influx experiments using 86Rb+ were performed. oshkt2;1 mutant, TosWT and WT plants, grown in 1 mM CaSO4 solution, were used for time-dependent Rb+ influx assays at 100 μM external Rb+. Only very small differences were detected between oshkt2;1 mutants and the WT lines, which showed a linear increase in Rb+ influx under the imposed condition (Figure 7E), in contrast to Na+ influx (Figure 7A). These results show that mutations in the OsHKT2;1 gene do not have a strong impact on high-affinity K+ (Rb+) uptake into intact rice roots. Small differences may be due to indirect effects of the oshkt2;1 mutations (e.g. on membrane potential).
Toxic sodium levels rapidly downregulate OsHKT2;1 transporter function in rice roots
To determine whether OsHKT2;1-mediated Na+ influx is affected by the amount of applied K+ or Na+ concentrations, oshkt2;1 mutant, TosWT and WT plants were grown in 1 mM CaSO4 solution including either 5 mM KCl or 30 mM NaCl. 22Na+ influx was monitored at 0.1 mM external Na+ (Figure 8A and Supplementary Figure 4B) and 1.0 mM external Na+ (Figure 8B and Supplementary Figure 4C). Interestingly, when the growth medium was supplemented with either 5 mM K+ or 30 mM Na+, Na+ influx into roots of TosWT and WT plants was dramatically repressed (Figure 8A and B). Comparisons with 22Na+ influx rates of oshkt2;1 knockout mutant plants demonstrated that the OsHKT2;1-mediated Na+ influx is tightly regulated by the level of external K+ and Na+ in the growth medium (Figure 8A, and B).
Figure 8.
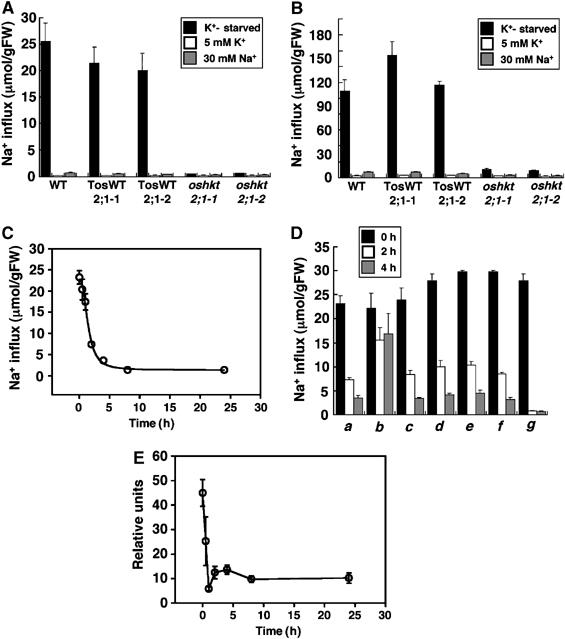
Sodium influx into K+-starved rice roots via OsHKT2;1 is subjected to posttranslational inactivation in response to high external Na+. (A, B) Sodium influx into roots of 10-day-old WT, TosWT2;1-1, TosWT2;1-2, oshkt2;1-1 and oshkt2;1-2, which were grown either in 1 mM CaSO4 solution (black bars), or the same solution supplemented with 5 mM KCl (white bars), or the 1 mM CaSO4 solution supplemented with 30 mM Na+ (gray bars). Sodium influx into roots was monitored at 0.1 mM external Na+ (A) and 1.0 mM external Na+ concentrations (B) (n=3; ±s.d.) 20 min after exposure of roots to the indicated solutions. (C) Time-dependent repression of Na+ influx into K+-starved rice roots via OsHKT2;1 in response to addition of 30 mM NaCl (n=3; ±s.d.). (D) Repression of OsHKT2;1-mediated Na+ influx in response to inhibitors. Sodium influx into roots of 10-day-old WT plants grown in 1 mM CaSO4 solution (0 h black bars) or plants incubated with the indicated test solutions for 2 h (white bars), or plants incubated with test solutions for 4 h (gray bars) are shown. Solutions contained (a) 30 mM NaCl, (b) 30 mM N-methyl-D-glucamine–Cl, (c) 30 mM NaCl and 0.1%DMSO, (d) 30 mM NaCl and the proteasome inhibitor MG132 (50 μM), (e) 30 mM NaCl and actinomycin D (20 μg/ml), (f) 30 mM NaCl and the translational inhibitor cycloheximide (100 μM), and (g) 30 mM NaCl and 5 μM K252a. Time-dependent sodium influx into roots shown in (C, D) was monitored at 0.1 mM external Na+ (n=3; ±s.d.). (E) Time-course expression analyses of the OsHKT2;1 mRNA in roots in response to 30 mM NaCl by quantitative real-time RT–PCR. Expression levels of OsHKT2;1 were monitored at 0, 0.5, 1, 2, 4, 8 and 24 h after the addition of 30 mM NaCl. The OsHKT2;1 expression level of plants grown in 1 mM CaSO4 solution supplemented with 30 mM KCl was also analyzed as a reference of basal OsHKT2;1 expression. All data were normalized to the level of the constitutively expressed OsSMT3 mRNA expression. OsHKT2;1 mRNA levels are plotted relative to the basal OsHKT2;1 expression level. Data at each time point show the average±s.e. of four independent experiments.
The time course of Na+ influx repression was further analyzed. Sodium influx in WT plants at 0.1 mM Na+ was monitored at six time points after the addition of 30 mM NaCl into the 1 mM CaSO4 growth solution. The 22Na+ influx time course demonstrated that OsHKT2;1-dependent Na+ influx is rapidly repressed in response to addition of 30 mM NaCl (Figure 8C; half time=1.45±0.11 h).
Several inhibitors were applied to gain initial insight into the biological mechanism(s) that mediates the rapid repression of the OsHKT2;1 transporter upon Na+ stress. A solution composed of N-methyl-D-glucamine (NMDG) was first tested as a source of a relatively membrane impermeable cation. The results showed that 30 mM NMDG solution was not effective at evoking the repression of the OsHKT2;1 compared with 30 mM Na+ (Figure 8D, b). The transcriptional inhibitor actinomycin D (20 μg/ml), the translational inhibitor cycloheximide (100 μM), as well as the proteasome inhibitor MG132 (50 μM), all did not greatly affect the 30 mM NaCl-induced rapid repression (Figure 8D, d–f). On the other hand, the serine/threonine protein kinase inhibitor K252a caused a more severe repression of Na+ influx than the effect of 30 mM NaCl (Figure 8D, g).
Quantitative real-time RT–PCR analyses of OsHKT2;1 mRNA upon the addition of 30 mM NaCl were subsequently performed. Interestingly, time-course analyses showed that the addition of NaCl caused a rapid reduction of OsHKT2;1 mRNA (Figure 8E; n=4 replicates). These data suggest that both posttranslational (Figure 8D) and transcript expression (Figure 8E) mechanisms control OsHKT2;1 activity in intact rice roots.
Discussion
Several Arabidopsis mutants such as sos1 (Wu et al, 1996; Shi et al, 2002), atnhx1 (Apse et al, 2003) and athkt1;1 (Rus et al, 2001; Mäser et al, 2002a; Berthomieu et al, 2003), which are disrupted or mutated in genes encoding Na+ transporters, have been characterized. These reports have demonstrated that the Na+ transporters in Arabidopsis, SOS1, AtNHX1 and AtHKT1;1, function to protect Arabidopsis plants from salt stress by different mechanisms (Zhu, 2002; Horie and Schroeder, 2004). HKT cDNAs have been isolated and characterized from many plant species (Fairbairn et al, 2000; Uozumi et al, 2000; Horie et al, 2001; Golldack et al, 2002; Garciadeblás et al, 2003; Su et al, 2003; Ren et al, 2005). TaHKT2;1-antisense wheat plants have been shown to exhibit an approximately 42% decrease in 22Na+ uptake at a high 100 mM external NaCl concentration, but no substantial difference in 22Na+ influx at 20 and 50 mM NaCl concentrations (Laurie et al, 2002). Despite these advances, no strong Na+ influx genetic mutant line has been identified in plants, indicating overlapping redundancies in Na+ transporters that mediate Na+ influx into plants (Schroeder et al, 1994). The present findings reveal that OsHKT2;1 mediates a major contribution to Na+ influx into K+-starved roots. An approximately 10–30% Na+ influx activity at millimolar concentrations remains in oshkt2;1-1 and oshkt2;1-2 mutant plants (Figures 7 and 8). Studies investigating whether the remaining Na+ influx in intact rice roots depends on other OsHKT transporters and/or different Na+ permeable channels/transporters will be an interesting subject for future inquiry. Genome analysis of rice cv. Nipponbare led to the identification of seven expressed OsHKT genes (Garciadeblás et al, 2003). OsHKT1;5 (OsHKT8 or SKC1) and its closest homologue in Arabidopsis, AtHKT1;1 (AtHKT1), have been shown to function in salt tolerance by reducing Na+ levels in the xylem sap (Ren et al, 2005; Sunarpi et al, 2005; Horie et al, 2006). More recently from initial mapping experiments, the TaHKT7 (TaHKT1;4) and TaHKT8 (TaHKT1;5) genes in durum wheat, close homologues of AtHKT1;1, have also been hypothesized to play a protective role in maintaining viability of wheat leaves under saline conditions (Huang et al, 2006; Byrt et al, 2007).
Genetic evidence that Na+ enhances growth of rice plants and a major role for OsHKT2;1 in nutritional Na+ uptake
Unlike K+, which is an essential macronutrient and the most abundant cation in plant cells, Na+ is not required as an essential nutrient by most higher plants, including glycophytes, despite the fact that the availability of Na+ and K+ is in general similar. On the other hand, however, classic plant physiological studies have shown that moderate amounts of Na+ enhance the growth of many plant species (Mengel and Kirkby, 1982; Flowers and Läuchli, 1983). Nutritional effects of Na+ have been found on the growth of beet and tomato plants (Woolley, 1957; Marschner, 1971; Besford, 1978). Despite these physiological observations, no genetic evidence for growth enhancement by Na+ has been obtained and genes mediating this process have not been identified.
oshkt2;1-1 and oshkt2;1-2 mutant plants were indistinguishable from WT plants under K+-rich nutrient conditions (Figure 2A and data not shown). Interestingly, however, oshkt2;1-1, oshkt2;1-2 and oshkt2;1-3 plants exhibited reduced growth, accompanied by leaf chlorosis under low Na+ and K+-limiting conditions (Figure 2C–E and Supplementary Figure 2B–D). These results, together with Na+ transport, accumulation and OsHKT2;1 localization analyses, provide first genetic evidence that Na+ enhances growth of rice plants under K+-starved condition, and that the plasma membrane transporter/channel OsHKT2;1 plays a central role in acquiring and distributing Na+ as a ‘nutrient'. Potassium depletion strongly induces OsHKT2;1 gene expression (Figure 4H) (Horie et al, 2001; Golldack et al, 2002; Garciadeblás et al, 2003) and OsHKT2;1-dependent Na+ influx in roots (Figure 8A and B). However, Na+ does not completely rescue rice plants from K+ deficiency (Figure 2A and E). Presumably, Na+ replaces only some of the roles of K+, which functions in many processes, including osmotic adjustment, enzyme activation, cell expansion, maintenance of membrane potential, protein synthesis and photosynthetic activity. Possible main functions of Na+ for cellular viability in K+-starved plants may be maintenance of turgor pressure and cell expansion.
K+ (Rb+) uptake and the OsHKT2;1 transporter in K+-starved rice roots
A Na+-selective has been detected in ion selectivity analyses of OsHKT2;1 expressed in heterologous expression systems (Horie et al, 2001; Mäser et al, 2002b; Garciadeblás et al, 2003), with an exception in which similar K+ and Na+ transport were observed (Golldack et al, 2002). To test whether dysfunctional mutations in the OsHKT2;1 gene affect net K+ influx in planta, we performed short-term 86Rb+ tracer influx analyses and compared the results between oshkt2;1 mutants and WT plants. Small reductions in 86Rb+ influx were detected in 86Rb+ influx assays performed at 0.1 mM external RbCl (Figure 7E). Two not mutually exclusive explanations may account for this difference. OsHKT2;1 may have a small permeability to K+. Alternatively, disruption of the OsHKT2;1 gene may affect other K+ influx transporters, for instance, by causing less hyperpolarized membrane potential in the Na+-free Rb+ influx buffers used for Rb+ influx assays. A slightly less hyperpolarized membrane potential in oshkt2;1 plants would indirectly result in decreasing net Rb+ (K+) influx in oshkt2;1 mutant roots compared to WT plants. The relatively small effect of oshkt2;1 disruption on Rb+ influx compared with Na+ influx shows that OsHKT2;1 mainly transports Na+ in planta under the imposed conditions.
Downregulation of OsHKT2;1 in response to high levels of sodium
Sodium over-accumulation in plant cells triggers diverse toxicities by perturbing vital cellular processes and reduces plant viability (Glenn et al, 1999; Zhu, 2002; Ward et al, 2003). Sodium influx into barley roots has been found to show multiple kinetic components (Rains and Epstein, 1967a), and an increase in the concentration of either K+ or Ca2+ in 50 mM Na+ solution led to reductions of low-affinity Na+ influx into K+-starved barley roots (Rains and Epstein, 1967b), and in a reduction in external Na+ depletion at 100 μM external Na+ (Haro et al, 2005). OsHKT2;1-expressing yeast cells showed Na+ hypersensitivity even under relatively mild stress conditions and OsHKT2;1 expressed in Xenopus oocytes yielded large Na+-selective inward currents at 100 mM external Na+ concentration (Horie et al, 2001). Furthermore, we found in this study that OsHKT2;1 mediates Na+ influx into rice roots, with substantial Vmax values of ≈2700 to 2900 nmol/gFW/min (Figures 7C and D, 8A; Table Ib). It cannot be excluded that a portion of the OsHKT2;1-dependent Na+ influx into intact roots (Figures 7, 8A and B) may be mediated by other transporters that are influenced by OsHKT2;1 activity (Schroeder et al, 1994). On the other hand, oshkt2;1 knockout would cause more hyperpolarized potentials in the presence of Na+, which can drive more Na+ influx via other transporters, and thus 22Na+ influx analyses of oshkt2;1 mutants may underestimate Na+ influx transported by OsHKT2;1.
OsHKT2:1 has been suggested to mediate high-affinity Na+ uptake in roots of rice plants (Garciadeblás et al, 2003). The determined Km values in this study differ to a degree from the Km (20 μM) values for OsHKT2;1 expressed in yeast (Garciadeblás et al, 2003). The reason for this difference may be several factors, including homologous versus heterologous expression of OsHKT2;1, different membrane potentials and possibly measurements of external Na+ depletion in yeast (Garciadeblás et al, 2003) compared to 22Na+ influx here.
At high Na+ concentrations of more than 25 mM, an apoplastic Na+ influx pathway contributes to Na+ uptake into rice roots (Yeo et al, 1987). The present study shows a major contribution of OsHKT2;1 to Na+ influx at a wide concentration range. The maximum Na+ influx rates of low-affinity Na+ uptake components in K+-starved barley roots, determined in the presence of 0.5–50 mM NaCl, can be estimated to approximately 4–5 times smaller than the Vmax values of OsHKT2;1-dependent Na+ influx (Rains and Epstein, 1967a) (Table Ib). Maximum OsHKT2;1-dependent Na+ influx rates with 1 mM Ca2+ in the influx buffer (Figure 7D) were approximately twice as high as the Na+ influx rate of Arabidopsis roots exposed to 50 mM NaCl and mM Ca2+ (Essah et al, 2003). These findings and comparisons show that OsHKT2;1 can mediate substantial Na+ influx rates in intact plants and suggest that OsHKT2;1-mediated Na+ influx would potentially cause Na+ toxicity in K+-starved rice plants. In contrast, however, oshkt2;1-1 and oshkt2;1-2 plants did not show any difference in salt sensitivity when treated with several high concentrations of NaCl, even under K+-starved conditions (data not shown).
To investigate whether OsHKT2;1 contributes to toxic Na+ influx, we performed tracer influx experiments, adding high levels of Na+ to K+-starved roots. Interestingly, OsHKT2;1-dependent Na+ influx was rapidly repressed upon the addition of 30 mM NaCl to K+-starved roots (Figure 8C). OsHKT2;1 mRNA was also found to undergo rapid reduction in response to the addition of NaCl (Figure 8E). Use of proteasome, transcriptional, translational and protein kinase inhibitors suggests that rapid downregulation of OsHKT2;1-dependent Na+ influx might not be directly and solely mediated by ubiquitination-related protein degradation, (Figure 8D, d–g). The rapid downregulation process correlates with OsHKT2;1 mRNA degradation (Figure 8C, D (g), E). However, it should be noted that a relatively large and measurable level of OsHKT2;1 mRNA remains even after the OsHKT2;1-dependent Na+ influx is almost completely shut down (Figure 8C and E), suggesting that mechanisms such as phosphorylation-dependent posttranslational modifications could be contributing to the downregulation of OsHKT2;1-dependent Na+ influx (Figure 8D, g).
In summary, the present study reports the identification of genetic mutants, oshkt2;1-1 and oshkt2;1-2, that strongly reduce Na+ influx into plant roots. Moreover, our findings provide robust genetic evidence that a moderate amount of Na+ enhances the growth of rice plants as a nutrient and the OsHKT2;1 gene is responsible for a major portion of nutritional Na+ uptake and distribution in K+-starved rice plants. OsHKT2;1 functions in Na+-selective influx relative to K+ in planta. Our findings also reveal that OsHKT2;1-induced Na+ influx is rapidly downregulated by posttranslational and transcriptional mechanisms, which monitor and restrict the amount of Na+ influx via OsHKT2;1 to prevent Na+ over-accumulation and thus Na+ toxicity. A complete elucidation of the molecular mechanisms that mediate rapid inactivation of OsHKT2;1 under high Na+ concentrations will be an important subject for understanding mechanisms controlling nutritional Na+ uptake and salinity resistance.
Materials and methods
Plant materials and growth conditions
Rice seeds were sterilized as described previously (Horie et al, 2001). Sterilized seeds were then planted on a plastic screen (Caisson labs, Sugar City, ID, USA) that was subsequently placed in a sterilized vessel (Caisson labs, Sugar City, ID, USA) filled with 1 mM CaSO4 solution or the same solution supplemented with KCl or NaCl. Seedlings were grown under light/dark cycles of 16/8 h and a temperature regime of 28/26°C. Hydroponic cultures were conducted using a modified basic nutrient medium (Mäser et al, 2002a) composed of 1.25 mM KNO3, 0.5 mM NH4H2PO4, 0.5 mM MgSO4, 0.5 mM Ca(NO3)2, 50 μM Fe2(SO4)3, 26 μM H3BO3, 5 μM ZnSO4 and 40 μM MnSO4. For 0.5 mM Na+-containing medium, KNO3 was substituted with NaNO3 and NMDG neutralized with HNO3. pH was brought to 5.5 by NMDG. Seedlings grown in 1 mM CaSO4 solution were transferred onto each modified medium using plugs (Jaece Industries Inc., North Tonawanda, NY, USA). Solutions were changed every 3 days.
Tobacco (Nicotiana tabacum cv. SR1) seeds were surface sterilized and sown on one-half Murashige and Skoog agar medium (Murashige and Skoog, 1962) containing 15 g/l sucrose. Tobacco plants were grown at 25°C and maintained by in vitro sterile shoot culture under light/dark cycles of 16/8 h in a growth chamber. Arabidopsis (A. thaliana ecotype Columbia) plants were grown on sterile soil for 20 days at 22°C under light/dark cycles of 16/8 h in a growth chamber.
Nucleic acid analyses and transgenic plants
Standard methods for nucleic acid isolations and analyses, for reporter genes analyses and for plant transformations are given in Supplementary data.
Ion content determinations
Ten-day-old plants grown in 1 mM CaSO4 solution were transferred onto 0.5 mM Na+-containing basic nutrient medium and hydroponically grown 9 further days. Ion contents of shoots and roots were measured by ICP-OES.
Ten-day-old plants grown in 1 mM CaSO4 solution were exposed to 0.5 mM Na+-containing basic nutrient medium for 2 days. Xylem sap samples were collected as described previously (Ma et al, 2004). Xylem sap samples were diluted in 5% HNO3 and Na+ contents of the samples were measured by ICP-OES.
Transient gene expression analyses
The EGFP DNA fragment was amplified by PCR and subcloned into the pBI221 vector (Clontech, Mountain View, CA, USA). The coding region of the OsHKT2;1 cDNA was also amplified and fused in frame to the 3′ end of the EGFP DNA. EGFP-OsHKT2;1 and control constructs were introduced into protoplasts of leaf mesophyll cells of tobacco (Nicotiana tabacum) cv. SR1 by polyethylene–glycol-mediated transformation, as described previously (Sparla et al, 2006). Before confocal microscopy, FM4-64 (Invitrogen Molecular probes, Eugene, OR, USA) was added to protoplast incubation solutions at a final concentration of 17 μM and incubated for 2 min at room temperature. Fluorescence was analyzed by confocal microscopy (Eclipse TE2000-U; Nikon, Tokyo, Japan), using 488 nm excitation and either 510 nm emission filters for EGFP or 600 nm emission filters for FM4-64.
EGFP-OsHKT2;1 and control constructs were also introduced into leaf epidermal cells of Arabidopsis (Columbia). Abaxial sides of WT plants grown on soil for 20 days were transfected by bombardment (PDS-000/He; Bio-Rad, Hercules, CA, USA) (Arnim et al, 1998). After 1 day, GFP expression was examined by confocal microscopy (Eclipse TE2000-U, Nikon, Tokyo, Japan).
Tracer influx analyses
Rice plants were grown in 1 mM CaSO4 solution or the same solution supplemented with the indicated concentrations of KCl or NaCl. Influx assays were performed as described previously (Gierth et al, 2005), with minor modifications described below. The influx buffer was composed of 2 mM MES-1,3 bis[tris(hydroxymethyl9-methylamino)]propane (BTP) pH5.5, 1 mM CaCl2 supplemented with either cold NaCl and 22NaCl (Amersham Bioscience, Pittsburgh, PA, USA) in Na+ influx assays or RbCl and 86RbCl (Amersham Bioscience, Pittsburgh, PA, USA) in Rb+ uptake assays. The same solution, excluding radioisotope, was used as a washing buffer. K+-starved intact plants were pretreated with the indicated concentrations of each inhibitor in the presence of 1 mM CaSO4 for 1 h. Plants were incubated for 20 min except for time-dependent influx assays. Excised roots were placed into a liquid scintillation vial and radioactivity was measured with a scintillation counter (LS6500 Beckman, Fullerton, CA, USA).
Supplementary Material
Supplementary Figures
Supplementary Information and Figure Legends
Acknowledgments
We thank Drs Markus Gierth (University of Cologne, Germany) and Mamoru Okamoto (University of California, San Diego) for helpful discussions regarding tracer influx experiments and quantitative real-time RT–PCR analyses. This work was supported by grants from the US Department of Energy (DOE-DE-FG02-03ER15449), the National Institutes of Health (ES010337-GM060396) and the National Science Foundation (MCB0417118) to JIS and the Crop Functional Genomic Center of the 21st Century Frontier Program (Grant CG1111) to GA.
References
- Aharon GS, Apse MP, Duan S, Hua X, Blumwald E (2003) Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil 253: 245–256 [Google Scholar]
- Amtmann A, Laurie S, Leigh R, Sanders D (1997) Multiple inward channels provide flexibility in K+/Na+ discrimination at the plasma membrane of barley suspension culture cells. J Exp Bot 48: 431–440 [DOI] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285: 1256–1258 [DOI] [PubMed] [Google Scholar]
- Apse MP, Sottosanto JB, Blumwald E (2003) Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J 36: 229–239 [DOI] [PubMed] [Google Scholar]
- Arnim AG, Deng XW, Stacey MG (1998) Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221: 35–43 [DOI] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, Savio C, Uozumi N, Oiki S, Yamada K, Cellier F, Gosti F, Simonneau T, Essah PA, Tester M, Very AA, Sentenac H, Casse F (2003) Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO J 22: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besford RT (1978) Effect of replacing nutrient potassium by sodium in uptake and distribution of sodium in tomato plants. Plant Soil 50: 399–409 [Google Scholar]
- Blumwald E, Poole R (1985) Na+/H+-antiport in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol 78: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Poole RJ (1987) Salt tolerance in suspension cultures of sugar beet: induction of Na+/H+ antiport activity at the tonoplast by growth in salt. Plant Physiol 83: 884–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Talbot C, Boutte Y, Catrice O, Read ND, Satiat-Jeunemaitre B (2004) FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J Microsc 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Buschmann PH, Vaidyanathan R, Gassmann W, Schroeder JI (2000) Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol 122: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, Dennis ES, Tester M, Munns R (2007) HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol 143: 1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport RJ, Tester M (2000) A weakly voltage-dependent, nonselective cation channel mediates toxic sodium influx in wheat. Plant Physiol 122: 823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essah PA, Davenport R, Tester M (2003) Sodium influx and accumulation in Arabidopsis. Plant Physiol 133: 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbairn DJ, Liu W, Schachtman DP, Gomez-Gallego S, Day SR, Teasdale RD (2000) Characterisation of two distinct HKT1-like potassium transporters from Eucalyptus camaldulensis. Plant Mol Biol 43: 515–525 [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Läuchli A (1983) Sodium versus potassium: substitution and compartmentation. Inorganic Plant Nutrition 15b: 651–681 [Google Scholar]
- Garciadeblás B, Senn ME, Bañuelos MA, Rodríguez-Navarro A (2003) Sodium transport and HKT transporters: the rice model. Plant J 34: 788–801 [DOI] [PubMed] [Google Scholar]
- Gassmann W, Rubio F, Schroeder JI (1996) Alkali cation selectivity of the wheat root high-affinity potassium transporter HKT1. Plant J 10: 852–869 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR (1999) The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA 96: 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierth M, Mäser P, Schroeder JI (2005) The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiol 137: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn E, Brown J, Blumwald E (1999) Salt tolerance and crop potential of halophytes. Crit Rev Plant Sci 18: 227–256 [Google Scholar]
- Gobert A, Park G, Amtmann A, Sanders D, Maathuis FJ (2006) Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J Exp Bot 57: 791–800 [DOI] [PubMed] [Google Scholar]
- Golldack D, Su H, Quigley F, Kamasani UR, Munoz-Garay C, Balderas E, Popova OV, Bennett J, Bohnert HJ, Pantoja O (2002) Characterization of a HKT-type transporter in rice as a general alkali cation transporter. Plant J 31: 529–542 [DOI] [PubMed] [Google Scholar]
- Gong JM, Waner DA, Horie T, Li SL, Horie R, Abid KB, Schroeder JI (2004) Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 15404–15409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro R, Bañuelos MA, Senn ME, Barrero-Gil J, Rodríguez-Navarro A (2005) HKT1 mediates sodium uniport in roots. Pitfalls in the expression of HKT1 in yeast. Plant Physiol 139: 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H (1997) Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol 35: 231–240 [PubMed] [Google Scholar]
- Hirochika H (2001) Contribution of the Tos17 retrotransposon to rice functional genomics. Curr Opin Plant Biol 4: 118–122 [DOI] [PubMed] [Google Scholar]
- Horie T, Horie R, Chan WY, Leung HY, Schroeder JI (2006) Calcium regulation of sodium hypersensitivities of sos3 and athkt1 mutants. Plant Cell Physiol 47: 622–633 [DOI] [PubMed] [Google Scholar]
- Horie T, Schroeder JI (2004) Sodium transporters in plants. Diverse genes and physiological functions. Plant Physiol 136: 2457–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Yoshida K, Nakayama H, Yamada K, Oiki S, Shinmyo A (2001) Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa. Plant J 27: 129–138 [DOI] [PubMed] [Google Scholar]
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, Munns R (2006) A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 142: 1718–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie S, Feeney KA, Maathuis FJ, Heard PJ, Brown SJ, Leigh RA (2002) A role for HKT1 in sodium uptake by wheat roots. Plant J 32: 139–149 [DOI] [PubMed] [Google Scholar]
- Ma JF, Mitani N, Nagao S, Konishi S, Tamai K, Iwashita T, Yano M (2004) Characterization of the silicon uptake system and molecular mapping of the silicon transporter gene in rice. Plant Physiol 136: 3284–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, Sanders D (2001) Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol 127: 1617–1625 [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1971) Why can sodium replace potassium in plants?. Proceedings of the 8th Colloquium of the International Potash Institute, Uppsala, Sweden, 1971. Int. Potash Institute: Berne
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, Kubo M, Yamagami M, Yamaguchi K, Nishimura M, Uozumi N, Robertson W, Sussman MR, Schroeder JI (2002a) Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett 531: 157–161 [DOI] [PubMed] [Google Scholar]
- Mäser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, Schroeder JI, Uozumi N (2002b) Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants. Proc Natl Acad Sci USA 99: 6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel K, Kirkby EA (1982) Principles of Plant Nutrition, 3rd edn. Worblaufen-Bern, Switzerland: International Potash Institute pp 425–426 [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H (2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays in tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Murguía JR, Bellés JM, Serrano R (1995) A salt-sensitive 3′(2′),5′-bisphosphate nucleotidase involved in sulfate activation. Science 267: 232–234 [DOI] [PubMed] [Google Scholar]
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, Fairbairn DJ, Horie T, Leigh RA, Lin HX, Luan S, Maser P, Pantoja O, Rodriguez-Navarro A, Schachtman DP, Schroeder JI, Sentenac H, Uozumi N, Very AA, Zhu JK, Dennis ES, Tester M (2006) Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci 11: 372–374 [DOI] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1965) Transport of sodium in plant tissue. Science 148: 1611. [DOI] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1967a) Sodium absorption by barley roots: role of the dual mechanisms of alkali cation transport. Plant Physiol 42: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rains DW, Epstein E (1967b) Sodium absorption by barley roots: its mediation by mechanism 2 of alkali cation transport. Plant Physiol 42: 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX (2005) A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet 37: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Roberts S, Tester M (1997) A patch clamp study of Na+ transport in maize roots. J Exp Bot 48: 431–440 [DOI] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI (1995) Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270: 1660–1663 [DOI] [PubMed] [Google Scholar]
- Rus A, Yokoi S, Sharkhuu A, Reddy M, Lee BH, Matsumoto TK, Koiwa H, Zhu JK, Bressan RA, Hasegawa PM (2001) AtHKT1 is a salt tolerance determinant that controls Na+ entry into plant roots. Proc Natl Acad Sci USA 98: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI (1994) Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370: 655–658 [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Ward JM, Gassmann W (1994) Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: biophysical implications for K+ uptake. Annu Rev Biophys Biomol Struct 23: 441–471 [DOI] [PubMed] [Google Scholar]
- Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci USA 97: 6896–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu JK (2002) The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P (2006) Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis. Plant Physiol 141: 840–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Balderas E, Vera-Estrella R, Golldack D, Quigley F, Zhao C, Pantoja O, Bohnert HJ (2003) Expression of the cation transporter McHKT1 in a halophyte. Plant Mol Biol 52: 967–980 [DOI] [PubMed] [Google Scholar]
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, Yoda K, Horie R, Chan WY, Leung HY, Hattori K, Konomi M, Osumi M, Yamagami M, Schroeder JI, Uozumi N (2005) Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Swarup R, Kargul J, Marchant A, Zadik D, Rahman A, Mills R, Yemm A, May S, Williams L, Millner P, Tsurumi S, Moore I, Napier R, Kerr ID, Bennett MJ (2004) Structure–function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholema N, Vor der Bruggen M, Maser P, Nakamura T, Schroeder JI, Kobayashi H, Uozumi N, Bakker EP (2005) All four putative selectivity filter glycine residues in KtrB are essential for high affinity and selective K+ uptake by the KtrAB system from Vibrio alginolyticus. J Biol Chem 280: 41146–41154 [DOI] [PubMed] [Google Scholar]
- Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H (1999) A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11: 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman S, Skerrett M, Garrill A, Findlay G, Leigh R (1997) Pathways for the permeation of Na+ and Cl− into protoplasts derived from the cortex of wheat roots. J Exp Bot 48: 459–480 [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI (2000) The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol 122: 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JM, Hirschi KD, Sze H (2003) Plants pass the salt. Trends Plant Sci 8: 200–201 [DOI] [PubMed] [Google Scholar]
- Woolley TJ (1957) Sodium and silicon as nutrient for the tomato plant. Plant Physiol 32: 317–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJ, Ding L, Zhu JK (1996) SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 8: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Tsugawa H, Miyao A, Yano M, Wu J, Yamamoto S, Matsumoto T, Sasaki T, Hirochika H (2001) The rice retrotransposon Tos17 prefers low-copy-number sequences as integration targets. Mol Genet Genomics 265: 336–344 [DOI] [PubMed] [Google Scholar]
- Yeo AR, Yeo ME, Flowers TJ (1987) The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J Exp Bot 38: 1141–1153 [Google Scholar]
- Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM (2002) Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. Plant J 30: 529–539 [DOI] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Information and Figure Legends


