Abstract
Germline mutations in BRCA2 predispose to hereditary breast cancers. BRCA2 protein regulates recombinational repair by interaction with RAD51 via a series of degenerate BRC repeat motifs encoded by exon 11 (BRCA2996–2113), and an unrelated C-terminal domain (BRCA23265–3330). BRCA2 is also required for meiotic recombination. Here, we show that human BRCA2 binds the meiosis-specific recombinase DMC1 and define the primary DMC1 interaction site to a 26 amino-acid region (BRCA22386–2411). This region is highly conserved in BRCA2 proteins from a variety of mammalian species, but is absent in BRCA2 from Arabidopsis thaliana, Caenorhabditis elegans, and other eukaryotes. We demonstrate the critical importance of Phe2406, Pro2408, and Pro2409 at the conserved motif 2404KVFVPPFK2411. This interaction domain, defined as the PhePP motif, promotes specific interactions between BRCA2 and DMC1, but not with RAD51. Thus, the RAD51 and DMC1 interaction domains on BRCA2 are distinct from each other, allowing coordinated interactions of the two recombinases with BRCA2 at meiosis. These results lead us to suggest that BRCA2 is a universal regulator of RAD51/DMC1 recombinase actions.
Keywords: chromosome synapsis, genome instability, meiosis-defective, sterility, RAD51
Introduction
The BRCA2 tumour suppressor protein is involved in maintaining the integrity of our genome by its participation in error-free double-strand break (DSB) repair via homologous recombination (HR; Venkitaraman, 2002). It does so by regulating the actions of RAD51 protein. Recombinases belonging to the RecA/RadA/RAD51 family play a key role in HR by binding to single-stranded DNA generated at the site of a DSB, to form helical nucleoprotein filaments that interact with homologous DNA and promote strand exchange (West, 2003). BRCA2 and RAD51 interact through a series of eight degenerative motifs within the exon 11 region of BRCA2, which are called the BRC repeats (Bignell et al, 1997; Wong et al, 1997; Chen et al, 1998b). In addition, there is an unrelated RAD51-binding site located close to the C terminus of BRCA2 (Mizuta et al, 1997; Sharan et al, 1997), which is regulated in a cell cycle- and DNA damage-dependent manner (Esashi et al, 2005). In response to DNA damage, RAD51 localises to nuclear foci with BRCA2 (Scully et al, 1997; Sharan et al, 1997; Chen et al, 1998a), by a mechanism that is BRCA2-dependent (Yuan et al, 1999). Consistent with a role for BRCA2 in the regulation of HR, BRCA2-deficient cells are defective in DSB repair (Moynahan et al, 2001; Xia et al, 2001) and exhibit spontaneous gross chromosomal rearrangements (Yu et al, 2000).
HR has a dual cellular function: while it maintains the integrity of DNA in somatic cells, it creates genetic variability by directing DNA exchanges in meiotic cells that result in random combinations of alleles and traits. BRCA2 appears to play an important role in meiotic recombination in mammalian cells, as it is highly expressed during spermatogenesis in mice (Connor et al, 1997), and localises to meiotic chromosomes during early prophase I when homologous chromosomes undergo pairing (Chen et al, 1998a). Although targeted disruption of the BRCA2 gene in mouse leads to embryonic lethality (Ludwig et al, 1997; Sharan et al, 1997; Suzuki et al, 1997), an analysis of the role of BRCA2 in meiosis has been made possible through the generation of a viable Brca2 null mouse carrying a bacterial artificial chromosome containing the human BRCA2 gene (Sharan et al, 2004). These mice, which exhibit low levels of expression of human BRCA2 protein in the gonads, are infertile because spermatocytes fail to progress beyond the early prophase I stage of meiosis. A role for BRCA2 in meiotic recombination has also been demonstrated in Arabidopsis thaliana where siRNA against BRCA2 leads to meiotic defects and partial sterility (Siaud et al, 2004).
In addition to RAD51, meiotic recombination requires the functions of DMC1, a meiosis-specific paralog of RAD51 (Bishop et al, 1992; Yoshida et al, 1998). Like RAD51, recombinant human DMC1 protein forms octameric rings that are capable of binding DNA and forming helical nucleoprotein filaments that catalyse strand exchange (Masson et al, 1999; Passy et al, 1999; Kinebuchi et al, 2004; Sehorn et al, 2004; Bugreev et al, 2005). The two recombinases colocalise to nuclear foci during meiotic recombination (Bishop, 1994; Chen et al, 1998a; Tarsounas et al, 1999). Consistent with these observations, mouse Dmc1 knockouts are infertile due to defects in chromosome synapsis during meiotic recombination (Pittman et al, 1998; Yoshida et al, 1998). However, it is not known how these two recombinases function during meiosis, whether they play independent roles or act in concert, or how their activities are regulated and coordinated.
A potential role for BRCA2 in coordinating the activities of RAD51 and DMC1 was implied by studies showing that meiotic chromosomes from mouse spermatocytes exhibiting impaired expression of BRCA2 had reduced numbers of RAD51 and DMC1 foci (Sharan et al, 2004). Also, in plants the AtBRCA2 protein has been shown to interact with AtDMC1, an interaction mediated by one of the four BRC repeats (BRC2) found in AtBRCA2 (Siaud et al, 2004; Dray et al, 2006). To date, however, there is no information relating to interactions between mammalian BRCA2 and DMC1 proteins.
In this study, we define the interactions between human BRCA2 and DMC1. Surprisingly, we found that the primary DMC1 interaction domain of BRCA2 maps to a region of BRCA2 that is distinct from the BRC repeats that facilitate interactions with RAD51. These results provide the first insight into understanding the coordinating role that BRCA2 provides in mammalian meiotic recombination.
Results
In vivo interactions of BRCA2 with DMC1
The interaction of human BRCA2 and DMC1 in vivo was investigated by using rabbit polyclonal antibodies to immunoprecipitate BRCA2 complexes from extracts prepared from human 293T cells transiently expressing Myc-tagged DMC1. The BRCA2 pull-downs were analysed by Western blotting using mouse monoclonal antibodies specific for BRCA2 (Figure 1, top panel), Myc-DMC1 (middle panel), and RAD51 (lower panel). We found that the BRCA2-specific antibodies pulled down both DMC1 and RAD51 (lane f), whereas IgG control reactions failed to pull down either of these proteins (lane e). Control reactions, using extracts from mock-transfected 293T cells, rather than Myc-DMC1-transfected 293T cells, showed the absence of DMC1 from the BRCA2-RAD51 immunoprecipitates, as expected since DMC1 is not normally expressed in somatic cells (lane d). Interestingly, the amounts of endogenous RAD51 present in the BRCA2 immunoprecipitates from Myc-DMC1 or mock-transfected cells were similar, indicating that while DMC1 can interact with BRCA2 in vivo, its overexpression does not significantly affect the binding of RAD51 to BRCA2.
Figure 1.

DMC1 interacts with BRCA2 in human cells. Human 293T cells were either mock-transfected (input, lane a) or transfected with Myc-DMC1 (input, lane b). Post-transfection, cells were harvested, lysed, and protein complexes were immunoprecipitated with polyclonal BRCA2 antibodies or nonspecific rabbit IgG, as indicated. The immunoprecipitates were then analysed by Western blotting for BRCA2 (upper panel), Myc-DMC1 (middle panel), or RAD51 (lower panel).
Mapping the DMC1 interaction sites in BRCA2
Studies of the interaction between BRCA2 and DMC1 in A. thaliana indicate that DMC1, like RAD51, interacts with BRCA2 through the BRC repeat sequences (Dray et al, 2006). To map the interaction domains between the human BRCA2 and DMC1 proteins, we used a series of nine overlapping glutathione S-transferase (GST) fusion proteins, designated B2-1 to B2-9 (Lee et al, 2004), that span the entire coding region of BRCA2 (Figure 2A). Lysates from Escherichia coli expressing each of the nine GST fusion proteins (Figure 2B, lanes b–j), and GST alone (lane a), were incubated with glutathione beads, and purified recombinant human DMC1 or RAD51. Complexes bound to the beads were analysed by Western blotting for either DMC1 (Figure 2B, top panel) or RAD51 (Figure 2B, middle panel). Similar amounts of GST-tagged BRCA2 fragments were observed in each pull-down (Figure 2B, bottom panel). As reported previously (Esashi et al, 2005), RAD51 interacted specifically with B2-3 (lane d) and B2-4 (lane e), the fragments containing BRC repeats 1–5, and with B2-9 (lane j), which corresponds to the C-terminal region of BRCA2. In the case of DMC1, however, we failed to observe interactions between DMC1 and any of the BRCA2 GST fragments containing BRC repeats (Figure 2B, top panel, lanes d–f). Instead, we found that DMC1 interacted specifically with B2-6 (lane g) and with the C-terminal region B2-9 (lane j).
Figure 2.

Interactions of BRCA2 with RAD51 and DMC1. (A) Schematic representation of human BRCA2 protein divided into nine GST-tagged fusion proteins. BRC repeats 1–8 (dark blue), the helical domain (green), OB folds (yellow), and nuclear localisation signals (black) are indicated. The boundaries of the nine fragments are as follows: B2-1 is BRCA21–454; B2-2 is BRCA2420–906; B2-3 is BRCA2885–1362 and contains BRC1 and BRC2; B2-4 is BRCA21338–1781 and contains BRC3, BRC4, and BRC5; B2-5 is BRCA21744–2115 and contains BRC6, BRC7, and BRC8; B2-6 is BRCA22106–2472; B2-7 is BRCA22438–2824 and contains the helical domain and one OB fold; B2-8 is BRCA22780–3197 and contains two OB folds; B2-9 is BRCA23189–3418 and contains two nuclear localisation signals. (B) Interactions of recombinant DMC1 or RAD51 with the nine GST-tagged BRCA2 fusion proteins. Lysates of E. coli cells expressing the indicated GST fusions were mixed with glutathione beads, and purified human RAD51 or DMC1 was subsequently added. After extensive washing, complexes bound to beads were analysed by Western probing for DMC1 (upper panel), RAD51 (middle panel), or the BRCA2 fragments (lower panel).
A region of BRCA2 that interacts with DMC1 but not RAD51
Presently, there is very little information relating to the function of the region of BRCA2 defined by B2-6 (amino acids 2106–2472). To map the DMC1 interaction domain in greater detail, we subdivided the region into a series of smaller GST fusion proteins, depicted in Figure 3A, designated B2-6.1 to B2-6.5. These fusion proteins were then expressed and used in DMC1 pull-down assays (Figure 3B). We found that DMC1 interacted strongly with B2-6.5 (Figure 3B, lane g), corresponding to BRCA22340–2472, but did not interact with any of the other fusion proteins (lanes c–f).
Figure 3.

Identification of a DMC1 interaction domain in an internal region of BRCA2. (A) Schematic representation of BRCA2 B2-6, subdivided into five smaller GST fusion proteins. B2-6.1 is BRCA22106–2190; B2-6.2 is BRCA22106–2218; B2-6.3 is BRCA22190–2260; B2-6.4 is BRCA22218–2340; B2-6.5 is BRCA22340–2472. (B) The indicated GST fusions were expressed in E. coli as described in Figure 2 legend and their ability to bind purified DMC1 was analysed as described. (C) Competition assay to analyse interactions between DMC1 or RAD51 and B2-6.5 fusion protein. B2-6.5 was mixed with DMC1 (200 ng) and increasing amounts of RAD51 (0, 200, 400, or 2 μg, respectively). Proteins bound to glutathione beads were analysed by Western blotting.
To ensure that the B2-6.5 region of BRCA2 interacts specifically with DMC1, competition experiments were carried out between RAD51 and DMC1 for binding to B2-6.5. We found that a 10-fold excess of purified RAD51 had no effect on the interaction between DMC1 and B2-6.5 (Figure 3C, compare lanes a–d). In these reactions, RAD51 could not be detected in the B2-6.5 pull-down by Western blotting (data not shown). These experiments indicate that there are no direct interactions between the two recombinases and show that BRCA22340–2472 contains a novel domain that binds specifically to DMC1 but does not interact with RAD51.
The interaction of this novel BRCA2 domain with DMC1 was confirmed by yeast two-hybrid analysis, a system used previously to evaluate BRCA2-RAD51 interactions (Wong et al, 1997; Chen et al, 1998b). It was also the primary method used to map interactions between a BRC repeat of AtBRCA2 and AtDMC1 (Dray et al, 2006). For the yeast two-hybrid analysis, DMC1 and RAD51 were fused to the GAL4 activation domain, while a variety of BRCA2 fragments were fused to the GAL4 DNA-binding domain. We used the eight BRC repeats, B2-6 and B2-6.5 (both identified in the above GST pull-down experiments to interact specifically with DMC1), and the C-terminal BRCA2 fragments B2-9 and TR2 (previously shown to be the C-terminal RAD51 interaction domain (Esashi et al, 2005)). We also expressed a large BRCA2 fragment, BRCA2996–2113, which contains all eight BRC repeats. We found that BRCA2-DMC1 interactions were observed with the two constructs containing the newly identified DMC1 interaction domain BRCA22340–2472 (Figure 4, B2-6.5 and B2-6). Neither of these BRCA2 fragments resulted in growth when combined with RAD51. These results confirm the finding that BRCA22340–2472 contains a domain that interacts specifically with DMC1.
Figure 4.
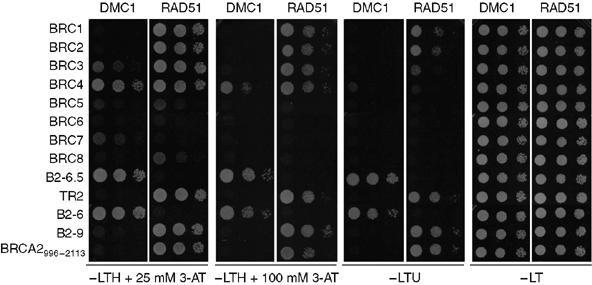
Yeast two-hybrid analysis of interactions between DMC1 or RAD51 and a series of BRCA2 fragments. Two-hybrid analysis was carried out as described in Methods, and diluted cells were plated on permissive −Leu/−Trp plates (−LT), or selection plates: −Leu/−Trp/−His + 25 mM 3-AT (−LTH+25 mM 3-AT); −Leu/−Trp/−His +100 mM 3-AT (−LTH+100 mM 3-AT); or −Leu/−Trp/−Ura (−LTU). The BRCA2 fragment boundaries are as follows: BRC1 (BRCA2996–1084); BRC2 (BRCA21206–1274); BRC3 (BRCA21415–1483); BRC4 (BRCA21511–1579); BRC5 (BRCA21658–1726); BRC6 (BRCA21831–1899); BRC7 (BRCA21965–2033); BRC8 (BRCA22045–2113); B2-6 (BRCA22106–2472); B2-6.5 (BRCA22340–2472); B2-9 (BRCA23189–3418); TR2 (BRCA23265–3330). A large BRCA2 fragment, BRCA2996–2113, containing all eight BRC repeats was also analysed.
As expected, RAD51 was found to interact with the C-terminal part of BRCA2, several of the individual BRC repeats, and with the BRCA2996–2113 fragment containing all BRC repeats. In the two-hybrid assay, we also observed an interaction between DMC1 and BRC repeat 4 (BRC4), but this interaction was relatively weak in comparison with those of B2-6 or B2-6.5, as indicated by the lack of growth in the absence of uracil (−LTU). Importantly, growth was not observed when DMC1 was expressed with BRCA2996–2113, which contains all eight BRC repeats. The weakness of the BRC4-DMC1 interaction, coupled with the absence of similar interactions in the GST pull-down experiments, may question the physiological relevance of this interaction.
Interactions of RAD51 and DMC1 with the C-terminal region of BRCA2
The two-hybrid analysis confirmed interactions between the C-terminal region of BRCA2 (TR2 and B2-9) with RAD51, but failed to support the pull-down experiments indicating an interaction of this region with DMC1. Further experiments were therefore carried out to establish that both RAD51 and DMC1 could interact with this domain of BRCA2. First, we used three small GST-tagged BRCA2 fragments, TR1, TR2, and TR3 (Figure 5A), to map the interaction of DMC1 with the C-terminal region of BRCA2 in greater detail. GST pull-downs showed that both DMC1 and RAD51 bind to the TR2 fusion protein (BRCA23265–3330), but not to either TR1 (BRCA23189–3274) or TR3 (BRCA23325–3418) (Figure 5B). Next, we carried out competition experiments to determine which of the two proteins exhibited the highest binding affinity for this site in BRCA2. For this analysis, TR2 bound to glutathione beads was mixed with constant amounts of DMC1 and increasing amounts of RAD51 and, after extensive washing, the complexes bound to beads were analysed by Western blotting. We found that the presence of RAD51, even at sub-molar concentrations compared with DMC1, efficiently competed away the binding of DMC1 to the TR2 region of BRCA2 (Figure 5C, lanes a–d). In contrast, DMC1 failed to compete with RAD51 for interaction with TR2 (lanes e–h). These results indicate that the DMC1-TR2 interaction is relatively weak compared to RAD51-TR2 complex formation, helping to explain why these interactions were not seen in the yeast two-hybrid studies.
Figure 5.

Characterisation of interactions between DMC1 and the C-terminal region of BRCA2. (A) Schematic representation of BRCA2 B2-9, subdivided into three smaller GST fusion proteins. TR1 is BRCA23189–3274 and contains one nuclear localisation signal (black); TR2 is BRCA23265–3330 and contains both nuclear localisation signals; TR3 is BRCA23325–3418. (B) Interaction of DMC1 or RAD51 with the three GST-tagged BRCA2 TR-fragments. Complexes bound to glutathione beads were analysed by Western probing for DMC1, RAD51, and BRCA2 fragments, as described in Figure 2b. (C) Competition between RAD51 and DMC1 for TR2 binding. TR2 was mixed with DMC1 and increasing amounts of RAD51 (lanes a–d), or RAD51 was mixed with increasing amounts of DMC1 (lanes e–h). Complexes bound to glutathione beads were analysed by Western blotting. (D) Schematic representation of the TR2 region of BRCA2. Red circles correspond to putative phosphorylation sites and blue circles to sites that have been phosphorylated in synthetic biotinylated peptides. The S3291 is known to be phosphorylated in vivo. (E) Interactions between RAD51 or DMC1 and the series of phosphopeptides shown in (d). Synthetic biotinylated phosphopeptides (300 ng) were mixed with either DMC1 or RAD51 (60 ng), and complexes bound to Streptavidin beads were analysed by Western blotting. (F) Effect of S3291 phosphorylation on the competition between RAD51 and DMC1 for TR2. TR2 peptide or S3291Phospho-TR2 peptide (300 ng) was mixed with DMC1 (60 ng) and increasing amounts of RAD51 (0, 30, 60, or 180 ng). Complexes bound to Streptavidin beads were analysed by Western blotting. Biotinylated peptides were visualised by HRP-conjugated Streptavidin.
Previously, it was shown that CDK-dependent phosphorylation of serine 3291 in the TR2 region of BRCA2 inhibits RAD51 interaction with this site and that this modification may serve as a regulatory mechanism for recombinational repair (Esashi et al, 2005). We therefore investigated the effect of TR2 phosphorylation on the binding of DMC1 to this region. Using biotinylated TR2 peptides phosphorylated at S3291, or a series of other control sites (Figure 5D), we found that peptides containing phosphorylated S3291 failed to bind RAD51 but were capable of interacting normally with DMC1 (Figure 5E, lanes c, e and f). These results were confirmed by competition studies in which biotinylated peptides corresponding to TR2, unphosphorylated, or phosphorylated at S3291, were incubated with DMC1 in the presence of increasing amounts of RAD51. As observed with TR2, we found that RAD51 could easily compete for DMC1 binding to TR2 (Figure 5F, lanes a–d), whereas it failed as a competitor with S3291-phosphorylated TR2 (lanes e–h). Thus, the phosphorylation of S3291 can modulate interactions with RAD51, but not DMC1, and may provide a mechanism for the differential regulation of BRCA2-RAD51 and BRCA2-DMC1 interactions.
Identification of a conserved domain in BRCA2, essential for DMC1 interaction
Since yeast two-hybrid and GST pull-down analyses had both confirmed the presence of a specific DMC1 interacting region in B2-6.5. (BRCA22340–2472), we wished to define this domain in greater detail. First, we generated four overlapping biotinylated synthetic peptides covering the region (Figure 6A, IR1–IR4), and used them in streptavidin pull-down experiments to show that the DMC1 interaction domain was contained within IR2 (BRCA22350–2417) and IR3 (BRCA22371–2438) (Figure 6B). Second, to narrow down this interaction domain even further, we synthesised a 30-mer peptide array that covered the entire BRCA22350–2417 region. On this membrane, which was subsequently probed with DMC1, each peptide overlapped the next by 29 amino acids, leading to a change of just one amino acid per peptide. We found that DMC1 interacted strongly with six to seven consecutive peptides, all containing the region corresponding to BRCA22386–2411 (Figure 6C, lower panel). This region is highly conserved in mammalian BRCA2 from a variety of species (Figure 6D), and is also found in chicken BRCA2. In contrast, the region has diverged in the BRCA2 homologues found in A. thaliana, Caenorhabditis elegans, Ustilago maydis, and Trypanosoma cruzi.
Figure 6.

Identification of the DMC1 interacting domain PhePP. (A) Schematic representation of four overlapping peptides corresponding to B2-6.5. IR1 is BRCA22340–2407; IR2 is BRCA22350–2417; IR3 is BRCA22371–2438; IR4 is BRCA22405–2472. (B) Interactions between the biotinylated IR peptides (300 ng) and DMC1 (60 ng) were assayed by streptavidin pull-downs and detected by Western blotting. Biotinylated TR2 was used as a control. (C) Peptide array with overlapping 30 mers of BRCA22350–2417 spotted on a membrane (see text for detail), probed with DMC1, and analysed by Western blotting. Subsequently, the membrane was stained with Ponceau to reveal the exact location of the spotted peptides. (D) Sequence alignment of the PhePP motif BRCA2 from a variety of organisms. At (A): A. thaliana BRCA2 homologue A. Ce: C. elegans. Um: Ustilago maydis. Tc: Trypanosoma cruzi. Amino acids indicated in blue are fully conserved in mammalian species, green indicate conservation of strong groups, whereas purple indicates conservation of weaker groups. (E) Analysis of essential amino acids required for interactions between BRCA22382–2411 and DMC1. The amino-acid substitution peptide array contains 600 30-amino acid long peptides spotted onto a membrane. Each peptide has one amino acid of BRCA22382–2411 replaced with one of the 20 common amino acids (see text for details). The membrane was probed with DMC1 followed by Western blotting.
Finally, to define precisely which amino acids are essential for the BRCA2-DMC1 interaction, we generated a mutant peptide array of 30-mer peptides containing the conserved region of BRCA2 important for DMC1 interaction. In this array, the 30 amino acids corresponding to BRCA22382–2411 were sequentially substituted (one at a time) by the 20 common amino acids (each substitute is indicated at the right of the membrane in Figure 6E). Hence, on this membrane, the first phenylalanine (F2382) is substituted in the first column, in the second column the tyrosine (Y2383) has been substituted, and so on. When this membrane was probed for DMC1 interaction, we observed that substitution of Phe2406 with any amino acid, except the two other aromatic amino acids, abolished interaction with DMC1. Similarly, substitution of Pro2408 or Pro2409 led to a loss of interaction with DMC1. All three amino acids were found to be located in the highly conserved 2404KVFVPPFK2411 region of BRCA2.
Discussion
In this work, we have demonstrated that human BRCA2 is capable of binding the meiosis-specific recombinase DMC1 via two different binding sites. The first is located at the C terminus of BRCA2 and corresponds to the region also bound by RAD51 protein. The second, however, represents the primary DMC1 interaction domain located within the relatively uncharacterised central part of BRCA2, contained within B2-6. Using yeast two-hybrid assays, pull-down experiments, and peptide arrays, we defined this interaction domain to the region corresponding to BRCA22386–2411. Within this region, we showed the critical importance of Phe2406, Pro2408, and Pro2409 located in the conserved motif KVFVPPFK, and for simplicity will refer to this DMC1 interaction domain as the PhePP motif.
The PhePP motif of BRCA2 interacts specifically with DMC1, and not with RAD51. The importance of the region is indicated by its conservation in different mammalian species and in chicken BRCA2. This DMC1 interaction domain, however, is divergent in BRCA2 homologues from other eukaryotes such as A. thaliana and C. elegans. When our studies were initiated, we were surprised to identify this novel interaction domain, as it was known that AtBRCA2 and AtDMC1 can interact through one of the four degenerate plant BRC repeats (Dray et al, 2006). However, modelling studies based on the crystal structure of a RAD51-BRC4 fusion protein suggest that DMC1 has polar or charged residues at the hydrophobic RAD51-binding patch for BRC4, which is likely to preclude its interaction with this BRC repeat (Pellegrini et al, 2002; Lo et al, 2003). It might also be significant that the AtBRCA2 proteins (there are two AtBRCA2 isoforms at 1151 or 1155 amino acids) are considerably shorter than the human counterpart (3418 amino acids), such that two of the plant BRC repeats (BRC4 and BRC2, respectively) have evolved independently to interact with either RAD51 or DMC1 (Dray et al, 2006). It remains to be established whether the diverged PhePP motif in A. thaliana can interact with AtDMC1. The divergence of the PhePP domain in C. elegans is less surprising given the absence of DMC1 in this organism, which promotes meiotic recombination through the high-level expression of RAD51 alone (Takanami et al, 2000).
In addition to the specific DMC1 interaction domain in BRCA2, we found that DMC1, like RAD51, could interact with the C-terminal region of BRCA2 known as TR2 (BRCA23265–3330). In comparison to the RAD51-TR2 interaction, the association of DMC1 with TR2 appears to be relatively weak, and it was found that RAD51 could easily compete with DMC1 for the C-terminal interaction site. However, it is known that interactions between RAD51 and the C terminus of BRCA2 are blocked by S3291 phosphorylation, a modification event that has little or no impact upon DMC1 binding. Thus, circumstances might exist that would favour DMC1, rather than RAD51, interaction with the BRCA2 C terminus. Our attempts to demonstrate phosphorylation of S3291 during meiotic recombination, which might favour interactions with DMC1 have so far proven to be unsuccessful, so the relevance of this potential binding switch upon phosphorylation is presently unknown. However, the lack of meiotic defects in mice harbouring deletions of the BRCA2 C terminus may well indicate that interactions between the BRCA2 C-terminal region and RAD51/DMC1 may not be so critical for the functions of BRCA2 during meiotic recombination (Atanassov et al, 2005).
The precise role played by BRCA2 in regulating homologous recombinational repair of DNA DSBs has yet to be fully defined. However, we are now beginning to understand the key role played by this protein in binding RAD51 and facilitating its relocalisation in somatic cells in response to DNA breaks. A similar situation exists in meiotic cells, as RAD51 and DMC1 colocalise to Spo11-induced DSBs during meiotic recombination (Bishop, 1994; Tarsounas et al, 1999). Our observations of a novel DMC1 interaction site at BRCA22386–2411, located away from the BRC repeats that mediate the primary interactions with RAD51, lead us to speculate that BRCA2 is capable of coordinately binding RAD51 and DMC1 to form a BRCA2-RAD51-DMC1 complex, which may facilitate the localisation of both recombinases to DNA break sites at the initiation of meiotic recombination. There, as in mitotic cells, it is likely that BRCA2 may participate in nucleoprotein filament formation leading to subsequent repair.
Materials and methods
Plasmids
The GST-tagged BRCA2 constructs B2-1 to B2-9 (Lee et al, 2004) and TR1–TR3 (Esashi et al, 2005) have been described previously. The GST-tagged BRCA2 constructs B2-6.1 to B2-6.5 were cloned into the SalI/NotI sites of pGEX-4T3 (Amersham) using PCR amplification. The mammalian expression vector pMyc-DMC1 was created by PCR amplification and subsequent insertion of the DMC1 sequence at the BamHI/SalI sites in the pCMV-Tag3 vector (Stratagene). The yeast two-hybrid plasmids were all cloned by PCR into the Gateway pDONR221 vector (Invitrogen) and subsequently transferred into pDEST22 (RAD51 and DMC1), or pDEST32 (all the BRCA2 constructs) (Invitrogen). All PCR-amplified sequences were verified by sequencing.
Proteins and peptides
Recombinant His10-DMC1 and RAD51 were prepared as described (Baumann et al, 1997; Masson et al, 1999). Peptides were made by solid-phase synthesis and purified by high-performance liquid chromatography, and their sequences were verified by mass spectroscopy. All peptides contained an N-terminal biotin group with an aminohexanoic spacer. GST-tagged BRCA2 constructs were expressed in E. coli strain BL21 RIL codon plus (Stratagene) by IPTG induction (0.1 mM) for 2 h at 18°C.
In vivo protein interactions
Human 293T cells were cultured in E4 medium supplemented with 10% FBS and antibiotics. Transfection with pMyc-DMC1 was performed using Fugene 6 (Roche) according to the manufacturer's instructions. Twenty-four hours post-transfection, cell were collected and lysed using a modified RIPA buffer (50 mM Tris–HCl, pH 7.5, 0.2% NP-40, 0.1% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 1 mM NaF, and Complete Protease Inhibitor Cocktail (Roche)). The lysates were added to 20 μl protein A magnetic beads (Invitrogen), which had been preincubated with 20 μg nonspecific rabbit IgG (Santa Cruz Biotechnology) or anti-BRCA2 rabbit polyclonal antibodies (Ab-2, Calbiochem) in RIPA buffer containing 2 mg/ml BSA. After incubation at 4°C, the beads were washed extensively, boiled in SDS loading buffer, and the precipitates analysed by Western blotting using either BRCA2 mouse monoclonal antibodies (Ab-1, Calbiochem), c-Myc mouse monoclonal antibodies (9E10, CRUK), or RAD51 monoclonal antibodies (14B4, Abcam).
In vitro GST pull-downs
E. coli cells expressing BRCA2 fragments were lysed in IP buffer (50 mM Tris–HCl, pH 7.5, 250 mM KCl, 5% glycerol, 0.5% NP-40, 2 mM EDTA, 1 mM DTT, 1 mM Pefabloc SC, Complete Protease Inhibitor Cocktail (Roche), and 25 U/ml Benzonase Nuclease). Saturating amounts of lysates were mixed with 15 μl glutathione Sepharose 4B beads (Amersham), and incubated with 200 ng DMC1 or RAD51 in IP buffer containing 2 mg/ml BSA. After incubation and extensive washing with IP buffer, the precipitates were analysed by Western blotting using HRP-conjugated GST mouse monoclonal antibodies (B14, Santa Cruz Biotechnology), 6 × His mouse monoclonal antibodies (Clontech) for detection of DMC1, and RAD51 monoclonal antibodies (14B4, Abcam). For DMC1/RAD51 competition, the procedure was as above, adding 200 ng DMC1 or RAD51 alone or in combination with the designated amounts of competitor.
In vitro peptide interaction studies
Biotinylated peptides (300 ng) were mixed with 6 μl magnetic Streptavidin Beads (Invitrogen) in IP buffer containing 2 mg/ml BSA and washed, before addition of 60 ng DMC1 or RAD51 in IP buffer containing 2 mg/ml BSA. After incubation and extensive washing with IP buffer, the precipitates were analysed by Western blotting using HRP-conjugated Streptavidin for detection of the biotinylated peptides (Abcam), 6 × His mouse monoclonal antibodies (Clontech) for detection of DMC1, and RAD51 monoclonal antibodies (14B4, Abcam). For DMC1/RAD51 competition experiments, the procedure was similar to that described except that the reactions contain 60 ng DMC1 alone or in combination with the designated amounts of RAD51.
Peptide arrays
For the peptide array studies, peptides were synthesised and spotted onto cellulose membranes. The membrane used in Figure 6C contained peptides, 30 amino acids in length, corresponding to BRCA22350–2417. The first peptide in this array corresponds to BRCA22350–2379, the next to BRCA22351–2380, the third to BRCA22352–2381, and so on. The membrane was activated in 50% methanol before blocking in 5% milk in TBS+0.1% Tween 20 (TBS-T). The membrane was then incubated with His-tagged DMC1 in 5% milk in TBS-T overnight, and bound DMC1 was detected by incubation with anti-His monoclonal antibodies and chemiluminescence. Subsequently, the membrane was stained with Ponceau, to reveal the exact location of each peptide. The mutant peptide array in Figure 6E was generated by synthesising 600 peptides corresponding to BRCA22382–2411, where each peptide sequentially had one of the amino acids substituted with each of the 20 common amino acids. The membrane was organised so that each of the 30 columns had the same amino acids in the 30-mer substituted. Each of the 20 rows on the membrane contains the identical amino-acid substitution.
Yeast two-hybrid assays
The ProQuest Two-Hybrid System (Invitrogen) was used for the analysis according to the manufacturer's instructions. DMC1 and RAD51 were fused to the GAL4 activation domain, while the BRCA2 fragments were fused to the GAL4 DNA-binding domain. After transforming the MaV203 strain with the different combinations of constructs by the lithium-acetate method, colonies were picked and grown in SD-medium minus Leu and Trp (Clontech) to stationary phase. The cells were then diluted and 10 000, 1000, or 100 cells, respectively, were plated onto selective plates, which were subsequently incubated at 30°C for 3 days. Interactions were assessed by analysing transcription of two different reporter genes, URA3 and HIS3, which resulted in grown on agar plates lacking uracil and histidine, respectively. Furthermore, on plates lacking histidine, two different concentrations of the HIS3 inhibitor 3-Amino-1,2,4-triazole (3-AT) was used, resulting in different stringency of these plates. Control experiments showed that growth was not observed on selective plates when the cells were transfected either with the GAL4 activation- or with DNA binding-domains.
Acknowledgments
We thank Nicola O'Reilly and Dhira Joshi for peptide synthesis, and our laboratory colleagues for suggestions and support. This work was supported by Cancer Research UK, the EU DNA Repair Consortium, the Breast Cancer Campaign, and the Jeantet Foundation. TT is supported by a post-doctoral fellowship from the Carlsberg Foundation (grant no 2005-1-216). FE is a recipient of a post-doctoral fellowship from the Human Frontiers Science Program and the Japanese Society for the promotion of Science.
References
- Atanassov BS, Barrett JC, Davis BJ (2005) Homozygous germ line mutation in exon 27 of murine BRCA2 disrupts the FANCD2-BRCA2 pathway in the homologous recombination-mediated DNA interstrand cross-links' repair but does not affect meiosis. Genes Chromosomes Cancer 44: 429–437 [DOI] [PubMed] [Google Scholar]
- Baumann P, Benson FE, Hajibagheri N, West SC (1997) Purification of human RAD51 protein by selective spermidine precipitation. Mut Res DNA Repair 384: 65–72 [DOI] [PubMed] [Google Scholar]
- Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R (1997) The BRC repeats are conserved in mammalian BRCA2 proteins. Human Mol Genet 6: 53–58 [DOI] [PubMed] [Google Scholar]
- Bishop DK (1994) RecA homologs DMC1 and RAD51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu LZ, Kleckner N (1992) DMC1: a meiosis-specific yeast homolog of E. coli RecA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69: 439–456 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Golub EI, Stasiak AZ, Stasiak A, Mazin AV (2005) Activation of human meiosis-specific recombinase DMC1 by Ca2+. J Biol Chem 280: 26886–26895 [DOI] [PubMed] [Google Scholar]
- Chen JJ, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R (1998a) Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328 [DOI] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen YM, Xiao J, Sharp ZD, Lee WH (1998b) The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci USA 95: 5287–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor F, Bertwistle D, Mee PJ, Ross GM, Swift S, Grigorieva E, Tybulewicz VLJ, Ashworth A (1997) Tumorigenesis and a DNA-repair defect in mice with a truncating BRCA2 mutation. Nat Genet 17: 423–430 [DOI] [PubMed] [Google Scholar]
- Dray E, Siaud N, Dubois E, Doutriaux MP (2006) Interaction between Arabidopsis BRCA2 and its partners RAD51, DMC1 and DSS1. Plant Physiol 140: 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC (2005) CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434: 598–604 [DOI] [PubMed] [Google Scholar]
- Kinebuchi T, Kagawa W, Enomoto R, Tanaka K, Miyagawa K, Shibata T, Kurumizaka H, Yokoyama S (2004) Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein DMC1. Mol Cell 14: 363–374 [DOI] [PubMed] [Google Scholar]
- Lee M, Daniels MJ, Venkitaraman AR (2004) Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene 23: 865–872 [DOI] [PubMed] [Google Scholar]
- Lo T, Pellegrini L, Venkitaraman AR, Blundell TL (2003) Sequence fingerprints in BRCA2 and RAD51: implications for DNA repair and cancer. DNA Repair 2: 1015–1028 [DOI] [PubMed] [Google Scholar]
- Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A (1997) Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of BRCA1, BRCA2, BRCA1/BRCA2, BRCA1/p53, and BRCA2/p53 null zygous embryos. Genes Dev 11: 1226–1241 [DOI] [PubMed] [Google Scholar]
- Masson J-Y, Davies AA, Hajibagheri N, Van Dyck E, Benson FE, Stasiak AZ, Stasiak A, West SC (1999) The meiosis-specific recombinase hDMC1 forms rings structures and interacts with hRAD51. EMBO J 18: 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta R, Lasalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW (1997) Rab22 and Rab163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci USA 94: 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–272 [DOI] [PubMed] [Google Scholar]
- Passy SI, Yu X, Li Z, Radding CM, Masson J-Y, West SC, Egelman EH (1999) Human DMC1 protein binds DNA as an octameric ring. Proc Natl Acad Sci USA 96: 10684–10688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee MY, Blundell TL, Venkitaraman AR (2002) Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature 420: 287–293 [DOI] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for DMC1, a germline-specific RecA homolog. Mol Cell 1: 697–705 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM (1997) Association of BRCA1 with RAD51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P (2004) Human meiotic recombinase DMC1 promotes ATP-dependent homologous DNA strand exchange. Nature 429: 433–437 [DOI] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim SS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A (1997) Embryonic lethality and radiation hypersensitivity mediated by RAD51 in mice lacking BRCA2. Nature 386: 804–810 [DOI] [PubMed] [Google Scholar]
- Sharan SK, Pyle A, Coppola V, Babus J, Swaminathan S, Benedict J, Swing D, Martin BK, Tessarollo L, Evans JP, Flaws JA, Handel MA (2004) BRCA2 deficiency in mice leads to meiotic impairment and infertility. Development 131: 131–142 [DOI] [PubMed] [Google Scholar]
- Siaud N, Dray E, Gy I, Gerard E, Takvorian N, Doutriaux MP (2004) BRCA2 is involved in meiosis in Arabidopsis thaliana as suggested by its interaction with DMC1. EMBO J 23: 1392–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Delapompa JL, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, Koo W, Billia P, Ho A, Fukumoto M, Hui CC, Mak TW (1997) BRCA2 is required for embryonic cellular proliferation in the mouse. Genes Dev 11: 1242–1252 [DOI] [PubMed] [Google Scholar]
- Takanami T, Mori A, Takahashi H, Higashitani A (2000) Hyper-resistance of meiotic cells to radiation due to a strong expression of a single recA-like gene in Caenorhabditis elegans. Nucl Acids Res 28: 4232–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsounas M, Morita T, Pearlman RE, Moens PB (1999) RAD51 and DMC1 form mixed complexes associated with mouse meiotic chromosome cores and synaptonemal complexes. J Cell Biol 147: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108: 171–182 [DOI] [PubMed] [Google Scholar]
- West SC (2003) Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445 [DOI] [PubMed] [Google Scholar]
- Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL (1997) RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J Biol Chem 272: 31941–31944 [DOI] [PubMed] [Google Scholar]
- Xia F, Taghian DG, DeFrank JS, Zeng ZC, Willers H, Iliakis G, Powell SN (2001) Deficiency of human BRCA2 leads to impaired homologous recombination but maintains normal nonhomologous end joining. Proc Natl Acad Sci USA 98: 8644–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T (1998) The mouse recA-like gene DMC1 is required for homologous chromosome synapsis during meiosis. Mol Cell 1: 707–718 [DOI] [PubMed] [Google Scholar]
- Yu VPCC, Köehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR (2000) Gross chromosomal rearrangements and genetic exchange between non-homologous chromosomes following BRCA2 inactivation. Genes Dev 14: 1400–1406 [PMC free article] [PubMed] [Google Scholar]
- Yuan S-SF, Lee S-Y, Chen G, Song M, Tomlinson GE, Lee EY (1999) BRCA2 is required for ionizing radiation-induced assembly of RAD51 complex in vivo. Cancer Res 59: 3547–3551 [PubMed] [Google Scholar]


