Abstract
The human cytomegalovirus UL82 gene encodes a protein (pp71) that is localized in the tegument domain of the virus particle. The UL82 gene product is delivered to the nucleus at the time of infection, and it is believed to function in gene activation. We have constructed a human cytomegalovirus mutant, ADsubUL82, that lacks a substantial portion of the UL82 coding region. It was propagated on human diploid fibroblasts expressing the UL82 gene product, and it was possible to produce a mutant virus lacking the UL82 protein by passaging virus stocks for one cycle of growth on normal, noncomplementing fibroblasts. The UL82-deficient mutant displays a multiplicity-dependent growth defect in normal human fibroblasts. The growth of ADsubUL82 is severely restricted at low input multiplicities (0.01–0.1 plaque-forming units per cell), producing a yield that is reduced by a factor of about 105 in comparison to wild-type virus. At higher input multiplicities (10 plaque-forming units per cell), ADsubUL82 grew nearly as well as the wild-type virus. By using a human cytomegalovirus gene array, we demonstrated that UL82 functions to facilitate virus mRNA accumulation very early during the human cytomegalovirus replication cycle. The growth phenotype associated with the UL82 mutant seems to result from its inability to efficiently activate human cytomegalovirus immediate early genes.
Human cytomegalovirus (HCMV) is a ubiquitous human pathogen. Although HCMV infection is usually asymptomatic in healthy individuals, HCMV infection can result in severe disease in immunocompromised individuals and newborn infants (1).
Like all herpesviruses, the HCMV virion contains a region between the capsid and lipid envelope called the tegument. The tegument of HCMV consists of ≈20–25 virally encoded proteins (2–5). On fusion of the HCMV virion with the host cell membrane, the constituents of the tegument are delivered to the infected cell. The UL82 gene of HCMV encodes one of the tegument proteins, which is termed the UL82 protein or pp71. The UL82 protein is a phosphoprotein that localizes to the nucleus immediately after infection (6–8). Transfection experiments have demonstrated that the UL82 protein can enhance the infectivity of HCMV DNA (9) and transactivate a number of viral promoters either alone or in conjunction with other viral proteins (10–14).
To elucidate the function of UL82 during HCMV replication, we have characterized a UL82 deletion mutant, ADsubUL82. UL82 is essential for HCMV replication when human fibrobasts are infected at a relatively low input multiplicity. However, at higher input multiplicities, the defect is substantially overcome, and the mutant replicates to near wild-type (wt) levels. By using an HCMV gene array, we demonstrate that the growth defect associated with ADsubUL82 results at least in part from the inability of the mutant to activate its immediate-early (IE) genes efficiently.
Materials and Methods
Cells and Viruses.
Human diploid foreskin fibroblasts at passage 11–16 (each passage is a 1:4 split of a recently confluent monolayer) and WF28-71-HA cells (15) were cultured in DMEM with 10% (vol/vol) FCS/100 units/ml penicillin/100 μg/ml streptomycin in a 37°C incubator with a 5% CO2 atmosphere. WF28-71-HA cells were derived from human fibroblasts with a lifespan that was extended (15) by the introduction of the catalytic subunit of telomerase. These cells contain the HCMV UL82 gene controlled by the HCMV late UL99 promoter (15). Consequently, the promoter is activated, and UL82 is expressed most abundantly during the late phase of the viral growth cycle.
The generation of the recombinant virus ADsubUL82 has been described (15). It is derived from the AD169 strain (16) of HCMV, and it contains a substitution mutation comprised of a deletion spanning nucleotides 117,648–119,185 of the viral genome plus an inserted marker gene cassette. The marker cassette (17) is expressed under control of the simian virus 40 early promoter and contains the enhanced green fluorescent protein coding region followed by an internal ribosomal entry site followed by the puromycin-resistance gene coding region. The substitution's insertion point and its flanking regions in ADsubUL82 (nucleotides 116,631–120,311) were sequenced to ensure that no additional mutations were introduced into neighboring genes during the construction of the mutant. A revertant virus, designated ADrevUL82, was generated by homologous recombination with infectious ADsubUL82 DNA and a construct containing the UL82 coding region and flanking regions corresponding to nucleotides 116,631 and 120,311 of the AD169 genome. To distinguish ADrevUL82 from wt AD169, a mutation within the UL82 coding region at nucleotide 119,133 was introduced that eliminated a PshAI cleavage site but did not alter the UL82 amino acid sequence. ADrevUL82 was isolated by three rounds of plaque purification on noncomplementing human fibroblasts. Revertant and wt virus stocks were prepared as described (17, 18). ADsubUL82 stocks were generated by infecting WF28-71-HA cells that express UL82 (15). ADsubUL82 virus propagated on WF28-71-HA cells contains UL82 protein within the tegument, and the virus is designated ADsubUL82+UL82. ADsubUL82 propagated on normal, noncomplementing human fibroblasts for one passage is devoid of UL82 within the tegument and is designated ADsubUL82−UL82. In some experiments, partially purified virus (2, 18) was used. The infectious titers of all virus stocks were determined by plaque assay on WF28-71-HA cells.
Analysis of Gene Expression.
HCMV gene arrays on membranes have been described (19). To analyze mRNA levels with a viral gene array, human fibroblasts were infected and harvested as specified for different experiments. For experiments in which cells were treated with cycloheximide, mRNA was isolated from total RNA with an Oligotex mRNA isolation kit (Qiagen, Chatsworth, CA), and 250 ng of mRNA was subjected to reverse transcription by using oligo(dT) and 3′ gene specific primers (19) in the presence of [32P]dCTP. RNA was degraded; unincorporated nucleotides were removed; and the labeled cDNA was then hybridized to the membrane-bound array. Total RNA (15 μg) was used for the reverse transcription reaction in all experiments that did not employ a drug block.
Southern, Northern, and Western blot analyses were performed as described (17, 18, 20).
Results
Propagation of ADsubUL82.
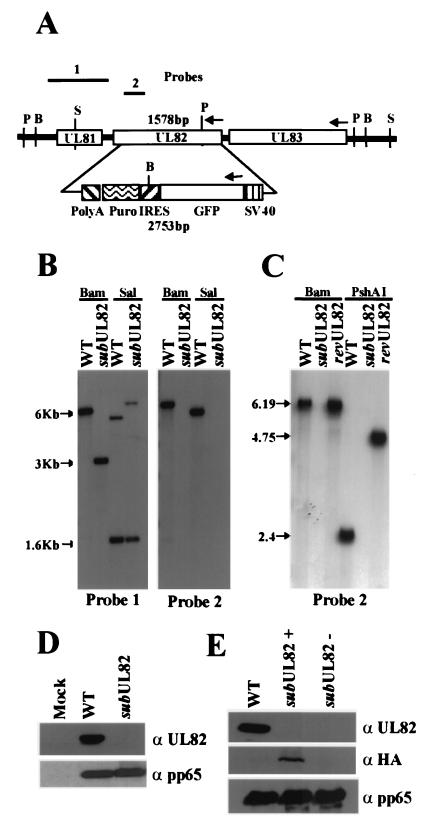
A substitution mutant containing a marker cassette encoding the enhanced green fluorescent protein and puromycin resistance in place of the UL82 coding region was produced by homologous recombination within human fibroblasts (Fig. 1A) as reported (15). The substitution mutant, termed ADsubUL82, is defective for growth and therefore was propagated and purified on a complementing cell line, called WF28-71-HA, which expresses UL82 protein (15). Southern blot analysis of the mutant viral DNA digested with BamHI or SalI confirmed that ADsubUL82 lacks the UL82 coding region and that the marker cassette had recombined properly within the viral genome (Fig. 1B). A revertant virus, termed ADrevUL82, was generated by using infectious viral DNA isolated from ADsubUL82 and a construct containing the nucleotide sequences 116,631–120,311 from the AD169 genome. Southern blot analysis of ADrevUL82 DNA digested with BamHI or PshAI revealed that ADrevUL82 was identical to wt AD169 except for a single point mutation at nucleotide 119,133 within the PshAI site that was introduced to tag the revertant (Fig. 1C). To confirm that we had not introduced a secondary mutation within the flanking regions of ADsubUL82 or ADrevUL82, the flanking sequences corresponding to nucleotides 116,631–117,649 and 119,184–120,311 were sequenced together with the wt virus. The flanking sequences were identical in the three viruses (data not shown).
Figure 1.
Characteristics of ADsubUL82. (A) Schematic representation of the UL81–UL83 region of the HCMV genome, the green fluorescent protein (GFP)/internal ribosomal entry site (IRES)/puromycin (Puro) cassette controlled by the simian virus 40 promoter and poly(A) site substituted for the UL82 ORF, and the probes used to characterize the mutant virus. BamHI (B), SalI (S), and PshAI (P) restriction sites are shown. (B) Southern blot analysis of wt AD169 (WT) and ADsubUL82 (subUL82) viral DNA. Viral DNA was digested with either BamHI or SalI, separated by electrophoresis, transferred to nitrocellulose, and analyzed with 32P-labeled probe 1 or probe 2. (C) Southern blot analysis of wt, ADsubUL82, and ADrevUL82 viral DNA. Viral DNA was digested with either BamHI or PshAI and analyzed with 32P-labeled probe 2, which encompasses a portion of the UL82 ORF. (D) Western blot analysis of UL82 (α UL82) expression at 120 h after infection of human fibroblasts with wt or ADsubUL82. As a control, expression of pp65 (α pp65) was also measured. (E) Detection of UL82 protein packaged within virus particles. Equal amounts of partially purified wt, ADsubUL82+UL82, or ADsubUL82−UL82 virus particles were assayed for the presence of UL82 protein with either a UL82-specific antibody (α UL82) or a hemagglutinin-specific (α HA) antibody directed against an epitope contained within the UL82 protein expressed from WF28-71-HA cells. Incorporation of the tegument protein pp65 (α pp65) was also measured.
To confirm that ADsubUL82 was unable to express UL82 protein, human fibroblasts were mock-infected or infected with wt or ADsubUL82 virus at a multiplicity of 1.0 plaque-forming unit (pfu) per cell. Cell lysates were harvested 120 h after infection and assayed by Western blot for UL82 expression (Fig. 1D). UL82 was abundantly expressed in cells infected with wt virus, whereas it was absent from cells infected with ADsubUL82. Another tegument protein, UL83 (pp65), was expressed at similar levels in cells infected with either wt or ADsubUL82. It is possible to generate two types of ADsubUL82 viral stocks. Virus cultured on WF28-71-HA cells can package into virions UL82 protein expressed from the cell line, giving rise to a stock of virus lacking the coding region but containing UL82 protein packaged within the tegument of the virus particle. We have designated this type of viral stock ADsubUL82+UL82. A virus stock that is devoid of both the UL82 coding region and protein was produced by infecting normal, noncomplementing human fibroblasts with ADsubUL82+UL82 virus at a high input multiplicity. This type of virus stock is designated ADsubUL82−UL82. To confirm that UL82 was packaged within virions cultured on complementing cells and was absent in virions grown on normal human fibroblasts, equal amounts of wt and mutant virus particles were purified and assayed by Western blotting for the presence of UL82 (Fig. 1E). By using an antibody directed against UL82, we could detect the protein packaged within wt virus particles, but we were unable to detect UL82 packaged within ADsubUL82+UL82 or ADsubUL82−UL82 particles. However, by using a more sensitive antibody directed at the hemagglutinin epitope that is fused to the UL82 protein expressed in the complementing cells (15), we could detect UL82 packaged within ADsubUL82+UL82 virions but, as expected, not within ADsubUL82-UL82 particles. In a control experiment, the three types of virus particles were found to contain equal amounts of the tegument protein pp65. These results demonstrate that a small quantity of UL82 protein is packaged within ADsubUL82+UL82 particles, and none of the protein is detected in ADsubUL82−UL82 stocks.
Multiplicity-Dependent Growth of ADsubUL82 on Noncomplementing Cells.
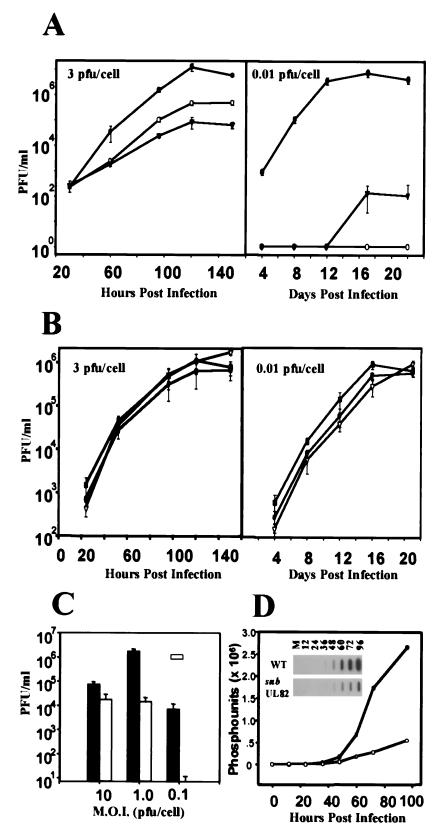
The growth kinetics of the ADsubUL82 mutant on noncomplementing human fibroblasts was examined at two multiplicities of infection (Fig. 2A). At a multiplicity of 3 pfu per cell, the yield of ADsubUL82−UL82 was reduced by a factor of 100 compared with that of the wt virus, whereas the growth of ADsubUL82+UL82 was reduced by a factor of 10. We interpret these results to indicate that the UL82 protein packaged within the virion of the ADsubUL82+UL82 virus is functional and provides a growth advantage over the ADsubUL82−UL82 virus. The residual defect observed for ADsubUL82+UL82 might result from the reduced amount of UL82 protein in virions, or it could reflect a second, later function for the gene product that requires its expression from the infecting genome. The growth defect observed at a multiplicity of 0.01 pfu per cell is much more profound. Both ADsubUL82+UL82 and ADsubUL82−UL82 produced a >105-fold reduced yield in comparison to the wt. The growth defect at a low multiplicity can be overcome if either mutant virus is cultured on complementing WF28-71-HA cells (15). These results indicate that UL82 is required for efficient virus replication when human fibroblasts are infected at a low input multiplicity.
Figure 2.
Growth kinetics of wt and mutant viruses. (A) Human fibroblasts were infected (3 or 0.01 pfu per cell) with wt (●), ADsubUL82+UL82 (○), or ADsubUL82−UL82 (▾) virus. Cultures were harvested at the indicated times after infection, and infectious virus was quantified by plaque assay on WF28-71-HA cells. (B) Human fibroblasts were infected (3 or 0.01 pfu per cell) with wt (●), ADrevUL82#1 (■), or ADrevUL82#2 (▿) virus. Viruses designated 1 and 2 are two independent isolates of the revertant virus. Cultures were harvested at the indicated times after infection, and virus was quantified by plaque assay on human fibroblasts. (C) Human fibroblasts were infected (10, 1.0, or 0.1 pfu per cell) with wt (black bars) or ADsubUL82−UL82 (open bars) virus. Cultures were harvested at 144 h after infection, and virus was quantified by plaque assay on WF28-71-HA cells. M.O.I., multiplicity of infection. (D) Accumulation of wt and ADsubUL82 viral DNA in human fibroblasts infected at a multiplicity of 2 pfu per cell. DNA was isolated at the indicated times after infection, and viral DNA was quantified by slot blot analysis with an HCMV-specific probe.
To demonstrate that the growth phenotype associated with ADsubUL82 was a direct consequence of deleting the UL82 gene and not the result of a secondary mutation somewhere else in the genome, the growth of the UL82 revertant virus, ADrevUL82, was examined (Fig. 2B). Two independent isolates were used to infect human fibroblasts at either a multiplicity of 3 pfu per cell or 0.01 pfu per cell. Both revertant isolates grew like wt virus, demonstrating that the growth defect of ADsubUL82 is a direct result of the UL82 deletion and not a secondary mutation elsewhere in the genome.
The results shown in Fig. 2A indicate that the growth of ADsubUL82−UL82 on noncomplementing cells is multiplicity dependent. To examine this dependence in greater detail, human fibroblasts were infected at a multiplicity of 10, 1.0, or 0.1 pfu per cell with either wt or ADsubUL82−UL82 virus (Fig. 2C). Virus was harvested at 144 h after infection, and yields were determined by plaque assay on WF28-71-HA cells. When used to infect cells at a multiplicity of 10 pfu per cell, the mutant showed only a 5-fold decrease in viral titers compared with wt. However, at a multiplicity of 0.1 pfu per cell, the mutant exhibited a 104-fold decrease compared with wt virus, confirming the multiplicity-dependent growth of ADsubUL82−UL82 on noncomplementing cells.
UL82 Is Required for Efficient Expression of HCMV IE Genes.
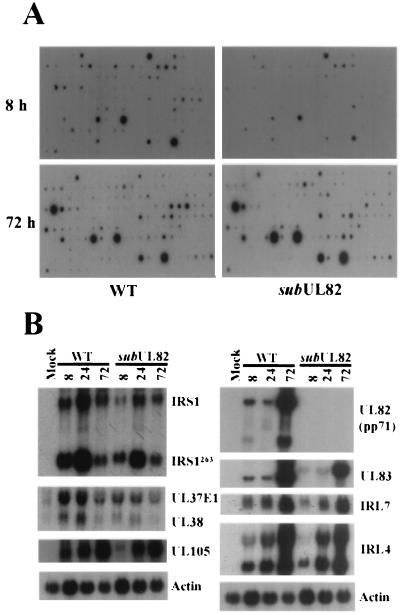
Viral DNA accumulation was monitored by slot blot analysis after infection of human fibroblasts with ADsubUL82−UL82 or wt virus at a multiplicity of 2 pfu per cell (Fig. 2D). A substantial decrease in viral DNA accumulation was observed in cells infected with the mutant virus as compared with wt virus, suggesting that UL82 functions before or during viral DNA synthesis. To search for an effect of the mutation on mRNA accumulation, which could in turn cause the defect in DNA accumulation, a DNA array analysis was performed. RNA was prepared from cells infected at a multiplicity of 2 pfu per cell with mutant or wt virus, and 32P-labeled cDNAs were prepared. These cDNAs were then hybridized to membrane-bound arrays of viral DNA fragments corresponding to HCMV ORFs ≥300 bp in size (Fig. 3A). At 8 h after infection, many mRNAs had accumulated to a reduced level in cells infected with ADsubUL82−UL82 as compared with cells infected with wt virus. By 72 h, there were many fewer differences in the transcriptional profiles of the two viruses. Northern blot assays for a variety of HCMV genes confirmed the deficiency in mRNA accumulation for the mutant at 8 h but not at 72 h (Fig. 3B). Taken together, the DNA and mRNA accumulation experiments suggest that a defect in cells infected with ADsubUL82−UL82 occurs very early in the replication cycle.
Figure 3.
HCMV mRNA accumulation after infection with wt or mutant viruses at a relatively high multiplicity. (A) Human fibroblasts were infected with wt or ADsubUL82−UL82 virus (2 pfu per cell). RNA was isolated 8 and 72 h after infection and reverse transcribed in the presence of [32P]dCTP. Quantities of 32P-labeled cDNA copied from RNA that was prepared from the same number of cells infected with wt or mutant virus was used as probe. The arrays contained an actin control DNA that was visible after longer exposure times than are displayed here. The intensity of the actin spot was similar for each preparation of probe. (B) Human fibroblasts were infected with wt or ADsubUL82−UL82 virus (2 pfu per cell). Total RNA was isolated at 8, 24, and 72 h after infection and assayed for HCMV gene expression by Northern blotting with 32P-labeled gene-specific probes. Actin was included as an internal loading control.
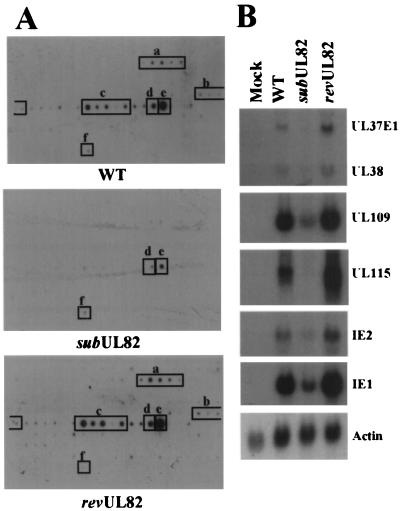
To test whether UL82 protein delivered to the host cell as a constituent of the virion tegument is involved in the activation of HCMV IE genes, a gene array experiment was performed to monitor mRNA accumulation in cycloheximide-treated cells infected with wt virus, ADsubUL82-UL82, or ADrevUL82 (Fig. 4). In the presence of the drug, no proteins are produced from the infecting viral genome, and only the IE class of viral genes are transcribed by wt virus (21, 22). Cells were harvested 8 h after infection at a multiplicity of 1 pfu per cell; 32P-labeled cDNAs were prepared from polyadenylated mRNAs and used to probe viral gene arrays (Fig. 4A). HCMV IE gene expression was greatly decreased in cells infected with ADsubUL82 relative to the wt or revertant viruses. The gene array results were confirmed by using Northern blot analysis to compare the expression of several IE genes in the presence of cycloheximide (Fig. 4B). A number of IE genes involved in transactivation of other viral promoters (IE1, IE2, UL37, and UL38) were dramatically decreased in mutant-virus-infected cells. IE gene expression was nearly identical in cells infected with wt or ADrevUL82 virus, which demonstrates that the transcriptional defect observed with ADsubUL82 is a direct result of the UL82 mutation (Fig. 4 A and B).
Figure 4.
HCMV IE mRNA accumulation after infection with wt or mutant viruses in the presence of cycloheximide. (A) Human fibroblasts were infected with wt, ADsubUL82−UL82, or ADrevUL82 virus (1 pfu per cell) in the presence of cycloheximide (100 μg/ml). At 8 h after infection, mRNA was isolated and reverse transcribed in the presence of [32P]dCTP. The labeled cDNA was then used to probe HCMV gene arrays on membranes. Transcripts expressed after infection with the wt or revertant virus correspond to the following open reading frames: UL36–UL38, a; UL106–UL109, b; UL115–UL119, c; UL122, d; UL123, e; and actin, f. Actin was used to confirm that equal quantities of each sample were analyzed, as described in the legend for Fig. 3A. (B) Human fibroblasts were infected with wt, ADsubUL82−UL82, or ADrevUL82 virus (1 pfu per cell) in the presence of cycloheximide (100 μg/ml). Total RNA was isolated at 8 h after infection and assayed for HCMV IE gene expression by Northern blotting with 32P-labeled gene-specific probes.
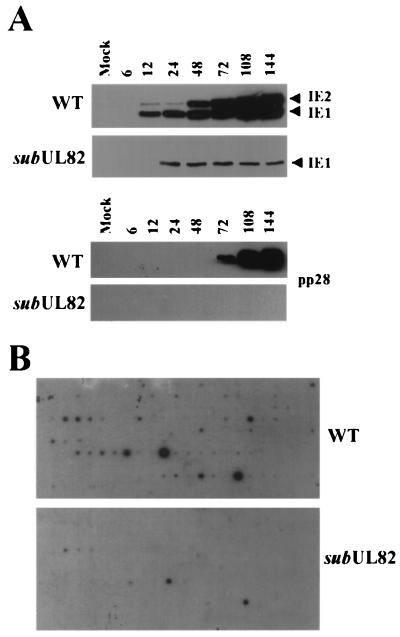
If UL82 helps to activate HCMV IE mRNA accumulation, one would predict that there would be a marked difference in the expression of this class of genes in cells infected with wt virus as compared with mutant viruses at a low multiplicity in the absence of cycloheximide. Accordingly, the abundance of the IE1 and IE2 proteins was examined in human fibroblasts infected with either wt or ADsubUL82−UL82 at a multiplicity of 0.1 pfu per cell. As shown in Fig. 5A, there is a dramatic decrease in the accumulation of IE1 and IE2 proteins in cells infected with the mutant as compared with those infected with the wt virus. Although detectable on very long exposures, IE2 protein was expressed at severely reduced levels in cells infected with ADsubUL82. Expression of the true late protein UL99 (pp28) was completely absent in cells infected with ADsubUL82−UL82 virus, which suggests that the mutant did not induce viral DNA synthesis or expression of late viral transcripts.
Figure 5.
HCMV mRNA accumulation after infection with wt or mutant viruses at a relatively low multiplicity. (A) Human fibroblasts were infected with wt or ADsubUL82−UL82 virus (0.1 pfu per cell). Cell lysates were prepared at the indicated times after infection and assayed for IE1, IE2, and pp28 protein expression by Western blotting. (B) Human fibroblasts were infected with wt or ADsubUL82−UL82 virus at a multiplicity of 0.1 pfu per cell. At 144 h after infection, RNA was isolated and reverse transcribed in the presence of [32P]dCTP. The labeled cDNA was then used to probe HCMV gene arrays. Actin was used to confirm that equal quantities of each sample were analyzed, as described in the legend for Fig. 3A.
The IE1 and IE2 proteins are potent transcriptional activators. Given the defect in their accumulation, one would anticipate that viral mRNA accumulation would be severely depressed after infection with ADsubUL82−UL82 as compared with its wt parent at a multiplicity of 0.1 pfu per cell. Gene array analysis confirmed this prediction, revealing a global transcriptional defect at 144 h after infection with the mutant virus (Fig. 5B). This result is consistent with the markedly reduced yield of infectious virus observed at 144 h after infection with ADsubUL82−UL82 at this input multiplicity (Fig. 2C), and it reinforces the conclusion that virion-associated UL82 protein facilitates the activation of IE genes whose products then induce the expression of later classes of viral genes.
Discussion
We describe here the phenotype associated with a substitution mutation (Fig. 1) within the UL82 gene of HCMV. The mutant exhibits a multiplicity-dependent growth defect in cultured human fibroblasts (Fig. 2). It replicates nearly to wt levels when used to infect cells at a relatively high multiplicity (10 pfu per cell), but it is severely growth restricted at low multiplicities of infection (<0.1 pfu per cell). A similar multiplicity-dependent growth phenotype has been observed for an independently generated UL82-null virus (M. J. Bentham and R. F. Greaves, personal communication). By using gene arrays, we demonstrate that UL82 is required for the efficient accumulation of HCMV IE mRNAs (Figs. 3–5). This observation is consistent with previous work demonstrating that UL82 can activate several HCMV promoters that control expression of reporter genes. The activation potential of UL82 has been demonstrated in transfection assays where the protein can act either alone or in conjunction with other viral proteins (10–12, 14) and in infected cells where UL82 is expressed from a recombinant herpes simplex virus genome (13). It might also directly facilitate expression of HCMV genes in later kinetic classes; however, it is not possible to be certain, because these genes depend on the prior expression of IE genes that are not properly induced in the absence of UL82.
Two observations argue that UL82 protein present in the virus particle, rather than newly synthesized protein, acts to facilitate IE gene expression and viral replication. First, in contrast to wt virus, ADsubUL82−UL82 fails to efficiently activate IE genes in the presence of cycloheximide, which blocks expression of proteins from the virus genome (Fig. 4). UL82 protein delivered by virions to the infected cell must contribute to the activation observed for wt virus in the presence of the drug. Second, growth of the UL82 mutant in complementing WF28-71-HA cells generates a virus stock (ADsubUL82+UL82) with a small amount of UL82 protein in virions, whereas propagation of the mutant for a single cycle in normal fibroblasts infected at a high input multiplicity generates a virus stock (ADsubUL82−UL82) lacking the protein (Fig. 1E). The mutant particles lacking the protein are more crippled for growth than particles containing a small quantity of the protein (Fig. 2A left). Consequently, UL82 protein in the virion must enhance replication of the virus.
One of the promoters that UL82 has been shown to activate in transfection assays is the major IE promoter, which controls expression of two major viral transcriptional regulatory proteins, IE1 and IE2 (12, 13). These IE proteins regulate expression from their own promoter as well as the expression of early and late viral genes (23–26). Interestingly, a mutant carrying a deletion within the IE1 coding region exhibits a similar multiplicity-dependent phenotype as that observed with the UL82 mutant (27). As has been shown for the UL82 protein in transfection assays (12), cells infected with herpes simplex virus (13), and cells infected with ADsubUL82−UL82 (Figs. 4 and 5), the IE1 protein enhances expression from the major IE promoter, the promoter that is responsible for its own synthesis (24, 27–29). Perhaps UL82 and IE1 are both required, when cells are infected at a relatively low multiplicity, to activate the major IE promoter and allow sufficient accumulation of the IE1 and IE2 proteins. However, at higher input multiplicities with the resulting higher promoter copy number, UL82 or IE1 alone might sufficiently activate expression from the major IE promoter. Indeed, high multiplicity infection of cells with either an IE1 mutant (27) or the UL82 mutant (data not shown) results in near wt levels of IE2 protein expression, which plays a central role in the activation of later kinetic classes of viral genes.
It is unlikely that the phenotype associated with the UL82 mutant is due to only its inability to activate the major IE promoter. We have shown that cotransfection with a plasmid expressing UL82 significantly enhances the infectivity of HCMV DNA, whereas cotransfection with plasmids expressing IE1 and IE2 has only a marginal effect on infectivity (9). This work predicted that UL82 does something to enhance the infectivity of viral DNA in addition to activating the major IE promoter and allowing expression of the IE1 and IE2 proteins. Here, we show that UL82 not only regulates IE1 and IE2 expression but also facilitates mRNA accumulation from other IE genes, including UL37exonI, UL38, UL106–UL109, and UL115–UL119 (Fig. 4). Several of these proteins have been shown to function as transcription factors (30–33). UL37 and UL38 have also been shown to be required for ori-Lyt HCMV DNA replication in a transient transfection assay (34–36), and UL37exon1 also functions as an antiapoptotic protein (37). Presumably, UL82-mediated activation of the major IE promoter in conjunction with the activation of additional IE genes is required for efficient HCMV replication.
Acknowledgments
We thank F. Ferrari for constructing the shuttle vector plasmid used in the production of ADsubUL82 and L. Enquist for thoughtful comments on the manuscript. This work was supported by National Cancer Institute Grant CA82396. W.A.B. received support from National Institute of Allergy and Infectious Disease Fellowship AI10448.
Abbreviations
- HCMV
human cytomegalovirus
- wt
wild type
- pfu
plaque-forming unit
References
- 1.Alford C A, Britt W J. In: Fields Virology. Fields B N, Knipe D M, Howely P M, editors. New York: Lippincott-Raven; 1995. pp. 2493–2543. [Google Scholar]
- 2.Baldick C J, Jr, Shenk T. J Virol. 1996;70:6097–6105. doi: 10.1128/jvi.70.9.6097-6105.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gibson W. Virology. 1981;111:516–537. doi: 10.1016/0042-6822(81)90354-8. [DOI] [PubMed] [Google Scholar]
- 4.Gibson W. Virology. 1983;128:391–406. doi: 10.1016/0042-6822(83)90265-9. [DOI] [PubMed] [Google Scholar]
- 5.Spaete R R, Gehrz R C, Landini M P. J Gen Virol. 1994;75:3287–3308. doi: 10.1099/0022-1317-75-12-3287. [DOI] [PubMed] [Google Scholar]
- 6.Hensel G M, Meyer H H, Buchmann I, Pommerehne D, Schmolke S, Plachter B, Radsak K, Kern H F. J Gen Virol. 1996;77:3087–3097. doi: 10.1099/0022-1317-77-12-3087. [DOI] [PubMed] [Google Scholar]
- 7.Roby C, Gibson W. J Virol. 1986;59:714–727. doi: 10.1128/jvi.59.3.714-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruger B, Klages S, Walla B, Albrecht J, Fleckenstein B, Tomlinson P, Barrell B. J Virol. 1987;61:446–453. doi: 10.1128/jvi.61.2.446-453.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldick C J, Jr, Marchini A, Patterson C E, Shenk T. J Virol. 1997;71:4400–4408. doi: 10.1128/jvi.71.6.4400-4408.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler M, Schmolke S, Plachter B, Stamminger T. Scand J Infect Dis Suppl. 1995;99:8–9. [Google Scholar]
- 11.Winkler M, Rice S A, Stamminger T. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu B, Stinski M F. J Virol. 1992;66:4434–4444. doi: 10.1128/jvi.66.7.4434-4444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homer E G, Rinaldi A, Nicholl M J, Preston C M. J Virol. 1999;73:8512–8518. doi: 10.1128/jvi.73.10.8512-8518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chau N H, Vanson C D, Kerry J A. J Virol. 1999;73:863–870. doi: 10.1128/jvi.73.2.863-870.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bresnahan W A, Shenk T. J Virol. 2000;74:10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowe W P, Hartely J W, Waterman S, Turner H C, Huebner R J. Proc Soc Exp Biol Med. 1956;92:418–424. [PubMed] [Google Scholar]
- 17.Patterson C E, Shenk T. J Virol. 1999;73:7126–7131. doi: 10.1128/jvi.73.9.7126-7131.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch A J, Shenk T. J Virol. 1999;73:404–410. doi: 10.1128/jvi.73.1.404-410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bresnahan W A, Shenk T. Science. 2000;288:2373–2376. doi: 10.1126/science.288.5475.2373. [DOI] [PubMed] [Google Scholar]
- 20.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 21.Wathen M W, Stinski M F. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wathen M W, Thomsen D R, Stinski M F. J Virol. 1981;38:446–459. doi: 10.1128/jvi.38.2.446-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spector D H. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 24.Stenberg R M, Stinski M F. J Virol. 1985;56:676–682. doi: 10.1128/jvi.56.3.676-682.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stenberg R M, Depto A S, Fortney J, Nelson J A. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greaves R F, Mocarski E S. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski E S, Kemble G W, Lyle J M, Greaves R F. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambucetti L C, Cherrington J M, Wilkinson G W, Mocarski E S. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherrington J M, Mocarski E S. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colberg-Poley A M, Huang L, Soltero V E, Iskenderian A C, Schumacher R F, Anders D G. Virology. 1998;246:400–408. doi: 10.1006/viro.1998.9212. [DOI] [PubMed] [Google Scholar]
- 32.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang H, al-Barazi H O, Colberg-Poley A M. Virology. 1996;223:292–302. doi: 10.1006/viro.1996.0481. [DOI] [PubMed] [Google Scholar]
- 34.Pari G S, Anders D G. J Virol. 1993;67:6979–6988. doi: 10.1128/jvi.67.12.6979-6988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pari G S, Kacica M A, Anders D G. J Virol. 1993;67:2575–2582. doi: 10.1128/jvi.67.5.2575-2582.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J A, Pari G S. J Virol. 1995;69:1925–1931. doi: 10.1128/jvi.69.3.1925-1931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldmacher V S, Bartle L M, Skaletskaya A, Dionne C A, Kedersha N L, Vater C A, Han J, Lutz R J, Watanabe S, McFarland E D, et al. Proc Natl Acad Sci USA. 1999;96:12536–12541. doi: 10.1073/pnas.96.22.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]