SUMMARY
The plasmid R1162 encodes proteins that enable its conjugative mobilization between bacterial cells. It can transfer between many different species and is one of the most promiscuous of the mobilizable plasmids. The plasmid-encoded protein MobA, which has both nicking and priming activities on single-stranded DNA, is essential for mobilization. The nicking, or relaxase, activity has been localized to the 186 residue N-terminal domain, called minMobA. We present here the 2.1 Å X-ray structure of minMobA. The fold is similar to that seen for two other relaxases, TraI and TrwC. The similarity in fold, and action, suggests these enzymes are evolutionary homologs, despite the lack of any significant amino acid similarity. MinMobA has a well- defined target DNA called oriT. The active site metal is observed near Tyr 25, which is known to form a phosphotyrosine adduct with the substrate. A model of the oriT substrate complexed with minMobA has been made, based on observed substrate binding to TrwC and TraI. The model is consistent with observations of substrate base specificity, and provides a rationalization for elements of the likely enzyme mechanism.
Keywords: MobA, X-ray structure, DNA binding, drug resistance, relaxases
Resistance to antibiotics by bacterial pathogens is a major health concern throughout the world. Bacteria acquire resistance to antibiotics most commonly by the conjugative transfer of plasmids and transposable elements. There are two classes of plasmids involved in conjugative transfer. One of these consists of large (greater than 30 Kb) self-transmissible elements 1 and the other smaller (~5-10 Kb) mobilizable plasmids 2 3. Both groups encode the proteins required to process the DNA for transfer, but only the self-transmissible plasmids encode as well the complex, type IV secretion system (T4SS) required to transport the DNA into a new cell. The mobilizable plasmids use a T4SS that is generally provided by a co-resident, self-transmissible plasmid 4, but can also take advantage of type IV systems involved in the transport of effector proteins active in pathogenesis 5. An important characteristic of T4SS is that it can transport both protein and DNA across species barriers 6, 7 and even into cells of different kingdoms 8; 9. In addition, some groups of plasmids are maintained in a large number of different bacterial species 10. These two characteristics have together resulted in the dramatic spread of antibiotic resistance among a wide range of microorganisms.
Among the best studied mobilizable plasmids are the nearly identical R1162 and RSF1010, which have been independently isolated from Pseudomonas aeruginosa and E. coli, respectively 11; 12. Along with R300b, from Salmonella enterica 11, these IncQ plasmids are probably all the same element, widely disseminated because of its broad host-range. These plasmids encode resistance to the antibiotics streptomycin (strA and strB) and sulfonamide (sulII). They also contain an origin of transfer (oriT), and three genes, mobA, B, and C, which encode proteins for conjugative mobilization. The Mob proteins assemble at oriT 13 to form a complex called the relaxosome. One of these proteins, MobA (78 kDa, 709 residues), has three domains and two known enzymatic activities. The largest of these domains (43 kDa), is a DNA primase located at the C-terminal end. The N-terminal domain is a “relaxase” or “nickase” that locally unwinds DNA and catalyzes cleavage of one of the DNA strands within oriT 14; 15. The cleavage site in oriT has been identified 13; 16, and the Tyr 25 residue of MobA has been shown to be essential for activity 17. This residue carries out a nucleophilic attack on the GC phosphodiester bond at the nick site (nic), forming a covalent adduct to the 5’ phosphate of the C and releasing the 3’ hydroxyl of the G residue. The transesterification is readily reversible, and appears to take place in the cell as an idling reaction within the relaxosome 13. During transfer, the cleaved strand is unwound from its complement and transferred, in the 5’ to 3’ direction 18, through the intercellular pore formed by the T4SS. Circular plasmid DNA is then re-formed in the target cell, presumably because the covalently-bound MobA catalyzes the rejoining of the two ends of the strand by the reverse transesterification reaction, with release of the free protein.
Ilyina and Koonin 19 used sequence analysis to categorize two large families of DNA strand-transferases, or nicking enzymes: rolling circle replicases (RCR or Rep) and mobilization proteins (Mob). Both families contain two signature sequences. One sequence is an HUH motif (His-hydrophobic residue-His) which is thought to bind active site metals. This motif was expanded to include a third histidine in the Mob family (typically HxDxxxxHUH). The other signature includes one or two Tyr residues (frequently YXXXY in the RCR family and NXY in the Mob family). Each of these conserved tyrosines may form a phosphotyrosine ester with DNA. In the RCR family, the two tyrosines may participate alternately as the rolling circle synthesis proceeds 20.
The RCR family differs from the Mob family in that secondary structural elements appeared to be circularly permuted so that the ordering of the two signature motifs is reversed 19. This proved to be true when X-ray structures became available. The first super-family member solved was the catalytic domain of the RCR from adeno-associated virus 21. Next the relaxase from the self-transmissible F factor plasmid, TraI, was solved 22. The relaxase had the same overall structural features as the replicase although the order of these was permuted as predicted. For example, in the central β sheet the strand order for the viral replicase was 41325 and was 13542 in the relaxase 23.
Structural data for relaxase enzymes is presently rather modest. In addition to the native form of TraI 22, the structure of an active site variant in complex with a 10 base oligonucleotide has been solved 24. The structure of the 293-residue relaxase TrwC from self-transmissible IncW family plasmid R388 has also been solved as a complex with DNA 25; 26. TrwC is a clear homologue of TraI with 42% sequence identity; the corresponding Cα atoms of the two structures superimpose with an rms value of 1.3 Å. Although MobA, TraI, and TrwC contain the signature HUH and catalytic tyrosine sequence motifs, there is no statistically significant amino acid sequence homology between MobA and either TraI or TrwC.
The 321 residue MobA relaxase domain has been expressed independently as MobA*, and has been shown to be competent for conjugative transfer between cells 27. Using fragments generated from a partial enzymatic digest, Becker and Meyer showed that a 184 residue amino terminal fragment is capable of cleaving oriT at nic 27.
In this paper, we report the crystal structure of the 186 residue N-terminal fragment of MobA (minMobA) from the drug resistance plasmid R1162, at 2.1 Å resolution and compare it to the structures of TraI and TrwC.
RESULTS AND DISCUSSION
X-ray structure determination
Crystals of minMobA formed in space group P41212 with cell constants, a =b= 48.0, c = 165.9 Å. There is one molecule per asymmetric unit, giving a Vm of 2.3 Å3 / dalton.
Crystallographic data for minMobA are summarized in Table 1. A Ramachandran plot shows 88.5% of residues to be in the most favorable region, 10.8% in additional allowed space and 0.6% in generously allowed space. The refined structure includes 3 bound Mn2+ ions and 112 solvent molecules.
Table 1.
Crystallographic Data
|
Native |
Se |
|||
|---|---|---|---|---|
| Space group | P41212 | P41212 | ||
| Cell dimensions (Å) | a=b=47.967, c=165.927 | a=b=47.802, c=164.116 | ||
| Inflection | Peak | Remote | ||
| Wavelength (Å) | 1.0076 | 0.97924 | 0.97900 | 0.93925 |
| Resolution (Å) (last shell) | 20.−2.1 (2.17–2.10) | 20.–2.2 (2.28–2.20) | 20.–2.2 (2.28–2.20) | 20.–2.2 (2.28–2.20) |
| Rmerge (%) (last shell) | 4.6 (17.0) | 7.9 (29.0) | 9.7 (31.3) | 8.8 (31.3) |
| <I/σI> (last shell) | 17.1 (10.5) | 9.1 (4.0) | 8.1 (3.6) | 8.1 (3.6) |
| Completeness (%) (last shell) | 97.8 (92.9) | 93.4 (78.3) | 93.6 (83.1) | 91.1 (76.3) |
| Unique reflections | 11,903 | 9709 | 9774 | 9510 |
| Redundancy | 9.7 | 10.4 | 10.0 | 10.6 |
| Phasing FOM (20.–3.0 Å) | 0.67 | |||
| Phasing power (20.–3.0 Å) | 1.1 | |||
| Number of residues | 185 | |||
| Number of Protein atoms | 1442 | |||
| Number of metal atoms | 3 | |||
| Number of bound waters | 112 | |||
| Rworking (last shell) | 0.238 (0.255) | |||
| Rfree (last shell) | 0.287 (0.277) | |||
| Average B factor for protein atoms (Å2) | 21.8 | |||
| Average B factor for metal atoms (Å2) | 43.1 | |||
| Average B factor for bound waters (Å2) | 27.3 | |||
| rms deviation from ideality | ||||
| bonds | 0.006 | |||
| angles | 1.205 | |||
The final structure does not include the N-terminal Met residue, which is likely processed away from the expressed protein, or residues 13–14. The missing residues in this loop are not visible in the electron density maps and are most likely disordered. Sections of the original MAD-phased electron density map and a map phased from the final refined model are shown in Figure 1.
Figure 1.
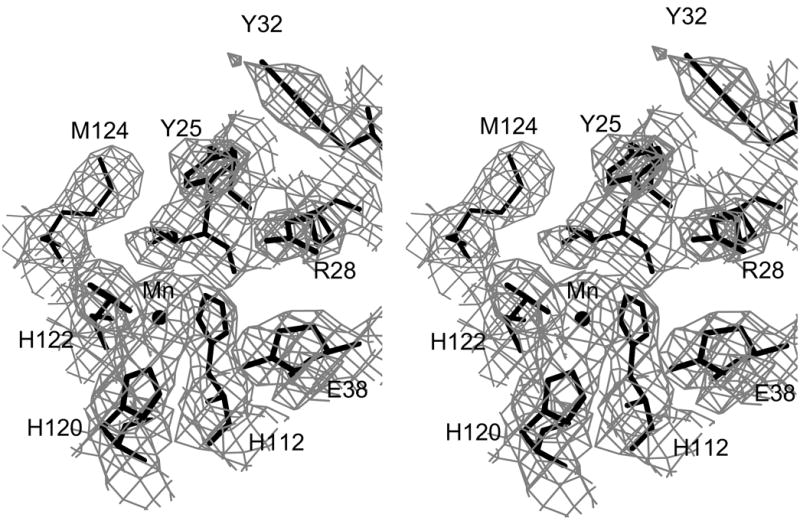
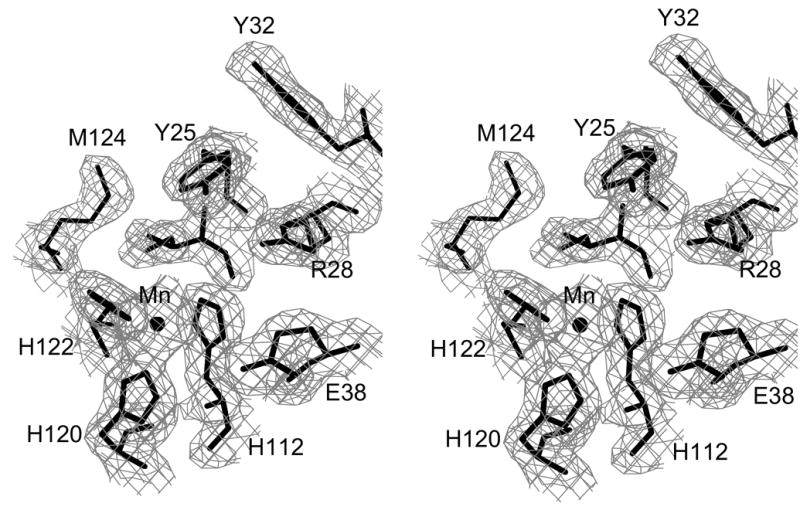
Electron density for minMobA protein. These stereo pictures display several active site residues with (a) a 3.0 Å MAD-phased Fo electron density map and (b) a 2.1 Å SIGMAA58-weighted 2Fo–Fc electron density map; these maps are contoured at 0.8 σ. The active site Mn atom is shown, bound by three histidine residues, along with the catalytic Tyr 25 and other nearby residues.
Structure of minMobA
The structure of minMobA can be described as a 5-strand antiparallel β−sheet with the helices connecting the strands lying on both sides of the sheet as shown in Figure 2. The first β−strand (a) lies on one edge of the sheet and is connected by helix A to strand b on the opposite edge. Helix B connects strand b to strand c, which lies adjacent and antiparallel to strand a. Helices A and B cross the sheet on the "front" side; that is, the side which contains the active site. Strand c is connected to strand d, which lies adjacent and antiparallel to strand b, by helix C across the back side of the sheet. Strand d is connected to the central strand e by a short loop. Strand e is followed by a short helix D, which lies above and in front of the sheet. These helices are followed by helix E across the back side of the sheet. Tyr 25, which has been shown to be involved in catalysis 13; 16, lies on helix A on the front side of the molecule and close to an apparent metal ion, which is bound on the front side of the β sheet, chelated by His 112 from strand d and histidines 120 and 122 from strand e.
Figure 2.
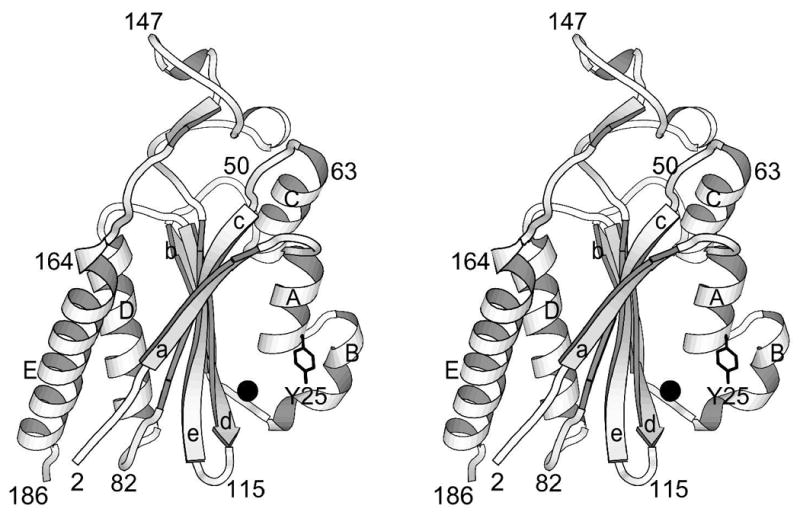
Ribbon drawing of minMobA. This stereo picture illustrates the fold of the protein and displays its secondary structural elements. Residues along the backbone are labeled to aid in following the polypeptide path. The active site Mn atom and Tyr side chain are also shown.
Previously, Scherzinger et al. found that catalysis of the nicking reaction by MobA in vitro depended on the presence of a divalent cation, such as Mg2+, Mn2+, Ca2+, or Ba2+13. Similarly, the relaxase TrwC has been shown to nick DNA in vitro with the addition of Mg2+, Mn2+, Ca2+, Ni2+, Cu2+, or Co2+, with much higher concentrations being required for Mg or Ca-catalyzed cleavage 26. The active site metal in relaxases TraI and TrwC has been modeled as Mg2+, Zn2+, and Ni2+22; 25. In recent soaking experiments with crystals of TrwC, it was shown that Zn2+, Ni2+, and Cu2+ bind in the active site whereas Mn2+ and Ca2+ do not 26. The tetrahedral geometry with three histidine residues and a water molecule is also more appropriate for Zn2+, Ni2+, or Cu2+.
An anomalous difference Fourier from the native minMobA crystal diffraction data and the experimental MAD phases indicated that there are three metal ions bound. As the minMobA crystallization condition included 10 mM MnCl2, it seems reasonable that the two metal ions outside the active site are Mn2+. To investigate the identity of the active site metal ion, two anomalous difference Fouriers were calculated using the MAD phases, one with native data collected from CuKα radiation (wavelength 1.5418 Å) and the other with data collected from synchrotron radiation at wavelength 1.0076 Å. The ratio of anomalous scattering for Mn is nearly 1:1 at these two wavelengths, while the ratio for all other plausible metals differs significantly. For example the ratio for Zn is 1:4. The experimental data, that is the rank ordering of anomalous peaks heights at the two wavelengths, are consistent with all three metals being Mn. These results indicate that Mn2+ is likely occupying the active site metal position in the minMobA crystal, probably having displaced the ion usually present under physiological conditions.
The refined B factors of the metal ions indicate that the active site ion, with a B factor of 24.0 Å2, is the most tightly bound. A second Mn2+ ion has a B factor of 35.4 Å2 and is chelated by the side chains of H46 and Q97. A third Mn2+ ion appears to be loosely bound with a B factor of 69.7 Å2; it is bound by H98 and by E52 of a symmetry-related molecule.
Comparison with TraI and TrwC
A Dali search 28 of the Protein Data Bank with the minMobA coordinates showed the strongest structural correlation to be with the relaxase domains of TraI and its homolog TrwC, Z = 8.5 in both cases. TraI and TrwC are the relaxases involved in conjugative cell-to-cell transfer of plasmids F factor and R388, respectively 29; 30. MinMobA, TraI, and TrwC all share a common fold as shown with the superposition of minMobA and TrwC in Figure 3a. The superposition of minMobA and TraI gives an rms distance between 106 equivalent Cα atoms of 2.3 Å. Similarly, a superposition of minMobA and TrwC gives an rms distance between 105 equivalent Cα atoms of 2.4 Å. TraI and TrwC are much more closely related to each other as a superposition of those two proteins gives an rms distance between 227 Cα atoms of 1.3 Å. A structure-based sequence alignment of minMobA, TraI, and TrwC is shown in Figure 4. There are only 12% and 10% sequence identities of minMobA with TraI and TrwC, respectively. However, the sequence identity between TraI and TrwC is 42%. Because of the apparent conservation of fold and function, it appears likely that the three proteins have descended from a common ancestor. However, the lack of sequence homology of MobA with the other two indicates that its evolutionary relationship is very distant. Comparison of the structure-aligned minMobA, TraI, and TrwC sequences reveals several conserved hydrophobic residues, which are likely critical to the formation of the common fold of MobA, TraI, and TrwC. These conserved residues are highlighted in dark gray in Figure 4. A previous analysis of several glycohydrolases that contained a common structural core fold showed a conservation of hydrophobic core residues despite there being no significant sequence homology 31.
Figure 3.
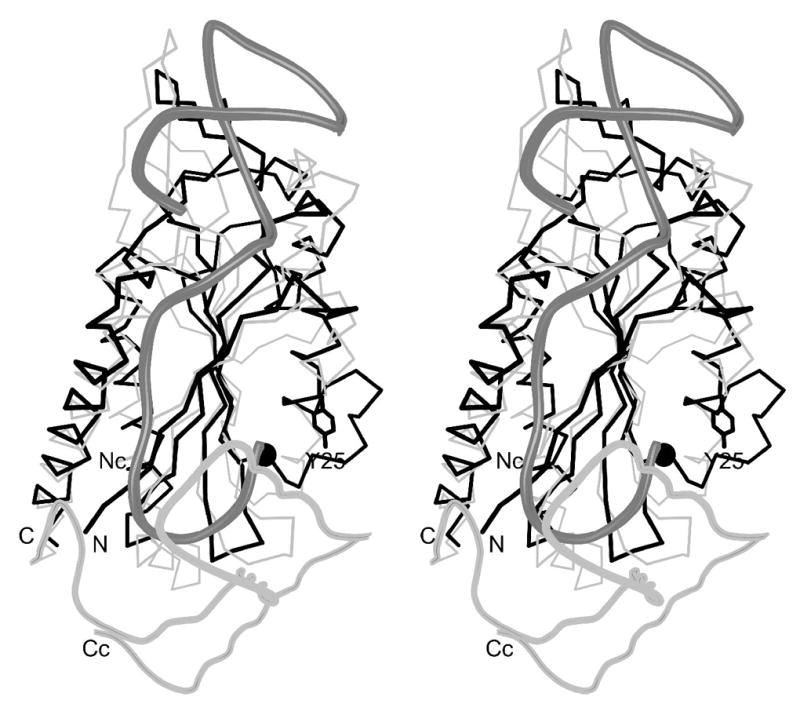
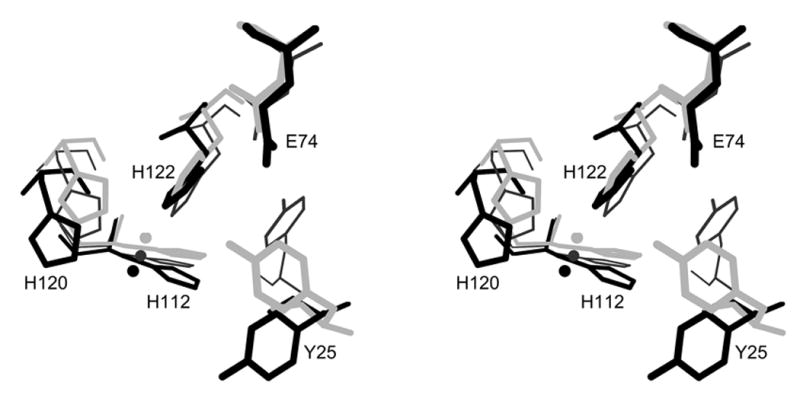
(a) Superposition of minMobA and TrwC. This stereo picture shows the least squares superposition of minMobA and TrwC complexed with DNA. The Cα trace of minMobA is shown in black bonds. The N and C termini are labeled. The active site metal atom and Tyr 25 side chain are also shown. The N-terminal region of TrwC, which has a similar fold to minMobA, is shown in thin, gray bonds. The N-terminus is labeled Nc. The C-terminal region of TrwC, for which there is no counterpart in minMobA, is shown in thick, gray bonds and helps enclose the single-stranded stretch of DNA. The phosphate backbone of the 25mer oligonucleotide complexed with TrwC is shown with thick, dark gray bonds. (b) Superposition of active site residues and metal. This stereo picture shows the superposition of the bound metal atom and several active site residues of minMobA with their counterparts in TrwC and TraI. Residues of minMobA are shown with thick, black bonds and are labeled whereas TraI residues are shown with thin, black bonds and TrwC with gray. Shown are the metal atom and its three histidine ligands and the catalytic tyrosine (Y18 for TrwC and Y16 for TraI). Also shown is a conserved acidic residue (D85 for TrwC and D81 for TraI) that has been hypothesized to act as the catalytic base.
Figure 4.

The structure-based sequence alignment of minMobA with TraI and TrwC. The sequence numbering and secondary structural elements of minMobA are shown on top. The secondary structural elements of TrwC and TraI are shown at bottom. The conserved catalytic tyrosine and metal-binding histidines are highlighted with black. Conserved hydrophobic core residues are highlighted in dark gray. A conserved arginine, involved in DNA binding, and a putative catalytic acidic residue are highlighted in light gray.
When compared with minMobA, TraI and TrwC have additional helices immediately following strands b and c. They have a more extended loop between helix B and strand c and an extended β-loop between helix C and strand d. Both of these loops are observed to interact with the double-stranded region of the 25mer oligonucleotide complexed with TrwC 25. TraI and TrwC lack the short helix D seen in minMobA and, in its place, have a β−loop. Following the C-terminus of minMobA, TraI and TrwC have a β-loop and three helices which contact the single-stranded region of oligonucleotides bound to both TraI and TrwC 24; 25. These, of course, are not part of the minMobA structure but may be part of the 321 residue MobA*, the domain of MobA fully active in transfer.
All three proteins have both a catalytic tyrosine residue and an active site metal ion bound by three histidine residues. The geometric arrangement of these active site moieties, shown in Figure 3b, is very similar. Interestingly, the sequence position of the TrwC catalytic tyrosine (Y18) differs from that of MobA and perhaps TraI by one residue (Figures 3b and 4).
Model of minMobA bound to oriT
The oriT of R1162 consists of no more than 38 base pairs 32. It is characterized by an inverted repeat, referred to as the outer and inner arms, and a core composed of an AT-rich region and adjacent, GC-rich segment containing the nic site (Figure 5a) 13; 17. The inverted repeat can form a 10 base-pair hairpin with one mismatch. The sequence of the core region is highly conserved in the oriTs of other plasmids 33. In vitro, MobA binds tightly to the oriT strand that is conjugally transferred and poorly to both the complementary strand and linear, duplex oriT 34. Tight binding to single-stranded DNA requires both the complete inverted repeat and adjacent TAA within the core 35; 36. The core and inner arm of the inverted repeat are sufficient for initiation but not termination 37. This indicates that, at initiation, MobA likely binds the inner arm in normal duplex form. At termination, the hairpin formed by the inner and outer arms recreates the required duplex DNA.
Figure 5.
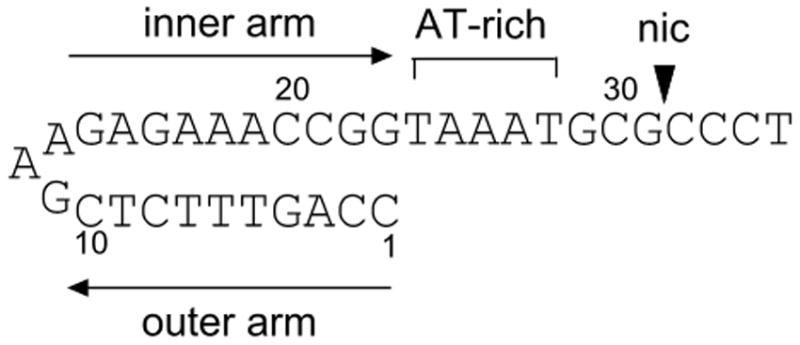
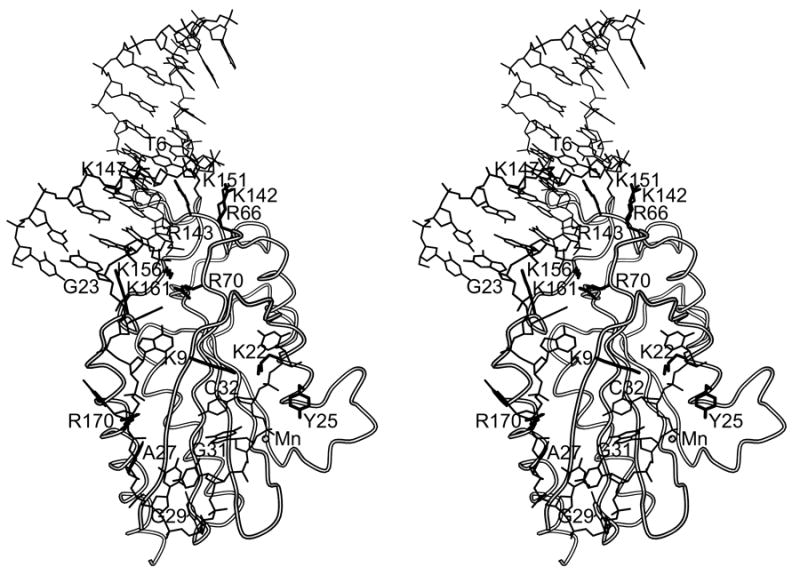
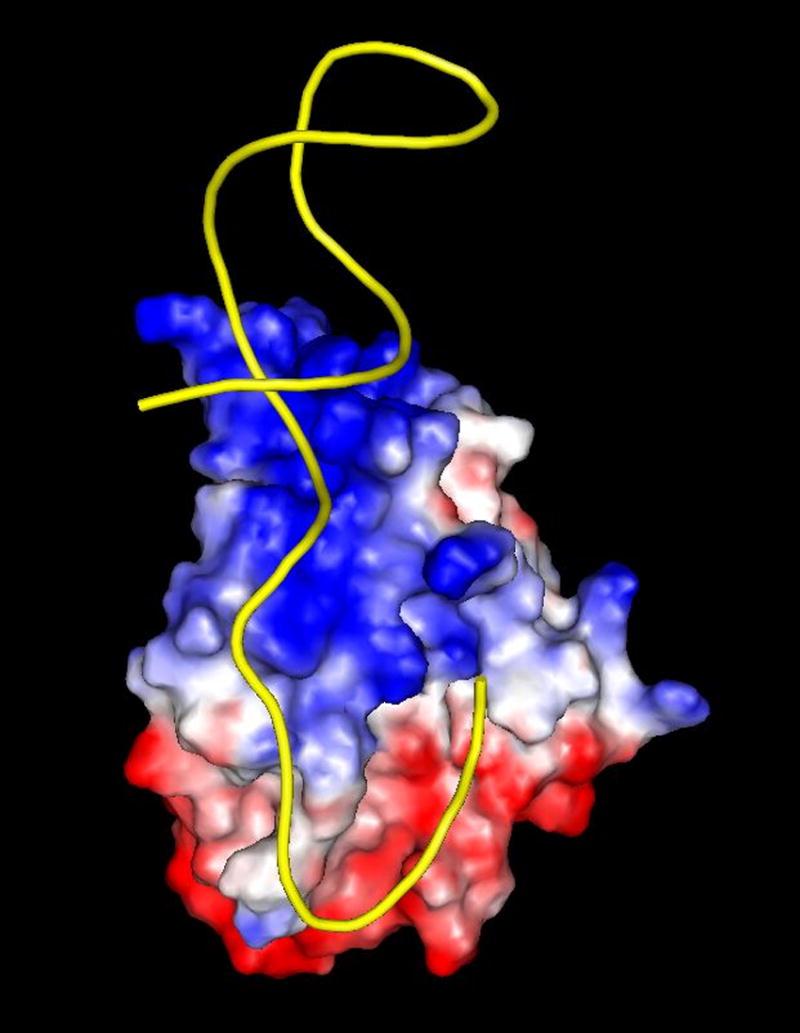
(a) R1162 oriT (b) Drawing of minMobA-33mer model complex. This stereo picture shows the Cα trace of minMobA in a model complex with a 33mer oligonucleotide. The bound Mn atom and catalytic Tyr 25 residue are shown and labeled along with several protein side chains that interact with the DNA in the model complex. Nucleotides mentioned in the narrative are also labeled. (c) Surface electrostatic potential of minMobA. Electronegative charges are shown in red, and positive charges are shown in blue. The bound DNA of the model complex is represented as a trace of the phosphate backbone.
In view of these observations, we have made a model of minMobA bound to single-stranded oriT DNA in the hairpin conformation (Figure 5b). The 33mer DNA in the model contains a 23 base double-stranded hairpin loop followed by a 10 residue single-stranded region. The docking of the double-stranded region of the 33mer to minMobA is modeled after the observed binding of double-stranded DNA to TrwC 25. Similarly, the single-stranded region of the 33mer is docked to minMobA based on the observed complex of a 10mer to the TraI Y16F variant 24. The 33mer contains the sequence of the first 33 nucleotides of plasmid R1162 oriT, with the nic site lying between nucleotides 31 and 32 (G and C). This model therefore represents oriT bound to MobA following termination of transfer and the strand rejoining at the nic site, and not the binding to double-stranded DNA at initiation.
In the surface electrostatic potential of the minMobA molecule shown in Figure 5c, it is the striking, positively charged area that contacts the DNA making up the stem loop. In contrast, the protein surface area that contacts the single-stranded core shows more variation in charge. These observations indicate that binding to the duplex region might involve primarily an interaction with the phosphate backbone, whereas MobA makes more specific contacts with the bases in the conserved core. This is consistent with the work of Scherzinger and co-workers 17 that showed the MobA recognition sequence to lie within an 11-residue oligonucleotide beginning with base 25.
In the oriT core region, the protein catalytic Y25 and the metal atom lie near the scissile P-O bond of the nic site, and the complex may be stabilized by the interaction with the phosphate backbone by positively charged side chains K9, K22, and R170. As observed in the TraI-10mer complex, the bases of nucleotides 27 and 29 sit in pockets of the protein, facilitating a sharp turn between bases 27 and 30 in the single-stranded core.
The core sequence is conserved in the oriTs of numerous plasmids 33. Genetic data have shown that the inverted repeat and the adjacent TAA (bases 24-26) in the core region are sufficient for strong binding by MobA. In addition, the dinucleotide YG (bases 28-29) determines the site of strand cleavage 36. In our model complex with minMobA, we see interactions of the thymidine at position 24 with residue R70. As shown in Figure 4, R70 is conserved in all three relaxases and may play a role in stabilizing the juncture of the double- and single-stranded regions. We do not see specific interactions with the adenosine bases at positions 25-26 or with the CG bases at positions 28 and 29. However in the observed TraI-10mer complex, specific interactions with those bases come from residues 255-266 24. These binding sites have no counterpart in the truncated minMobA structure, but might be present in MobA*. It might be that minMobA has a less stringent requirement for a specific sequence in the core region than MobA*.
As expected from Figure 5c, the phosphate backbone of the stem loop DNA is stabilized by interactions with numerous positively charged amino acids. K147 and K156 interact with the inner arm while the outer arm backbone is contacted by R66, K142, and K151. Our model also displays characteristics of the interaction between MobA and the DNA stem loop that are in agreement with those deduced from genetic studies. First, the inverted repeat region of the R1162 oriT, which contains 10 base pairs, is longer than that of plasmid R388 (only 5 base pairs). Thus, in our model the double-stranded hairpin loop of the 33mer extends well beyond that observed with the 25mer bound to TrwC, and only the first 9 nucleotides at the base of the stem interact with protein. This is consistent with mutational studies that predicted that only 7 bases of the inner arm of oriT (bases 17–23 in Figure 5) make specific interactions with MobA 38.
Mutational analysis of the oriT inner arm has shown that adenosine bases at positions 17 and 18 are highly preferred for MobA activity 38, and previous mutational analysis of the inner and outer arms showed that AT to TA transversions in the AAA triplet (bases 17–19) within the hairpin stem are well-tolerated by the protein 35. This tolerance of TA transversions is consistent with binding to the minor groove, where hydrogen-bonding contacts for the protein are not greatly altered by an AT-to-TA change 39, and it was proposed that MobA interacts with the DNA in the minor groove at positions 17 and 18 38. In our complex model, the side chain of R143 interacts with the thymidine bases 6 and 7, the pairing bases of A17 and 18, in the DNA minor groove, consistent with this proposal.
Mutational analysis of the oriT inner arm also showed that a guanosine at position 23 is preferred for MobA activity 38. There are no contacts in our complex model between the protein and G23 with K161 being the closest residue at a distance of ~5 Å. This base is near the juncture between the double-stranded and single-stranded regions, and it may well be that the conformation of our model in this area is not quite right. In the mutational studies, base 23 was found to be only G or A, consistent with a long contact with the protein. Mutational analysis also indicated that MobA tolerates changes at positions 20–22 38. In agreement with this, there are no close contacts between the protein and the DNA at these locations. It is interesting to note that the homologous MobA from plasmid pSC101 has a very stringent requirement for the base sequence at the corresponding locations of its oriT 38. How this is reflected in the structure of the pSC101 MobA is now being determined.
Mechanism of transesterification reaction
The action of MobA, and other relaxases, consists of the catalysis of a transesterification reaction involving a tyrosine residue in both the forward and reverse directions. The forward and reverse transesterification reactions are separated temporally by the transport of the protein-DNA covalent complex into the target cell. The action of type IA and IB DNA topoisomerases also involves the formation of a covalent 5' phosphotyrosine intermediate, and a mechanism has been proposed for those enzymes in which the scissile phosphate is held in position by positively-charged arginine and lysine side chains while a base abstracts a proton from the catalytic tyrosine, activating it for attack 40; 41; 42. A similar mechanism has been proposed for the transesterification reaction of TrwC involving its Tyr 18, Asp 85, and Arg 154 active site residues as well as the bound metal ion Zn2+26. In that proposed mechanism, the scissile phosphate is held by the positively-charged metal ion and arginine side chain (R154); and the side chain of the acidic residue (D85) acts as the general base, abstracting a proton from the side chain of the catalytic Y18. Asp 85 was implicated because of its proximity to the catalytic tyrosine and because a plasmid containing the D85A mutation was shown to have very low transfer frequency 25.
From the structure-based sequence alignment (Figure 4), Glu 74 in MobA corresponds in position to D85 of TrwC, and this residue is therefore a candidate to serve as the general base in an acid-base mechanism for transesterification. In the minMobA structure, the side chain of the catalytic Y25 is oriented away from E74; but rotation of the side chain about the Cα–Cβ bond can bring the tyrosine Oη atom within 2.5 Å of the E74 Oε2. To help assess the significance of E74 to catalysis, we carried out DNA nicking assays with formation of a radiolabeled covalent adduct. Figure 6 shows the time course of adduct formation. Conversion of E74 to Ala or Gln decreases adduct formation in one hour to 50% and 70%, respectively, of wild type activity. The double mutant E74AE76A reduces activity 10 fold. This suggests E74 plays only the most minor role in catalysis. Consistent with this notion is the observation that the frequency of mobilization of R1162 is virtually unaffected when an E74A change is introduced into MobA (Table 2). Another potential base in the active site is E38, which is observed to form a hydrogen bond both with the water molecule that is the fourth ligand of the bound metal. However, rotation of side chains can bring the E38 Oε1 and Y25 Oη atoms only to within ~5 Å. Changing this residue to alanine, or changing E76, the third acid residue in the vicinity of the active tyrosine, also has virtually no effect on DNA transfer (Table 2).
Figure 6.
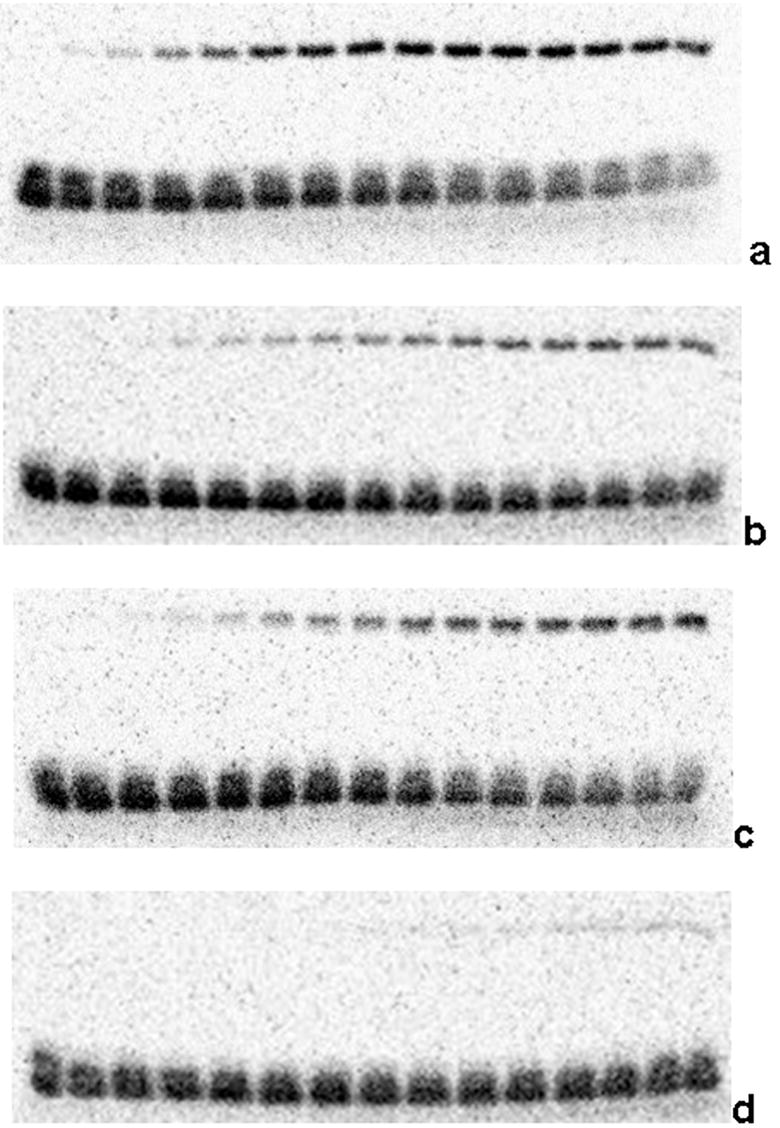
In vitro cleavage of 35mer oligonucleotide by minMobA variants. Cleavage reactions were begun by mixing protein with radiolabeled 35mer oligonucleotide containing the R1162 oriT sequence. Reactions were stopped at various time points by addition of EDTA, and the reaction mixtures were separated on 12% SDS-PAGE gel. In each gel, each lane contains the separated components from the reaction stopped after a time ranging from 0 (left) to 60 (right) minutes. The bottom bands correspond to the unreacted oligonucleotide, and the top bands represent the protein-oligonucleotide adduct. Each gel shows the reaction time course with a particular minMobA variant: (a) wild-type, (b) E74A, (c) E74Q, (d) E74AE76A.
Table 2.
Conjugal transfer frequency of R1162 derivatives containing mutations in mobA
| Change in MobA | Relative mobility |
|---|---|
| none | (100) |
| Y25F | 0.39 |
| E74A | 48 |
| E76A | 113 |
| E74AE76A | 35 |
| E38A | 104 |
| Y32F | 588 |
E74 does not appear to be critical to the cleavage mechanism, and probably does not act directly as a general base in an acid-base mechanism. That role may be filled by solvent molecule, perhaps binding to a carboxylate. However, it may very well be that activating the MobA tyrosine nucleophile is not mechanistically important. In vitro experiments have shown that the rate of adduct formation is very slow 17, and it can be seen in Figure 6 that the reaction occurs on the scale of hours. MobA does not effectively turnover as an enzyme, but instead produces a stable protein-DNA adduct. Compared with transfer, the nicking reaction is not the rate-limiting step, and there is no biological reason to enhance the rate of transesterification.
Although E74 does not appear to be important to the cleavage reaction, it may play a role in stabilizing the active site. We have observed that the E74AE76A variant of MobA supports transfer at a reduced frequency (Table 2). The in vitro rate of cleavage by the E74AE76A variant is also clearly reduced (Figure 6). In the minMobA structure, the Oε2 atom of E74 is within hydrogen bonding distance of the His 122 Nδ1 atom. With residue 74 changed to alanine, the side chain of E76 can rotate to hydrogen bond H122. Without E74 and E76, the positioning of the H122 side chain may be less stable.
Question of a second catalytic tyrosine
The initiation proteins of plasmids and single-stranded phage DNAs that undergo rolling circle replication have common functional motifs with the relaxases 19, and both groups of proteins belong to the same superfamily. For both rolling-circle replication and conjugative transfer, an initiator protein binds a specific site on the double-stranded DNA and locally unwinds the DNA at that site. This is followed by the protein nicking the DNA within the locally single-stranded region and with the formation of the covalent phosphotyrosine adduct. In the case of rolling-circle replication, the ensuing unwinding of the double-stranded DNA is coupled with leading strand replication by extension from the free 3’ OH end, to form a new target for cleavage by the initiation protein. A round of replication is completed as the newly-generated origin on the displaced, single-stranded DNA is cleaved to form a free monomer, which is then recircularized by the reverse of the cleavage reaction. In conjugative strand transfer, the mobilized strand is also unwound, and it is reasonable to suggest that rolling-circle replication might be occurring in the donor cell, as a means of strand replacement. In agreement with this, the R1162 relaxase can efficiently initiate transfer at one oriT and terminate it at a second oriT within the same DNA molecule 18.
The F TraI relaxase has two tyrosines in the active site, Y16 and 17 24, both of which are required for optimal cleavage of oligonucleotides in vitro 43. TrwC also has two adjacent active sites tyrosines, 18 and 19, but only 18 appears to be necessary for cleavage and for strand transfer 44. R1162 MobA lacks adjacent tyrosines, but, in addition to Y25, does have a second tyrosine (Y32) in the active site area. However, as for TrwC, changing this residue to phenylalanine does not decrease the frequency of transfer (Table 2). Parker and Meyer have suggested that R1162 lacks the characteristics of a rolling circle system, and so it is unlikely that a second tyrosine plays any known role in the activity of MobA 45.
MATERIALS AND METHODS
Protein purification
MinMobA, a polypeptide consisting of the first 186 amino acids of MobA, was purified by first amplifying the DNA with primers CAGCGAGGCCATATGGCGATTTATCACCTTACG and CGGGGCGGGCCACGTGGCCAG. The product was digested with NdeI and PmlI restriction endonucleases, and cloned into pTYB2 plasmid DNA (New England Biolabs Inc., Ipswich, MA) cleaved with NdeI and SmaI. The integrity of the cloned DNA was confirmed by DNA sequencing. The cloning fuses the C-terminal histidine of the MobA fragment to the Sce VMA1 intein and chitin-binding domain of the vector. Thiol-induced intein cleavage releases the MobA fragment with an addition C-terminal glycine due to the cloning. E. coli K-12 strain ER2566 (New England Biolabs) containing the plasmid was grown overnight in broth medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl) containing ampicillin (100 μg/ml) at 37 ºC. The cells were then diluted 1:80 in 2 x 2 L medium and grown to a cell density of approximately 4x108 per ml. IPTG was added to 0.3mM and the cells then grown overnight at 25 C. Protein was purified essentially according to a procedure developed by New England Biolabs. Briefly, cells were collected by centrifugation, resuspended in 18 ml column buffer (CB: 20 mM Tris, pH 8.0, 500 mM NaCl, 1 mM EDTA) and disrupted by sonication. Cellular debris was pelleted by centrifugation at 17,000xg for 30 min at 4 ºC. The supernatant was applied to a column containing chitin beads (18 ml bed volume, equilibrated with CB), and washed with 180 ml CB. Intein cleavage was initiated by rapidly flushing the column with 30 ml CB containing 40 mM DTT, and the reaction allowed to proceed for 48–60 hrs at 4 ºC. Released polypeptide was eluted from the column by washing the beads with CB and collecting 15 three-ml fractions. About three samples or 9 ml contained protein. The protein was concentrated to 1–2 ml by using an Amicon bioseparator fitted with a 25 mm YM-3 membrane and applying 60-70 psi N2 for 7–10 hrs at 4 ºC. The sample was washed with 8–9 ml 5 mM Tris HCl, pH 10.47, 500 mM NaCl and reconcentrated. This step was then repeated twice. The amount of protein was estimated following 15% PAGE-SDS electrophoresis with lysozyme as a standard. Typically, preparations contained about 8 mg/ml protein; a sample of 13 μg contained the MobA polypeptide and no other visible bands after Coomassie Blue staining. The protein was tested for cleavage of an oriT oligonucleotide as described previously 36.
MobA containing selenomethionine was prepared generally following the procedure of Hoffman et al. 46 Incorporation of selenomethionine was confirmed by electro-spray ionization mass spectroscopic analysis at the Mass Spectrometry Facility, Department of Chemistry and Biochemistry, University of Texas at Austin.
Construction of plasmid mutations
For mutagenesis of mobA, R1162 plasmid DNA was isolated by the Qiagen miniprep method (Qiagen Inc., Valencia, CA) and a DNA fragment containing the desired mutation was generated by the two-step PCR method described by Horton et al 47, except for the Y32F mutation, which was prepared according to the method of Kunkel 48. In all cases, either Taq polymerase or KOD polymerase (Novagen, San Diego, CA) was used for DNA amplification. The PCR product (or phage RF1 in the case of Y32F construction) was digested with EcoO109 and AflIII, and used to replace the same fragment in R1162. The presence of the desired mutation was confirmed by DNA sequencing.
Assays for frequency of plasmid mobilization
Mating experiments were carried out as previously described 14. Donors were the C600 derivative MV10 49 containing the mobilizing vector R751 50 and R1162 or the mutated derivative. The recipient was a C600 derivative resistant to nalidixic acid 51. The transfer frequency was calculated as the number of transconjugants per donor cell (average of two matings).
Construction of minMobA variants
The mutations were introduced by site-directed mutagenesis, according to the Stratagene protocol (Stratagene, La Jolla, CA). Briefly, approximately 50 ng of plasmid and 200 ng of each primer were combined with reaction buffer (20 mM Tris-HCl, pH 7.5, 8 mM MgCl2, 7.5 mM DTT, 50 μg/ml BSA), 150 μM dNTP mix, 2 U KOD Hot Start DNA Polymerase (Novagen), and sterile deionized water to 50 μl. Reaction were cycled with a denaturing temperature of 95 °C for 50 seconds, a reannealing temperature of 65 °C for 50 seconds, and an extension temperature of 72 °C for 9 min for 18 cycles on a GeneAmp 2400 thermocycler (Perkin-Elmer, Norwalk, CT). DpnI (10U) was added to each reaction and incubated at 37 °C for overnight to digest the methylated parental strand. One μl of reaction mixture was then added to 50 μl of E. coli DH5α competent cells and placed on ice for 30 minutes. Cells were heat shocked for 30 seconds at 42 °C, placed on ice for 1 minute and then combined with 200 μl LB media. Cells were shaken for one hour, and all 250 μl of solution plated on LB+ ampicillin plates. Plates were incubated at 37 ºC overnight and screened for colonies the following morning. The presence of the expected minMobA mutations was confirmed by DNA sequencing.
Cleavage assay
A 35mer oligonucleotide (Integrated DNA Technologies, Coralville, IA) containing the R1162 oriT sequence CCAGTTTCTGAAGAGAAACCGGTAAATGCGCCCT was 3’-33P-labeled with terminal transferase, according to the manufacturer’s instructions (New England Biolabs). The reaction was terminated by incubation at 65 °C for 20 minutes, and excess nucleotide was removed using a Micro Spin G25 column (GE Healthcare, Piscataway, NJ). Cleavage assay by minMobA was performed in reaction mixtures containing 40 mM Tris-HCl, pH 8.0, 50 mM MgCl2, and 0.5 mM EDTA. The reactions were terminated by addition of EDTA to 40 mM after 0, 1, 2, 4, 6, 9, 11, 15, 20, 25, 28, 36, 45, 55, and 60 minutes. Reaction products were separated by 12% SDS-PAGE and imaged on a Molecular Imager FX system (BioRad Laboratories, Inc., Hercules, CA).
Crystallization and data collection
MinMobA was crystallized at 4 ºC using the sitting drop method from 21–27% PEG4000, 0.1 M CAPS, pH10.0, 0.2 M NaCl, 10 mM MnCl2.
Prior to data collection, crystals were treated with cryoprotectant by transferring to 40% PEG4000, 0.1 M CAPS, pH 10.0, 0.2 M NaCl, 10 mM MnCl2, for 1–5 seconds. Crystals, mounted in a cryoloop (Hampton Research, Laguna Niguel, CA), were frozen by dipping in liquid nitrogen and placed in the cold stream on the goniostat.
Native crystal data and MAD data from the selenomethionine derivative crystal were collected at 100 K on the Gulf Coast Protein Crystallography Consortium (GCPCC) Beamline at the Center for Advanced Microstructures and Devices (CAMD). The MAD data were collected by Henry Bellamy as part of the mail-in program at the GCPCC Beamline. Diffraction images were processed and data reduced using HKL2000 52.
Structure determination and analysis
Three selenium sites were determined from the selenomethionine derivative MAD data (20–3.0 Å resolution) using SOLVE (Version 2.09) 53. The resulting MAD phases were submitted to RESOLVE (Version 2.09) for density modification and initial chain tracing 54. P41212 was determined to be the correct enantiomorphic space group as density modification using MAD phases in that space group yielded an interpretable electron density map. RESOLVE successfully traced 113 of the 186 residues of MinMobA, correctly identifying the side chains of 76 residues. The density-modified map was skeletonized with MAPMAN 55. The remaining backbone of the protein was traced from the density-modified map using the skeleton as a guide. The remaining side chains were also identified and built into appropriate electron density. Model building was done on a Gateway Select SB computer (Poway, CA) using O 56.
Models were refined with the Crystallography and NMR System (CNS) (Version 1.1) suite, using the slow-cooling protocol 57. There were several rounds of refinement followed by rebuilding of the model. To facilitate manual rebuilding of the model, a difference map and a 2Fo-Fc map, SIGMAA-weighted to eliminate bias from the model 58, were prepared. 5% of the diffraction data were set aside throughout refinement for cross-validation 59. PROCHECK 60 was used to determine areas of poor geometry. CNS was used to select solvent peaks. Candidates were positive peaks on a difference map 3.5 standard deviations above the mean, and within 3.5 Å of a protein nitrogen or oxygen atom. O was used to manually view and accept water sites. Computations were done on an HP Pavilion a510n computer (Hewlett-Packard Co., Palo Alto, CA).
Structural searches of the Protein Data Bank 61 were done using Dali 62. Superpositions of protein molecules were done with O.
Model pictures were made using MOLSCRIPT 63, BOBSCRIPT 64, and PyMOL (DeLano Scientific, San Carlos, CA).
DNA modeling
A model of a 33 base DNA oligonucleotide (sequence CCAGTTTCTGAAGAGAAACCGGTAAATGCGCC) was made to emulate the 38 base-pair oriT of plasmid R1162 32. The double-stranded stem with a 3-base loop (residues 1–23) was modeled after the NMR structure observed for an 11mer containing such a loop (PDB entry 1BHJ) 65. The single-stranded 3' terminus (residues 24–33) was taken from the 10-base oligonucleotide observed bound to the TraI Y16F variant (PDB entry 2A0I) 24. The nucleotide sequence of the model 33mer was changed to that of the R1162 oriT region using Xfit 66 and O. Initial docking of the model 33mer with the minMobA structure was made by homology with the observed binding of a 25-base double- and single-stranded oligonucleotide to TrwC (PDB entry 1QX0) 25 and a 10mer to the TraI Y16F mutant. The model of the minMobA-33mer complex was energy-minimized by the conjugate gradient method 67 using CNS, with stereochemical, van der Waals, and electrostatic terms being minimized. The resulting model had good stereochemical properties with an rms deviation from standard bond lengths of 0.002 Å and an rms deviation from standard bond angles of 0.91º.
Atomic coordinates
Coordinates of the refined model of minMobA have been deposited in the Protein Data Bank with entry code 2NS6.
Acknowledgments
We thank Sarah Jandle for technical assistance in the preparation of minMobA and Mehdi Moini (Department of Chemistry and Biochemistry, University of Texas at Austin) and Klaus Linse (Institute for Cellular and Molecular Biology, University of Texas at Austin) for assistance with the mass spectroscopic analysis. We thank Henry Bellamy and David Neau for technical assistance and H.B. for data collection at the GCPCC Beamline at CAMD; the beamline is supported by NSF grant DBI-9871464 with co-funding from the National Institute for General Medical Sciences. This work was supported by grants to J.D.R. from NIH (GM 63593) and the Robert A. Welch Foundation, by support for the Center for Structural Biology from the College of Natural Sciences and in part by a grant from NIH (GM37462) to R.J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zechner EL, de la Cruz F, Eisenbrandt R, Grahn AM, Koraimann G, Lanka E, Muth G, Pansegrau W, Thomas CM, Wilkins BM, Zatyka M. Conjugative DNA Transfer Processes. In: Thomas CM, editor. The Horizontal Gene Pool: Bacterial Plasmids and Gene Spread. Harwood Academic Publishers; Amsterdam: 2000. pp. 87–174. [Google Scholar]
- 2.Rawlings DE, Tietze E. Comparative biology of IncQ and IncQ-like plasmids. Microbiol Mol Biol Rev. 2001;65:481–496. doi: 10.1128/MMBR.65.4.481-496.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francia MV, Varsaki A, Garcillan-Barcia MP, Latorre A, Drainas C, de la Cruz F. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–83. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 5.Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–6. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 6.Mazodier P, Davies J. Gene transfer between distantly related bacteria. Annu Rev Genet. 1991;25:147–71. doi: 10.1146/annurev.ge.25.120191.001051. [DOI] [PubMed] [Google Scholar]
- 7.Courvalin P. Transfer of antibiotic resistance genes between gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 1994;38:1447–51. doi: 10.1128/aac.38.7.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A. 2004;101:841–6. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinemann JA, Sprague GF., Jr Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–9. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 10.Gormley EP, Davies J. Transfer of plasmid RSF1010 by conjugation from Escherichia coli to Streptomyces lividans and Mycobacterium smegmatis. J Bacteriol. 1991;173:6705–8. doi: 10.1128/jb.173.21.6705-6708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barth PT, Grinter NJ. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974;120:618–30. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerry P, van Embden J, Falkow S. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol. 1974;117:619–30. doi: 10.1128/jb.117.2.619-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherzinger E, Lurz R, Otto S, Dobrinski B. In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res. 1992;20:41–8. doi: 10.1093/nar/20.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasch MA, Meyer RJ. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J Bacteriol. 1986;167:703–10. doi: 10.1128/jb.167.2.703-710.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholz P, Haring V, Wittmann-Liebold B, Ashman K, Bagdasarian M, Scherzinger E. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene. 1989;75:271–88. doi: 10.1016/0378-1119(89)90273-4. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharjee MK, Meyer RJ. A segment of a plasmid gene required for conjugal transfer encodes a site-specific, single-strand DNA endonuclease and ligase. Nucleic Acids Res. 1991;19:1129–37. doi: 10.1093/nar/19.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherzinger E, Kruft V, Otto S. Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur J Biochem. 1993;217:929–38. doi: 10.1111/j.1432-1033.1993.tb18323.x. [DOI] [PubMed] [Google Scholar]
- 18.Kim K, Meyer RJ. Unidirectional transfer of broad host-range plasmid R1162 during conjugative mobilization. Evidence for genetically distinct events at oriT. J Mol Biol. 1989;208:501–5. doi: 10.1016/0022-2836(89)90513-5. [DOI] [PubMed] [Google Scholar]
- 19.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–85. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Mansfeld AD, van Teeffelen HA, Baas PD, Jansz HS. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 1986;14:4229–38. doi: 10.1093/nar/14.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickman AB, Ronning DR, Kotin RM, Dyda F. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol Cell. 2002;10:327–37. doi: 10.1016/s1097-2765(02)00592-0. [DOI] [PubMed] [Google Scholar]
- 22.Datta S, Larkin C, Schildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure (Camb) 2003;11:1369–79. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Dyda F, Hickman AB. A mob of reps. Structure (Camb) 2003;11:1310–1. doi: 10.1016/j.str.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure. 2005;13 :1533–44. doi: 10.1016/j.str.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat Struct Biol. 2003;10:1002–10. doi: 10.1038/nsb1017. [DOI] [PubMed] [Google Scholar]
- 26.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F. Unveiling the molecular mechanism of a conjugative relaxase: The structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J Mol Biol. 2006;358:857–69. doi: 10.1016/j.jmb.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Becker EC, Meyer RJ. MobA, the DNA strand transferase of plasmid R1162: the minimal domain required for DNA processing at the origin of transfer. J Biol Chem. 2002;277:14575–80. doi: 10.1074/jbc.M110759200. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–38. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 29.Matson SW, Morton BS. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J Biol Chem. 1991;266:16232–7. [PubMed] [Google Scholar]
- 30.Llosa M, Grandoso G, Hernando MA, de la Cruz F. Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J Mol Biol. 1996;264:56–67. doi: 10.1006/jmbi.1996.0623. [DOI] [PubMed] [Google Scholar]
- 31.Monzingo AF, Marcotte EM, Hart PJ, Robertus JD. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996;3:133–40. doi: 10.1038/nsb0296-133. [DOI] [PubMed] [Google Scholar]
- 32.Brasch MA, Meyer RJ. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987;198:361–9. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- 33.Becker EC, Meyer RJ. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J Bacteriol. 2003;185:3538–46. doi: 10.1128/JB.185.12.3538-3546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharjee MK, Meyer RJ. Specific binding of MobA, a plasmid-encoded protein involved in the initiation and termination of conjugal DNA transfer, to single-stranded oriT DNA. Nucleic Acids Res. 1993;21:4563–8. doi: 10.1093/nar/21.19.4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee M, Rao XM, Meyer RJ. Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J Bacteriol. 1992;174:6659–65. doi: 10.1128/jb.174.20.6659-6665.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Becker EC, Meyer RJ. Recognition of oriT for DNA processing at termination of a round of conjugal transfer. J Mol Biol. 2000;300:1067–77. doi: 10.1006/jmbi.2000.3902. [DOI] [PubMed] [Google Scholar]
- 37.Kim YJ, Lin LS, Meyer RJ. Two domains at the origin are required for replication and maintenance of broad-host-range plasmid R1162. J Bacteriol. 1987;169:5870–2. doi: 10.1128/jb.169.12.5870-5872.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jandle S, Meyer R. Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J Bacteriol. 2006;188:499–506. doi: 10.1128/JB.188.2.499-506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73 :804–8. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–81. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 41.Krogh BO, Shuman S. Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J Biol Chem. 2002;277:5711–4. doi: 10.1074/jbc.C100681200. [DOI] [PubMed] [Google Scholar]
- 42.Sherratt DJ, Wigley DB. Conserved themes but novel activities in recombinases and topoisomerases. Cell. 1998;93:149–52. doi: 10.1016/s0092-8674(00)81566-4. [DOI] [PubMed] [Google Scholar]
- 43.Street LM, Harley MJ, Stern JC, Larkin C, Williams SL, Miller DL, Dohm JA, Rodgers ME, Schildbach JF. Subdomain organization and catalytic residues of the F factor TraI relaxase domain. Biochim Biophys Acta. 2003;1646:86–99. doi: 10.1016/s1570-9639(02)00553-8. [DOI] [PubMed] [Google Scholar]
- 44.Grandoso G, Avila P, Cayon A, Hernando MA, Llosa M, de la Cruz F. Two active-site tyrosyl residues of protein TrwC act sequentially at the origin of transfer during plasmid R388 conjugation. J Mol Biol. 2000;295:1163–72. doi: 10.1006/jmbi.1999.3425. [DOI] [PubMed] [Google Scholar]
- 45.Parker C, Meyer R. Mechanisms of strand replacement synthesis for plasmid DNA transferred by conjugation. J Bacteriol. 2005;187:3400–6. doi: 10.1128/JB.187.10.3400-3406.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffman DW, Davies C, Gerchman SE, Kycia JH, Porter SJ, White SW, Ramakrishnan V. Crystal structure of prokaryotic ribosomal protein L9: a bi-lobed RNA-binding protein. Embo J. 1994;13:205–12. doi: 10.1002/j.1460-2075.1994.tb06250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–35. [PubMed] [Google Scholar]
- 48.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985;82:488–92. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hershfield V, Boyer HW, Yanofsky C, Lovett MA, Helinski DR. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974;71:3455–9. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Willetts N, Crowther C. Mobilization of the non-conjugative IncQ plasmid RSF1010. Genet Res. 1981;37:311–6. doi: 10.1017/s0016672300020310. [DOI] [PubMed] [Google Scholar]
- 51.Figurski D, Meyer R, Miller DS, Helinski DR. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1:107–19. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- 52.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;27:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 53.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55 (Pt 4):849–61. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56 (Pt 8):965–72. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kleywegt GJ, Jones TA. xdlMAPMAN and xdlDATAMAN - programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr D Biol Crystallogr. 1996;52:826–8. doi: 10.1107/S0907444995014983. [DOI] [PubMed] [Google Scholar]
- 56.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47 (Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 57.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54 (Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 58.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallog. sect. A. 1986;42:140–149. [Google Scholar]
- 59.Brunger AT. Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr D Biol Crystallogr. 1993;49:24–36. doi: 10.1107/S0907444992007352. [DOI] [PubMed] [Google Scholar]
- 60.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J App Cryst. 1993;26:283–291. [Google Scholar]
- 61.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holm L, Sander C. Searching protein structure databases has come of age. Proteins. 1994;19:165–73. doi: 10.1002/prot.340190302. [DOI] [PubMed] [Google Scholar]
- 63.Kraulis PJ. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structure. J Appl Cryst. 1991;2:946–950. [Google Scholar]
- 64.Esnouf RM. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr D Biol Crystallogr. 1999;55 (Pt 4):938–40. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 65.Chou SH, Zhu L, Gao Z, Cheng JW, Reid BR. Hairpin loops consisting of single adenine residues closed by sheared A.A and G.G pairs formed by the DNA triplets AAA and GAG: solution structure of the d(GTACAAAGTAC) hairpin. J Mol Biol. 1996;264:981–1001. doi: 10.1006/jmbi.1996.0691. [DOI] [PubMed] [Google Scholar]
- 66.McRee DE. A visual protein crystallographic software system for X11/XView. J Molecular Graphics. 1992;10:44–46. [Google Scholar]
- 67.Powell MJD. Restart Procedures for the Conjugate Gradient Method. Mathematical Programming. 1977;12:117–132. [Google Scholar]


