Abstract
Control of poly(A) tail length can affect translation and stability of eukaryotic mRNAs. Although well established for individual cases, it was not known to what extent this type of adjustable gene control is used to shape expression of eukaryotic transcriptomes. Here we report on microarray-based measurements of mRNA poly(A) tail lengths and association with the poly(A)-binding protein Pab1 in S. cerevisiae, revealing extensive correlation between tail length and other physical and functional mRNA characteristics. Gene ontology analyses and further directed experiments indicate coregulation of tail length on functionally and cytotopically related mRNAs to coordinate cell-cycle progression, ribosome biogenesis, and retrotransposon expression. We show that the 3′-untranslated region drives transcript-specific adenylation control and translational efficiency of multiple mRNAs. Our findings suggest a wide-spread interdependence between 3′-untranslated region-mediated poly(A) tail length control, Pab1 binding, and mRNA translation in budding yeast. They further provide a molecular explanation for deadenylase function in the cell cycle and suggest additional cellular processes that depend on control of mRNA polyadenylation.
Keywords: polyadenylation, microarray, mRNA translation, cell cycle, 3′-untranslated region
INTRODUCTION
In the nuclei of eukaryotic cells, a 5′ m7G(5′)ppp(5′)N cap structure and a 3′ poly(A) tail are added to nascent (pre-)mRNA concurrent with gene transcription (Proudfoot et al. 2002). Both cap and tail are known to influence mRNA splicing, transport, translation, and stability. In Saccharomyces cerevisiae, the coupled processes of mRNA cleavage, polyadenylation, and trimming to a mature length of ∼ 70–90 adenosines are instructed by sequence elements near the 3′ end of the transcript and carried out by a multiprotein complex (Minvielle-Sebastia and Keller 1999; Proudfoot et al. 2002). In a longstanding model, mRNA cleavage and onset of polyadenylation by the nuclear poly(A) polymerase Pap1 lead to the recruitment of multiple molecules of the canonical poly(A)-binding protein Pab1 to the emerging poly(A) tail. Pab1 stimulates the deadenylase PAN (a dimer of catalytic Pan2 and regulatory Pan3 subunits) to trim the tail to its mature length (Brown and Sachs 1998; Mangus et al. 2004; Dunn et al. 2005). In the cytoplasm, the poly(A) tail bound by Pab1 cooperates with the cap structure and its cognate binding protein eukaryotic initiation factor (eIF) 4E during translation initiation. As first discovered in yeast, the two ends of the mRNA are brought in close proximity by a bridging interaction of eIF4G with both eIF4E and Pab1. This closed-loop conformation of the mRNA is critical for efficient translation initiation (Sachs and Varani 2000; Mangus et al. 2003; Hentze et al. 2006), which in turn antagonizes mRNA degradation (Schwartz and Parker 1999). A major mRNA decay pathway in eukaryotes begins with a shortening of the poly(A) tail by the CCR4-NOT complex (catalytic subunit Ccr4), followed by decapping and degradation of the mRNA body (Meyer et al. 2004; Parker and Song 2004; Yamashita et al. 2005). The CCR4-NOT complex is inhibited by Pab1 bound to the poly(A) tail (Tucker et al. 2002). Thus, while PAN engages in initial trimming of the poly(A) tail, CCR4-NOT is considered the predominant deadenylase complex involved in mRNA deadenylation (Daugeron et al. 2001; Tucker et al. 2001).
Much evidence supports a significant role for poly(A) tail length control in gene regulation. microRNAs interfere with poly(A) tail function (Humphreys et al. 2005) and cause mRNA deadenylation (Giraldez et al. 2006; Wu et al. 2006). Best understood is the example of “stock-piled,” underadenylated maternal mRNAs in arrested Xenopus oocytes. Controlled by cytoplasmic polyadenylation elements (CPEs) in their 3′-untranslated region (UTR), these mRNA become readenylated and translationally active during oocyte maturation and early embryonic divisions (Mendez and Richter 2001; Read and Norbury 2002; Colegrove-Otero et al. 2005). Drosophila ccr4 mutants have specific defects in the female germ line (Benoit et al. 2005; Morris et al. 2005), and male mice deficient for the CCR4-associated factor Caf1 (Pop2 in yeast) exhibit disruption of spermatogenesis (Berthet et al. 2004). S. cerevisiae mutants of the core deadenylase subunits Pan2, Pan3, Ccr4, or Pop2 display many genetic interactions indicative of their importance in cell-cycle control (Liu et al. 1997; Hata et al. 1998; Hammet et al. 2002; Westmoreland et al. 2004; Traven et al. 2005; Pan et al. 2006; Woolstencroft et al. 2006).
In this study, we sought to identify the scope of poly(A) tail length control in a eukaryotic transcriptome to explore its cellular function(s) and the molecular mechanisms that establish it. We devised a method, termed polyadenylation state array (PASTA) analysis, which combines separation of cellular mRNA on poly(U) Sepharose with subsequent microarray analyses. PASTA surveys of the S. cerevisiae transcriptome identified mRNA groups with tendencies toward either long or short tails at steady state (∼ 700 each). Microarray analyses of mRNA fractions purified from an affinity-tagged Pab1 strain demonstrated a positive correlation between tail length and Pab1 interaction. Tail length correlated positively with ribosome density, and negatively with open reading frame (ORF) and UTR lengths. We further found extensive coregulation of poly(A) tail length on functionally or cytotopically related mRNAs, suggesting pervasive regulation of gene expression through poly(A) tail length control. Finally, expression studies with multiple chimeric green fluorescent protein (GFP) reporter constructs showed that mRNA 3′ regions autonomously mediate gene-specific poly(A) length control.
RESULTS
Separation of mRNA by poly(A) tail length
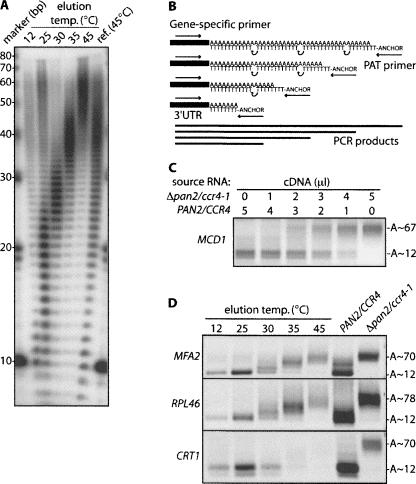
Binding to poly(U) Sepharose at low temperature, followed by stepwise thermal elution, can be used to fractionate cellular mRNAs by the length of their poly(A) tail (Palatnik et al. 1979; Binder et al. 1994). We adapted this approach for S. cerevisiae, where mature mRNA poly(A) tail lengths are ∼ 70–90 adenosines. Figure 1A shows the bulk tail length distribution in fractions from a typical poly(U)100 chromatography run with RNA from exponentially growing wild-type BY4741 cells. Each fraction is enriched for a distinct population of tail lengths (peak sizes [nucleotides]: 12°C, ∼ 10; 25°C, ∼ 20; 30°C, ∼ 27; 35°C, ∼ 38; and 45°C ∼ 58). There was minor contamination of long-tailed mRNAs in the first two elution fractions. These mRNAs may have bound nonspecifically to the matrix or through internal poly(A) runs (note also that ethanol precipitation may introduce some bias against short oligo(A) fragments).
FIGURE 1.
Fractionation of mRNA by poly(U)-Sepharose chromatography. (A) Total RNA from BY4741 cells was fractionated on poly(U) Sepharose by stepwise temperature increases (12°C–45°C, as indicated above the panels). Separately, all bound mRNA was eluted from poly(U) Sepharose in a single step (reference [45°C]). Aliquots of each mRNA fraction were end labeled with [32P]-pCp, digested with RNases A and T1, and poly(A) fragments analyzed by denaturing PAGE. DNA markers are indicated on the left. (B) Schematic of the LM-PAT assay. (C) To test for a PCR bias toward shorter amplicons, total RNA was isolated from either wild-type cells (PAN2/CCR4) or a deadenylase mutant strain (Δpan2/ccr4-1). The cDNA synthesis step of the LM-PAT assay was carried out separately for each RNA sample. Both cDNA preparations were then mixed in proportions outlined above the panel, prior to PCR for MCD1 mRNA. (D) Temperature eluates were analyzed by LM-PAT with primers specific to the mRNAs indicated to the left of the panels. Parallel assays were done with RNA from wild-type (PAN2/CCR4) or mutant (Δpan2/ccr4-1) strains. PCR products were sized against a 100-bp DNA ladder to determine the range of poly(A) lengths for each mRNA (indicated on the right).
To measure poly(A) tail lengths of individual mRNAs, we used the ligation-mediated poly(A) test (LM-PAT) (Fig. 1B; Sallés and Strickland 1995). This RT-PCR-based assay yields product sizes that reflect the poly(A) tail lengths present on a specific mRNA. Note that the use of a oligo(dT)12–18 mix and (dT)12–Anchor primer introduces some “laddering” of PCR products. Specifically, the lowest “rung” in the LM-PAT ladder represents all short tails (∼ 7–22 adenosines, e.g., mRNA enriched in the 12°C and 25°C elution fractions) (Fig. 1A) that can accommodate a single (dT)12–Anchor but not an additional oligo(dT)12–18 primer. LM-PAT results can also be affected by alternative mRNA 3′-end processing. We have sequenced suspect bands in some cases and found evidence for alternative 3′-cleavage site usage in four cases (PWP1, SPA2, SMC3, IRR1) (data not shown). A further concern with the method is that a bias toward shorter poly(A) lengths may be introduced by incomplete saturation of tails with oligo(dT)12–18 prior to cDNA synthesis or a drift toward shorter amplicons during PCR. We have included control reactions with total RNA from a deadenylase double mutant Δpan2/ccr4-1 strain (Traven et al. 2005) throughout this study to guard against this problem. Δpan2/ccr4-1 cells are known to accumulate all mRNAs examined thus far, with a homogenous tail reflecting their initial endowment in the nucleus (Tucker et al. 2001, 2002). LM-PAT assays for numerous mRNAs showed only one prominent band corresponding to fully adenylated mRNA in the double mutant strain (additional data not shown; Fig. 3, lane 2 in all panels; Figs. 1D, 6B,C), demonstrating that there is no problem with incomplete saturation of tails with oligo(dT)12–18 prior to cDNA synthesis in the LM-PAT assay as used here. Similar results were obtained with artificial transcripts carrying a defined length of poly(A) tail, while reactions with a model transcript carrying a continuum of tail lengths returned the expected ladder of PCR products (data not shown; see also Fig. 7C in Preiss et al. 1998). We further mixed cDNA preparations from either wild-type cells (PAN2/CCR4) or a deadenylase mutant strain (Δpan2/ccr4-1) in different proportions, followed by PCR for MCD1 mRNA, which exhibits mostly short tails in wild-type cells. We observed approximately linear relationships between the amount of each cDNA added to the reaction and the amount of the corresponding PCR products, demonstrating that the LM-PAT assay as used here does not display a substantial bias toward shorter PCR amplicons (Fig. 1C).
FIGURE 3.
Confirmation of mRNA tail length distribution by LM-PAT assay. Shown are analyses of five PASTA–long candidates in A and five PASTA–short examples in B. For each case, assays were done in parallel with total RNA from wild-type, Δpan2/ccr4-1, Δpan2, and ccr4-1 strains (1, 3, 3, and 1 μg input, respectively). Range of poly(A)-tail lengths is shown to the right of the panels. The dashed box exemplifies quantification of PCR product size distribution by densitometry that was done for all lanes with wild-type RNA. A ratio of signal above to signal below the dashed line was calculated as an arbitrary measure of mRNA tail length distribution. This ratio was ≥1.5 for all examples in A and ≤0.5 for all cases in B.
FIGURE 6.
The regulation of poly(A) tail length within functional groups is linked to deadenylation kinetics. (A) The bar chart displays the distribution of PASTA assignments within indicated mRNA groups. (B, C) Total RNA was isolated from BY4147 cells in stationary phase and after replenishing with fresh media (time-points indicated above the panels). LM-PAT assays were done for six mRNAs as indicated within the panels. Controls with RNA from exponentially growing PAN2/CCR4 and Δpan2/ccr4-1 cells are also shown, and poly(A) tail lengths ranges are indicated to the right of the panels.
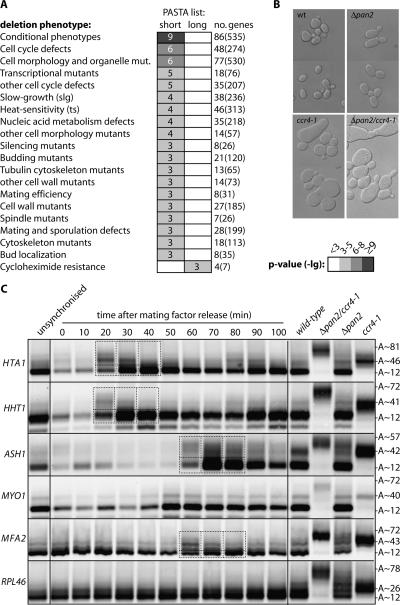
FIGURE 7.
poly(A) tail length control is important during the cell cycle. (A) PASTA lists were uploaded onto the FunSpec Web site (http://funspec.med.utoronto.ca/; default parameters; Robinson et al. 2002) and analyzed for enrichment of MIPS nonessential haploid deletion phenotype descriptors. The table lists all associations with a P-value of ∼ 10−3 or less; box shading indicates approximate P-value (see legend to the right of the table). No significant enrichment was found with the “unassigned” group. (B) DIC images (equal magnification) of the indicated yeast strains. Morphological changes are consistent with cell size-check point failure in ccr4-1 cells and a failure in the axial-to-isotropic growth switch in Δpan2 cells. Double mutants display a combination of these defects with increased cell size and body axis elongation. (C) LM-PAT assays were done on total RNA samples taken from wild-type cells before, and at indicated time-points after, release from mating factor synchronization. The HTA1, HHT1, ASH1, MYO1, and MFA1 mRNAs all fluctuate in transcript level during the time-course. LM-PAT data for RPL46(39) is shown in the bottom panel as a control with approximately constant mRNA level and adenylation state throughout the time-course. Although the MYO1 mRNA is clearly induced at 50 min after cell-cycle release, no long-tailed form is seen, possibly because deadenylation of MYO1 mRNA proceeds too quickly to be captured in our assays. Results with total RNA from unsynchronized wild-type, Δpan2/ccr4-1, Δpan2 and ccr4-1 strains (as in Fig. 3) and estimated poly(A) tail lengths are further shown to the right of the panels. The dashed boxes frame time-points where mRNA deadenylation following transcript induction is seen. Signals within the dashed boxes were quantified and partitioned into intensity derived from mRNA with an elongated or a minimal poly(A) tail (cut-off indicated by the dashed horizontal lines). This yielded the following long/short intensity ratios: (20, 30, 40 min) HTA1, 2.171, 0.901, 0.489; HHT1, 0.923, 0.312, 0.208; (60, 70, 80 min) ASH1, 1.413, 0.579, 0.573; MFA2, 2.287, 1.390, 0.991.
To further test the performance of the poly(U) chromatography method, we carried out LM-PAT assays for three mRNAs of known poly(A) tail length: MFA2 (long), RPL46/39 (intermediate; Brown and Sachs 1998), and CRT1 mRNA (short; Woolstencroft et al. 2006). RNA from each poly(U) chromatography eluate was analyzed (Fig. 1C), alongside total RNA controls from a PAN2/CCR4 and a Δpan2/ccr4-1 strain (for strain details, see Materials and Methods). As expected, MFA2 and RPL46(39) mRNAs with increasing tail lengths are present throughout all elution steps, whereas CRT1 is detected as a short-tailed mRNA eluting only in the low-temperature fractions. Δpan2/ccr4-1 cells accumulate all mRNAs with a homogenous tail reflecting their initial endowment in the nucleus (Tucker et al. 2001). These results demonstrate that poly(U) chromatography faithfully resolves bulk as well as individual mRNAs into fractions of distinct poly(A) tail lengths.
PASTA analysis
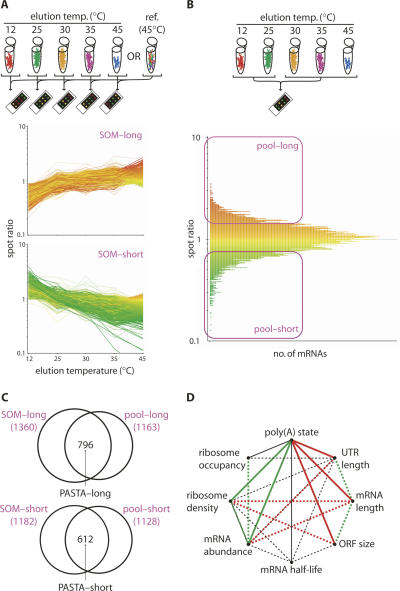
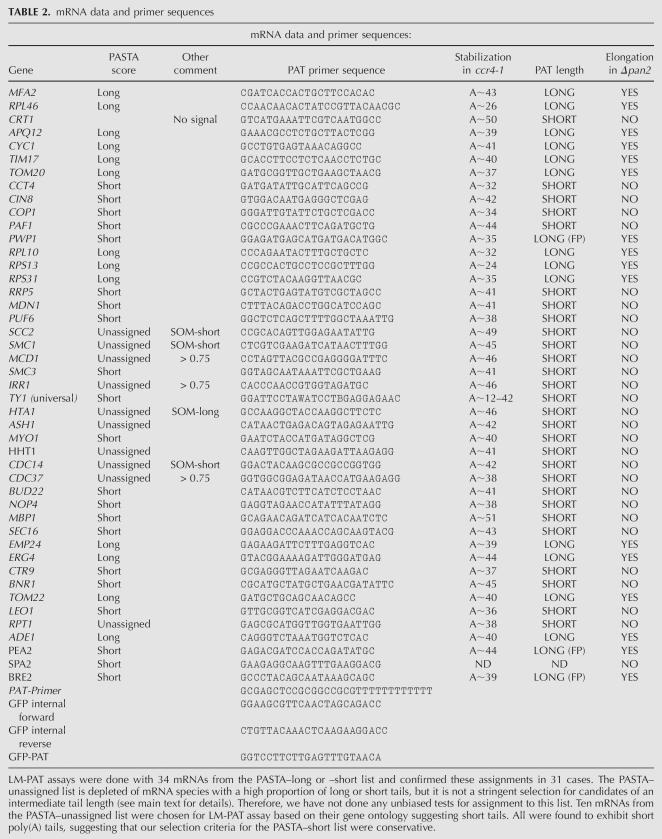
Next, we devised two strategies for microarray analysis of poly(U) eluates. First, we used five separate arrays to compare each eluate against reference mRNA (Fig. 2A). To obtain this reference, bound mRNA was eluted from beads in one batch at 45°C (reference [45°C]). Three biological repeat experiments were performed, and array data were normalized, replicate averaged, and further processed as detailed in the Materials and Methods. Each mRNA elution profile is normalized against the average elution pattern of all mRNAs, represented by a spot ratio of one on each array. Second, we compared pooled low-temperature fractions against a pool of high-temperature eluates on a single microarray (three biological repeats, data analysis as above; Fig. 2B). Here, poly(A) status is represented by a single spot ratio, which is greater than one for an mRNA with a higher proportion of long-tailed molecules than average and less than one when this proportion is below average. The two PASTA data sets correlated well with each other (Spearman rank correlation coefficient r = 0.6921, P < 0.0001) (data not shown). We then classified the high-resolution elution profile data using a self-organizing-map (SOM) algorithm-based approach, similar to that applied to polysome profiles in previous work (Preiss et al. 2003). Inspection of the SOM classes (data not shown) suggested two groups of ∼ 1250 mRNAs each, as candidates for a high proportion of short- or long-tailed mRNA molecules, respectively (Fig. 2A, bottom half). For the pool comparison, we chose spot ratio cutoffs of > 1.33 and < 0.75 (Fig. 2B, bottom half) to generate pools of roughly equal size to those derived from the SOM selection. To suppress experimental noise, we then required an equivalent tail length assignment in both experimental approaches, yielding a combined PASTA–long list of 796 and a combined PASTA–short list of 612 mRNAs (Fig. 2C). All detectable unassigned mRNAs were entered into a third list (PASTA–unassigned, 4446 mRNAs). To independently verify our PASTA selections, we performed LM-PAT assays. In wild-type cells, candidates from the PASTA–long list typically exhibited noticeable proportions of longer-tailed molecules, while cases chosen from the PASTA–short list had mostly short tails (Fig. 3A,B, left-most lanes). Estimates by densitometry (dashed box) suggest a shift of at least threefold in the long/short ratio between examples from the different PASTA lists. LM-PAT assays were done with 34 mRNAs from either the PASTA–long or –short lists and confirmed these assignments in 31 cases (Figs. 3, 6, 7; see Table 2 below). These data indicate that our PASTA–long and –short lists are highly enriched for “true” cases.
FIGURE 2.
Polyadenylation state array (PASTA) analysis. mRNA was fractionated as detailed in Figure 1A. (A) For high-resolution PASTA analyses, each temperature eluate (reverse-transcribed into Cy5-labeled cDNA) was compared against reference mRNA (ref. 45°C; Cy3-labeled cDNA). The lower panels show the mRNA elution profiles of candidate mRNA groups (SOM–short or SOM–long; averaged data from three biological repeat experiments). Profiles are colored by spot ratio for the 45°C elution. (B) For low-resolution PASTA analyses, pools of elution fractions were compared to each other by microarray (30°C, 35°C, and 45°C [Cy5-labeled cDNA] versus 12°C and 25°C [Cy3-labeled cDNA]). The graph displays the resulting data (averaged spot ratios from three biological repeats against mRNA frequency; high over low temperature pool). Outlined in purple are mRNA groupings chosen for further analysis (pool–short or pool–long). (C) The diagrams illustrate the selection of mRNAs on the basis of coherent tail length assignment in A and B. (D) Statistically significant relationships between PASTA assignment and characteristics of mRNAs in S. cerevisiae taken from Table 1 are visualized here in simplified form. Each parameter is represented as a black dot with connecting lines indicating relationships as follows: Solid lines represent correlations exposed by this study; dashed lines are correlations determined by previous studies (Graber et al. 2002; Wang et al. 2002; Arava et al. 2003; Hurowitz and Brown 2003; Beyer et al. 2004; MacKay et al. 2004); green lines represent positive correlations; red lines represent negative correlations; and black lines indicate no simple correlation. (Multiple connections indicate conflicting published information.)
TABLE 2.
mRNA data and primer sequences
LM-PAT assays throughout this study were also done with RNA from deadenylase mutant strains. These data indicate that prevalence of short-tailed forms in wild-type cells for any of the 44 mRNAs tested is not due to diminished initial polyadenylation, as all had long tails (> 55 adenosines) in the Δpan2/ccr4-1 strain (e.g., Figs. 1D, 3, 6B,C). The ccr4-1 strain exhibited accumulation of all mRNAs tested with tails of an intermediate length (∼ 25–45 adenosines) (Figs. 3, 7C; see Table 2 below). These results mirror changes in bulk poly(A) tail length in these mutants (data not shown). Results in the Δpan2 strain revealed that mRNA from the PASTA–long list showed increases in long-tailed molecules in the absence of Pan2, whereas those with a PASTA–short assignment remained unchanged (Figs. 3, 7C; Table 2 below). This suggests a stronger dependence on Ccr4 for mRNAs in the PASTA–short list.
Relationship of PASTA assignments to established genome-wide trends
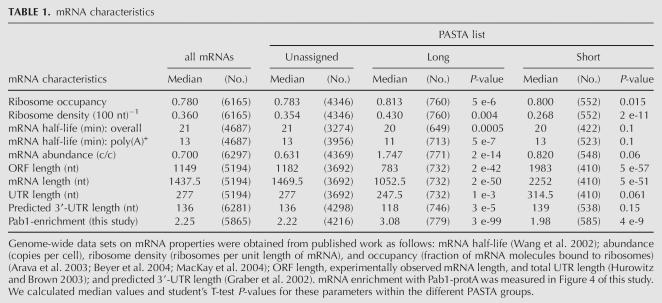
To detect global relationships between our PASTA assignments and other mRNA characteristics in S. cerevisiae, we obtained microarray or serial analysis of gene expression (SAGE) data sets of mRNA half-life (Wang et al. 2002), abundance, as well as ribosome density (ribosomes per unit length of mRNA) and occupancy (fraction of mRNA molecules bound to ribosomes) (Arava et al. 2003; Beyer et al. 2004; MacKay et al. 2004), observed mRNA length and total UTR length (Hurowitz and Brown 2003), and predicted 3′-UTR length (Graber et al. 2002). After calculating the median and student's T-test P-value for all these parameters within each PASTA list (Table 1), we found a positive correlation of poly(A) tail length with ribosome density on the mRNA. Negative correlations were observed between poly(A) tail length and ORF size, mRNA–, and (3′) UTR length. Median mRNA abundance was strongly increased in the PASTA–long list and only marginally increased in the PASTA–short list. The latter probably reflects a small bias in the microarray data toward more reliably detectable, abundant mRNAs. Thus, we tentatively conclude a positive correlation between poly(A) tail length and mRNA abundance. Microarray data are available for overall mRNA half-life (based on random-primed cDNA synthesis) as well as poly(A)+ mRNA half-life (based on oligo[dT] priming) (Wang et al. 2002). We found no clear correlation of PASTA assignments with overall mRNA half-lives, while there was a significantly reduced median poly(A)+ mRNA half-life in the PASTA–long list. The graph in Figure 2D summarizes all identified global relationships between poly(A) tail length and other mRNA characteristics. We note that new and established relationships within this correlative network are largely compatible with each other, suggesting that poly(A) tail length control is an important part of the gene regulation program of S. cerevisiae.
TABLE 1.
mRNA characteristics
Poly(A) tail length positively correlates with Pab1 binding
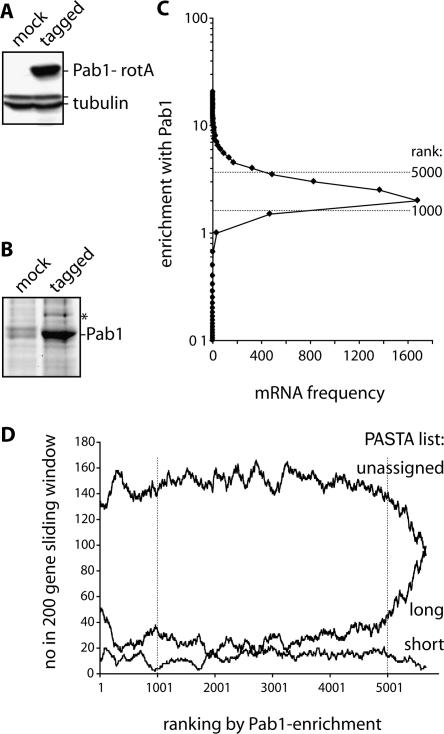
Integral to models of poly(A) tail function is that a longer tail will bind Pab1 in larger numbers and/or with increased affinity. To test this tenet on a genome-wide scale, we generated a strain carrying protein A–tagged Pab1. Pab1–RNA complexes were purified from cell lysates on IgG Sepharose and eluted by TEV protease cleavage (Fig. 4A,B; Gerber et al. 2004). We prepared RNA from Pab1 purifications (three biological repeats) or mock purifications (two biological repeats) as well as aliquots of total RNA from each of the yeast cultures. Each purified RNA sample was then compared with the corresponding total RNA by microarray. The array data were processed to generate a measure of mRNA enrichment with Pab1 as detailed in the Materials and Methods. Median enrichment was ∼ 2.3-fold across all mRNAs (Fig. 4C; Table 1), consistent with Pab1 binding to most mRNAs. Figure 4D plots the frequency of members of the different PASTA lists occurring within the tagged Pab1 enrichment ranking, illustrating a positive correlation between tail length and extent of Pab1 association. Median values of Pab1 enrichment within the PASTA lists confirm this trend (Table 1). These data support a functional relationship between poly(A) tail length and Pab1 binding on a genome-wide level and add physiological relevance to results obtained by the PASTA approach.
FIGURE 4.
Poly(A) tail length positively correlates with Pab1 binding. (A) Western blot of cell extracts from the control (mock) or protein A-tagged Pab1 strain (tagged) probed with β-tubulin antibodies (Pab1-protA and tubulin bands indicated on the right). (B) Material eluted from IgG Sepharose by TEV protease cleavage was separated by 10% SDS-PAGE, followed by deep purple staining (positions of cleaved Pab1 and a copurifying protein [asterisk] are marked on the right). (C) RNA was isolated from Pab1-protA or mock purifications (Cy5-labeled cDNA) and compared with total RNA isolated from the corresponding yeast cultures (Cy3-labeled cDNA) by microarray analysis. mRNA enrichment, Pab1-protA over mock (derived from several biological repeats), is plotted against mRNA frequency. Dashed lines indicate positions of the 1000th and 5000th mRNA in enrichment ranking. (D) The graph displays the frequency of members of different PASTA groups occurring within the Pab1-protA enrichment ranking using a 200-gene sliding window.
Poly(A) tail length coregulation of functionally and cytotopically related mRNAs
Next, we assessed any statistically significant enrichment of gene annotations within the PASTA–short or –long lists using GOstat (http://gostat.wehi.edu.au/; Beissbarth and Speed 2004). We found highly significant association of a number of interrelated gene ontology (GO) terms from all three categories—molecular function, biological process, and cellular component—with either the PASTA–short or –long lists (for a full display, see Fig. 5). The GO terms DNA/Ty element transposition (biological process) and retrotransposon nucleocapsid (cellular component) associated with the PASTA–short assignment and were the most highly enriched terms (∼ 55% of all mRNAs with that annotation were in the PASTA–short list, with P-values ≤ 10−55). LM-PAT using a primer universal to the TY1 3′ long terminal repeat (LTR) confirmed that these transcripts are severely underadenylated in wild-type cells, while they are fully adenylated (∼ 57 adenosines) in the Δpan2/ccr4-1 strain (data not shown), suggesting that deadenylation of retrotransposon transcripts may limit their expression.
FIGURE 5.
Gene ontology term enrichment within the PASTA lists. The PASTA—long and—short lists were uploaded into the GOstat tool, available at http://gostat.wehi.edu.au/ (Beissbarth and Speed 2004). GOstat was run with SGD as the gene ontology database, a requirement for a minimal GO path of three, and correction for multiple testing by false discovery rate (Benjamini). Over- or underrepresented GO terms are displayed only if the associated P-values were in the order of ≤10−4. Diagrams summarize association with terms from (A) molecular function, (B) cellular component, and (C) biological process ontologies. Box shading indicates approximate P-value (see legend in the center of figure).
Ribosome biogenesis-related terms also featured prominently in these analyses. High representation in the PASTA–long list of mRNAs encoding proteins of both subunits of the cytoplasmic ribosome (65 of 136 detected by PASTA) (Fig. 6A) drove overrepresentation of several relevant GO terms (structural constituent of the ribosome [molecular function], P-value ∼ 10−27, cytosolic ribosome –small/–large subunit [cellular component], P-values ∼ 10−10–10−26) with that assignment. By contrast, mRNAs encoding nucleolar proteins involved in ribosome biogenesis were enriched in the PASTA–short list, driving overrepresentation of GO terms such as ribosome biogenesis and assembly, rRNA processing (biological process), P-values ∼ 10−12–10−13, nucleolus (cellular component), P-value ∼ 10−8, with that list. A group of ∼ 220 mRNAs, termed the RiBi regulon, are coregulated with cytoplasmic ribosomal protein (RP) mRNAs in transcriptome profiling studies across a variety of cellular conditions (Gasch et al. 2000; Beer and Tavazoie 2004; Grigull et al. 2004; Jorgensen et al. 2004). Most RiBi genes encode proteins involved in ribosome biogenesis, but other functions are also represented. We find that the RiBi regulon is enriched for the PASTA–short assignment (55 of 220 detected by PASTA); however, this does not apply uniformly to all constituent functional subgroups (Fig. 6A). For instance, the ribosome biogenesis subgroup in isolation is more highly enriched for the PASTA–short assignment (45 of 135), while RiBi members involved in ribonucleotide metabolism show the opposite tendency (six of 22). The latter tendency also persisted beyond the RiBi regulon in that a group of GO-Biological Process terms related to ribonucleotide metabolism were enriched in the PASTA–long list, with P-values ranging from ∼ 10−4 to 10−5 (Fig. 5C). Thus, PASTA analysis indicates that a large cohort of transcriptionally coregulated genes involved in ribosome biogenesis is partitioned into distinct subgroups at the post-transcriptional level along functional (e.g., ribosomal protein versus ribosome biogenesis) and cytotopic lines (cytoplasmic versus nucleolar protein localization).
Furthermore, coregulation of mRNAs encoding proteins destined to mitochondria and the endoplasmic reticulum was suggested by enrichment of corresponding GO terms (cellular component) within the PASTA–long list (Fig. 5B). This trend is exemplified by the mRNAs encoding components of the mitochondrial import machinery (11 of 29) (data not shown). Enrichment of several additional nuclear functionalities in the PASTA–short list was also apparent (GO terms relating to chromosome condensation, chromatin remodeling and modification, DNA replication, and RNA transcription and processing) (Fig. 5). Finally, we noticed a propensity of short-tailed mRNAs to encode cell-cycle-related polarity functions, suggested for instance, by enrichment of the GO terms cytoskeleton, cell cortex, bud neck (cellular component), P-values ∼ 10−4–10−7, or establishment of cell polarity, cell budding (biological process), P-values ∼ 10−4–10−5. Collectively, the enrichment of multiple GO terms in either the PASTA–long or –short lists indicate a widespread and purposeful use of differential poly(A) tail length control in the transcriptome of S. cerevisiae.
A corollary of the above is that mRNAs encoding members of a protein complex tended to be enriched in the same PASTA group. This is illustrated by the example of cohesin, a complex required for cohesion between sister chromatids. mRNAs for three of the six subunits of cohesin (SMC3, IRR1, and REC8) were in the PASTA–short list. We confirmed a short tail length for two of these by LM-PAT assay and established that the three “unassigned” cohesin mRNAs (SCC2, SMC1, MCD1) did in fact also have very short poly(A) tails at steady state (data not shown). Thus, mRNAs encoding all members of a protein complex can display equivalent poly(A) tail length tendencies.
Origin of different mRNA polyadenylation states
Yeast cells reentering the cell cycle from stationary phase are known to activate transcription of over 2500 genes within minutes of nutrient repletion, including the RP and RiBi genes (Radonjic et al. 2005). Thus, we employed this readily accessible experimental system to investigate how distinct adenylation states of transcriptionally coregulated mRNAs are established. We followed three RP mRNAs (RPL10, RPS13, and RPS31), from the PASTA–long list (Fig. 6B), and three mRNAs from the PASTA–short list (Fig. 6C), which encode ribosome biogenesis factors (RRP5, MDN1) and/or are part of the RiBi regulon (RRP5, PUF6). LM-PAT assays were done on RNA isolated from stationary phase cells at the indicated time points after suspension in to fresh media; controls from exponentially growing PAN2/CCR4 and Δpan2/ccr4-1 cells were also included. The results confirm the PASTA assignment for all six mRNAs (see PAN2/CCR4 lanes; in total we confirmed the PASTA assignment of four RP mRNAs and four RiBi mRNAs) (Table 2). Importantly, an initial burst of transcripts with long poly(A) tails is clearly seen for the mRNAs with a PASTA–long assignment, which become slowly deadenylated toward a steady-state length distribution. By contrast, the mRNAs with PASTA–short assignment accumulate few, if any, molecules with long tails, detectable only at 10 min. All six mRNAs display long poly(A) tails in Δpan2/ccr4-1 cells, indicating that differences in their tail length distribution are predominantly established through different rates of deadenylation.
Role of tail length control during the cell cycle
Our GO analyses showed that mRNAs encoding cell-cycle-related functions are enriched in the PASTA–short list. This is consistent with observations that both CCR4 and PAN2 interact genetically with cell cycle and DNA damage checkpoint regulators (e.g. CRT1 [Woolstencroft et al. 2006], RAD5 [Westmoreland et al. 2004], and DUN1 [Hammet et al. 2002]). Thus, we predicted that genes enriched in the PASTA–short list may, when deregulated, generate cell-cycle-related or cell-morphology-related phenotypes. This was indeed born out by a statistically significant enrichment of such phenotype descriptors (from the MIPS nonessential haploid deletion set) in the PASTA–short list (Fig. 7A, produced on the FunSpec Web site; Robinson et al. 2002). Further, microscopy of deadenylase mutant cells revealed morphological changes consistent with cell size check point failure in cells lacking functional Ccr4 and a failure in the axial-to-isotropic growth switch in deletants of PAN2. Double mutants display a combination of these defects with both an increased cell size and elongation of the body axis (Fig. 7B). Similar observations were made for Δccr4 cells in a genomic scale screen (Zhang et al. 2002). We also monitored the induction/deadenylation profiles of five cell-cycle-regulated genes (HTA1, HTT1, ASH1, MYO1, and MFA2) in α-factor synchronized cells (Fig. 7C). This showed accumulation of polyadenylated molecules in a temporal “spike” that is much tighter than overall fluctuation in mRNA level for four of these. Given the link between poly(A) tail length and translation efficiency (Figs. 2D, 8; Table 1), translation of these mRNAs will only be efficient during the spike period. Taken together, these observations suggest that mRNA-specific control of deadenylation is important for cell-cycle progression, by delimiting the expression of many mRNAs in the PASTA–short list to shorter intervals than suggested by changes in mRNA expression level.
FIGURE 8.
The 3′-UTR drives adenylation state and protein expression level. GAL1-regulated GFP constructs harboring the indicated 3′-UTRs were maintained in BY4741 cells. Transcription of GFP was induced with galactose at time zero and repressed by addition of glucose at 10 min. (A) GFP-mRNA analyses at various time-points by LM-PAT assay (top) and semi-quantitative RT-PCR reaction using internal GFP primers (middle). The bottom panel shows input total RNA. (B) Western blot detection of GFP and GAPDH proteins by infrared fluorescent imaging. (C) Graph of corresponding GFP/GAPDH protein ratios over time (ratio for GFP-3′RPL10 at 40 min set to one).
3′-UTRs drive differences in mRNA polyadenylation state and protein synthesis
Translational fitness and decay of many individual mRNAs have been linked to their 3′-UTRs (Muhlrad and Parker 1992; Wickens et al. 2002; Colegrove-Otero et al. 2005; Hentze et al. 2006). To test for a 3′-UTR involvement in establishing mRNA-specific tail length as seen here, we engineered and constitutively expressed a series of GFP reporter plasmids in wild-type cells. The plasmids contained immediately after the GFP stop codon, 500 base pairs (bp) of 3′ genomic sequence taken from 10 candidate genes, as well as two TY1 element LTRs. LM-PAT assays revealed that in all cases the 3′ sequences conferred poly(A) tail length distribution to the GFP reporter mRNA, as seen with the endogenous transcripts under the same conditions. GFP protein expression from these constructs furthermore positively correlated with the tail-length (data not shown).
Next, we introduced the inducible GAL1 promoter into our GFP plasmids and followed mRNA deadenylation and protein expression over time after generating a transcriptional pulse using a galactose induction/ glucose repression regimen (Decker and Parker 1993). Shown in the upper panel of Figure 8A are LM-PAT assays of GFP transcripts terminated by 3′-UTRs derived from the RPL10, MDN1, or MCD1 genes. While the GFP-RPL10 mRNA is deadenylated slowly, GFP-MDN1 and GFP-MCD1 are deadenylated rapidly, with short-tailed forms predominating by 15 min after glucose repression. These patterns match the deadenylation kinetics and steady-state tail distributions of the corresponding endogenous transcripts (data not shown; Fig. 6C). We also performed semiquantitative PCR with primers amplifying an internal portion of the GFP transcripts (middle panel in Fig. 8A). The results suggest some increases in transcript turnover at later time points (∼ 25–30 min) for GFP-MDN1 and GFP-MCD1 compared with GFP-RPL10, and they reveal a minor resurgence of transcription at the 40-min time point. This imperfection was previously seen in this inducible system (Decker and Parker 1993) but does not affect interpretation of the experiment, which is based on observations at earlier time points. Next, we measured GFP protein expression in the same cultures by quantitative Western blotting (Fig. 8B, representative of four independent experiments). There was some lag time between mRNA induction (at 5 min) and an visible increase in GFP protein (at 10–15 min). Quantitation of the ratio of GFP signal over the GAPDH loading control (Fig. 8C) shows that, from the earliest time point onward, GFP expression correlates positively with mRNA poly(A) tail length. Similar trends were observed with GFP-chimera exhibiting the 3′-UTRs of the RPL46(39) or the CRT1 mRNA (specifying slow or fast deadenylation, respectively) (data not shown). Collectively, these results demonstrate that multiple 3′-UTR sequences can drive gene-specific differences in mRNA adenylation state outside their normal genomic and mRNA sequence context. These differences are set up through differences in mRNA deadenylation rates and correlate positively with protein synthesis from the mRNA.
DISCUSSION
poly(A) tail lengths and other mRNA characteristics
The surveys of poly(A) tail length and function in the transcriptome of S. cerevisiae reported here document the importance of poly(A) tail length control to eukaryotic gene regulation. The multiple correlative relationships of tail length and several other physical and functional characteristics of the S. cerevisiae transcriptome (Fig. 2D; Table 1) suggest a remarkable level of integration in gene expression. Comprehensive genome-wide analyses in Schizosaccharomyces pombe further demonstrate evolutionary conservation of these relationships between the distantly related yeasts (Lackner et al. 2007). Any global correlation between mRNA characteristics could imply mechanistic relationships or reflect independent co-opting of parameters to suit an overall goal in gene expression. Definitive statements in this regard are not possible on the basis of global data alone, but our evidence is indicative of particular scenarios as follows. We find a positive correlation of tail length with an ability to interact with Pab1p and mRNA translation efficiency (ribosome density) on the mRNA. This strongly suggests that the translational functions of Pab1 and poly(A) tail length identified in single gene studies are important for the expression of substantial proportions of the transcriptome.
The lack of a positive correlation between poly(A) length and mRNA half-life observed here was a surprise. Although this may be due to poor microarray data compatibility between separate studies and laboratories, we also find no such correlation in fully coherent data obtained for S. pombe (Lackner et al. 2007). Genome-wide studies have found global mRNA half-lives to be significantly longer than poly(A)+ half-lives (Wang et al. 2002), indicating that the model of poly(A) shortening as a prerequisite for further mRNA decay holds true on a transcriptome-wide level. Work with exemplary mRNAs has described poly(A) shortening to an oligo(A)7–16 tail, and the decay of this oligoadenylated intermediate as kinetically distinct and mechanistically separable phases of turnover (Decker and Parker 1993). Together with the prevalence of oligo(A)-tailed mRNA observed in our LM-PAT assays, this suggests as a testable scenario that processes acting on the oligo(A)-tailed intermediate (e.g., terminal deadenylation, decapping) may limit the decay rate of a large enough number of yeast mRNAs to preclude detection of a clear genome-wide trend. The negative correlation of ORF length and mRNA length with poly(A) tail length is reminiscent of the inverse relationship between ribosome density and ORF length found in S. cerevisiae (Arava et al. 2003) as well as S. pombe (Lackner et al. 2007). The length of an ORF is subject to constraints in terms of the encoded protein function and thus this may represent an example of co-opted parameters (see also Arava et al. 2003). Finally, the negative correlation between poly(A) tail- and 3′-UTR length is suggestive of regulatory elements residing in the 3′-UTR (see also below).
Cellular roles of poly(A) tail length control
Our PASTA analyses demonstrate a striking coregulation of poly(A) tail lengths on functionally and cytotopically related mRNAs. This is consistent with early observations that protein coding potential was not distributed evenly between cellular mRNA fractions of different poly(A) tail length in Dictyostelium discoideum (Palatnik et al. 1979), and with recent studies to identify polyadenylation-controlled neuronal mRNAs (Du and Richter 2005) and post-transcriptionally regulated Xenopus tropicalis maternal mRNAs (Graindorge et al. 2006). Our data further broadly support the “post-transcriptional operon” model, which posits that eukaryotes coregulate related mRNAs based on their functional organization in ribonucleoprotein (RNP) complexes (Keene and Lager 2005). We chose to analyze this issue further for the mRNAs encoding proteins of the cytoplasmic ribosome and ribosome biogenesis factors. Our data indicate that cells adjust the expression of RP mRNAs to be sustained and efficient by keeping the tail long, while ribosome biogenesis mRNA expression is under stricter control through more avid deadenylation. This demonstrates that poly(A) tail length control divides this group of transcriptionally coregulated mRNAs into distinct subclasses, suggesting a previously unrecognized mode of regulation of ribosome biogenesis. The latter may have interesting implications for cell-cycle control, as the rate of ribosome synthesis was proposed to set the critical cell size threshold at the G1/S phase transition of the budding yeast cell cycle (Jorgensen et al. 2004) and connections have been made between nucleolar function and DNA replication, positioning of the mitotic spindle, mitotic exit, or cytokinesis (Dez and Tollervey 2004). Further links to the cell cycle abound among the GO terms and MIPS deletion phenotypes enriched with the PASTA–short list and our deadenylase mutant strains display cell-cycle-related phenotypes. This indicates that many mRNAs encoding cell-cycle-relevant functions are present with mostly short tails in a mixed cell population, and suggests that appropriate (temporal) limitation of their expression during the cell cycle depends on proper deadenylase function (see Fig. 7C). This provides an attractive explanation for the reported genetic interactions of the deadenylases with cell-cycle regulators (Hammet et al. 2002; Westmoreland et al. 2004; Woolstencroft et al. 2006).
S. cerevisiae appears to lack cytoplasmic poly(A) polymerases to readenylate mRNAs and thus establishes any waves or spikes of polyadenylated mRNAs mainly through a combination of transcriptional control and differential deadenylation rates. S. pombe and higher eukaryotic cells possess cytoplasmic poly(A) polymerases, and ample evidence in these organisms documents the importance of cytoplasmic polyadenylation during the meiotic and mitotic cell cycle (see Introduction; Groisman et al. 2002). This suggests deep evolutionary conservation of poly(A) tail length control in the context of the eukaryotic cell cycle, which can be further explored with the PASTA method described here.
Mechanism of poly(A) tail length regulation
The genome-wide studies reported here establish global positive correlations between poly(A) tail length, Pab1 binding, and efficiency of mRNA translation. They further suggest a mechanistic involvement of the 3′-UTR, which is substantiated by experiments with GFP-3′-UTR fusion constructs. This is fully consistent with models established previously for individual examples; however, since our observations are based on genome-wide surveys, they establish these principles for a substantial portion of the yeast transcriptome. Pulse-chase analyses with the chimeric GFP mRNAs demonstrate a tight temporal correlation between mRNA tail length and translation and highlight the accumulation of an oligoadenylated mRNA intermediate prior to decay of the mRNA body. The experiments demonstrate that the 3′-UTR drives these mRNA-specific differences and suggest that changes in translation efficiency are primarily responsible for differences in protein expression. Our conclusions are valid for exponentially growing cells, but it is an attractive possibility that cells may disrupt the link between poly(A) tail length and translation in response to physiological cues, as recently described for environmental stress (Hilgers et al. 2006).
Detailed studies for each individual mRNA species will be required to determine the exact mechanistic relationships between 3′-UTR function, poly(A) tail length, translation, and mRNA decay. An example is set by the HO mRNA, which is regulated at the post-transcriptional level by binding of Puf5 and Puf4 to its 3′-UTR. Both PUF proteins interact with Pop2 and recruit the Ccr4 deadenylase and associated proteins (Tadauchi et al. 2001; Goldstrohm et al. 2006), including the translational repressor Dhh1 (Coller and Parker 2005). This enhances HO mRNA deadenylation, destabilization, and possibly also translational silencing (Goldstrohm et al. 2006). Specific mRNA targets have been identified for all six S. cerevisiae PUF proteins. Each PUF target group exhibits common themes in the functions and subcellular localization of the proteins they encode (Gerber et al. 2004), which broadly match those that we find enriched in our PASTA lists. Furthermore, genome-wide studies have shown that Ccr4, Pan2, and Puf4 all contribute to the decay of mRNAs encoding ribosomal proteins and ribosome biogenesis factors, while another RNA-binding protein, Pub1, preferentially stabilized RP mRNAs (Grigull et al. 2004). Puf6 represses translation of ASH1 mRNA and possibly several additional mRNAs that localize to the distal bud tip (Gu et al. 2004).
Oligoadenylated mRNA may represent a state of translational silencing or imminent decay, suggesting that inclusion of an mRNA in our PASTA–short list may reflect on the removal of the mRNA from polysomes toward the repressive environment of processing (P-) bodies. This notion is supported by the presence of Ty retrotransposon-derived transcripts in our PASTA–short list. Virus-like retrotransposon particle assembly takes place at or near P-bodies. As P-bodies are sites of mRNA degradation and storage (Brengues et al. 2005), it has been suggested that P-body–associated translational repression promotes assembly of retrotransposon RNA into virus-like particles (Beliakova-Bethell et al. 2006). Our observations suggest poly(A) tail length control as another aspect of this switch from translation to packaging of retrotransposon RNA. P-bodies fluctuate during the cell cycle (Lian et al. 2006), possibly tying them in with the cellular functions of poly(A) tail length control described here. Further, the P-body–resident Dhh1 binds Pop2 and DHH1 interacts genetically with CCR4 and POP2 (Hata et al. 1998; Coller et al. 2001). Thus, in current work we address the possible involvement of the PUF proteins and P-bodies in dynamically establishing transcriptome-wide poly(A) tail length patterns.
MATERIALS AND METHODS
Yeast strains and culture
To generate the protein A-tagged PAB1 strain, primers designed with 40 nucleotides (nt) of PAB1 sequence directly upstream and downstream of the native stop codon were used in PCR with pYM8 cassette (Knop et al. 1999) as a template. The integration cassette was transformed into W303-1A cells (MATa ade2-1 can1-100 his3-11,15 leu2-3112 trp1-1 ura3-1) and correct integration determined by PCR and Western blotting. Integration of this tag had no impact on cell growth under normal conditions. The CCR4/PAN2 wild-type (Y136=MATa trp11 ura3–52 gcn4 leu2∷PET56), and ccr4-1 (Y369=Y136 but ccr4-E556A) strains were gifts from Jörg Heierhorst, St. Vincent's Institute, Melbourne, Australia (Traven et al. 2005). To generate Δpan2 mutants, the disruption cassette was amplified from the Euroscarf (BY4741) deletion series and transformed into Y139 and Y369 to generate the isogenic Δpan2 (YTP1) and Δpan2/ccr4-1 (YTP2) strains.
For PASTA analyses, BY4741 cells were grown in YPAD to an OD600 of 0.5–0.8. In refeeding experiments, BY4741 cells were grown to saturation overnight, 50 OD units were harvested, washed and resuspended in 50 mL fresh media, and incubated at 30°C. Samples (5 mL) were taken at indicated time intervals into tubes containing 100 μL 10% aqueous sodium azide, pelleted, washed in dH2O, and snap-frozen for RNA analysis. For cell-cycle experiments, 50 OD units of BY4741 cells (OD600 ∼ 0.5) were harvested and resuspended in to 250 mL fresh media containing 300 μg/mL of mating factor (Sigma T-6901) and incubated for ∼ 80 min at 30°C. Cells were monitored by microscopy until > 90% were large unbudded cells (Amon 2002). Synchronized cells were harvested, washed once, and resuspended in 100 mL fresh media. Samples (∼ 5 OD units) were taken at 10-min intervals, washed in dH2O, and snap frozen for RNA analysis. DIC images were captured using a Zeiss Axioplan microscope.
GFP reporter constructs
We amplified 500 bp of 3′ sequence from within ∼ 20 bp of the gene's native stop codon with primers that include a BamHI or BglII site immediately followed by a stop codon at the 5′ end, and a KpnI site at the 3′ end to facilitate the replacement of the original CYC1 terminator of the pMET25 GFP S65T plasmid (Egan et al. 1999). Inclusion of the stop codon in the primer ensured that the same carboxy terminus was present on GFP in all cases. For steady-state RNA expression studies (data not shown), BY4741 cells transformed with the reporter constructs were grown to OD600 0.5–0.8 in synthetic glucose media supplemented with CSM-URA (Bio-101 Systems).
Plasmids for galactose transcriptional pulse chase experiments were generated by replacing the MET25 promotor (flanked by SacI and SpeI) from plasmids detailed above by the 820bp GAL1 promoter (a gift from Gabriel Perrone, UNSW, Sydney, Australia).
Poly(A) tail measurements
To measure poly(A) tail lengths of multiple individual mRNAs, we used the Ligation-Mediated poly(A) Test (LM-PAT) assay (see schematic in Fig. 1B; Sallés and Strickland 1995); 1–3 μg total RNA, or 10% of each fraction eluted from poly(U) chromatography, served as input for the LM-PAT assays. Briefly, RNA was first incubated with oligo(dT)12–18 primers in the presence of T4-DNA ligase at 42°C. This creates a poly(dT) copy of each mRNA's poly(A) tail within the sample. Addition of an excess of (dT)12–anchor primer and further incubation at 12°C favors annealing of the anchor primer to unpaired poly(A) ends and ligation to the poly(dT) stretch. This assembly is then used to prime synthesis of first-strand cDNA with Superscript II (Invitrogen). Aliquots of this cDNA were then used as templates in PCR reactions with primers specific to the mRNA 3′-UTR of choice and the anchor region. Fast-Start polymerase (Roche) was used to avoid preferential amplification of shorter amplicons. LM-PAT PCR products were visualized by 2% high-resolution agarose gel electrophoresis (Agarose 1000, Invitrogen) and scans on a FLA-5100 imager and MultiGauge software (Fuji).
For bulk poly(A) tail length analysis, 90% of each poly(U) chromatography fraction was 3′-end labeled with [32P]-pCp and digested with RNAses A and T1 as described (Minvielle-Sebastia et al. 1991). Poly(A) tracts were resolved by denaturing (urea) 16% PAGE and analyzed by autoradiography or phosphorimaging using the Fuji FLA-5100 imager.
Poly(U) Sepharose chromatography
The procedure was based on published protocols (Binder et al. 1994), with modifications. Dry poly(U)100 Sepharose 4B (GE Healthcare) was hydrated and washed three times in 0.1 M NaCl and 10 mM Tris-HCl (pH 7.4) at room temperature. Then followed washes in elution buffer (EB; 0.1 M NaCl, 0.01 M EDTA, 0.5 M Tris-HCl at pH 7.4, 0.2% SDS, 25% formamide) and high salt binding buffer (HSBB; 0.7 M NaCl, 0.01 M EDTA, 0.5 M Tris-HCl at pH 7.4, 0.2% lauryl sarcosine, 12% formamide), one time each for 5 min at 70°C. Total cellular RNA was purified using the Hot Phenol method. For each run, 600 μL of HSBB, ∼ 150 μL of wet gel volume, and 150 μg of total yeast RNA were denatured for 5 min at 70°C in 1.5 mL tubes. For binding, tubes were incubated in an Eppendorf Thermomixer (1100 rpm in a cold room), first for 10 min at 35°C, followed by cooling to 12°C for 90 min (including ∼ 25 min ramp-down time). The matrix was washed four times with HSBB at 12°C for 5 min. Each thermal elution step was performed in the Thermomixer, by resuspending the matrix in 600 μL EB at the specified temperature for 5 min. The supernatant from a 1/2 min microcentrifuge spin at 6000 rpm containing the eluted mRNA was removed with an insulin syringe, respun and precipitated in 2 vol ethanol and 1/10 vol of 5 M NaCl, and coprecipitant (glycogen or Pellet Paint, Novagen). mRNA was resuspended in H2O and desalted using BIO-RAD Micro Bio-Spin6 columns.
For high-resolution PASTA analyses, corresponding elution fractions from three parallel poly(U) chromatography runs were reverse-transcribed and labeled with Cy5. As a reference sample, three single-step eluates at 45°C were pooled and then split into five aliquots for reverse transcription and Cy3 labeling. For low-resolution PASTA analysis, the 30°C, 35°C, and 45°C fractions (Cy5-labeling) were pooled and reverse-transcribed, as were the 12°C and 25°C fractions (Cy3-labeling).
RNP affinity purification
Purification of protein A–tagged Pab1/mRNA complexes was performed as described (Gerber et al. 2004). RNA was isolated from tagged Pab1 purifications or from mock purifications using an isogenic untagged strain. In parallel, total RNA was isolated from an aliquot of each of the yeast cultures. Cy5-labeled cDNA was made from each purified RNA sample and compared on a microarray to Cy3-labeled cDNA prepared from corresponding total RNA. Microarray methods were as detailed below except that a 1:1 mixture of random decamer and anchored dT20VN was used for reverse transcription.
Microarray methods
Reverse transcription and dye coupling were performed as described in the TIGR protocol (http://pga.tigr.org/sop/M004_1a.pdf). Aliquots of spike-in RNAs (Lucidea ScoreCard, GE Healthcare) were added to samples where required. Labeled cDNAs were hybridized to spotted oligonucleotide microarrays (MWG Biotech set of 40-mer probes for 6250 yeast ORFs; UNSW Center for Gene Function Analysis). Hybridization and wash conditions were as previously described (Gelling et al. 2004). Images were recorded on an Axon GenePix 4000A scanner with software GenePix 5.0. GPR files were installed in GeneSpring 7.2 for Lowess normalization and further data processing. Microarray data are available through ArrayExpress.
For high-resolution PASTA, three biological repeat experiments were performed and replicate averaged. Weak and/or low quality signals were removed by accepting only spots flagged “present” and applying filters on minimal intensity (> 15 in both channels) and number of replicate measurements (two or more replicates in at least three of five conditions). Additional removal of 11 nonsensical elution profiles left 5772 elution profiles for further analysis. For low-resolution PASTA, the same criteria for spot quality and intensity were applied, qualifying spot ratios of 6121 mRNAs for further analysis.
RNA preparations from the protA-Pab1 and mock purifications received aliquots of spike-in RNAs (Lucidea ScoreCard, GE Healthcare) prior to reverse transcription. Array data from the protA-Pab1 and mock purifications were normalized against ScoreCard signals, replicate data averaged, and minimal spot intensity filters applied as detailed above. Each mRNA's ratio from the protA-Pab1 microarrays was finally divided by that mRNA's ratio in the mock microarrays to generate a measure of mRNA-enrichment with Pab1.
Pulse chase analysis of GFP reporter mRNAs
BY4741 cells were transformed with Gal1-driven plasmids harboring the GFP coding region followed by 500 bp of sequence taken from regions immediately 3′ of the stop codon of different genes. Cells were grown in synthetic CSM-URA (Bio-101 Systems) with 2% raffinose to an OD600 of 0.7–0.9. Two percent galactose was added to activate transcription. Repression was achieved by the addition of 4% glucose. Aliquots of cells (5 OD600 units for RNA or 1 OD600 unit for protein analysis) were harvested directly into prechilled tubes containing 1/100 volume of 10% aqueous sodium azide at each time point. Cells for RNA extraction were flash-frozen and treated as above. Total cellular protein was extracted using the NaOH/TCA method (Yaffe and Schatz 1984). Quantitative Western blot was performed using the LI-COR Odyssey Infrared Imager. Anti-GFP and -GAPDH antibodies were gifts form Trevor Lithgow, University of Melbourne, Melbourne, Australia. Fluorescent secondary antibodies were anti-mouse/alexa-680 and anti-rabbit/alexa-750 conjugated antibodies (Invitrogen).
ACKNOWLEDGMENTS
We thank Jörg Heierhorst, Gabriel Perrone, Chris Norbury, and Trevor Lithgow for gifts of reagents. Jürg Bähler is acknowledged for sharing results prior to publication, and Roy Parker for valuable comments on the manuscript. Matthias Hentze is thanked for his support of early exploratory experiments. This work was funded by grants from the Australian Research Council, the UNSW Goldstar Programme, and the Sylvia & Charles Viertel Charitable Foundation to T.P. T.H.B. was the recipient of a National Health and Medical Research Council Howard Florey Fellowship.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.569407.
REFERENCES
- Amon, A. Synchronization procedures. Methods Enzymol. 2002;351:457–467. doi: 10.1016/s0076-6879(02)51864-4. [DOI] [PubMed] [Google Scholar]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer, M.A., Tavazoie, S. Predicting gene expression from sequence. Cell. 2004;117:185–198. doi: 10.1016/s0092-8674(04)00304-6. [DOI] [PubMed] [Google Scholar]
- Beissbarth, T., Speed, T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Beliakova-Bethell, N., Beckham, C., Giddings T.H., Jr, Winey, M., Parker, R., Sandmeyer, S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit, B., Mitou, G., Chartier, A., Temme, C., Zaessinger, S., Wahle, E., Busseau, I., Simonelig, M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila . Dev. Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Berthet, C., Morera, A.M., Asensio, M.J., Chauvin, M.A., Morel, A.P., Dijoud, F., Magaud, J.P., Durand, P., Rouault, J.P. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol. Cell. Biol. 2004;24:5808–5820. doi: 10.1128/MCB.24.13.5808-5820.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer, A., Hollunder, J., Nasheuer, H.P., Wilhelm, T. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics. 2004;3:1083–1092. doi: 10.1074/mcp.M400099-MCP200. [DOI] [PubMed] [Google Scholar]
- Binder, R., Horowitz, J.A., Basilion, J.P., Koeller, D.M., Klausner, R.D., Harford, J.B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′-UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, C.E., Sachs, A.B. Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol. Cell. Biol. 1998;18:6548–6559. doi: 10.1128/mcb.18.11.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colegrove-Otero, L.J., Minshall, N., Standart, N. RNA-binding proteins in early development. Crit. Rev. Biochem. Mol. Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Coller, J., Parker, R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron, M.C., Mauxion, F., Seraphin, B. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001;29:2448–2455. doi: 10.1093/nar/29.12.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dez, C., Tollervey, D. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 2004;7:631–637. doi: 10.1016/j.mib.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Du, L., Richter, J.D. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, E.F., Hammell, C.M., Hodge, C.A., Cole, C.N. Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Genes & Dev. 2005;19:90–103. doi: 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, B., Beilharz, T., George, R., Isenmann, S., Gratzer, S., Wattenberg, B., Lithgow, T. Targeting of tail-anchored proteins to yeast mitochondria in vivo. FEBS Lett. 1999;451:243–248. doi: 10.1016/s0014-5793(99)00581-5. [DOI] [PubMed] [Google Scholar]
- Gasch, A.P., Spellman, P.T., Kao, C.M., Carmel-Harel, O., Eisen, M.B., Storz, G., Botstein, D., Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelling, C.L., Piper, M.D., Hong, S.P., Kornfeld, G.D., Dawes, I.W. Identification of a novel one-carbon metabolism regulon in Saccharomyces cerevisiae . J. Biol. Chem. 2004;279:7072–7081. doi: 10.1074/jbc.M309178200. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:E79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez, A.J., Mishima, Y., Rihel, J., Grocock, R.J., Van Dongen, S., Inoue, K., Enright, A.J., Schier, A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goldstrohm, A.C., Hook, B.A., Seay, D.J., Wickens, M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 2006;13:533–539. doi: 10.1038/nsmb1100. [DOI] [PubMed] [Google Scholar]
- Graber, J.H., McAllister, G.D., Smith, T.F. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge, A., Thuret, R., Pollet, N., Osborne, H.B., Audic, Y. Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 2006;34:986–995. doi: 10.1093/nar/gkj492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigull, J., Mnaimneh, S., Pootoolal, J., Robinson, M.D., Hughes, T.R. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals post-transcriptional control of ribosome biogenesis factors. Mol. Cell. Biol. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman, I., Jung, M.Y., Sarkissian, M., Cao, Q., Richter, J.D. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- Gu, W., Deng, Y., Zenklusen, D., Singer, R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammet, A., Pike, B.L., Heierhorst, J. Posttranscriptional regulation of the RAD5 DNA repair gene by the Dun1 kinase and the Pan2-Pan3 poly(A)-nuclease complex contributes to survival of replication blocks. J. Biol. Chem. 2002;277:22469–22474. doi: 10.1074/jbc.M202473200. [DOI] [PubMed] [Google Scholar]
- Hata, H., Mitsui, H., Liu, H., Bai, Y., Denis, C.L., Shimizu, Y., Sakai, A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae . Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze, M.W., Gebauer, F., Preiss, T. Cis-regulatory sequences and trans-acting factors in translational control. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 273–299. [Google Scholar]
- Hilgers, V., Teixeira, D., Parker, R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae . RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys, D.T., Westman, B.J., Martin, D.I., Preiss, T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurowitz, E.H., Brown, P.O. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae . Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., Rupes, I., Sharom, J.R., Schneper, L., Broach, J.R., Tyers, M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes & Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D., Lager, P.J. Post-transcriptional operons and regulons coordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Knop, M., Siegers, K., Pereira, G., Zachariae, W., Winsor, B., Nasmyth, K., Schiebel, E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Lackner, D.H., Beilharz, T.H., Marguerat, S., Mata, J., Watt, S., Schubert, F., Preiss, T., Bahler, J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol. Cell. 2007;26:145–155. doi: 10.1016/j.molcel.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian, S., Jakymiw, A., Eystathioy, T., Hamel, J.C., Fritzler, M.J., Chan, E.K. GW bodies, MicroRNAs and the cell cycle. Cancer Biol. Ther. 2006;5:242–245. doi: 10.4161/cc.5.3.2410. [DOI] [PubMed] [Google Scholar]
- Liu, H.Y., Toyn, J.H., Chiang, Y.C., Draper, M.P., Johnston, L.H., Denis, C.L. DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J. 1997;16:5289–5298. doi: 10.1093/emboj/16.17.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay, V.L., Li, X., Flory, M.R., Turcott, E., Law, G.L., Serikawa, K.A., Xu, X.L., Lee, H., Goodlett, D.R., Aebersold, R., et al. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: Response of yeast to mating pheromone. Mol. Cell. Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., Jacobson, A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Smith, M.M., McSweeney, J.M., Jacobson, A. Identification of factors regulating poly(A) tail synthesis and maturation. Mol. Cell. Biol. 2004;24:4196–4206. doi: 10.1128/MCB.24.10.4196-4206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, R., Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Keller, W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- Minvielle-Sebastia, L., Winsor, B., Bonneaud, N., Lacroute, F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate: Sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.Z., Hong, A., Lilly, M.A., Lehmann, R. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005;132:1165–1174. doi: 10.1242/dev.01672. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes & Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- Palatnik, C.M., Storti, R.V., Jacobson, A. Fractionation and functional analysis of newly synthesized and decaying messenger RNAs from vegetative cells of Dictyostelium discoideum . J. Mol. Biol. 1979;128:371–395. doi: 10.1016/0022-2836(79)90093-7. [DOI] [PubMed] [Google Scholar]
- Pan, X., Ye, P., Yuan, D.S., Wang, X., Bader, J.S., Boeke, J.D. A DNA integrity network in the yeast Saccharomyces cerevisiae . Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Parker, R., Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Preiss, T., Muckenthaler, M., Hentze, M.W. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA. 1998;4:1321–1331. doi: 10.1017/s1355838298980669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss, T., Baron-Benhamou, J., Ansorge, W., Hentze, M.W. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat. Struct. Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., Dye, M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Radonjic, M., Andrau, J.C., Lijnzaad, P., Kemmeren, P., Kockelkorn, T.T., van Leenen, D., van Berkum, N.L., Holstege, F.C. Genome-wide analyses reveal RNA polymerase II located upstream of genes poised for rapid response upon S. cerevisiae stationary phase exit. Mol. Cell. 2005;18:171–183. doi: 10.1016/j.molcel.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Read, R.L., Norbury, C.J. Roles for cytoplasmic polyadenylation in cell cycle regulation. J. Cell. Biochem. 2002;87:258–265. doi: 10.1002/jcb.10300. [DOI] [PubMed] [Google Scholar]
- Robinson, M.D., Grigull, J., Mohammad, N., Hughes, T.R. FunSpec: A web-based cluster interpreter for yeast. BMC Bioinformatics. 2002;3:35. doi: 10.1186/1471-2105-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B., Varani, G. Eukaryotic translation initiation: There are (at least) two sides to every story. Nat. Struct. Biol. 2000;7:356–361. doi: 10.1038/75120. [DOI] [PubMed] [Google Scholar]
- Sallés, F.J., Strickland, S. Rapid and sensitive analysis of mRNA polyadenylation states by PCR. PCR Methods Appl. 1995;4:317–321. doi: 10.1101/gr.4.6.317. [DOI] [PubMed] [Google Scholar]
- Schwartz, D.C., Parker, R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi, T., Matsumoto, K., Herskowitz, I., Irie, K. Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 2001;20:552–561. doi: 10.1093/emboj/20.3.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven, A., Hammet, A., Tenis, N., Denis, C.L., Heierhorst, J. Ccr4-not complex mRNA deadenylase activity contributes to DNA damage responses in Saccharomyces cerevisiae . Genetics. 2005;169:65–75. doi: 10.1534/genetics.104.030940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Tucker, M., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D., Parker, R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae . EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Liu, C.L., Storey, J.D., Tibshirani, R.J., Herschlag, D., Brown, P.O. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland, T.J., Marks, J.R., Olson J.A., Jr, Thompson, E.M., Resnick, M.A., Bennett, C.B. Cell cycle progression in G1 and S phases is CCR4 dependent following ionizing radiation or replication stress in Saccharomyces cerevisiae . Eukaryot. Cell. 2004;3:430–446. doi: 10.1128/EC.3.2.430-446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens, M., Bernstein, D.S., Kimble, J., Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Woolstencroft, R.N., Beilharz, T.H., Cook, M.A., Preiss, T., Durocher, D., Tyers, M. Ccr4 contributes to tolerance of replication stress through control of CRT1 mRNA poly(A) tail length. J. Cell Sci. 2006;119:5178–5192. doi: 10.1242/jcs.03221. [DOI] [PubMed] [Google Scholar]
- Wu, L., Fan, J., Belasco, J.G. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe, M.P., Schatz, G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, A., Chang, T.C., Yamashita, Y., Zhu, W., Zhong, Z., Chen, C.Y., Shyu, A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Schneider, C., Ottmers, L., Rodriguez, R., Day, A., Markwardt, J., Schneider, B.L. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae . Curr. Biol. 2002;12:1992–2001. doi: 10.1016/s0960-9822(02)01305-2. [DOI] [PubMed] [Google Scholar]