Abstract
Two cyclooxygenase (COX) enzymes, COX-1 and COX-2, are present in human cells. While COX-1 is constitutively expressed, COX-2 is inducible and up-regulated in response to many signals. Since increased transcriptional activity accounts for only part of COX-2 up-regulation, we chose to explore other RNA processing mechanisms in the regulation of this gene. Previously, we showed that COX-2 is regulated by alternative polyadenylation, and that the COX-2 proximal polyadenylation signal contains auxiliary upstream sequence elements (USEs) that are very important in efficient polyadenylation. To explore trans-acting protein factors interacting with these cis-acting RNA elements, we performed pull-down assays with HeLa nuclear extract and biotinylated RNA oligonucleotides representing COX-2 USEs. We identified PSF, p54nrb, PTB, and U1A as proteins specifically bound to the COX-2 USEs. We further explored their participation in polyadenylation using MS2 phage coat protein-MS2 RNA binding site tethering assays, and found that tethering any of these four proteins to the COX-2 USE mutant RNA can compensate for these cis-acting elements. Finally, we suggest that these proteins (p54nrb, PTB, PSF, and U1A) may interact as a complex since immunoprecipitations of the transfected MS2 fusion proteins coprecipitate the other proteins.
Keywords: 3′-untranslated region (UTR), polyadenylation, cyclooxygenase-2 (COX-2)
INTRODUCTION
Cyclooxygenase isoenzymes 1 and 2 (COX-1 and -2) both catalyze oxygenation and peroxidation of arachidonic acid. This is the rate-limiting step in prostanoid biosynthesis and results in production of important biological mediators such as prostaglandins, thromboxanes, and prostacyclins. The two COX enzymes are ∼60% identical in humans and the same holds true within other species. Moreover, the exon–intron arrangement and amino acid sequence of COX-1 and COX-2 are very similar. Interestingly, the 3′-untranslated region (3′-UTR) of COX-2 is highly divergent from that of COX-1 (Xie et al. 1991; Hla and Neilson 1992; O'Banion et al. 1992; Appleby et al. 1994; Garavito and Mulichak 2003, and references therein). While COX-1 is a constitutive enzyme that makes prostaglandins for homeostatic physiological functions, COX-2 is induced by a wide variety of stimuli (Hla and Maciag 1991; Feng et al. 1993; Bazan et al. 1994; Ristimaki et al. 1994; O'Banion et al. 1996; Hla et al. 1999; Bazan 2001, and references therein). Exaggerated COX-2 protein expression is associated with pathological conditions; misregulated expression of COX-2 has been observed in rheumatoid arthritis and in cancers such as colorectal, breast, gastric, cervical, ovarian, and nonsmall cell lung cancers (San et al. 1995; Kutchera et al. 1996; Hwang et al. 1998; Williams et al. 1999; Prescott 2000; Prescott and Fitzpatrick 2000; Bishop-Bailey et al. 2002; Landen et al. 2003).
One common cellular pathway of increasing a protein's expression is to increase transcription of its mRNA. However, transcriptional activation of COX-2 mRNA by such mediators as interleukin-1β cannot alone account for the induced expression of COX-2 protein, suggesting a post-transcriptional mechanism of regulation (Ristimaki et al. 1994). It is now evident that the stability of the mRNA plays one role in the regulation of COX-2 (Ristimaki et al. 1996; Gou et al. 1998; Cok and Morrison 2001; Dixon et al. 2000, 2001, 2003). In addition, alternative COX-2 transcripts differing only in their 3′-UTRs have been found in a number of cell and tissue types (Ristimaki et al. 1996; Bracken et al. 1997; Lukiw and Bazan 1997; Newton et al. 1997, 1998; Dixon et al. 2001; Schmidt et al. 2003). Therefore, RNA stability and alternative 3′-end processing are two important regulatory mechanisms that can contribute to the expression control of COX-2.
3′-End formation of most mRNAs is accomplished by the process of polyadenylation (for review, see Colgan and Manley 1997; Wahle and Kuhn 1997; Wickens et al. 1997; Zhao et al. 1999; Proudfoot 2000; Edmonds 2002, and references therein). Although polyadenylation is distinct from other RNA processing events, it is now accepted that all RNA processing events are cotranscriptional and interdependent (for review, see Maniatis and Reed 2002; Proudfoot et al. 2002; Bentley 2005, and references therein). Identification of polyadenylation signals is accomplished by recognition of two core sequence elements, usually a hexamer (AAUAAA) and a U/GU-rich downstream element. However, the mechanism for selecting among alternative polyadenylation signals, that is, more than one polyadenylation signal within an mRNA, is still obscure. Accordingly, use of an alternative polyadenylation signal can alter the structure of the 3′-UTR and can impact the final amount of protein product (Edwalds-Gilbert et al. 1997). Recent bioinformatic studies concluded that ∼54% of human genes and ∼32% of mouse genes have multiple polyadenylation signals (Tian et al. 2005). This comprehensive study shed light on the need to better understand alternative polyadenylation.
It is also unclear how polyadenylation signals that vary from the consensus elements, that is, “suboptimal” core elements, are recognized. It may be that auxiliary cis-acting motifs or elements (that is, RNA elements found on the same RNA molecule) aid in use of such suboptimal polyadenylation signals. Bioinformatic examination of sequences flanking the poly(A) addition site identified not only core elements (AAUAAA hexamer and U/GU-rich downstream element) but also found additional, overrepresented cis-acting element motifs that may be involved in the regulation of polyadenylation (Hu et al. 2005). The cis-acting RNA auxiliary elements identified include elements upstream of the polyadenylation signal that are reminiscent of known upstream sequence elements (USEs), previously shown to influence regulation of polyadenylation signal usage, including Simian Virus 40 (SV40), C2 complement, and collagen polyadenylation signals (Carswell and Alwine 1989; Moreira et al. 1995, 1998; Natalizio et al. 2002). These USEs may interact with trans-acting protein factors that, in turn, influence mRNA processing and may be involved in enhancing polyadenylation at alternative or suboptimal signals.
We have previously investigated alternative polyadenylation of the human COX-2 mRNA (Hall-Pogar et al. 2005). COX-2 has two predominant polyadenylation signals, which we called proximal and distal, the use of which results in mRNAs of ∼2.8 kb and ∼4.6 kb, respectively. We also identified auxiliary cis-acting USEs upstream of the proximal COX-2 polyadenylation signal. These USEs directly influenced usage of the COX-2 proximal polyadenylation signal, as their mutation significantly diminished polyadenylation in in vivo polyadenylation assays (Hall-Pogar et al. 2005).
Our previous studies have shown that the COX-2 USE sequence, when added as a oligonucleotide competitor to in vitro polyadenylation reactions, can decrease polyadenylation, presumably by titrating away or sequestering cellular factors important to polyadenylation (Hall-Pogar et al. 2005). It was therefore important to identify the trans-acting protein factors that specifically interact with COX-2 USEs to better understand the mechanism and regulation of COX-2 alternative polyadenylation. The results presented here demonstrate that U1A, p54nrb, PSF, and PTB proteins specifically interact with COX-2 USEs. These proteins were able to bind specifically to USEs as demonstrated by affinity purification and mass spectrometry identification. Extracts depleted of these factors did not polyadenylate substrate RNA despite the presence of core polyadenylation factors. Finally, the need for cis-acting sequence elements for in vivo polyadenylation of these reporter mRNAs was abrogated by phage MS2 coat protein-MS2 RNA binding site tethering assays in which the trans-acting protein factors were tethered to a USE-mutated COX-2 polyadenylation signal. These data suggest that these USE-specific factors may stabilize or promote assembly of the polyadenylation machinery on the polyadenylation signal.
RESULTS
Specific proteins interact with the COX-2 USEs
We have previously identified three USEs upstream of the COX-2 proximal polyadenylation signal that affect the efficiency of polyadenylation at that signal (Hall-Pogar et al. 2005). This polyadenylation signal was shown to be suboptimal, and we suggested then that the presence of the USEs enhanced usage of this polyadenylation signal (Hall-Pogar et al. 2005). We showed that in vitro polyadenylation reactions containing a SVL substrate RNA could be specifically inhibited using an oligoribonucleotide representing the COX-2 USEs in a dose-dependent manner, suggestive of titratable protein factors (Hall-Pogar et al. 2005). We now wanted to define the trans-acting protein factors that interact with these specific sequence elements and study overall implications of their impact on polyadenylation.
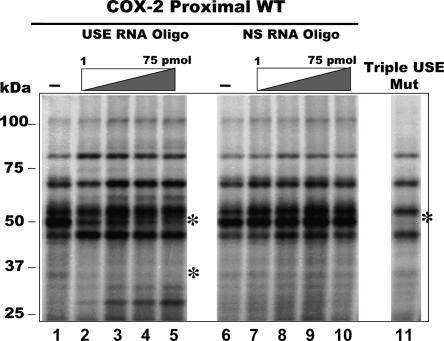
To visualize the RNA-binding proteins that interact with the COX-2 proximal polyadenylation signal, we performed UV cross-linking reactions with radiolabeled RNA representing the COX-2 proximal polyadenylation signal. These experiments used HeLa nuclear extract and 32P-labeled in vitro transcribed pGEM4-COX-2-Proximal RNAs (see Hall-Pogar et al. 2005 for a detailed description of the COX-2 RNA). As shown in Figure 1, lanes 1 and 6, a specific pattern was detected for radiolabeled proteins that specifically bound to and associated with the COX-2 proximal RNA.
FIGURE 1.
Analysis of proteins which are UV-cross-linked and competed by specific USE oligoribonucleotides. Radiolabeled RNA transcript representing wild-type COX-2 proximal polyadenylation signal was incubated with HeLa nuclear extract and UV cross-linked in the absence of any oligoribonucleotides (lanes 1,6), in the presence of increasing amounts of a COX-2 USE oligoribonucleotide (1, 10, 25, 50 nmol oligo, lanes 2–5, respectively), or in the presence of increasing amounts of a nonspecific oligoribonucleotide (1, 10, 25, 50 nmol oligo, lanes 7–10, respectively). (Lane 11) RNA representing the COX-2 proximal polyadenylation signal with mutations in all three COX-2 USEs was used in a UV cross-linking reaction, called Triple USE mut (Hall-Pogar et al. 2005).
To further define and identify the proteins that are specifically interacting with the USE elements of the COX-2 RNA, we used oligoribonucleotides representing COX-2 USE #3 and a nonspecific sequence as competitors in the UV-cross-linking reactions (Hall-Pogar et al. 2005). Each COX-2 USE is similar; USE #3 was used previously in competitions, and mutation of this particular USE caused the largest decrease in in vivo polyadenylation assays (Hall-Pogar et al. 2005). With addition of increasing amounts of the specific COX-2 USE competitor oligo, several specific protein bands began to disappear or be greatly reduced in intensity, as shown in Figure 1, lanes 2–5. The differences in the UV cross-linking patterns suggested that the competitor oligo was competing for binding to specific trans-acting factors that were bound to the USE sequence on the labeled substrate RNA. Addition of nonspecific oligo competitor did not result in any significant changes in the pattern of proteins associated with the COX-2 proximal RNA (Fig. 1, lanes 7–10). The protein–RNA interactions could not be competed further with additional amounts of competitor (data not shown).
We next tested the effect of USE mutations on the UV cross-linking pattern. When UV cross-linking reactions were performed using HeLa nuclear extract and 32P-labeled substrate RNA representing the COX-2 proximal polyadenylation signal with all three USEs mutated (Triple USE mut; Hall-Pogar et al. 2005), there was a concurrent loss in specific protein bands of a molecular weight similar to those seen in the USE oligo competition (Fig. 1, cf. lanes 5 and 11). This indicates that by either mutating the cis-acting RNA elements or by competing for binding to the trans-acting protein factors away with USE oligo similar loss of protein association was observed. Therefore, we conclude the USE elements of the COX-2 proximal polyadenylation signal interact with specific trans-acting factors.
Depletion with the USE sequence removes specific proteins and inhibits polyadenylation
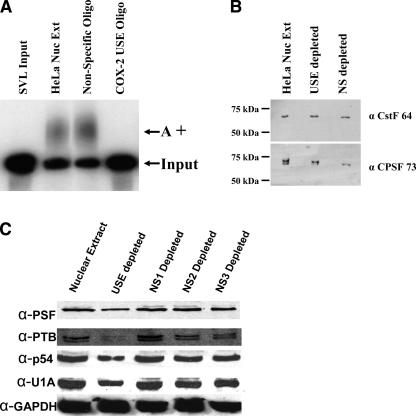
We next investigated the role(s) of the trans-acting protein factors that interact with COX-2 USEs. Previously we have shown that titration of these possible trans-acting protein factors from COX-2 USE sequences can affect polyadenylation of an SV40 late polyadenylation substrate RNA (SVL) in an in vitro polyadenylation assay (Hall-Pogar et al. 2005). COX-2 USEs are very similar in sequence and position relative to the AUUAAA core upstream polyadenylation element as compared to SVL USEs (Lutz and Alwine 1994; Hall-Pogar et al. 2005). To further confirm this observation, we used biotinylated oligoribonucleotides representing USEs to deplete HeLa nuclear extract of proteins that specifically associate with the USE. These RNA sequences were the same as the oligo competitors used in Figure 1. After isolation of biotin RNA-bound proteins on streptavidin agarose, the supernatant was used in in vitro polyadenylation reactions using an SVL polyadenylation substrate RNA. Polyadenylation was nearly abolished after the HeLa nuclear extract was depleted using the COX-2 USE oligo (Fig. 2A). There was no notable change in the amount of polyadenylation after HeLa nuclear extract was depleted using the nonspecific oligo (Fig. 2A).
FIGURE 2.
COX-2 USE depleted HeLa nuclear extracts are unable to polyadenylate substrate RNAs in vitro, yet the core polyadenylation factors remain. Biotinylated COX-2 USE or nonspecific oligoribonucleotides were used to deplete HeLa nuclear extract. Depleted supernatants were then used in in vitro polyadenylation reactions with SVL substrate RNA. (A) Results of in vitro polyadenylation reactions. SVL input, unreacted substrate RNA; HeLa Nuc Ext, regular undepleted nuclear extract; nonspecific oligo, nonspecific oligo depleted extract; COX-2 USE oligo, COX-2 USE oligo depleted extract; A+, polyadenylated product. (B) Western blot of depleted and undepleted extracts using antibodies to two core polyadenylation factors, CstF-64 and CPSF-73, as indicated at right. HeLa Nuc Ext, regular undepleted nuclear extract; USE depleted, COX-2 USE oligo depleted extract; NS depleted, nonspecific oligo depleted extract. Molecular weight markers are indicated at left. (C) Western blot of depleted and nondepleted extracts using specific antibodies as indicated to the left. Nuclear extract, HeLa undepleted nuclear extract; USE depleted, supernatant of biotinylated COX-2 USE oligo depleted HeLa nuclear extract; NS1 depleted, supernatant of biotinylated NS1 oligo depleted HeLa nuclear extract; NS2 depleted, supernatant of biotinylated NS2 oligo depleted HeLa nuclear extract; NS3 depleted, supernatant of biotinylated NS3 oligo depleted HeLa nuclear extract. See Materials and Methods for sequences of the oligos.
To determine whether the core polyadenylation factors were still present in the USE-depleted nuclear extract, we performed Western blot analysis on the depleted extracts. CstF-64 and CPSF-73 were chosen to be examined because CstF-64 is the critical factor that binds to the core downstream polyadenylation element (Wilusz et al. 1990), and CPSF-73 has been reported to be the endonuclease (Ryan et al. 2004). We found that both CstF-64 and CPSF-73 were still present in the extracts after either depletion with the COX-2 USE sequence or the nonspecific sequence (Fig. 2B). This indicates that proteins that associated with the COX-2 USE oligo did not deplete the core components of the polyadenylation complexes.
The COX-2 proximal USEs interact with PSF, p54nrb, PTB, and U1A
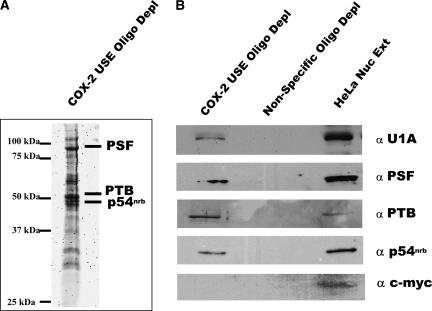
We next wanted to identify trans-acting factors that specifically associated with COX-2 USEs. In order to accomplish this, we used a biotinylated pull-down assay with the biotinylated COX-2 USE oligo and HeLa nuclear extract. The COX-2 USE biotinylated RNA oligo and bound proteins were isolated on streptavidin agarose and, after extensive washing, the proteins were separated by gel electrophoresis on a 12% SDS-PAGE gel. This gel was stained with Sypro Ruby for visualization, and bands specific to the USE oligo were excised and digested with trypsin, and tryptic peptides were analyzed by mass spectrometry. MALDI-TOF/TOF analysis revealed that polypyrimidine tract binding protein associated splicing factor (PSF), polypyrimidine tract binding protein (PTB), and p54 nonO-related binding protein (p54nrb) proteins specifically associated with the COX-2 USE sequence (Fig. 3A). These proteins are multifunctional nuclear proteins (Peng et al. 2006) and are further discussed later. The proteins were assigned by comparison with masses calculated from amino acid sequences followed by analysis with a local MASCOT search engine (V.1.9) against data present in NCBI. Peptides recognized as belonging to specific proteins with a confidence interval of 95% or greater were considered to be identified. These specific proteins were not found when the biotinylated nonspecific oligo was used (data not shown).
FIGURE 3.
Specific proteins interact with the COX-2 USE oligoribonucleotide. (A) Sypro Ruby stained gel from which protein bands were excised and submitted for MALDI TOF/TOF analysis. Identified proteins are indicated on the right, molecular weight markers at left. (B) Western blot analyses confirm that the identified proteins do interact specifically with the COX-2 USE oligoribonucleotide and not a nonspecific oligo. Proteins bound to biotinylated COX-2 USE oligo or a nonspecific oligo were isolated using streptavidin agarose followed by extensive washing, then bound proteins were separated on a 12% SDS-PAGE gel and Western blotted with antibodies as indicated to the right. HeLa nuclear extract (HeLa Nuc Ext) and anti-c-myc antibody were used as controls.
To confirm the association of the PSF, PTB, and p54nrb proteins with the COX-2 USE, the biotinylated RNA oligo pull-down assay was repeated with HeLa nuclear extract using the USE and nonspecific oligos. Bound proteins were separated on a 12% SDS-PAGE gel, which was transferred to nitrocellulose. Western blot analysis was then performed using antibodies specific for each individual protein. PSF, PTB, and p54nrb all specifically coprecipitated with the USE (Fig. 3B). At the same time, none of the proteins interacted with the nonspecific sequence (Fig. 3B).
Previously, it was shown that U1A can interact with the USEs of SV40 (Lutz and Alwine 1994). Mass spectrometry was not able to conclusively identify the U1A protein in our sample, but there were bands present in the 32-kDa range. In order to address the possibility that one of these bands might be U1A, we utilized Western blotting. The biotinylated pull-down was repeated, and the gel was then Western blotted. Antibodies specific for U1A were able to specifically detect the presence of the U1A protein bound to the COX-2 USE oligo (Fig. 3B), while U1A did not coprecipitate with the nonspecific oligo. Therefore, we concluded that U1A, PTB, PSF, and p54nrb proteins specifically interact with the COX-2 USEs.
We have also used three additional nonspecific biotinylated oligos of 19 nucleotides (nt) each, in addition to the biotinylated USE oligo in HeLa nuclear extract pull-down assays (Fig. 2C). We found that while the biotinylated COX-2 USE oligo showed diminished PSF, p54nrb, U1A, and PTB proteins in the depleted extract as measured by Western blotting, the nonspecific oligos had no effect on the abundance of these proteins (Fig. 2C). GAPDH was not affected by any biotinylated oligo treatment. Also, these new nonspecific oligos did not affect in vitro polyadenylation (data not shown). Taken together, these data suggest that the USE oligo specifically depletes PSF, p54nrb, PTB, and U1A proteins, and that polyadenylation activity in vitro is diminished in depleted extracts.
A novel interaction between U1A and PTB indicates that these proteins may interact together at the COX-2 USEs
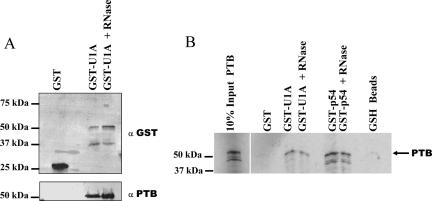
We next wanted to address protein–protein interactions of these proteins that we found associated with the COX-2 USE. Previously, we have shown that U1A is able to interact directly with both PSF (Lutz et al. 1998) and p54nrb (Liang and Lutz 2006). These data suggested that these protein interactions can directly affect polyadenylation reactions. Furthermore, PTB has been found to interact with the USEs of lamin B2 (Brackenridge and Proudfoot 2000) and complement C2 (Moreira et al. 1998) and to affect the efficiency of polyadenylation. To investigate a possible interaction between U1A and PTB proteins, an in vitro GST pull-down experiment was performed using GST-U1A fusion protein and HeLa nuclear extract. The GST-U1A fusion protein was prepared as previously described (Lutz et al. 1996). After isolation using glutathione-sepharose beads and extensive washing, the complexes were analyzed on a 10% SDS-polyacrylamide gel. The gel was transferred to nitrocellulose and the resulting Western blot was probed with anti-GST antibody and anti-PTB antibodies (Fig. 4A). The GST-U1A fusion protein was able to coprecipitate the PTB protein (Fig. 4A). Additionally, this interaction was not affected by the presence of RNase A, suggesting that the interaction is direct and not mediated by RNA bridging.
FIGURE 4.
PTB interacts directly with U1A protein. (A) HeLa nuclear extracts were incubated with the GST moiety alone (GST) or with GST-full length U1A protein. Bound proteins were separated on a 12% SDS-PAGE gel and were assayed using Western blotting with anti-GST antibodies (top) or with anti-PTB antibodies (bottom). Addition of RNase A did not affect the U1A-PTB interaction (GST U1A + RNase). Molecular weight markers are indicated to the left. (B) GST pull-down assays were performed in the presence of in vitro transcribed and translated 35S methionine labeled PTB protein. GST fusion proteins used in this experiment were the GST moiety alone (GST), GST full-length U1A (GST-U1A), or GST-p54nrb (GST-p54). After incubation of the GST fusion proteins to the 35S PTB and subsequently to the glutathione beads, extensive washing took place, and the bound proteins were separated on a 12% SDS-PAGE gel. Any proteins bound to the glutathione-sepharose beads when incubated alone with the 35S PTB were shown in the last lane (GSH beads). Proteins were visualized by autoradiography.
To further establish an interaction between U1A and PTB we analyzed protein–protein interactions using the GST-U1A fusion protein and an in vitro transcribed and translated 35S-labeled PTB protein. The glutathione-binding moiety (GST) alone was used as a negative control. After incubation, the complexes containing the beads, GST-U1A, and 35S-labeled PTB were washed extensively and were analyzed on a 10% SDS-polyacrylamide gel and visualized by autoradiography. Figure 4B shows that PTB bound specifically to the GST-U1A and not to the GST alone. The addition of RNase to the reaction did not affect this protein–protein interaction, indicating that RNA does not mediate the U1A and PTB interaction. GST-p54 also demonstrates an interaction between p54nrb and PTB that is not sensitive to RNase A (Fig. 4B). Although interactions between U1A, PSF, and p54nrb have been previously described (Liang and Lutz 2006), this is the first evidence of a direct protein–protein interaction between U1A and PTB proteins.
USE binding proteins can enhance the polyadenylation at the COX-2 proximal polyadenylation signal by interacting with the USEs
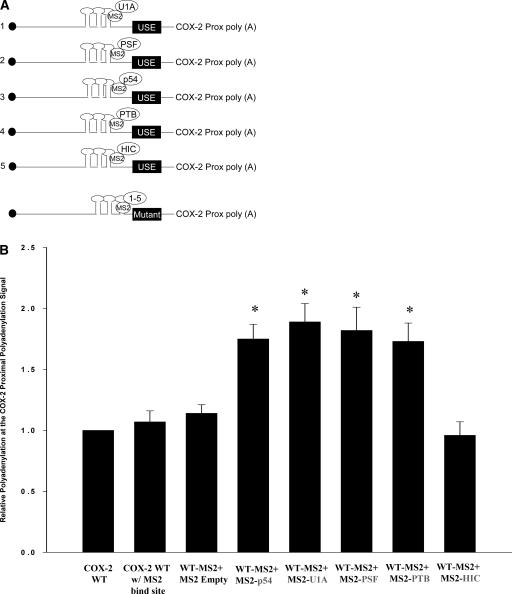
In order to determine the influence of the PSF, p54nrb, PTB, and U1A proteins in vivo at the COX-2 USEs, we employed a system whereby we could specifically tether these proteins to the COX-2 proximal polyadenylation signal. We utilized the highly specific binding of the bacteriophage MS2 19-nt RNA stem–loop structure to a version of the MS2 coat protein that binds the stem–loop but is no longer able to dimerize (Lykke-Andersen et al. 2000). These researchers found that six copies of the MS2 stem–loops worked well (J. Lykke-Andersen, pers. comm.). We specifically engineered the placement of these six MS2 stem–loops upstream of the COX-2 proximal polyadenylation signal in our tandem polyadenylation reporter vector, pCßS-COX-2 proximal (Fig. 5A; see also Hall-Pogar et al. 2005). Constructs that expressed MS2 fusion proteins were then prepared using the PSF, p54nrb, PTB, U1A, and HIC proteins, to direct tethered binding of these trans-acting protein factors to the COX-2 proximal polyadenylation signal. HIC is a nuclear RNA binding protein that binds specifically to the 3′-UTR of a protein called I-mfa (Young et al. 2003) and was used as a negative control. Fusion proteins of the expected sizes were easily detected by immunoblotting with anti-FLAG tag antibody (data not shown).
FIGURE 5.
MS2 tethering assays show that each protein can enhance polyadenylation. (A) Schematic of each protein fused to MS2 coat protein and cotransfected with a reporter vector containing wild-type COX-2 proximal polyadenylation signal with MS2 stem-loop structures inserted upstream. Only three stem-loops are depicted; actually six were inserted. (Bottom) Each individual MS2 coat protein fusion was also cotransfected with a similar COX-2 construct that had the three USE elements mutated. (B) Quantification of RNase protection assays using cotransfections of COX-2 wild-type reporter vector containing MS2 binding sites and individual MS2 coat-protein fusion proteins. Results are shown as relative polyadenylation at the COX-2 polyadenylation signal, with usage of the COX-2 proximal polyadenylation signal set as a value of one. COX-2 WT w/MS2 bind site, COX-2 proximal polyadenylation signal with six MS2 stem–loops inserted; WT-MS2 + MS2 empty, cotransfected with MS2 coat protein vector alone; WT-MS2 + MS2 p54, cotransfected with MS2-p54nrb; WT-MS2 + U1A, cotransfected with MS2-U1A; WT-MS2 + PSF, cotransfected with MS2-PSF; WT + MS2 PTB, cotransfected with MS2-PTB; WT-MS2 + HIC, cotransfected with MS2-HIC as a negative control. Bars represent means ±SD, n = 7. Values were significantly different (two-tailed, two-sample t-test assuming unequal variances) from COX-2 proximal WT. *P < 0.001467.
Transient transfections were performed in order to assess utilization of the wild-type COX-2 proximal polyadenylation signal in the presence of the MS2-PSF, -p54nrb, -PTB and -U1A fusion proteins. HeLa cells were cotransfected with each individual MS2 fusion protein expression plasmid and the COX-2 WT-MS2 reporter plasmid. Total RNA was extracted from the cells after 24 h, and RNase protection assays were performed using the COX-2 proximal antisense probe previously described (Hall-Pogar et al. 2005). We measured the utilization of the COX-2 proximal signal relative to the BGH polyadenylation signal as an internal control. Previously we have shown that the relative utilization of the COX-2 proximal signal in this experimental system is ∼25% in HeLa cell transfections (Hall-Pogar et al. 2005). In this experiment we set the utilization of the COX-2 proximal polyadenylation signal as a value of 1. Quantification of multiple independent experiments is shown in Figure 5B. Transfection of the WT-MS2 reporter plasmid alone did not change the utilization of the COX-2 proximal polyadenylation signal. This indicated that the six MS2 stem–loops did not influence 3′-end processing at the COX-2 polyadenylation signal. Cotransfection with the MS2 coat protein empty vector likewise did not affect polyadenylation at the COX-2 proximal polyadenylation signal. Cotransfection with MS2-PSF, p54nrb, PTB, and U1A fusion proteins individually did increase the utilization of the COX-2 proximal polyadenylation signal (Fig. 5B). Therefore, each of the identified proteins that interact with the COX-2 USE was able to enhance polyadenylation. The MS2-HIC fusion protein, which served as a negative control, was unable to affect the utilization of the COX-2 proximal polyadenylation signal (Fig. 5B; Young et al. 2003).
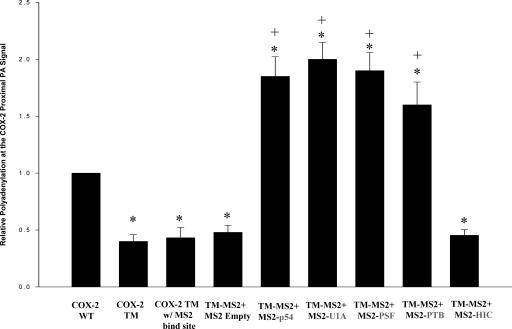
Next, we placed the MS2 stem–loops upstream of the COX-2 polyadenylation signal that contained mutations in each of the three previously described USE sequence elements (Triple mutant [TM]) (Fig. 5A; see also Hall-Pogar et al. 2005). This TM-MS2 reporter plasmid was also cotransfected with the fusion protein expression plasmids as described above. In this experiment, utilization of the COX-2 TM proximal polyadenylation was ∼60% less than that of COX-2 wild-type (WT) proximal polyadenylation signal, as expected from our previous results (Hall-Pogar et al. 2005). The MS2 binding sites in this tethering assay should compensate for the absence of the cis-acting elements in this experiment. We observed an approximately fourfold increase in polyadenylation at the mutant COX-2 proximal signal when MS2-PSF, p54nrb, PTB, and U1A fusion proteins were individually cotransfected with the USE mutant reporter (Fig. 6). Indeed, the tethered proteins were able to increase polyadenylation at the mutant COX-2 proximal signal to levels slightly higher than those seen with the wild-type reporter. We concluded that tethering these proteins to the USE mutant COX-2 polyadenylation signal abrogated the need for the USE sequence elements.
FIGURE 6.
MS2 tethering abrogates the need for the cis-acting USE elements. Same as Figure 4B but the COX-2 proximal polyadenylation signal reporter contained mutations in all three COX-2 USEs (Triple mutation [TM]) so that each USE was mutated (n = 6; Hall-Pogar et al. 2005). Values were significantly different (two-tailed, two-sample t-test assuming unequal variances) from COX-2 proximal WT. *P < 0.02915 and from COX-2 TM, +P < 0.001076.
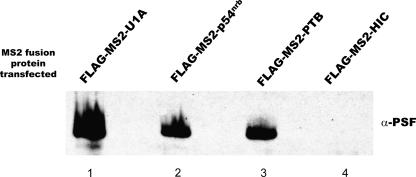
Presumably, if these proteins act as a complex, then transfection of one fusion protein should be able to recruit other binding partners to interact at the USE. We next performed transfections of each of the MS2 fusion proteins in HeLa cells, followed by immunoprecipitation using the FLAG tag present at the N-terminal end of all the MS2 fusion proteins. After extensive washing of the coprecipitated proteins, these proteins were separated by SDS-PAGE analysis and transferred to nitrocellulose. The Western blot was then probed with an antibody to PSF. As shown in Figure 7, the MS2-U1A, p54nrb, and PTB fusion proteins coprecipitated with PSF, but the MS2-HIC fusion protein did not. We have also performed this experiment in other combinations and obtained similar results (data not shown). These data suggest that U1A, p54nrb, PTB, and PSF may interact at the USE as a complex of proteins in order to affect polyadenylation at the suboptimal COX-2 proximal polyadenylation signal.
FIGURE 7.
Immunoprecipitation of individual transfected FLAG-tagged proteins coprecipitate PSF. Individual transfections were performed using FLAG-tagged MS2 protein constructs as indicated at the top. Immunoprecipitations were performed using a FLAG-tag antibody, and following separation of precipitated proteins on a 12% SDS-PAGE gel, the gel was Western blotted using anti-PSF antibodies.
DISCUSSION
We have previously shown that the COX-2 proximal polyadenylation signal contains auxiliary cis-acting RNA sequence elements upstream of the suboptimal AUUAAA core upstream polyadenylation element, and that these auxiliary sequence elements promote efficient utilization of this polyadenylation signal (Hall-Pogar et al. 2005). We have now shown that trans-acting protein factors can specifically associate with these cis-acting RNA elements. We have also demonstrated that tethering each of these RNA binding proteins to the region upstream of the COX-2 proximal polyadenylation signal abrogates the need for the cis-acting RNA sequence elements since the tethering assays in the USE mutant construct restored the same level of polyadenylation signal use. This suggests that the cis-acting USEs function to enhance polyadenylation of suboptimal poly(A) addition signals by recruiting trans-acting protein factors that promote RNA cleavage and polyadenylation.
The PTB-U1A protein–protein interaction shown here is intriguing, especially since both proteins have previously been shown to interact with USE-like sequence elements. U1A has previously been shown to interact with the USEs of SV40 (Lutz and Alwine 1994), while PTB has been shown to interact with the USEs of the C2 complement and lamin B2 genes and enhance 3′-end formation (Moreira et al. 1995, 1998; Brackenridge and Proudfoot 2000). A USE found in the early polyadenylation signal of human papillomavirus 16 was shown to interact with PTB as demonstrated by UV-cross-linking assays (Zhao et al. 2005). In addition, PTB has also been shown to compete with CstF for binding to the core downstream element (Castelo-Branco et al. 2004). When PTB binds in this fashion it is able to inhibit the polyadenylation and cleavage reaction, presumably because CstF can no longer bind (Castelo-Branco et al. 2004). Finally, coordinate expression of U1A and PTB has previously been reported in a number of human tissues, suggesting that these two proteins may regulate polyadenylation across cell types (Zhang et al. 2005).
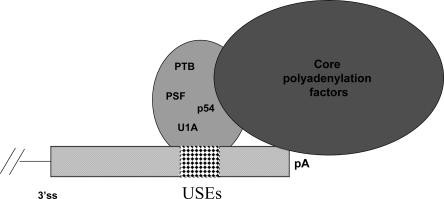
On the basis of the binding of PSF, p54nrb, PTB, and U1A proteins to the USEs of the COX-2 proximal polyadenylation signal, and from our coimmunoprecipitation assays, we suggest that these proteins may work together as a complex to regulate the usage of this polyadenylation signal. The COX-2 3′-UTR is large and contains not only multiple polyadenylation signals, but also many stability elements. Therefore, this example of alternative polyadenylation could result in mRNAs with different stabilities. Because PTB and U1A have previously been shown to interact with USE-like sequences (Lutz and Alwine 1994; Brackenridge and Proudfoot 2000), and p54nrb and PSF play a key role in 3′-end processing events (Liang and Lutz 2006), it is quite possible that in this instance they interact to recruit or stabilize the polyadenylation factors on the suboptimal COX-2 proximal polyadenylation signal (Fig. 8). This may also explain why, in the nonmutant USE constructs, tethering each protein enhanced polyadenylation (Fig. 5B). Moreover, the binding of these proteins to the USEs may enhance the stability of CPSF and CstF complexes on the polyadenylation signal core elements. Therefore, if one or both elements of the core polyadenylation signal are nonconsensus in nature, the interaction of these proteins could compensate for or enhance cleavage and polyadenylation reactions.
FIGURE 8.
A model: proteins bound to USEs at the COX-2 proximal polyadenylation signal may enhance polyadenylation at that signal by recruiting or stabilizing core polyadenylation factors on the polyadenylation signal. The proteins we have found associated with the COX-2 USEs are listed inside a large oval; since each protein has been found to associate with each other and with USEs it is not clear which one(s) may be in direct association with the polyadenylation machinery.
This group of proteins acting as a complex to influence polyadenylation at the COX-2 USEs is reminiscent of the SF-A complex of proteins (O'Connor et al. 1997; Lutz et al. 1998; Liang and Lutz 2006). The SF-A complex has been shown to contain PSF and p54nrb (Lutz et al. 1998; Liang and Lutz 2006). In particular, p54nrb was shown to be critical, since immunodepletions followed by reconstitution experiments with recombinant p54nrb restored polyadenylation activity (Liang and Lutz 2006). PTB has not been demonstrated so far to be a member of the SF-A complex; however, its molecular size (55 kDa) is very close to that of p54nrb, and this fact may have hampered its isolation (Liang and Lutz 2006). Since all of these proteins are functionally linked to the polyadenylation process (Castelo-Branco et al. 2004; Liang and Lutz 2006), as well as play roles in mRNA splicing, an interplay between these RNA processing reactions could be envisioned, since it is appreciated that splicing and polyadenylation reactions are coupled in vivo (for review, see Maniatis and Reed 2002; Bentley 2005).
While this work was in progress, work from the Kulozik group found a similar group of proteins associated with upstream U-rich elements in the suboptimal polyadenylation signal of the prothrombin (F2) gene (Danckwardt et al. 2007). While this group found a larger number of proteins associated with their upstream U-rich element by a method similar to ours, we should point out that we occasionally also isolated additional proteins but these proteins were not reproducible in our assays (data not shown). Our groups both believe that perhaps this represents a more global mechanism to regulate or facilitate usage of suboptimal polyadenylation signals, and will provide rich ground for future studies on the regulation of such signals.
MATERIALS AND METHODS
Plasmids
A fragment containing six stem–loop binding sites for MS2 was obtained by cutting pcBwtB2–3MS2 (a gift from J. Lykke-Andersen, University of Colorado, Boulder) with XbaI and NotI. This fragment was blunted using an Epicentre End-It DNA repair Kit according to the manufacturer's protocol, and placed in the BamHI site of the pCßS-COX2-prox and pCßS-COX2-tripleUSEmut constructs (Hall-Pogar et al. 2005) to create pCßS-COX2prox-MS2 and pCßS –tripleUSEmut-MS2, respectively.
The pcNMS-2-FLAG constructs (a gift from J. Lykke-Andersen) were used for MS2 fusion protein construction. The FLAG tag is N-terminal to the MS2 moiety. U1A cDNA with BamHI restriction sites on both sides was PCR amplified from pGEX-2T-U1A (Lutz-Freyermuth et al. 1990); primers used were 1 and 2 (see Table 1). p54nrb cDNA with an XhoI restriction site on one side and a NotI restriction site on the other side was amplified by PCR using primers 3 and 4. The HIC cDNA fragment was digested from the XhoI site of pGEX-6p-1-HIC (a gift from M. Mathews, UMDNJ). Digested fragments were cloned into the corresponding sites of the pcNMS-2-FLAG vector. pcNMS2-FLAG-PSF was prepared by the following procedure: PCR-based site directed mutagenesis using a QuikChange kit (Stratagene) was performed on pcNMS2-FLAG with primers 5 and 6 to destroy the EcoRI site inside of the MS2 reading frame so that the remaining EcoRI site in the multiple cloning site was a unique EcoRI site. Then, PSF cDNA fragment was digested from pET28a-PSF (a gift from M. Konarska, Rockefeller University) at the EcoRI site and was cloned into the mutated pcNMS2-FLAG vector. To make PSF cDNA fragment in the right reading frame, another round of site directed mutagenesis was performed to add an extra “A” before the start codon of PSF cDNA. Primers used for site directed mutagenesis to create this extra “A” were primers 7 and 8. To make the PTB construct, PCR-based site directed mutagenesis (see below) was performed on the pcNMS2-FLAG vector using primers 9 and 10 to destroy the EcoRI site inside of the MS2 reading frame so that the remaining EcoRI site in the multiple cloning site is a unique EcoRI site. Then, PTB (gifts from L. Covey, Rutgers University and J. Patton, Vanderbilt University) was digested with EcoRI and cloned into the mutated pcNMS2-FLAG vector. To make PTB cDNA fragment in the right reading frame, another round of site directed mutagenesis was performed to delete an extra “C” (in the BamHI site). Primers used for this mutagenesis were primers 11 and 12.
TABLE 1.
Forward and reverse primers
The plasmid pGEM4 (Promega) was digested with BamHI and PstI. The pCßS-COX-2 proximal wild-type, nonspecific proximal mutant and triple USE mutant were digested with BamHI and PstI and then subsequently subcloned into digested pGEM4 vector. Digestion of these plasmids with PstI gave templates for transcription of RNAs used in UV-cross-linking assays. SV40 late polyadenylation signal construct (SVL) was used as previously described (Lutz and Alwine 1994).
In vitro transcription of RNA substrates
RNA transcripts for in vitro polyadenylation, in vivo polyadenylation assays, UV cross-linking, and RNase protection assays were synthesized as previously described (Hall-Pogar et al. 2005; Liang and Lutz 2006).
In vitro polyadenylation
HeLa nuclear extracts were prepared as described (Natalizio et al. 2002 and references therein) using HeLa cells purchased from the National Cell Culture Center or grown in our laboratory. In vitro polyadenylation assays using HeLa nuclear extract and labeled substrate RNAs were performed as described previously (Natalizio et al. 2002; Hall-Pogar et al. 2005).
Maintenance of mammalian cell culture
HeLa cells were maintained in DMEM (Sigma or Life Technologies) supplemented with 10% fetal bovine serum (Sigma) and 1% penicillin–streptomycin (Life Technologies).
MS2 tethering experiments
Transfection of HeLa cells was accomplished using 7 × 105 cells/well in 60-mm plates using TransIT-LT1 reagent (Mirus). The transfection cocktail consisted of 1.0 μg of pCßS-COX2prox-MS2 or pCßS–tripleUSEmut-MS2 and 0.5 μg of pMS2-fusion protein construct individually. Plasmid DNA was diluted in 180 μL of serum-free medium to which 6 μL of LT-1 was added and the mixture was incubated at room temperature for 20 min. Following addition of 1 mL complete medium to the transfection mixture, medium on the cells was removed and replaced with the entire transfection cocktail. After 24 h, cells were washed once with 1× phosphate-buffered saline (PBS), then were scraped and collected into 1 mL fresh PBS. Cells were then centrifuged at 7000 rpm for 5 min. PBS was aspirated and total RNA was extracted immediately from the cell pellet using Trizol following the manufacturer's protocol.
Total RNA isolation and RNase protection assays
We have employed our tandem in vivo polyadenylation signal reporter vector, pCßS, which contains the COX-2 proximal polyadenylation signal or the triple USE mutant COX-2 upstream of the bovine growth hormone (BGH) polyadenylation signal (Hall-Pogar et al. 2005). To determine polyadenylation signal use in vivo, total RNA from transfected cells was isolated and assayed by RNase protection as previously described (Natalizio et al. 2002; Hall-Pogar et al. 2005). The ratio of the COX-2 polyadenylation signal utilized relative to the internal control, the BGH polyadenylation signal, was quantified using a Typhoon PhosphorImager and ImageQuant software.
UV cross-linking assays
Each reaction was composed of HeLa nuclear extract and an in vitro transcribed RNA substrate and contained equivalent amounts of extract (50 μg), 1 × 105 cpm of labeled RNA, and 1 mM EDTA. Reaction volumes were equalized with 1× Buffer D (20 mM HEPES at pH 7.9, 20% [v/v] Glycerol, 100 mM KCl, 0.2 mM EDTA, 1 mM PMSF, 1 mM DTT). Reactions were incubated at 30°C for 15 min and then were irradiated with 254 nm UV light for 10 min. Ten micrograms of RNase A were added to each reaction and further incubated at 37°C for 15 min. Cross-linked proteins were separated on 10% denaturing SDS-PAGE gels. Visualization was performed using autoradiography or PhosphorImager analysis.
The COX-2 USE 3 RNA oligoribonucleotide used in the UV cross-linking competition assay was synthesized by Dharmacon Research, Inc. and had the sequence 5′-UUGUUUGAUUUCUUAAAGU-3′. The nonspecific RNA oligoribonucleotide used in Figures 1, 2A,B was described previously (Natalizio et al. 2002).
Western blot analysis
Cell lysates were prepared by incubating 1.5 × 106 cells on ice for 30 min in a solution containing 1% NP-40, 150 mM NaCl, and 50 mM Tris-HCl (pH 8.0), plus phenylmethylsulfonyl fluoride (50 μg/mL), leupeptin, aprotinin, and pepstatin A (each at 1 μg/mL). Proteins from the cleared lysate (75 μg) were separated by 10% SDS-PAGE, then transferred onto a nitrocellulose membrane using a semi-dry transfer apparatus. The blot was then blocked and probed as previously described (Liang and Lutz 2006). Visualization of bound antibodies was accomplished by chemiluminescence using an ECL kit (GE Healthcare) and autoradiography. Anti-PSF, anti-c-myc, anti-GST, and anti-p54nrb antibodies were used at dilutions of 1:1000; anti-PTB and anti-U1A antibodies were used at dilutions of 1:500; anti-CstF-64 and anti-CPSF-73 were used at dilutions of 1:100.
Antibodies
p54nrb antibody was purchased from BD Transduction laboratories (Pharmingen). PSF, anti-myc tagged 9E10, and FLAG tag antibodies were purchased from Sigma; PTB and GST antibodies were purchased from Santa Cruz Biotechnology. U1A antibody 310 was described previously (Lutz and Alwine 1994). CstF-64 and CPSF-73 antibodies were gifts of C. MacDonald (Texas Tech University) and D. Bentley (University of Colorado, Denver), respectively.
Immunoprecipitations followed by Western blotting
HeLa cells (7 × 105 cells/well) were transfected with 500 ng of the MS2 fusion protein constructs as described above. After 24 h, cells were washed with 1× PBS, scraped, and collected in 1 mL of 1× PBS. Centrifuged cell pellets were lysed using 300 μL mild lysis buffer (50 mM Tris, 150 mM NaCl, 0.5% NP40), plus phenylmethylsulfonyl fluoride (50 μg/mL), leupeptin, aprotinin, and pepstatin A (1 μg/mL each). Resulting extracts were precleared with protein G beads (Gammabind Plus Sepharose, GE Healthcare) by adding 25 μL of washed beads and incubating at 4°C for 1 h. After removal of beads, 1 μL of FLAG M2 antibody was added to the precleared extracts, which were then incubated at 4°C for 4 h Gammabind Plus Sepharose beads (25 μL) were added to the reactions and then further incubated for 1 h at 4°C. Extracts were washed three times with IP lysis buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 1% NP-40) and once in 1× PBS. Pellets were resuspended in Laemmli buffer and loaded on 12% SDS-PAGE gels. Western blot analysis was performed as described above.
Biotinylated RNA pull down assay
The biotinylated RNA–streptavidin binding assay was performed using the following reagents per reaction: 10 nmol biotin-RNA oligo (Dharmacon) representing either COX-2 USE #3 (same sequence as described above) or nonspecific oligo, 60 μL of HeLa nuclear extract (10 μg/μL), 40 μL streptavidin sepharose high performance beads (GE Healthcare), 1 μL RNasin, and 5 μg tRNA (to decrease nonspecific interactions). Streptavidin sepharose beads were washed with 200 μL cold 1× PBS three times and 50 μL Buffer D once and placed on ice until use. Twenty-five microliters of beads were used to preclear HeLa nuclear extract by gentle rocking for 15 min at room temperature. The extract was centrifuged at 5000 rpm for 30 sec to remove the beads and the supernatant was placed in a fresh tube. Next, the biotinylated RNA oligo was added to precleared nuclear extract and the reaction was incubated for 10–20 min at 30°C. A second set of prewashed streptavidin beads was added to the mixture of RNA-oligo-nuclear extract and the mixture was incubated on ice for an additional 15–30 min. The reaction mixture was centrifuged for 5 min at 2500 rpm and the supernatant was removed for polyadenylation and UV-cross-linking experiments as the depleted extract. The beads were washed with 200 μL PBS three times, and 30 μL of 2× Laemmli Buffer was added. The bead-bound protein mixtures were boiled for 5 min and loaded onto a denaturing SDS-PAGE gel.
Nonspecific biotinylated oligos used in Figure 2C had the following sequences: NS1, 5′-GGAUUAACUAAUUGAUACC-3′; NS2, 5′-UUGUAUGCAUAUCUGAUCU-3′; NS3, 5′-GUCACGUGUCACCUGUAUG-3′, and were synthesized by Dharmacon Research, Inc.
GST purification and pull-down assays
GST full-length U1A and full-length p54nrb fusion proteins were expressed and purified as previously described (Gruda et al. 1993; Lutz et al. 1996; Liang and Lutz 2006). GST pull-down assays were performed using HeLa nuclear extract or TNT product as previously described (Liang and Lutz 2006). Bound proteins were visualized by denaturing SDS-PAGE analysis and autoradiography.
Generation of in vitro transcribed/translated protein products
To generate in vitro radiolabeled protein, the Promega TNT Coupled Transcription/Translation System was used according to the manufacturer's protocol. The construct to make in vitro transcribed and translated PTB was a gift from M. Garcia- Blanco (Duke University).
Statistical analyses
Results are expressed as ± SD of the mean. Two-tailed, two-sample t-tests were also performed assuming unequal variances.
Mass spectrometry
Proteins were separated on SDS-PAGE gels and visualized using fluorescent SYPRO Ruby protein stain. Protein bands were excised, digested with trypsin, and analyzed by MALDI-TOF/TOF by the Center for Advanced Proteomics, UMDNJ–New Jersey Medical School.
ACKNOWLEDGMENTS
This work was supported by a grant from the National Science Foundation to C.S.L. (MCB-0426195). We thank David Bentley, Clint MacDonald, Lori Covey, Jim Patton, Magda Konarska, Mariano Garcia-Blanco, Michael Mathews, and Jens Lykke-Andersen for gifts of reagents. We thank Dr. Hong Li and members of the Center for Advanced Proteomics for their mass spectrometry work. We thank Bin Tian, Pat O'Connor, and Carol Wilusz for critical reading of the manuscript. Additionally, we thank all members of the Lutz laboratory for technical assistance, including Mr. Andrew Schulman, as well for helpful comments and experimental suggestions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.577707.
REFERENCES
- Appleby, S.B., Ristimaki, A., Neilson, K., Narko, K., Hla, T. Structure of the human cyclo-oxygenase-2 gene. Biochem. J. 1994;302:723–727. doi: 10.1042/bj3020723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan, N.G. COX-2 as a multifunctional neuronal modulator. Nat. Med. 2001;7:414–415. doi: 10.1038/86477. [DOI] [PubMed] [Google Scholar]
- Bazan, N.G., Fletcher, B.S., Herschmann, H.R., Mukherjee, P.K. Platelet-activating factor and retinoic acid synergistically activate the inducible prostaglandin synthase gene. Proc. Natl. Acad. Sci. 1994;91:5252–5256. doi: 10.1073/pnas.91.12.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley, D.L. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Bishop-Bailey, D., Calatayud, S., Warner, T.D., Hla, T., Mitchell, J.A. Prostaglandins and the regulation of tumor growth. J. Environ. Pathol. Toxicol. Oncol. 2002;21:93–101. [PubMed] [Google Scholar]
- Bracken, K.E., Elger, W., Jantke, I., Nanninga, A., Gellersen, B. Cloning of guinea pig cyclooxygenase-2 and 15-hydroxyprostaglandin dehydrogenase complementary deoxyribonucleic acids: Steroid-modulated gene expression correlates to prostaglandin F2 α secretion in cultured endometrial cells. Endocrinology. 1997;138:237–247. doi: 10.1210/endo.138.1.4889. [DOI] [PubMed] [Google Scholar]
- Brackenridge, S., Proudfoot, N.J. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol. Cell. Biol. 2000;20:2660–2669. doi: 10.1128/mcb.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell, S., Alwine, J.C. Efficiency of utilization of the Simian Virus 40 late polyadenylation site: Effects of upstream sequences. Mol. Cell. Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., Proudfoot, N.J. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol. Cell. Biol. 2004;24:4174–4183. doi: 10.1128/MCB.24.10.4174-4183.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cok, S.J., Morrison, A.R. The 3′-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J. Biol. Chem. 2001;276:23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- Colgan, D.F., Manley, J.L. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Danckwardt, S., Kaufmann, I., Gentzel, M., Foerstner, K.U., Gantzert, A.S., Gehring, N.H., Neu-Yilik, G., Bork, P., Keller, W., Wilm, M., et al. Splicing factors stimulate polyadenylation via USEs at noncanonical 3′ end formation signals. EMBO J. 2007 doi: 10.1038/sj.emboj.7601699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, D.A., Kaplan, C.D., McIntyre, T.M., Zimmerman, G.A., Prescott, S.M. Post-transcriptional control of cyclooxygenase gene expression J. Biol. Chem. 2000;275:11750–11757. doi: 10.1074/jbc.275.16.11750. [DOI] [PubMed] [Google Scholar]
- Dixon, D.A., Tolley, N.D., King, P.H., Nabors, L.B., McIntyre, T.M., Zimmerman, G.A., Prescott, S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, D.A., Balch, G.C., Kedersha, N., Anderson, P., Zimmerman, G.A., Beauchamp, R.D., Prescott, S.M. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 2003;198:475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds, M. A history of polyA sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert, G., Veraldi, K.L., Milcarek, C. Alternative poly(A) site selection in complex transcription units: Means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, L., Sun, W., Xia, Y., Tang, W.W., Chanmugam, P., Soyoola, E., Wilson, C.B., Hwang, D. Cloning two isoforms of rat cyclooxygenase: Differential regulation of their expression. Arch. Biochem. Biophys. 1993;307:361–368. doi: 10.1006/abbi.1993.1601. [DOI] [PubMed] [Google Scholar]
- Garavito, R., Mulichak, A. The structure of mammalian cyclooxygenases. Annu. Rev. Biophys Biomol. Struct. 2003;32:183–206. doi: 10.1146/annurev.biophys.32.110601.141906. [DOI] [PubMed] [Google Scholar]
- Gou, Q., Liu, C.H., Ben-Av, P., Hla, T. Dissociation of basal turnover and cytokine-induced transcript stabilization of the human cyclooxygenase-2 mRNA by mutagenesis of the 3′-untranslated region. Biochem. Biophys. Res. Commun. 1998;242:508–512. doi: 10.1006/bbrc.1997.7994. [DOI] [PubMed] [Google Scholar]
- Gruda, M.C., Zabolotny, J.M., Xiao, J.H., Davidson, I., Alwine, J.C. Transcriptional activation by simian virus 40 large T antigen: Interactions with multiple components of the transcription complex. Mol. Cell. Biol. 1993;13:961–969. doi: 10.1128/mcb.13.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar, T., Zhang, H., Tian, B., Lutz, C.S. Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res. 2005;33:2565–2579. doi: 10.1093/nar/gki544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hla, T., Maciag, T. Cyclooxygenase gene expression is down-regulated by heparin-binding (acidic fibroblast) growth factor-1 in human endothelial cells. J. Biol. Chem. 1991;266:24059–24063. [PubMed] [Google Scholar]
- Hla, T., Neilson, K. Human cyclooxygenase-2 cDNA. Proc. Natl. Acad. Sci. 1992;89:7384–7388. doi: 10.1073/pnas.89.16.7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hla, T., Bishop-Bailey, D., Liu, C.H., Schaefers, H.J., Trifan, O.C. Cyclooxygenase-1 and -2 isoenzymes. Int. J. Biochem. Cell Biol. 1999;31:551–557. doi: 10.1016/s1357-2725(98)00152-6. [DOI] [PubMed] [Google Scholar]
- Hu, J., Lutz, C.S., Wilusz, J., Tian, B. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA. 2005;11:1485–1493. doi: 10.1261/rna.2107305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, D., Scollard, D., Byrne, J., Levine, E. Expression of cyclooxygenase-1 and cyclooxygenase-2 in human breast cancer. J. Natl. Cancer Inst. 1998;90:455–460. doi: 10.1093/jnci/90.6.455. [DOI] [PubMed] [Google Scholar]
- Kutchera, W., Jones, D.A., Matsunami, N., Groden, J., McIntyre, T.M., Zimmerman, G.A., White, R.L., Prescott, S.M. Prostaglandin synthase 2 is abnormally expressed in human colon cancer: Evidence for a transcriptional effect. Proc. Natl. Acad. Sci. 1996;93:4816–4820. doi: 10.1073/pnas.93.10.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landen, C.N., Marthur, S.P., Richardson, M.S., Creasman, W.T. Expression of cyclooxygenase-2 in cervical, endometrial, and ovarian malignancies. Am. J. Obstet. Gynecol. 2003;188:1174–1176. doi: 10.1067/mob.2003.284. [DOI] [PubMed] [Google Scholar]
- Liang, S., Lutz, C.S. p54nrb is a component of the snRNP free U1A (SF-A) complex that promotes pre-mRNA cleavage during polyadenylation. RNA. 2006;12:111–121. doi: 10.1261/rna.2213506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw, W.J., Bazan, N.G. Cyclooxygenase 2 RNA message abundance, stability and hypervariability in sporadic Alzheimer neocortex. J. Neurosci. Res. 1997;50:937–945. doi: 10.1002/(SICI)1097-4547(19971215)50:6<937::AID-JNR4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S., Alwine, J.C. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes & Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S., Murthy, K.G.K., Schek, N., O'Connor, J.P., Manley, J.L., Alwine, J.C. Interaction between the U1snRNP-A protein and the 160-kDa subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes & Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- Lutz, C.S., Cooke, C., O'Connor, J.P., Kobayashi, R., Alwine, J.C. The snRNP-free U1A (SF-A) complex(es): Identification of the largest subunit as PSF, the polypyrimidine-tract binding protein-associated splicing factor. RNA. 1998;4:1493–1499. doi: 10.1017/s1355838298981183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz-Freyermuth, C., Query, C.C., Keene, J.D. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem–loop II of U1 RNA. Proc. Natl. Acad. Sci. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Reed, R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Moreira, A., Wollerton, M., Monks, J., Proudfoot, N.J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira, A., Takagaki, Y., Brackenridge, S., Wollerton, M., Manley, J.L., Proudfoot, N.J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′-end formation by two distinct mechanisms. Genes & Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio, B.J., Muniz, L.C., Arhin, G.K., Wilusz, J., Lutz, C.S. Upstream elements present in the 3′-UTR of collagen gene influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- Newton, R., Seybold, J., Liu, S.F., Barnes, P.J. Alternate COX-2 transcripts are differentially regulated: Implications for post-transcriptional control. Biochem. Biophys. Res. Commun. 1997;234:85–89. doi: 10.1006/bbrc.1997.6586. [DOI] [PubMed] [Google Scholar]
- Newton, R., Seybold, J., Kuitert, L.M.E., Bergmann, M., Barnes, P.J. Repression of cyclooxygenase-2 and prostaglandin E2 release by dexamethasone occurs by transcriptional and post-transcriptional mechanisms involving loss of polyadenylated mRNA. J. Biol. Chem. 1998;273:32312–32321. doi: 10.1074/jbc.273.48.32312. [DOI] [PubMed] [Google Scholar]
- O'Banion, M.K., Winn, V.D., Yong, D.A. cDNA cloning and functional activity of a glucocorticoid-regulated inflammatory cyclooxygenase. Proc. Natl. Acad. Sci. 1992;89:4888–4892. doi: 10.1073/pnas.89.11.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Banion, M.K., Miller, J.C., Chang, J.W., Kaplan, M.D., Coleman, P.D. Interleukin-1 β induces prostaglandin G/H synthase-2 (cyclooxygenase-2) in primary murine astrocyte cultures. J. Neurochem. 1996;66:2532–2540. doi: 10.1046/j.1471-4159.1996.66062532.x. [DOI] [PubMed] [Google Scholar]
- O'Connor, J.P., Alwine, J.C., Lutz, C.S. Identification of non-snRNP associated U1A protein in human cell nucleoplasm. RNA. 1997;3:1444–1455. [PMC free article] [PubMed] [Google Scholar]
- Peng, R., Hawkins, I., Link, A.J., Patton, J.G. The splicing factor PSF is part of a large complex that assembles in the absence of pre-mRNA and contains all five snRNPs. RNA Biol. 2006;3:69–76. doi: 10.4161/rna.3.2.3017. [DOI] [PubMed] [Google Scholar]
- Prescott, S.M. Is cyclooxygenase-2 the α and ω in cancer? J. Clin. Invest. 2000;105:1511–1513. doi: 10.1172/JCI10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott, S.M., Fitzpatrick, F.A. Cyclooxygenase-2 and carcinogenesis. Biochim. Biophys. Acta. 2000;1470:69–78. doi: 10.1016/s0304-419x(00)00006-8. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J., Furger, A., Dye, M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Ristimaki, A., Garfinkel, S., Wessendorf, J., Maciag, T., Hla, T. Induction of cyclooxygenase-2 by interleukin-1 α. Evidence for post-transcriptional regulation. J. Biol. Chem. 1994;269:11769–11775. [PubMed] [Google Scholar]
- Ristimaki, A., Narko, K., Hla, T. Down-regulation of cytokine-induced cyclooxygenase 2 transcript isoforms by dexamethasone: Evidence for post-transcriptional regulation. Biochem. J. 1996;318:325–331. doi: 10.1042/bj3180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K., Calvo, O., Manley, J.L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA. 2004;10:565–573. doi: 10.1261/rna.5214404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San, H., Kawahito, Y., Wilder, R.L., Hasiramoto, A., Mukai, S., Asai, K., Kimura, S., Kato, H., Kondo, M., Hla, T. Expression of cyclooxygenase-1 and -2 in human colorectal cancer. Cancer Res. 1995;55:3785–3789. [PubMed] [Google Scholar]
- Schmidt, C.M., Wang, Y., Wiesenauer, C. Novel combination of cyclooxygenase-2 and MEK inhibition in human hepatocellular carcinoma provides a synergistic increase in apoptosis. J. Gastrointest. Surg. 2003;7:1024–1033. doi: 10.1016/j.gassur.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Tian, B., Hu, J., Zhang, H., Lutz, C.S. A large scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle, E., Kuhn, U. The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog. Nucleic Acid Res. Mol. Biol. 1997;57:41–71. doi: 10.1016/s0079-6603(08)60277-9. [DOI] [PubMed] [Google Scholar]
- Wickens, M., Anderson, P., Jackson, R.J. Life and death in the cytoplasm: Messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- Williams, C.S., Mann, M., DuBois, R.N. The role of cyclooxygenases in inflammation, cancer and development. Oncogene. 1999;18:7908–7916. doi: 10.1038/sj.onc.1203286. [DOI] [PubMed] [Google Scholar]
- Wilusz, J., Shenk, T., Takagaki, Y., Manley, J.L. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 1990;10:1244–1248. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, W., Cipman, J.G., Robertson, D.L., Eirkson, R.L., Simmons, D.L. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc. Natl. Acad. Sci. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, T.M., Wang, Q., Pe'ery, T., Mathews, M.B. The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol. Cell. Biol. 2003;23:6373–6384. doi: 10.1128/MCB.23.18.6373-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Lee, J.Y., Tian, B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:1–13. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., Moore, C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulationm and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Oberg, D., Rush, M., Fay, J., Lambkin, H., Schwartz, S. A 57-nucleotide upstream early polyadenylation element in human papilloma virus type 16 interacts with hFip1, CstF64, hnRNP C1/C2, and polypyrimidine tract binding protein. J. Virol. 2005;79:4270–4288. doi: 10.1128/JVI.79.7.4270-4288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]