Abstract
We describe the first microarray analysis of a whole animal containing a mutation in the Dicer gene. We used adult Caenorhabditis elegans and, to distinguish among different roles of Dicer, we also performed microarray analyses of animals with mutations in rde-4 and rde-1, which are involved in silencing by siRNA, but not miRNA. Surprisingly, we find that the X chromosome is greatly enriched for genes regulated by Dicer. Comparison of all three microarray data sets indicates the majority of Dicer-regulated genes are not dependent on RDE-4 or RDE-1, including the X-linked genes. However, all three data sets are enriched in genes important for innate immunity and, specifically, show increased expression of innate immunity genes.
Keywords: Dicer, miRNA, RNAi, X chromosome, innate immunity
INTRODUCTION
The ribonuclease III enzyme Dicer processes microRNAs (miRNAs) and the small interfering RNAs (siRNAs) that function in RNA interference (RNAi) (Matzke and Birchler 2005; Sontheimer and Carthew 2005). While Dicer is central to the production of both types of small RNAs, other factors required for the two pathways are distinct. For example, in Caenorhabditis elegans, the double-stranded RNA (dsRNA) binding protein RDE-4 and the Argonaute protein RDE-1 are necessary for RNAi, but not the miRNA pathway. The miRNA pathway in C. elegans requires distinct Argonaute family members, and as in other organisms, in addition to Dicer, processing of miRNAs requires the ribonuclease III enzyme Drosha.
siRNAs and miRNAs also regulate mRNA expression by different mechanisms. siRNAs bind their target mRNA with perfect complementarity and promote its cleavage and subsequent degradation. miRNAs typically bind their target messages with imperfect complementarity, and while translation is inhibited, only in some cases has the mRNA been shown to be degraded (Bagga et al. 2005).
Many questions remain in regard to the biologic roles of small RNAs. Numerous miRNAs as well as endogenous siRNAs (endo-siRNAs) have been identified through cloning strategies (Lagos-Quintana et al. 2001; Lau et al. 2001; Ambros et al. 2003; Ruby et al. 2006), but few of the mRNA targets have been experimentally validated. One way to identify targets of small RNAs would be to analyze transcripts misregulated in organisms deficient for Dicer. A microarray analysis of a Schizosaccharomyces pombe Dicer mutant revealed few misregulated transcripts, but clearly indicated the importance of Dicer in generating small RNAs necessary for silencing centromeric repeats and retrotransposons (Hansen et al. 2005). However, S. pombe differs from higher organisms since it encodes a Dicer protein but lacks miRNAs. Microarray studies in higher organisms are difficult since miRNAs are necessary for proper development, and hence Dicer mutants rarely develop beyond the embryo stage. However, a microarray experiment has been performed in human HEK293 cells in which Dicer levels were reduced with an shRNA (Schmitter et al. 2006). These carefully performed experiments implicate miRNAs in the regulation of 5%–10% of the transcriptome, but the primary function of Dicer was not revealed.
C. elegans with a homozygous dcr-1 mutation, derived from heterozygote mothers, develop to adulthood, presumably due to maternally contributed dcr-1 mRNA. The viability of these animals allowed us to perform the first microarray analysis of a whole animal containing a homozygous mutation in the Dicer gene. To provide additional insight into the functions of Dicer in C. elegans, we also performed microarray analyses on animals defective for RNAi due to mutations in rde-4 and rde-1, which are not involved in miRNA processing. We find that genes misregulated in animals containing a mutation in the dcr-1 gene are enriched on the X chromosome, and this trend is not observed in animals defective for RDE-4 or RDE-1. Interestingly, all three microarray data sets are enriched for genes implicated in innate immunity.
RESULTS
Microarray analysis of C. elegans homozygous for a deletion in dcr-1
Our laboratory and others previously characterized recessive deletion alleles of the C. elegans dcr-1 gene (Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). As expected, animals homozygous for deletions in dcr-1 are defective for RNAi and miRNA processing. Mutant animals often exhibit a burst vulva phenotype, similar to the phenotype of animals with a mutation in the miRNA let-7, a miRNA involved in developmental timing (Reinhart et al. 2000). dcr-1 homozygote animals reach adulthood despite an impaired ability to process miRNAs, presumably due to the maternal contribution of DCR-1 from a heterozygote mother. However, homozygote animals are sterile, and the gonad of adult animals contains misshapen oocytes with an endomitotic phenotype. Due to these germline defects, dcr-1 mutants must be maintained in a heterozygous background.
To determine how mRNA levels are altered in C. elegans with mutations in dcr-1, we performed a genome-wide analysis using Affymetrix oligonucleotide microarrays. Isolation of dcr-1(ok247) homozygote animals was facilitated by marking ok247 with a recessive allele of unc-32(e189), which is <0.5 map units from dcr-1. unc-32(e189) homozygotes display a “coiler” phenotype and are easily distinguished from wild type (WT) and heterozygotes. dcr-1(ok247);unc-32(e189) homozygotes, hereafter referred to as dcr-1(−/−) animals, were collected by picking worms based on the coiler phenotype. unc-32(e189) homozygotes were isolated similarly for use in comparison. While maternal Dicer from heterozygote parents would undoubtedly contribute to embryogenesis of homozygote progeny, effects should be minimized at later stages, and thus we used adult stage animals for our analyses. Using RNA isolated from these animals, microarray hybridization experiments were performed based on the Affymetrix protocol for oligonucleotide microarrays (see Materials and Methods).
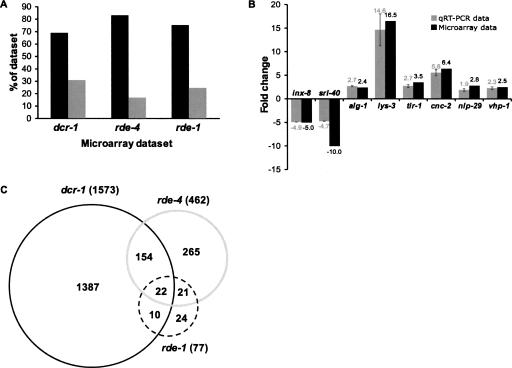
Not surprisingly, we observed a large number of misregulated genes in dcr-1(−/−) adult animals. Thus, a stringent statistical threshold was applied with a cutoff of P < 0.01, as determined by a multisample t-test and the Benjamini and Hochberg false discovery rate (FDR) correction (see Materials and Methods) (Benjamini and Hochberg 1995). The curated list of misregulated genes (Supplemental Table S1) showed 1573 transcripts that were misregulated at least 1.5-fold and, of these, 134 transcripts were misregulated at least fivefold. Thus, silencing by small RNAs generally affects transcript levels modestly but in some cases leads to dramatic changes in gene expression. As shown in Figure 1A, consistent with a role for DCR-1 in repressing gene expression, the majority of genes misregulated ≥1.5-fold were up-regulated (1085/1573, 69%).
FIGURE 1.
Summary of microarray analyses of dcr-1, rde-4, and rde-1 mutant C. elegans. (A) Consistent with a silencing role for small RNAs in C. elegans, the majority of transcripts in mutant animals were up-regulated (black bars) as opposed to down-regulated (gray bars). Data are plotted from curated lists for (up/down): dcr-1 (1085/488); rde-4 (384/78); and rde-1 (58/19). (B) Relative fold change of selected misregulated genes in dcr-1(−/−) animals compared to control unc-32(−/−) animals. Values determined by microarray analysis (black bars) were compared to those determined by qRT-PCR (gray bars). Three independent dcr-1(−/−) and control unc-32(−/−) RNA samples used in microarray analysis were pooled and assayed by qRT-PCR using at least two independent cDNA preparations and one technical replicate per cDNA preparation (n ≥ 4); error bars represent the standard error of the mean. (C) Venn diagram showing overlap between genes misregulated ≥ 1.5-fold in microarray analyses of dcr-1(−/−) (black circle), rde-4(−/−) (gray circle), and rde-1(−/−) (dotted circle) mutant animals. The intersection of data sets was far greater than the number expected based on random chance. The [observed/expected] values for the various intersections (∩) were: rde-4 ∩ dcr-1 [176/40], rde-4 ∩ rde-1 [43/2], rde-1 ∩ dcr-1 [32/7], rde-4 ∩ rde-1 ∩ dcr-1 [22/0] (for calculations see Materials and Methods). Curated lists of genes misregulated ≥1.5-fold and GeneSifter output are in Supplemental Table S1.
Given the germline defects in adult dcr-1(−/−) animals, we wondered whether misregulated genes were enriched for genes expressed in the germline. Thus, we compared our data set of genes misregulated ≥1.5-fold with a list of germline-enriched genes reported by Reinke and colleagues (WT versus glp-4TS) (Reinke et al. 2004). Of the 488 genes observed to be down-regulated in dcr-1(−/−) animals, 184 (38%) corresponded to genes reported to be germline-enriched. However, very few up-regulated genes in dcr-1(−/−) animals correlated with germline expression (14 genes, 1% of up-regulated genes). This analysis emphasized that DCR-1 plays a role in the C. elegans germline, but the large number of up-regulated genes was not simply a result of the germline defects.
To validate our microarray data, a subset of misregulated genes was analyzed by an independent method, quantitative RT-PCR (qRT-PCR). Genes were chosen to represent varying degrees of misregulation, including both up-regulated and down-regulated genes. There was strong agreement between the relative fold change of candidate genes using both qRT-PCR and microarray analysis (Fig. 1B). For sri-40 the fold change reported by the two methods was disparate, but both methods indicated sri-40 was down-regulated. This analysis indicates the microarray experiments accurately identified transcripts misregulated in dcr-1(−/−) animals, but in some cases the magnitude of misregulation was different from that observed using qRT-PCR.
Microarray analyses of rde-4(−/−) and rde-1(−/−) animals distinguish different roles of DCR-1
To delineate which small RNA pathways were responsible for the altered mRNA levels in dcr-1(−/−) animals, we performed additional microarray analyses using worms defective for rde-4 or rde-1. RDE-4 acts with Dicer to produce siRNAs (Parrish and Fire 2001; Parker et al. 2006), while RDE-1 is an Argonaute protein that functions in RNAi downstream from siRNA production (Tabara et al. 1999; Parrish and Fire 2001). Importantly, neither rde-4 nor rde-1 is involved in the miRNA pathway. For this analysis we isolated worms at the adult stage to allow comparison with the dcr-1(−/−) data, and as for the dcr-1 analysis the parental strain served as the reference strain, in this case Bristol N2 rather than unc-32(e189). To increase the stringency of our analysis, we used two independent alleles of rde-4(ne299, ne337) and rde-1(ne219, ne300). Two samples were obtained for each allele and together treated as quadruplicate samples in our microarray analysis; resulting data sets for the combined alleles are herein referred to as rde-4(−/−) and rde-1(−/−) (see Materials and Methods).
Microarray analyses showed 462 and 77 genes misregulated ≥1.5-fold for rde-4(−/−) and rde-1(−/−), respectively (Supplemental Table S1). Thus, fewer genes were misregulated in these animals compared to the dcr-1(−/−) animals. This is consistent with the idea that Dicer functions are more diverse. Of course, our use of two alleles for analyses of the rde mutants, while providing robust data sets, may underestimate the number of genes misregulated in these animals.
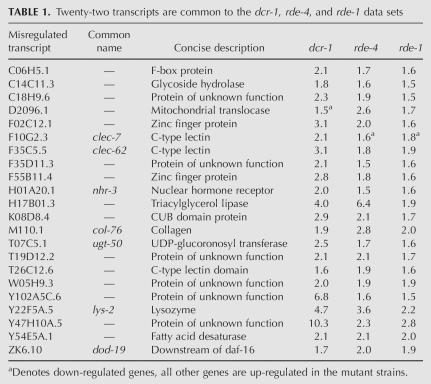
As observed for the dcr-1(−/−) data set, the majority of misregulated transcripts were up-regulated, for both rde-4 (384/462, 83%) and rde-1 (58/77, 75%) (Fig. 1A). Figure 1C presents a Venn diagram showing the overlap of genes determined to be misregulated in dcr-1(−/−), rde-4(−/−), and rde-1(−/−) animals (Supplemental Table S1, intersections). In C. elegans, a canonical RNAi response to an exogenous dsRNA “trigger” requires all three genes and, as shown in Table 1, we identified 22 transcripts that were misregulated ≥1.5-fold in all three mutants. There was good agreement in the magnitude and direction (up versus down) by which the common genes were misregulated in each strain and, interestingly, the 22 transcripts shared common functionality, as a number relate to innate immunity (see below).
TABLE 1.
Twenty-two transcripts are common to the dcr-1, rde-4, and rde-1 data sets
In addition to the 22 genes of the rde-4(−/−) data set that were common to all data sets, 154 genes overlapped with dcr-1(−/−) but not rde-1(−/−). A recent study indicates RDE-1 has functions specific to primary siRNAs generated in response to exogenous dsRNA (Yigit et al. 2006). However, C. elegans has 27 Argonaute genes, and this study also shows other family members have distinct roles in RNAi. Possibly, the 154 genes of the rde-4(−/−) data set that overlap with the dcr-1(−/−) data set, but not the rde-1(−/−) data set, reflect roles of RDE-4 that require these other Argonaute family members. We also observed that 265 genes of the rde-4(−/−) data set were unique to this data set, raising the possibility that RDE-4 has roles independent of both DCR-1 and RDE-1. Alternatively, a subset of these 265 genes could be DCR-1 dependent in the absence of maternal effects. This may be most applicable to genes whose silencing is initiated at early developmental stages where maternal DCR-1 is more abundant.
Finally, as apparent from the Venn diagram, the vast majority of misregulated genes in dcr-1(−/−) animals did not overlap with those misregulated in rde-4(−/−) and rde-1(−/−) animals. These are expected to include direct and indirect miRNA targets (see below).
dcr-1(−/−) up-regulated genes are enriched for predicted targets of miRNAs
Although miRNAs are generally thought to mediate post-transcriptional silencing by inhibiting translation, in some cases targeted message levels are also reduced (Bagga et al. 2005). Thus, we anticipated our microarray analysis of dcr-1(−/−) animals would generate a data set enriched for genes that were both direct and indirect targets of miRNA regulation.
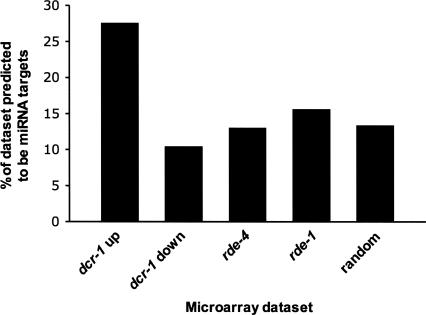
The discovery of miRNAs led to the development of numerous algorithms aimed at predicting mRNA targets of miRNAs. To determine if there was an enrichment for targets in the dcr-1(−/−) up-regulated list of genes, we focused on targets predicted by the PicTar algorithm (Lall et al. 2006). PicTar was chosen because it is a conservative algorithm, giving consideration to evolutionary conservation between related nematodes. The developmental profiles of most miRNAs analyzed in C. elegans show an appearance in embryos and a persistence through adulthood (Lau et al. 2001; Lee and Ambros 2001). The persistence of nearly all miRNAs into adulthood, combined with the inability to determine which of the miRNAs remain functionally active in adults, made it difficult to generate a list of adult-specific miRNAs. Therefore, we combined all targets predicted by PicTar into a single data set (cePicTar, Supplemental Table S2; see Materials and Methods). Next, we determined the percentage of genes in each microarray data set that were common with the cePicTar data set (Fig. 2; Supplemental Table S2). We observed that 27.6% of the dcr-1(−/−) up-regulated data set were predicted miRNA targets, a value that was twofold higher than that of the random control data sets (see Materials and Methods). There was no enrichment for targets in either the rde-4(−/−) or rde-1(−/−) data sets, as expected since neither gene is involved in the miRNA pathway. Interestingly, there was a slight underrepresentation of predicted targets in the dcr-1(−/−) down-regulated data set.
FIGURE 2.
dcr-1(−/−) up-regulated genes are enriched for predicted miRNA targets. The percentage of each microarray data set that were predicted miRNA targets according to the PicTar algorithm was plotted ([number of miRNA targets in data set/total genes in data set] × 100). Data for dcr-1(−/−) were analyzed after parsing into up-regulated and down-regulated genes while all misregulated genes for rde-4 and rde-1 were considered together. The random analysis is the average value using 10 independently generated random lists of genes (see Materials and Methods).
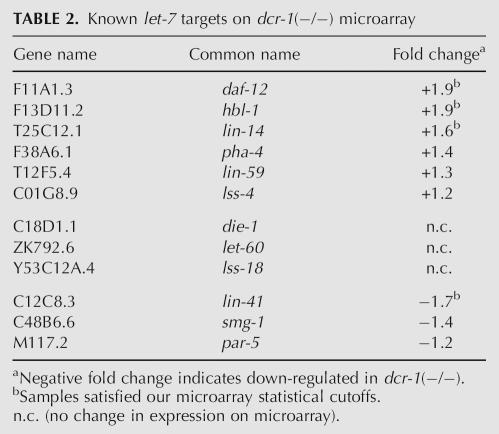
One of the best studied miRNAs in C. elegans, let-7, is temporally expressed, initiating in the L3 larval stage and continuing into adulthood (Reinhart et al. 2000). When we performed a similar analysis as shown in Figure 2, but limited the scope to let-7 targets predicted by the PicTar algorithm, there was an even higher enrichment (fourfold) in the dcr-1(−/−) up-regulated genes compared with random data sets (data not shown). Out of the 57 PicTar predicted let-7 targets, 11 were on our list of dcr-1(−/−) up-regulated genes (19%, see Supplemental Table S2). Finally, we considered only targets with corroborating experimental evidence for their consideration as let-7 targets (Reinhart et al. 2000; Grosshans et al. 2005). As shown in Table 2, 50% of these genes were up-regulated in the dcr-1(−/−) strain, including three that satisfied our rigorous statistical cutoff (daf-12, hbl-1, and lin-14). It is unclear why others, such as lin-41, are down-regulated or in other cases unchanged.
TABLE 2.
Known let-7 targets on dcr-1(−/−) microarray
As mentioned, another class of small RNAs in C. elegans is the endo-siRNAs. To determine if there was also an overrepresentation of endo-siRNA targets in our microarray data sets, we compared the list of endo-siRNA targets published by Ambros and colleagues (Lee et al. 2006) with our data sets (Supplemental Fig. S1). We did not see an enrichment of endo-siRNA targets among dcr-1(−/−) up-regulated genes. In fact, there was a higher percentage of endo-siRNA targets in the dcr-1(−/−) down-regulated genes than the up-regulated. We obtained similar results when we compared our data sets with the endo-siRNAs identified by Bartel and colleagues (data not shown; Ruby et al. 2006). The lists of genes up-regulated in rde-4(−/−) and rde-1(−/−) showed a 1.5–2.0-fold overrepresentation for endo-siRNA targets, but the overrepresentation of endo-siRNA targets was also observed in the list of genes down-regulated in rde-1.
In summary, our dcr-1(−/−) microarray data set was enriched for known and predicted miRNA targets. Thus, this data set will be beneficial to groups attempting to refine existing, or design future, miRNA target prediction algorithms. The lack of a correlation between the up-regulated genes in dcr-1(−/−) and predicted endo-siRNA targets is consistent with recent work indicating that many endo-siRNAs are generated in a DCR-1-independent pathway (Ruby et al. 2006; Pak and Fire 2007; Sijen et al. 2007).
Misregulated genes in dcr-1(−/−) animals are enriched on the X chromosome
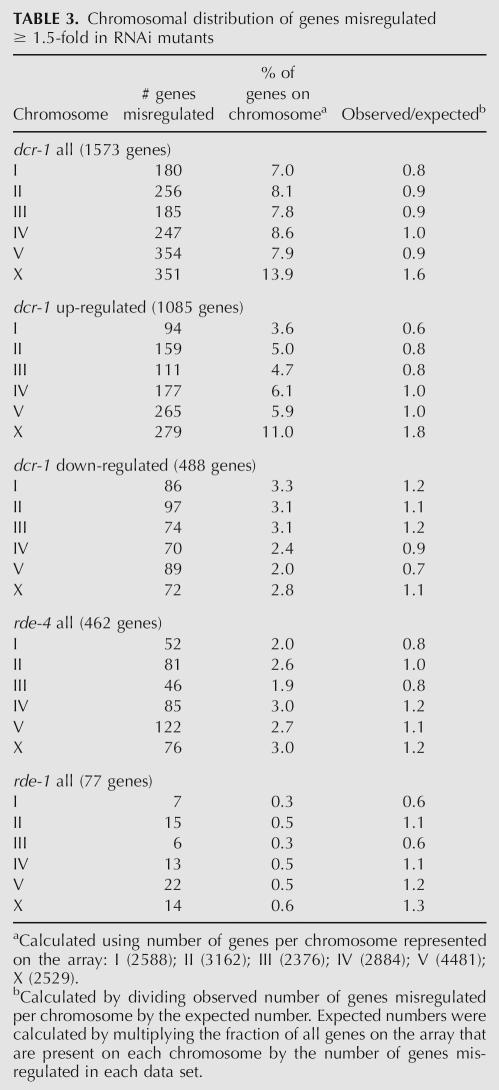
To determine the distribution of misregulated genes across the genome, we tabulated the number of genes misregulated on each chromosome (Table 3) and for dcr-1(−/−)depicted these data graphically according to chromosomal position (Supplemental Fig. S2). Chromosomes V and X had the highest number of misregulated genes in the dcr-1(−/−) animals (Table 3). However, Chromosome V is the longest chromosome and has more genes than other C. elegans chromosomes. When this was taken into account, by calculating the percentage of the genes on each chromosome that were misregulated, Chromosome V was similar to other autosomes, showing 7.9% of its genes misregulated in the dcr-1(−/−) animals. In contrast, almost twice the percentage (13.9%) of the genes on the X chromosome were misregulated in dcr-1(−/−) animals. The 351 transcripts misregulated on the X chromosome represent 1.6-fold more than the 221 expected for a random distribution. Upon analyzing the up-regulated and down-regulated genes separately, we found the correlation was specific to up-regulated genes (1.8-fold greater than chance) (Table 3). Neither the rde-4(−/−) nor rde-1(−/−) data sets showed an enrichment for X chromosome genes, suggesting that RNAi is not responsible for this bias.
TABLE 3.
Chromosomal distribution of genes misregulated ≥ 1.5-fold in RNAi mutants
Misregulated genes were associated with GO terms related to innate immunity
To gain insight into the biologic roles of dcr-1, rde-4, and rde-1, we analyzed gene ontology (GO) terms associated with genes misregulated ≥1.5-fold in the microarray data sets (Ashburner et al. 2000). A web-based application, FatiGO+ (http://www.babelomics.org), was used to evaluate whether misregulated genes exhibited an over- or underrepresentation of particular GO terms (Al-Shahrour et al. 2005). This software provides information about GO terms at different levels of the ontology hierarchy, from those describing general functions (e.g., level 3) to those that are quite specific (e.g., level 9).
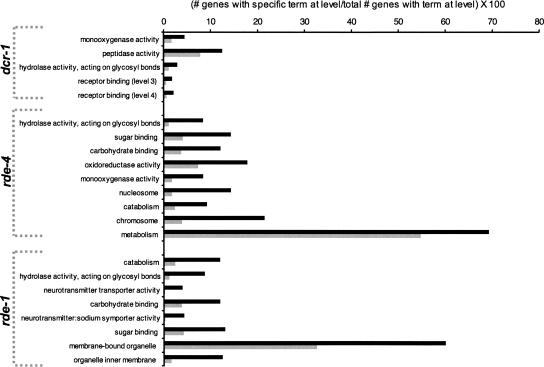
The three GO classifications, molecular function (MF), biological process (BP), and cellular component (CC), were evaluated separately by level (see Supplemental Table S5), but only significant terms at levels 3 and 4 of all ontologies are shown (Fig. 3). Bar height represents the percentage of all genes with GO annotations at the level that were associated with the specified GO term. Genes misregulated ≥1.5-fold (black bars) were compared with the remainder of the genes on the array (gray bars).
FIGURE 3.
Significant GO terms at levels 3 and 4 of the MF, BP, and CC ontologies. Overrepresented terms are shown for dcr-1(−/−) (P ≤ 0.01, unadjusted), rde-4(−/−) (P ≤ 0.01, unadjusted), and rde-1(−/−) (P ≤ 0.1, unadjusted); underrepresented terms using the same significance cutoff (Supplemental Fig. S3) and the complete analysis for all levels (Supplemental Table S5) are in Supplemental Data. For each mutant, GO terms are listed top to bottom from most to least significant according to the P-value, as determined from a Fishers exact test (unadjusted). Bar height represents the percentage of all genes with annotations at the level associated with the specified GO term; genes misregulated ≥ 1.5-fold (black bars) were compared with the remainder of the genes on the array (gray bars).
Several of the significant GO terms suggested a relationship to innate immunity. For example, the only GO term at level 3 or 4 that was common to analyses of all mutant animals, “hydrolase activity, acting on glycosyl bonds,” includes enzymes that hydrolyze pathogenic bacteria, such as the lysozyme genes (for review, see Schulenburg et al. 2004). Indeed, genes of the lysozyme family are misregulated in all three mutant animals. Similarly, for both rde-4(−/−) and rde-1(−/−), “sugar binding” and “carbohydrate binding” were significant GO terms, and the misregulated genes associated with these GO terms were almost exclusively of the C-lectin family. C. elegans C-type lectins are proposed to be pattern recognition receptors (PRRs), which function in innate immunity by recognizing pathogen-associated molecular pattern (PAMP) molecules on the pathogen to trigger phagocytosis and lysis (for review, see Schulenburg et al. 2004).
We also observed many GO terms that were underrepresented (Supplemental Fig. S3). Genes annotated as receptors were underrepresented in data sets for all mutant animals, and those annotated as kinases were underrepresented in the data set of dcr-1(−/−) animals. Similarly, GO terms related to post-embryonic and larval development, as well as those related to growth and metabolism, were underrepresented in data sets of all mutant animals (Supplemental Fig. S3; Supplemental Table S5). Some underrepresented annotations may relate to the fact that we analyzed adult animals, which would lack RNAs specifically expressed during later stages of embryogenesis and larvae.
Misregulated genes overlap with those altered during exposure to pathogen
Microarray analyses have revealed transcripts that are induced and/or repressed during infection of C. elegans by several pathogens (Mallo et al. 2002; Huffman et al. 2004; O'Rourke et al. 2006; Shapira et al. 2006). There is little overlap between the specific genes in the data sets of these studies, suggesting the innate immune system of C. elegans elicits tailored responses to specific pathogens. However, members of both the C-lectin and lysozyme gene families are induced following infection with either Microbacterium nematophilum, Pseudomonas aeruginosa, or Serratia marcescens. These are the same gene families that the GO analysis indicated are enriched in our RNAi mutants.
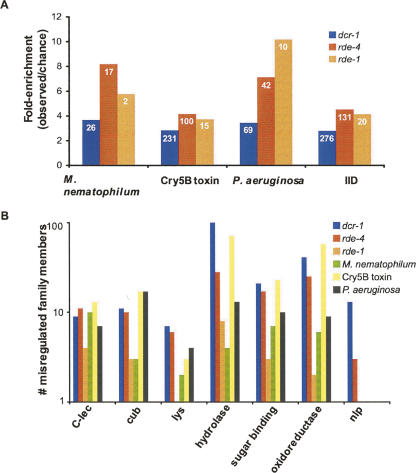
Although we observed elevated expression of innate immunity genes in the RNAi mutants even without exposure to pathogen, we wondered how much overlap existed between our data sets and those of published microarray analyses of worms exposed to M. nematophilum, P. aeruginosa (PA14), or the pore forming toxin Cry5B (Huffman et al. 2004; O'Rourke et al. 2006; Shapira et al. 2006). In addition, the three data sets were combined, and duplicate genes removed, to create a list of genes misregulated in response to infection with any of these pathogens (referred to as innate immunity data set or IID). Our data sets showed substantial overlap with all data sets that was considerably more than expected by random chance (Fig. 4A; Supplemental Table S3; see Materials and Methods for calculations of expected values). Furthermore, analysis of the direction in which the overlapping genes of the various data sets were misexpressed (up or down) displayed strong correlation (Supplemental Table S3). A breakdown of the three infection-related data sets and the dcr-1(−/−), rde-4(−/−), and rde-1(−/−) data sets by gene families involved in innate immunity revealed a similar number of misregulated family members in all data sets (Fig. 4B). The C-type lectins, CUB-like domain proteins, lysozyme proteins, and the antimicrobial neuropeptide-like proteins (nlp family) have been reported to be important in the innate immune system of C. elegans (Mallo et al. 2002; Couillault et al. 2004; Huffman et al. 2004; O'Rourke et al. 2006; Shapira et al. 2006; Troemel et al. 2006). We also analyzed the infection study data sets to determine how many misregulated genes had GO terms relating to hydrolase activity, sugar binding activity, and oxidoreductase activity, since these were enriched in our RNAi data sets (Fig. 3). Again, there were similar numbers of misregulated genes with these GO terms in the infection study data sets and our data sets (Fig. 4B).
FIGURE 4.
Microarray data sets of RNAi mutants overlap with those of C. elegans exposed to pathogen or toxin. (A) Overlap is expressed as number of genes in common/number of genes predicted by random chance, for comparisons of each RNAi mutant data set and data sets from animals exposed to M. nematophilum, Cry5B toxin, or P. aeruginosa. The IID data set combines all three of the infection-related microarrays (see Materials and Methods). The number of genes common between data sets is shown in white within each bar. (B) The number of gene family members that are misregulated in each of the microarray studies is presented. Lists of genes for each infection-related data set, as well as lists of family members, are provided in Supplemental Table S3.
Comparison of pathogen response in wild-type and RNAi mutant strains
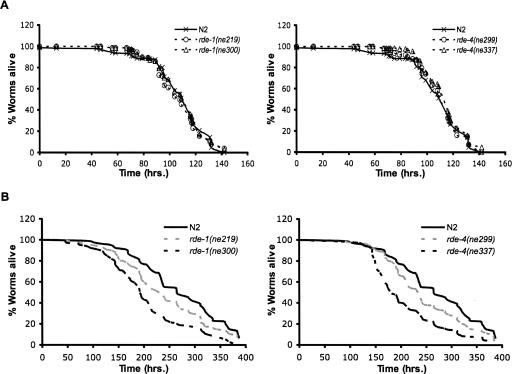
Since many genes involved in the C. elegans innate immune system were up-regulated in RNAi mutants, including the lysozyme family and C-lectins, we wondered if mutant animals would exhibit enhanced resistance to infection. Due to the sick phenotype of dcr-1 mutants, we used rde-4 and rde-1 mutants to test this hypothesis by performing killing assays with the C. elegans pathogen S. marcescens strain Db11. In this assay, L4 worms were grown on plates seeded with Db11 and survival was monitored every 8–12 h. The survival times of two alleles of rde-4(ne299, ne337), two alleles of rde-1(ne219, ne300), and wild-type (N2) worms are plotted in Figure 5A. Kaplan–Meier survival analyses including all data of each 140-h time course showed no significant difference in pathogen sensitivity between wild-type and RNAi mutants. Since wild-type animals induce the expression of innate immune effectors in response to pathogen, it is possible that this induction is rapid enough to negate any potential advantage of the constitutive overexpression of innate immunity effectors in our RNAi mutants (see Discussion).
FIGURE 5.
Analysis of pathogen response and longevity of RNAi mutants. (A) Kaplan–Meier survival analysis (Kaplan and Meier 1958) of animals fed S. marcescens (Db11). Left panel, rde-1(ne219) (open circles) and rde-1(ne300) (open triangles) compared to wild-type N2 (x). Right panel, rde-4(ne299) (open circles) and rde-4(ne337) (open triangles) compared to wild-type N2 (x). Curves represent combined results of three independent experiments (n ≥ 226 for each strain). (B) Longevity analysis for mutants as in A, during feeding with nonpathogenic OP50. Left panel, dashed lines represent rde-1(ne219) (gray) and rde-1(ne300) (black) compared to wild-type N2 (solid black line). Right panel, dashed lines represent rde-4(ne299) (gray) and rde-4(ne337) (black) compared to wild-type N2 (solid black line). Data are the combined results of three independent assays (n ≥ 149 for each strain). The decrease in survival of rde-4 and rde-1 compared to wild-type N2 is significant (P ≤ 0.01 one-sided log-rank test).
As a control for the killing assay we also performed longevity assays on the RNAi mutants under normal laboratory growth conditions in the absence of pathogen (Fig. 5B). Interestingly, all alleles of rde-4 and rde-1 animals had reduced lifespan compared to wild-type (N2) controls (P ≤ 0.01, one-sided log-rank test). While the molecular reason for the shorter lifespan is not yet known, it is possible that the elevated immune system of these animals has deleterious effects.
DISCUSSION
We describe a microarray analysis of adult C. elegans containing a homozygote deletion in the Dicer gene. We observed >8% of the protein-coding genes in these animals to be misregulated. Only a small subset of the misregulated genes overlapped with genes misregulated in the RNAi mutants, rde-4(−/−) and rde-1(−/−), suggesting that the majority of genes misregulated in dcr-1(−/−) animals relate to defective miRNA processing. Consistent with this idea, genes identified as misregulated in dcr-1(−/−) animals were enriched for predicted and actual targets of miRNAs. Surprisingly, in dcr-1(−/−) animals, but not rde-4(−/−) or rde-1(−/−) animals, misregulated genes showed a strong bias for the X chromosome. All animals showed increased expression of genes involved in innate immunity.
General trends and limitations of the analysis
Consistent with functions for dcr-1, rde-4, and rde-1 in gene silencing, the majority of misregulated genes in mutant animals were up-regulated (see Fig. 1A). However, some genes were down-regulated, and many of these may be downstream targets of genes regulated by small RNAs. Of the three mutant strains, Dicer has the most down-regulated genes. This may be due to the germline defects of dcr-1(−/−) animals, and, as mentioned earlier, 38% of these down-regulated genes are germline-enriched genes.
Our data sets are unlikely to represent all transcripts regulated by DCR-1, RDE-4, and RDE-1. For example, many small RNAs act in a stage-specific manner and would be missed in our analysis of adult worms. Of course, analyses of earlier developmental stages in dcr-1(−/−) animals are complicated by the presence of significant amounts of maternal DCR-1, and we analyzed adult worms to minimize this effect.
The dcr-1(−/−) animals used in our studies were in the background of unc-32(−/−) to facilitate isolation. Since unc-32(−/−) animals were used for comparison in our dcr-1(−/−) microarray analyses, we expect the majority of misregulated genes are true targets of DCR-1 and also misregulated in dcr-1(−/−) animals containing a wild-type unc-32 gene. Still, we cannot rule out the possibility that in some cases dcr-1 and unc-32 act synergistically to misregulate gene expression. unc-32 encodes the α subunit of a vacuolar ATPase that is expressed as six isoforms in C. elegans. The unc-32(e189) allele used in our study affects only two of the six isoforms and, furthermore, these two isoforms are only expressed in neurons (Pujol et al. 2001a). Thus, we expect synergistic effects to be minimal. Using qRT-PCR we compared expression of several members of the lysozyme gene family found in our microarray data set, using wild-type N2 animals and homozygote dcr-1 animals not in the unc-32 background (data not shown). These data are limited, but indicate induction of the lysozyme gene family in dcr-1(−/−) mutants is not dependent upon the unc-32 mutation. Regardless, this caveat suggests that researchers interested in long-term studies of specific genes in our data set should perform the relevant controls.
dcr-1(−/−) up-regulated genes are enriched for miRNA, but not siRNA, targets
We observed that 28% of the up-regulated genes in the dcr-1(−/−) microarray data set were miRNA targets as predicted by PicTar. The enrichment for miRNA targets was specific to the dcr-1(−/−) up-regulated data set, and neither the rde-4(−/−), rde-1(−/−), nor dcr-1(−/−) down-regulated data sets showed an enrichment compared to randomly generated lists of genes (see Fig. 2). There are several explanations for why we do not observe greater enrichment for miRNA targets. First, while some miRNAs down-regulate message levels, others may inhibit translation without affecting mRNA levels. The latter would not be identified by our experimental approach. Second, since our analyses were done on adult stage worms, miRNAs that act at earlier developmental stages would be missed. Finally, we note that our study was performed on whole animals, and thus miRNAs that act in a small subset of cells may also be missed, since changes in mRNA levels produced by these miRNAs may not be apparent in the background of mRNA derived from all cells.
We did not see a significant enrichment for previously published endo-siRNA targets within our dcr-1(−/−), rde-4(−/−), or rde-1(−/−) lists of up-regulated genes (Lee et al. 2006; Ruby et al. 2006). The endo-siRNAs from the Ambros and Bartel laboratories were isolated from mixed stage populations, while we analyzed adult animals, and this may explain the lack of overlap. Alternatively, the presence of a large number of direct and indirect miRNA targets within the dcr-1(−/−) data set may mask an enrichment for endo-siRNA targets if they are much less abundant in the population. However, it is also possible that most endo-siRNAs are produced in a pathway that does not require dcr-1, rde-4, or rde-1. In fact, recent work indicates that most endo-siRNAs are “secondary siRNAs” whose synthesis is independent of DCR-1 and instead requires a RNA-directed RNA polymerase, RdRP (Pak and Fire 2007; Sijen et al. 2007). Here we note that EGO-1, a putative germline RdRP, is responsible for a Dicer-independent silencing pathway in the meiotic germline of C. elegans (Maine et al. 2005).
Why does DCR-1 regulate so many genes on the X chromosome?
On average, 5% of the genes on each autosome were up-regulated in dcr-1(−/−) animals, while this number increased to 11% for the X chromosome (Table 3). Possibly DCR-1 has a role in an X chromosome regulatory process, such as dosage compensation. Dosage compensation in C. elegans occurs by down-regulating the expression of each X in an XX hermaphrodite by roughly half, to equal the expression of X-linked genes in the XO male (Meyer 2005). In support of the idea that DCR-1 is involved in dosage compensation, the enrichment for X chromosome genes in our dcr-1(−/−) data set most strongly correlated with gene expression changes around twofold (2.5-fold greater than average autosome); enrichment diminished outside of this range (1.3-fold greater than average autosome for genes up-regulated more than fivefold).
There are several mechanisms by which Dicer could affect dosage compensation. If a gene with an important role in dosage compensation was regulated by a miRNA, its expression might be altered in dcr-1(−/−) animals, thus affecting expression of all genes on the hermaphrodite X chromosome. Of 10 genes known to be important for dosage compensation in C. elegans (sdc-1, sdc-2, sdc-3, dpy-21, dpy-26, dpy-27, dpy-28, dpy-30, mix-1, and xol-1) (Meyer 2005), only sdc-3 was on the list of genes misregulated in dcr-1(−/−) animals. sdc-3 was up-regulated 1.9-fold in dcr-1(−/−) animals; it is unclear if this level of misregulation would cause the effects we observe. Alternatively, X chromosome genes subject to dosage compensation could be regulated directly by an miRNA or family of miRNAs.
Not only did we observe an enrichment of genes regulated by DCR-1 on the X chromosome, but there were more predicted miRNA targets on the X chromosome. Greater than 40% of the up-regulated genes on the X chromosome were predicted miRNA targets compared with 23% for the autosomal genes. Consistent with this observation, there is also an enrichment for miRNA targets residing on the X chromosome within the cePicTar data set (1.6-fold higher on X than the average autosome).
All of these observations indicate DCR-1 plays a special role in regulating expression of the X chromosome. Although our data are consistent with a role for DCR-1 in dosage compensation, we cannot rule out the possibility that miRNA binding sites are just easier to identify on the X chromosome, possibly because this chromosome is more conserved (e.g., see Lu and Wu 2005). Alternatively, miRNA-regulated genes may just be more abundant on the X chromosome. Of course, this raises the question as to why miRNA targets might be more abundant on the X chromosome. Perhaps the evolution of miRNA-mediated silencing coincided with the need to silence the X chromosome?
Finally, we note that it is possible that the observed X chromosome silencing involves some other RNA, for example, sense and antisense transcripts. However, while such noncoding RNAs participate in dosage compensation in other organisms, they have not been implicated in dosage compensation in C. elegans.
Does RNAi play a role in innate immunity?
Several signaling pathways have been identified in C. elegans that lead to the induction of innate immune effectors, including the TGF-β-like pathway, p38 MAPK pathway, DAF-2/DAF-16 insulin-like receptor pathway, and programmed cell death pathway (for review, see Schulenburg et al. 2004). We find that numerous immune effectors are up-regulated in dcr-1(−/−), rde-4(−/−), and rde-1(−/−) animals, and in future studies it will be of interest to determine which signaling pathways are involved in this induction. While molecules previously identified as effectors are enriched in our data sets, we also expect our lists to contain unrecognized effectors, as well as genes misregulated as a secondary consequence.
We did not see altered resistance to S. marcescens in rde-4 or rde-1 mutant animals and, thus, the biological significance of our microarray data awaits further experimentation. C. elegans responds to pathogen by inducing innate immunity genes (Mallo et al. 2002; Huffman et al. 2004; Kerry et al. 2006; O'Rourke et al. 2006; Shapira et al. 2006), and possibly the levels of immune effectors induced by infection of wild-type animals are too similar to those existing in RNAi mutants to show differences in the time frame of our assay. Further, while several studies have focused on the pathogen susceptibility of C. elegans lacking certain immune effectors (Mallo et al. 2002; Huffman et al. 2004; Kerry et al. 2006; O'Rourke et al. 2006; Shapira et al. 2006), effects of overexpression of multiple immune effectors, as we observe in the RNAi mutants, have not been studied. Some studies have monitored effects of overexpression of a single effector and observed little or no benefit in survival on pathogen (Mallo et al. 2002; Couillault et al. 2004).
It is important to note that since DCR-1 and RDE-4 act together to cleave dsRNA to siRNAs animals lacking these enzymes may have higher levels of uncleaved dsRNA. Thus, conceivably, dsRNA itself could lead to induction of innate immunity genes in RNAi mutant strains. However, while dsRNA clearly elicits an immune response in mammals (e.g., Kato et al. 2006), it is not thought to trigger such a response in C. elegans. Consistent with this, we assayed a subset of innate immunity genes after feeding C. elegans with bacteria expressing dsRNA and saw no changes in expression (unpublished data).
Several studies suggest roles for RNAi during viral infection (Schott et al. 2005; Wilkins et al. 2005; Galiana-Arnoux et al. 2006; van Rij et al. 2006; Wang et al. 2006). These studies indicate that during infection by RNA viruses Dicer actively participates in the antiviral response by cleaving viral dsRNA to produce siRNAs that target viral mRNA. So far, there is only one report implicating Dicer in the immune response to bacteria. In this case, a flagellin peptide of Pseudomonas syringae induces an Arabidopsis miRNA that down-regulates auxin signaling to promote resistance (Navarro et al. 2006).
We speculate that our data hint at a broader role for Dicer in innate immunity than previously recognized. Rather than simply promoting the degradation of the pathogenic agent (i.e., viral RNA), or processing a single miRNA that affects pathogen resistance, our studies raise the possibility that RNAi components act constitutively to silence the innate immune response, presumably until a pathogen is encountered. While it is possible that this function is mediated entirely through small RNAs, we offer an alternative model that is based on comparisons with innate immunity in vertebrates.
As mentioned, the innate immune system in all metazoa is based on the recognition of pattern-associated molecular patterns (often nucleic acid of the pathogen) by pathogen recognition receptors (PRRs). Two types of PRRs have been described: the Toll-like receptors that function at the membrane to intercept a pathogen before it enters the cytoplasm, and soluble cytoplasmic proteins that detect pathogens once they enter the cytoplasm (for review, see Decker et al. 2005; Kawai and Akira 2006; Meylan et al. 2006). All of these PRRs respond to pathogens by triggering a signaling pathway that ultimately leads to increased transcription of innate immunity genes, such as the Type-I interferons of vertebrates.
The cytoplasmic PRRs were more recently discovered, and thus less is known about these factors than the membrane bound Toll-like receptors. However, the RIG-1 and MDA5 cytoplasmic PRRs are characterized by a helicase domain with significant sequence similarity to the helicase domain of Dicer, as well as the C. elegans dicer-related helicases (DRH-1, DRH-2, DRH-3) found in a complex with DCR-1, RDE-1, and RDE-4 (Tabara et al. 2002; Duchaine et al. 2006). In fact, among all helicases in the nucleotide databases currently queried by BLAST, this group of helicases shows the highest scoring similarities (data not shown). Thus, while somewhat heretical, we are currently exploring the idea that the helicase domains of these RNAi factors play an active role in the induction of genes required for the immune response to a pathogen.
MATERIALS AND METHODS
Strains and maintenance
The following strains were maintained under standard laboratory conditions (Brenner 1974): Bristol strain N2, BB1 dcr-1(ok247) unc-32(e189)/++ III, CB189 unc-32(e189) III, BB9 rde-1(ne219) V, BB10 rde-1(ne300) V, BB11 rde-4(ne299) III, and BB12 rde-4(ne337) III.
RNA isolation
unc-32(−/−) and dcr-1(−/−) samples were harvested by picking adult worms based on the coiler phenotype. Other strains were synchronized by hypochlorite treatment (Emmons et al. 1979). Synchronized L1 worms were grown on NGM/OP50 plates and harvested at adulthood. Total RNA was isolated using Trizol (Invitrogen) as per manufacturer's protocol, treated with Turbo DNase (Ambion) followed by RNA cleanup using the RNeasy Mini Kit (Qiagen).
Affymetrix microarray analysis
Analyses were performed using Affymetrix Genechip Arrays for C. elegans as per manufacturer's protocol. Biological replicates were analyzed in triplicate for dcr-1(ok247); unc-32(e189) and unc-32(e189), and in quadruplicate for Bristol N2, rde-1(−/−), and rde-4(−/−). For rde-1(−/−), two samples derived from rde-1(ne219) and two from rde-1(ne300). Similarly, two samples were obtained using rde-4(ne299) and two from rde-4(ne337). Microarray analyses were performed with GeneSifter software (VizX Labs) using the GCRMA algorithm (Wu et al. 2003) and analyzed by applying a statistical t-test: P < 0.05 for rde-4(−/−) and rde-1(−/−) analyses; P < 0.01 for dcr-1(−/−) analysis with a threshold of 1.5-fold misregulation. The Benjamini and Hochberg FDR correction was applied for the dcr-1(−/−) analysis (Benjamini and Hochberg 1995). Gene lists were curated by cross-referencing with WormBase (http://www.wormbase.org, release WS157). When annotation indicated a single probe correlated with multiple genes, all such genes were excluded from our final lists. Finally, gene lists were curated to remove duplicate entries. Curated gene lists and raw GeneSifter output are included in Supplemental Table S1.
Determining expected overlap of different microarray data sets
Expected overlap was determined by multiplying the number of genes misregulated in data set 1 by the number misregulated in data set 2 and then dividing by the total number of genes represented on the microarray chips (18,020 genes).
Quantitative real-time PCR
cDNA was synthesized with 5 μg of total RNA per 20 μL reaction using random decamers (Ambion) and Superscript II (Invitrogen) according to the manufacturer's protocol. qRT-PCR was performed using a Lightcycler 2.0 instrument and Masterplus Sybr Green I kit (Roche). qRT-PCR primers spanned at least one exon boundary where possible. The quality of qRT-PCR products was assessed using melting curve analysis and gel electrophoresis. gpd-3 RNA levels were used to normalize.
miRNA and endo-siRNA analysis
A list of all PicTar miRNA targets was obtained by querying the PicTar website (http://pictar.bio.nyu.edu/cgi-bin/new_PicTar_nematode.cgi?species=nematode) with each available C. elegans miRNA (miRNA ID). Predicted targets were combined into a single file, duplicate entries removed, and isoforms treated as a single gene. The final data set included 2716 genes (see Supplemental Table S2). The endo-siRNA target data set was extracted from published supplemental data and included 1229 genes (Lee et al. 2006). Ten random lists of genes were used as control data sets, and the average of 10 independent comparisons is presented. These data sets were compared with lists of misregulated genes from microarray analyses to determine the number of overlapping genes.
GO analysis
Curated gene lists were used as input for FatiGO+ (http://www.babelomics.org) (Al-Shahrour et al. 2005). FatiGO+ version 2.0 was used, which relies on WormBase annotations of 1/28/2006 and Gene Ontology of 01/2006. Misregulated genes were compared to the remaining genes on the array (Supplemental Tables S4, S5). FatiGO software reports an unadjusted P-value based on a Fisher's exact text and an adjusted P-value calculated using the FDR procedure of Benjamini and Hochberg (Benjamini and Hochberg 1995).
Killing/longevity assays
Survival assays using S. marcescens (Db11) were as described (Pujol et al. 2001b). L4 stage worms were transferred to NGM plates seeded with S. marcescens (10–20 worms/plate) and cultivated at 23°C. Worms were moved daily to freshly seeded plates and assayed for viability every 8–12 h by response to touch. Kaplan–Meier survival analyses and one-sided log-rank tests were used to compare survival curves. Longevity assays were carried out similarly by growing worms at 20°C on OP50, and scoring viability every 8–12 h.
Accession numbers
Microarray data were deposited at ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under the accession numbers E-MEXP-956 (rde-1, rde-4) and E-MEXP-957 (dcr-1).
SUPPLEMENTAL DATA
Supplemental Data include three figures and five tables available at: http://www.biochem.utah.edu/bass/Publications/NWJHBB-RNA2007supp.htm.
ACKNOWLEDGMENTS
We thank the Caenorhabditis Genetics Center (CGC), Craig Mello, and Scott Knight for worm strains. We also thank Cyril-Leopold Kurz, previously of the Ewbank laboratory, for supplying S. marcescens (Db11), Rich Holubkov for advice regarding statistics, David Bartel and Janis Weiss for helpful discussions, and Alexander Kozik for sharing his Python source code for depicting the location of genes on chromosomes. This work was supported by funds to B.L.B. from the National Institutes of Health (GM067106); B.L.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.542107.
REFERENCES
- Al-Shahrour, F., Minguez, P., Vaquerizas, J.M., Conde, L., Dopazo, J. BABELOMICS: A suite of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., Jewell, D. MicroRNAs and other tiny endogenous RNAs in C. elegans . Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y., Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol). 1995;57:289–300. [Google Scholar]
- Brenner, S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J.F., Kohara, Y., Ewbank, J.J. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Decker, T., Muller, M., Stockinger, S. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- Duchaine, T.F., Wohlschlegel, J.A., Kennedy, S., Bei, Y., Conte D., Jr, Pang, K., Brownell, D.R., Harding, S., Mitani, S., Ruvkun, G., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Emmons, S.W., Klass, M.R., Hirsh, D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans . Proc. Natl. Acad. Sci. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J.A., Imler, J.L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila . Nat. Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Johnson, T., Reinert, K.L., Gerstein, M., Slack, F.J. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans . Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Hansen, K.R., Burns, G., Mata, J., Volpe, T.A., Martienssen, R.A., Bahler, J., Thon, G. Global effects on gene expression in fission yeast by silencing and RNA interference machineries. Mol. Cell. Biol. 2005;25:590–601. doi: 10.1128/MCB.25.2.590-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman, D.L., Abrami, L., Sasik, R., Corbeil, J., van der Goot, F.G., Aroian, R.V. Mitogen-activated protein kinase pathways defend against bacterial pore-forming toxins. Proc. Natl. Acad. Sci. 2004;101:10995–11000. doi: 10.1073/pnas.0404073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, E.L., Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–481. [Google Scholar]
- Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K.J., et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai, T., Akira, S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- Kerry, S., Tekippe, M., Gaddis, N.C., Aballay, A. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS ONE. 2006 doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans . Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, S.W., Bass, B.L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans . Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lall, S., Grun, D., Krek, A., Chen, K., Wang, Y.L., Dewey, C.N., Sood, P., Colombo, T., Bray, N., Macmenamin, P., et al. A genome-wide map of conserved microRNA targets in C. elegans . Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans . Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Hammell, C.M., Ambros, V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans . RNA. 2006;12:589–597. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J., Wu, C.I. Weak selection revealed by the whole-genome comparison of the X chromosome and autosomes of human and chimpanzee. Proc. Natl. Acad. Sci. 2005;102:4063–4067. doi: 10.1073/pnas.0500436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine, E.M., Hauth, J., Ratliff, T., Vought, V.E., She, X., Kelly, W.G. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr. Biol. 2005;15:1972–1978. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo, G.V., Kurz, C.L., Couillault, C., Pujol, N., Granjeaud, S., Kohara, Y., Ewbank, J.J. Inducible antibacterial defense system in C. elegans . Curr. Biol. 2002;12:1209–1214. doi: 10.1016/s0960-9822(02)00928-4. [DOI] [PubMed] [Google Scholar]
- Matzke, M.A., Birchler, J.A. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Meyer, B.J. X-chromosome dosage compensation. 2005 doi: 10.1895/wormbook.1.8.1. http://www.wormbook.org/chapters/www_dosagecomp/dosagecomp.html [DOI] [PMC free article] [PubMed]
- Meylan, E., Tschopp, J., Karin, M. Intracellular pattern recognition receptors in the host response. Nature. 2006;442:39–44. doi: 10.1038/nature04946. [DOI] [PubMed] [Google Scholar]
- Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., Voinnet, O., Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- O'Rourke, D., Baban, D., Demidova, M., Mott, R., Hodgkin, J. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum . Genome Res. 2006;16:1005–1016. doi: 10.1101/gr.50823006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, J., Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parker, G.S., Eckert, D.M., Bass, B.L. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, S., Fire, A. Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans . RNA. 2001;7:1397–1402. [PMC free article] [PubMed] [Google Scholar]
- Pujol, N., Bonnerot, C., Ewbank, J.J., Kohara, Y., Thierry-Mieg, D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J. Biol. Chem. 2001a;276:11913–11921. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- Pujol, N., Link, E.M., Liu, L.X., Kurz, C.L., Alloing, G., Tan, M.W., Ray, K.P., Solari, R., Johnson, C.D., Ewbank, J.J. A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans . Curr. Biol. 2001b;11:809–821. doi: 10.1016/s0960-9822(01)00241-x. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans . Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Reinke, V., Gil, I.S., Ward, S., Kazmer, K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans . Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Ruby, J.G., Jan, C., Player, C., Axtell, M.J., Lee, W., Nusbaum, C., Ge, H., Bartel, D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans . Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Schmitter, D., Filkowski, J., Sewer, A., Pillai, R.S., Oakeley, E.J., Zavolan, M., Svoboda, P., Filipowicz, W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott, D.H., Cureton, D.K., Whelan, S.P., Hunter, C.P. An antiviral role for the RNA interference machinery in Caenorhabditis elegans . Proc. Natl. Acad. Sci. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg, H., Kurz, C.L., Ewbank, J.J. Evolution of the innate immune system: The worm perspective. Immunol. Rev. 2004;198:36–58. doi: 10.1111/j.0105-2896.2004.0125.x. [DOI] [PubMed] [Google Scholar]
- Shapira, M., Hamlin, B.J., Rong, J., Chen, K., Ronen, M., Tan, M.W. A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. 2006;103:14086–14091. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen, T., Steiner, F.A., Thijssen, K.L., Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Carthew, R.W. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans . Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Yigit, E., Siomi, H., Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans . Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Troemel, E.R., Chu, S.W., Reinke, V., Lee, S.S., Ausubel, F.M., Kim, D.H. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans . PLoS Genet. 2006 doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij, R.P., Saleh, M.C., Berry, B., Foo, C., Houk, A., Antoniewski, C., Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes & Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.H., Aliyari, R., Li, W.X., Li, H.W., Kim, K., Carthew, R., Atkinson, P., Ding, S.W. RNA interference directs innate immunity against viruses in adult Drosophila . Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins, C., Dishongh, R., Moore, S.C., Whitt, M.A., Chow, M., Machaca, K. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans . Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Irizarry, R., Gentleman, R., Murillo, F., Spencer, F.A. Technical Report, Department of Biostatistics Working Papers, John Hopkins University; Baltimore, MD: 2003. “A model based background adjustment for oligonucleotide expression arrays.”. [Google Scholar]
- Yigit, E., Batista, P.J., Bei, Y., Pang, K.M., Chen, C.C., Tolia, N.H., Joshua-Tor, L., Mitani, S., Simard, M.J., Mello, C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]