Abstract
RNA silencing plays a key role in antiviral defense as well as in developmental processes in plants and insects. Negative strand RNA viruses such as the plant virus Rice hoja blanca tenuivirus (RHBV) replicate in plants and in their insect transmission vector. Like most plant-infecting viruses, RHBV encodes an RNA silencing suppressor, the NS3 protein, and here it is demonstrated that this protein is capable of suppressing RNA silencing in both plants and insect cells. Biochemical analyses showed that NS3 efficiently binds siRNA as well as miRNA molecules. Binding of NS3 is greatly influenced by the size of small RNA molecules, as 21 nucleotide (nt) siRNA molecules are bound > 100 times more efficiently than 26 nt species. Competition assays suggest that the activity of NS3 is based on binding to siRNAs prior to strand separation during the assembly of the RNA-induced silencing complex. In addition, NS3 has a high affinity for miRNA/miRNA* duplexes, indicating that its activity might also interfere with miRNA-regulated gene expression in both insects and plants.
Keywords: RNAi, silencing, virus, suppressor, miRNA, siRNA
INTRODUCTION
RNA silencing is an evolutionarily conserved mechanism in many, if not all, eukaryotes to target and degrade aberrant endogenous or exogenous RNA molecules (Sontheimer 2005; Tomari and Zamore 2005; Voinnet 2005). More recently, related processes were shown to be involved in eukaryotic gene regulation processes through host-encoded micro-RNAs (miRNAs) (for reviews, see Carrington and Ambros 2003; Bartel 2004; Herr 2005).
A common feature of all RNA silencing processes is the endonucleolytic cleavage of longer double-stranded (ds) RNA molecules into short interfering RNA (siRNA) or miRNA species. These small RNAs are 21–26 base pairs in size (Hamilton and Baulcombe 1999) and contain 2 nucleotide (nt) 3′-overhangs and 5′-phosphorylated termini, which are characteristic for their production by RNase-III-type enzymes from the Drosha/Dicer protein family (Bernstein et al. 2001; Lee et al. 2003). The siRNA guide strand or miRNA strand of the small RNA duplex programs a ribonucleoprotein complex, the RNA induced silencing complex (RISC), for sequence-specific recognition of RNA targets (Khvorova et al. 2003; Schwarz et al. 2003; Lee et al. 2004; Tomari and Zamore 2005). Enzymatic activity of members of the Argonaute (Ago) protein family enables RISC to slice complementary mRNAs or arrest their translation (Fagard et al. 2000; Hammond et al. 2000).
The core machinery of RNA silencing plays diverse and essential roles in regulation of gene expression by miRNAs, genome defense against transposons and viruses, and modification of chromatin structure (Mallory and Vaucheret 2006). MiRNAs originate from long noncoding single-stranded RNAs (ssRNAs) and negatively regulate complementary mRNAs by either guiding RNA slicing activity by RISC or translational repression (Bartel 2004). In plants, RNA silencing is an important antiviral defense initiated by structured viral RNAs, dsRNA replication intermediates of plant viruses, cytoplasmically replicating viruses, or dsRNA production by plant RNA-dependent RNA polymerase (RDR) action (RDR1 or RDR6). Subsequent processing of viral specific dsRNAs results in the accumulation of viral siRNAs (Silhavy and Burgyan 2004; Molnar et al. 2005). Interestingly, RNA silencing in plants can generate a second class of larger siRNAs (24–26 nt) that seem to be involved in systemic signaling and are proposed to travel to different plant organs in advance of the invading virus (Hamilton et al. 2002; Tang et al. 2003).
In insects the miRNA- and siRNA-mediated RNA silencing processes are initiated by Dcr-1 and Dcr-2, respectively (Lee et al. 2004). Dcr-1 function requires Ago1, whereas siRNA synthesis and active RISC complex formation is Ago2 dependent (Lee et al. 2004; Okamura et al. 2004). Cultured Drosophila cells or animals depleted of or lacking Dcr-2, R2D2, or Ago2 showed higher accumulation of the insect-infecting Flock house virus (FHV), Drosophila C virus (DCV), and Cricket paralysis virus (CrPV), suggesting also an antiviral role of RNA silencing in insects (Li et al. 2002, 2004; Galiana-Arnoux et al. 2006; van Rij et al. 2006; Wang et al. 2006). RISC complex formation is initiated by the R2D2/Dcr-2 complex, which is a sensor for siRNA strand loading (Pham et al. 2004; Tomari et al. 2004b). This complex associates with an Ago2-containing protein complex, and the passenger strand of the siRNA complex is released by the Ago2 slicer function (Matranga et al. 2005; Miyoshi et al. 2005; Rand et al. 2005) to give rise to the activated RISC complex, which catalyzes sequence-specific mRNA degradation (Tuschl et al. 1999).
To counteract antiviral RNA silencing, plant viruses were shown to encode specific proteins that were previously mainly associated with the enhancement of viral pathogenicity and accumulation. Over the years many RNA silencing suppressor proteins of plant viruses have been identified (Silhavy and Burgyan 2004; Voinnet 2005). Suppression of antiviral silencing can be accomplished by binding to ds siRNAs (Lakatos et al. 2006; Merai et al. 2006), masking long dsRNA molecules (Merai et al. 2005; van Rij et al. 2006), or inhibition of active RISC by physical interaction between the suppressor and the slicer component (Zhang et al. 2006). Besides suppression of siRNA-mediated antiviral silencing, several plant viral suppressors also induce developmental abnormalities in plants by interfering with the miRNA pathway (Chapman et al. 2004; Dunoyer et al. 2004).
The identification of RNA silencing suppressors has not remained limited to plant viruses, as the B2 protein of the insect-infecting FHV has been identified as a viral suppressor in insect cells. Cross-kingdom suppression of RNA silencing was observed for the FHV B2 protein in plants (Li et al. 2002). Also human-infecting viruses encode proteins that can act as suppressors of RNA silencing. The NS1 protein of Influenza virus A is active in insect cells as well as in plants (Bucher et al. 2004; Delgadillo et al. 2004; Li et al. 2004) and the NSs protein of La Crosse virus (LACV) shows RNA silencing inhibition in human cells (Soldan et al. 2005).
The Drosophila embryo extract in vitro RNA silencing system was employed for the molecular and biochemical characterization of the suppressor p19 of tombusvirus (Lakatos et al. 2004). Although members belonging to tombusviruses are often transmitted by insects, like many members belonging to positive (+) strand single-stranded (ss) RNA viruses, they replicate exclusively in their plant hosts and thus are unlikely to be involved in combating antiviral RNA silencing in insects. In previous studies we, and others, reported that two members of negative (−) strand RNA plant viruses also carry a suppressor of RNA silencing (Takeda et al. 2002; Bucher et al. 2003). In contrast to (+) strand RNA plant viruses, the (−) strand RNA plant viruses replicate in both insect vector and plant host (Wijkamp et al. 1993; Falk and Tsai 1998) and are therefore likely a target for antiviral silencing in plant host and insect vector. Rice hoja blanca virus (RHBV) of the genus Tenuivirus is such a (−) strand RNA virus (Ramirez et al. 1992, 1993), which is transmitted by and replicated in a plant hopper, Tagosodes orizicolus.
Since RHBV replicates both in insects and plants, we aspired to investigate whether the RHBV NS3 suppressor protein also suppresses RNA silencing in insects and furthermore examined the molecular mechanism of RNA silencing inhibition using the established insect in vitro embryo extract system of Drosophila. In addition, the ability of the NS3 protein to act on gene regulation through the miRNA pathway was examined by a biochemical approach.
RESULTS
NS3 is a functional RNA silencing suppressor in plants as well as in insects
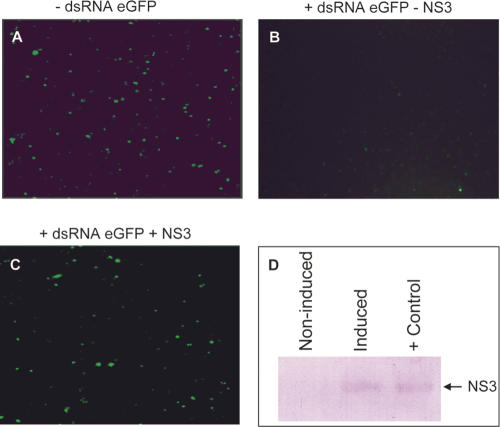
RHBV infects rice and is transmitted by plant hoppers in which it also replicates (Falk and Tsai 1998). It is therefore likely to be targeted by antiviral RNA silencing in both plants and insects, and the NS3 protein of RHBV was shown to be a suppressor of RNA silencing in plants (Bucher et al. 2003). In Drosophila, Dcr-2, R2D2, and Ago2 were shown to be involved in the antiviral response (Galiana-Arnoux et al. 2006; van Rij et al. 2006; Wang et al. 2006). To investigate the RNA silencing suppression activity of NS3 in insects, we expressed a reporter (eGFP) in cultured Drosophila S2 cells. Effective RNA silencing of eGFP was achieved by adding long GFP-specific dsRNA to the cell culture (Fig. 1A). After induction, the NS3 protein was detected by Western blotting (Fig. 1D), and GFP levels were notably higher compared to noninduced cells (Fig. 1, cf. B and C). These results show that NS3, next to being active in plants, is also able to suppress RNA silencing in insect cells.
FIGURE 1.
Suppression of RNA silencing by NS3 in cultured Drosophila cells. Cells were transfected with pAc-eGFP and empty pMK33 (A) and treated 1 h after transfection with dsRNA specific for eGFP. Cells in panels B and C were transfected with the same transfection mixture containing pAc-eGFP and pMK33-NS3. NS3 expression was induced with CuSO4, resulting in an increase of the eGFP signal (C) compared to the noninduced cells (B). Expression of NS3 was confirmed by Western blot analysis using bacterial expressed HIS-tagged NS3 as positive control (D).
The MBP–NS3 fusion protein is an active RNA silencing suppressor
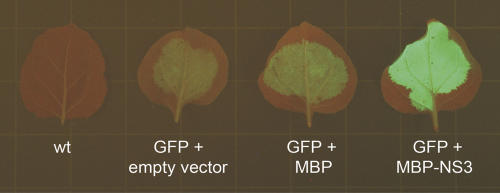
To address the question of how NS3 exerts its function as suppressor of RNA silencing, a biochemical approach was followed (Lakatos et al. 2004, 2006). First the NS3 protein was produced in bacteria, as a C-terminal fusion to the maltose binding protein for purification purposes. Suppressor activity of the MBP–NS3 fusion protein was verified in plants using the established Agrobacterium tumefaciens transient expression assay (ATTA) (Fig. 2; Bucher et al. 2003).
FIGURE 2.
GFP silencing suppression of MBP–NS3 in Agrobacterium-infiltrated Nicotiana benthamiana leaves visualized 5 d post-infiltration. From left to right: noninfiltrated wild-type and GFP expression constructs coinfiltrated with an empty binary vector, the MBP construct, and MBP–NS3 binary vector, respectively.
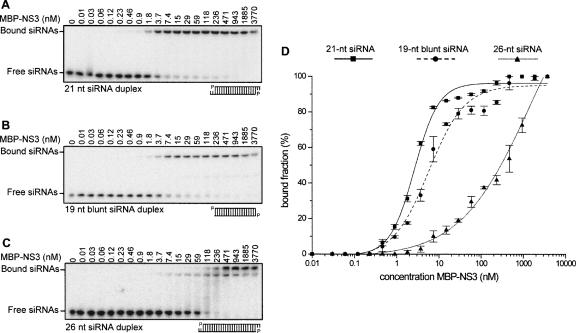
NS3 has high affinity for 21 nt but not for 26 nt siRNAs
Several other strong RNA silencing suppressors have been shown to bind small RNAs with varying affinities. Here it was investigated whether also NS3 is able to bind small RNAs and which size and structural features might influence the affinity of the interaction. The affinity of the NS3 protein to different small dsRNA molecules was determined by electrophoretic mobility shift assays (EMSA). Radiolabeled small RNA molecules were incubated with a dilution series of the NS3 protein and complexes were resolved by native gel electrophoresis. The dissociation constant (Kd) of NS3 for 21 nt siRNA was calculated to be 2.45 ± 0.26 nM (Fig. 3A,D), indicating a high binding affinity in the same order of magnitude as the well-studied suppressor p19 with a Kd of 0.17 ± 0.02 nM for 21 nt siRNA (Vargason et al. 2003). Only a slightly lower affinity (Kd 5.7 ± 0.80 nM) was observed for siRNAs lacking the 2 nt overhangs (Fig. 3B,D), whereas the Kd increased dramatically (> 300 nM) when 26 nt siRNAs were tested (Fig. 3C,D). The MBP expression tag by itself was shown not to bind to 21 nt siRNA molecules and had no effect in subsequent control experiments (data not shown), thereby excluding a role of the tag in the observed siRNA binding and further biochemical analyses. These experiments showed that NS3 binds short siRNA molecules with high affinity and that the 3′ 2 nt overhangs are not essential for NS3 binding.
FIGURE 3.
Affinity of MBP–NS3 for different RNA duplexes. A dilution series of MBP–NS3 (0.01–3770 nM) was incubated with 100 pM each of 32P-labeled 21 nt siRNA duplex (A), 19 nt blunt ended RNA duplex (B), or 26 nt siRNA duplex (C) for 20 min, then loaded onto a 5% native gel. The Kd was determined of MBP–NS3 for the different small RNA molecules by plotting the bound RNA fraction as a function of the MBP–NS3 concentration (D). In panels A, B, and C the first lane contains only siRNAs.
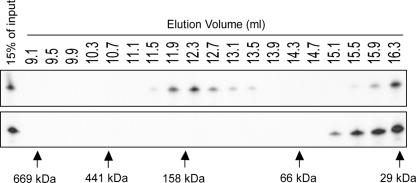
NS3 binds small RNAs as a dimer
To obtain more insight into the stoichiometry of siRNA binding by NS3, gel filtration experiments were carried out with purified NS3 protein and radiolabeled siRNA molecules. To do this, the EMSA reaction was scaled up and subsequently size separated. As a control, siRNA without NS3 was loaded at the same concentration onto the column. Gel filtration fractions were tested for the presence of siRNAs by denaturing gel electrophoresis. Our results showed that unbound siRNAs were found in the same fractions as control siRNAs. An additional peak, corresponding to the NS3–siRNA complex, was detected to be migrating at a size similar to 150 kDa (Fig. 4). This suggests that a single siRNA pair (∼ 14 kDa) is bound by two molecules of MBP–NS3 (∼ 66 kDa). Binding of RNA silencing suppressors as a dimer has been observed previously for the plant viral suppressor protein p19 (Vargason et al. 2003) and the insect viral B2 protein (Chao et al. 2005; Lingel et al. 2005).
FIGURE 4.
Gel filtration of the MBP–NS3–siRNA complex. MBP–NS3 was incubated with 32P-labeled siRNAs and size separated on a Superdex-200 column. Fractions were collected and tested for the presence of 32P-labeled siRNAs (top panel). As control 32P-labeled siRNAs were size separated in the absence of MBP–NS3 (lower panel). The elution position of protein molecular weight markers is indicated by arrows below the picture: 669 kDa, thyroglobulin (9.1 mL); 441 kDa, ferritin (10.5 mL); 158 kDa, aldolase (12.1 mL); 66 kDa, bovine serum albumin (14.3 mL); and 29 kDa, carbonic anhydrase (16.3 mL).
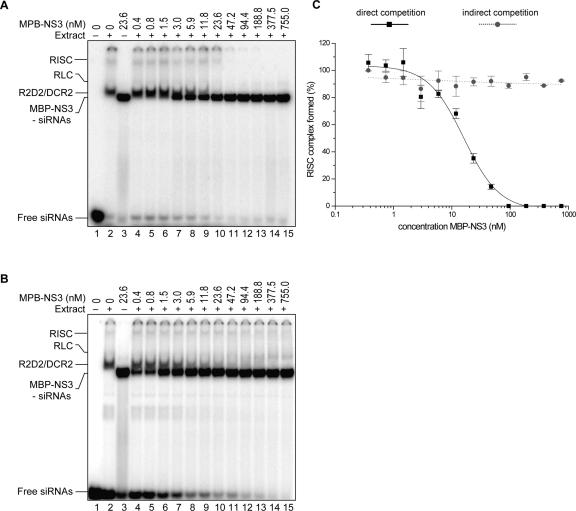
NS3 competes for siRNAs in the RISC assembly process
The biochemical analysis of RISC functionality and assembly is best studied in the in vitro Drosophila embryo extract RNA silencing system, where mature RISC and intermediate complexes can be visualized (Pham et al. 2004; Tomari et al. 2004a). RISC complexes are assembled in an organized manner (Pham et al. 2004; Tomari et al. 2004a). Initial complexes drive the assembly of the mature, active RISC complexes from the R2D2/Dcr-2 complex (R1) through a distinct intermediary complex, the RISC loading complex (RLC). The formation of active RISC complexes on siRNAs can be visualized by native gel electrophoresis and the influence on the complex formations studied by adding increasing concentration of the suppressor protein to the in vitro reactions. We used the system based on native gel electrophoresis using Drosophila embryo lysate (Pham et al. 2004) with modifications as described (Lakatos et al. 2006).
Labeled siRNAs, Drosophila extract, and recombinant NS3 protein were incubated simultaneously in direct competition assays, and formation of mature RISC decreased gradually with increasing NS3 concentration (Fig. 5A,C). Complete inhibition of RISC formation was observed at higher (> 90 nM) NS3 concentrations. In a second setup, the effect of NS3 on preassembled RISC complexes was tested. RISC was assembled by incubation of Drosophila extract with siRNAs and, after 30 min, different concentrations of NS3 were added to the mixtures. These results showed that in this case increasing NS3 amounts had no effect on the preassembled RISC (Fig. 5B,C). Though partially masked by the MBP–NS3 protein, Figure 5, A and B, suggests a decrease in the R1 complex upon elevation of MBP–NS3 concentrations, which might imply that NS3 is capable of extracting double-stranded siRNAs from R1 complexes prior to strand separation or sequestering free siRNAs during passive transient release of siRNAs by the R1 complex in RISC assembly. Control experiments using MBP alone indicated no inhibitory effect of the tag on the formation of silencing complexes (data not shown).
FIGURE 5.
Inhibition of RISC assembly by NS3 in vitro. (A) In direct competition experiments, RISC assembly was monitored by adding 32P-labeled siRNAs and MBP–NS3 (0.4–755.0 nM) to Drosophila embryo extract. (B) Indirect competition assay where RISC assembly was initiated by adding 32P-labeled siRNAs to embryo extract. MBP–NS3 (0.4–755.0 nM) was added to preincubated reactions after 30 min. (A,B) Lane 1 contains only free siRNAs, lane 2 32P-labeled siRNAs and embryo extract, and lane 3 32P-labeled siRNAs and 23.6 nM MBP–NS3. In lanes 4–15 the competition effect of MBP–NS3 on RISC assembly is shown. (C) For direct and indirect competition experiments the formation of the RISC complex as a function of MBP–NS3 concentration is plotted relative to the RISC formation in the absence of MBP–NS3 (lane 2).
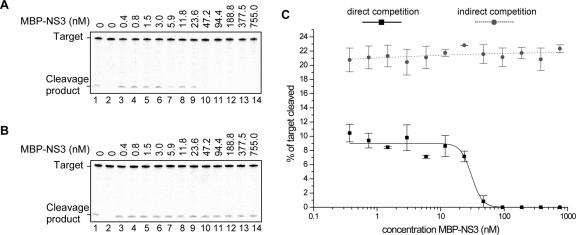
RISC-mediated cleavage is inhibited by NS3
RISC-mediated cleavage of a target is initiated by siRNAs with sequence complementarity to the target. With the observation that NS3 competes for siRNAs during RISC assembly in Drosophila extracts, RISC-mediated cleavage of an RNA target was tested, in a way described previously (Lakatos et al. 2006). The effect of NS3 on siRNA-mediated target cleavage by RISC was studied in direct and indirect competition assays. In direct competition, Drosophila extract, a target RNA (GFP mRNA), and synthetic siRNA-inducer molecules with a sequence homologous to the target were mixed with NS3. RISC-mediated cleavage was visualized by the 5′ cleavage product of the radiolabeled target and cleavage products were quantified at the different NS3 concentrations used. Indirect competition was established by preincubation of siRNA inducer and Drosophila extract for 30 min followed by the addition of target RNA and NS3. At the highest NS3 concentrations used, RISC-mediated target cleavage was inhibited in the direct competition assay (Fig. 6A,C). As could be expected from the RISC assembly studies, the activity of preassembled RISC was not inhibited by NS3, independent of the amount of NS3 added to the reactions (Fig. 6B,C).
FIGURE 6.
NS3 inhibits siRNA-mediated target cleavage in the Drosophila embryo extract in vitro RNA silencing system. (A) In direct competition assays, RISC-mediated target RNA (0.5 nM) cleavage was induced by siRNAs (5 nM) and MBP–NS3 (0.4–755.0 nM) simultaneously added to Drosophila embryo extracts. (B) In indirect competition, RISC was preassembled by adding siRNAs (5 nM) to embryo extract for 30 min and target RNA (0.5 nM) and MBP–NS3 (0.4–755.0 nM) subsequently added. (C) For direct and indirect competition experiments the percentage of cleaved target is plotted as a function of the MBP–NS3 concentration relative to the percentage of cleaved target in the absence of MBP–NS3. (A,B) Lanes 1 include siRNAs and lack MBP–NS3; lanes 2 lack inducer siRNA and MBP–NS3. We note that Drosophila embryo extract was used at the same concentration as we used for RISC assembly experiments (1 μg/μL in the test tube).
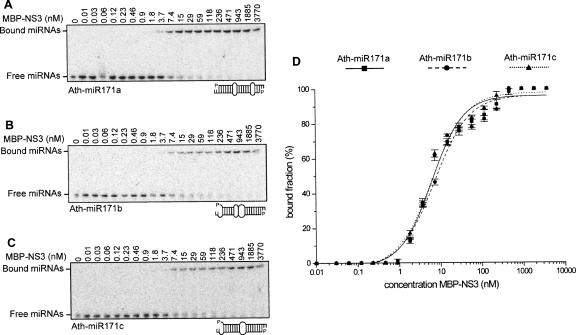
NS3 binds miRNAs in vitro
Besides a role in antiviral defense in plants, small RNAs play an essential role in the regulation of gene expression by miRNAs (Mallory and Vaucheret 2006). It has been reported that several plant viral suppressors also induce developmental abnormalities in plants by interfering with the miRNA pathway (Chapman et al. 2004; Dunoyer et al. 2004), possibly by interfering with the miRNA/miRNA* duplex unwinding. In order to investigate the base complementarity requirements of NS3 for small RNA complexes including miRNA/miRNA*, three miRNAs belonging to the Arabidopsis thaliana miRNA171 family were tested in a similar setup as was used for siRNAs. Members of the miR171 family show differences in structural features by containing two or three wobbles introduced by mismatches between the two strands of the miRNA duplex (Fig. 7A–C). The affinity of the NS3 protein to the miRNAs 171a (Kd 6.17 ± 0.80), 171b (Kd 7.19 ± 1.02), and 171c (Kd 6.26 ± 0.78) was compared (Fig. 7A–C). Despite their varying degree of base complementarity, all miRNA species were efficiently bound by NS3 (Fig. 7D), with an affinity in the same range as observed for the 21 nt and 19 nt blunt siRNA molecules. As indicated for the siRNAs molecules also for the tested miRNAs there was no binding to MBP observed (data not shown). This indicates that NS3, besides efficiently binding siRNAs, can interfere with the miRNA-regulated RNA silencing pathway by strongly binding miRNAs/miRNA* complexes before these can be incorporated into RISC. Considering that all three members of the miR171 family are bound at high affinity, it can be anticipated that many miRNA/miRNA* complexes can be subject to NS3 binding. Expression of this protein is therefore likely to influence host gene regulation in infected tissues of plants and insects (data not shown).
FIGURE 7.
Affinity of MBP–NS3 for different miRNA duplexes. A dilution series of MBP–NS3 (0.01–3770 nM) was incubated with 100 pM each of 32P-labeled Ath-miR171a (A), Ath-miR171b (B), or Ath-miR171c (C) for 20 min, then loaded onto a 5% native gel. The Kd was determined of MBP–NS3 for the different small RNA molecules by plotting the bound fraction as a function of the MBP–NS3 concentration (D). In panels A, B, and C the first lane contains only miRNAs.
DISCUSSION
NS3 is an RNA silencing suppressor in cultured insect cells
It has been reported that RNA silencing suppressors of several plant viruses also operate in an insect cell background (Li et al. 2002, 2004; Reavy et al. 2004). Additionally also true insect viruses, FHV and DCV, have been shown to specify such a suppressor, indicating that RNA silencing in insects also acts as an antiviral defense mechanism (Li et al. 2002; van Rij et al. 2006). Here we demonstrate that the NS3 RNA silencing suppressor protein of RHBV, a virus that replicates both in plants and insects, is not only operational in plant cells (Bucher et al. 2003) but also in insect cells. The mode of operation of NS3 was hitherto unknown, but based on these observations it must target a conserved part of the RNA silencing pathway.
NS3 suppresses RNA silencing by inhibiting RISC assembly in vitro
NS3 physically interacts with dsRNA molecules with size preference, showing the highest affinity for 21 nt siRNAs. Therefore NS3 is able to recognize the “standard” siRNA implicated in local silencing, but less efficiently to longer siRNA species that have been implicated in long-distance movement in plants (Hamilton et al. 2002; Tang et al. 2003). Our results showed that the NS3 protein is able to bind siRNAs with a high affinity in the presence or absence of the Drosophila extract, suggesting that no additional components are required for efficient siRNA binding.
The siRNAs play an important role by serving as the foundation for the ordered assembly of RISC complexes. In indirect competition experiments RISC complexes were preassembled, and it could be shown that NS3 was not able to interfere with sequence-specific target cleavage. Active RISC complexes contain ss siRNAs representing the guide strand of the initial ds siRNA complex that gives rise to the sequence specificity of RISC for the target. In additional experiments (results not shown) it was shown that NS3 does not efficiently bind to single-stranded siRNA. Consistent with this observation, no decrease in RISC complex formation was detected in the presence of increasing concentrations of NS3, because the ss siRNAs present in mature RISC complexes are no substrate for NS3. In contrast, a decrease in the formation of the R1 was observed with increasing concentration of NS3, suggesting that NS3, by having a higher affinity for ds siRNAs, can prevent the formation or maturation of the R1 complex. As the affinity of NS3 to larger dsRNA is low, our results suggest that NS3 action occurs after the cleavage of larger dsRNA complexes by Dcr-2 and before mature RISC formation.
As a result of RISC formation inhibition, NS3 was shown to inhibit cleavage of a target mRNA in the Drosophila embryo in vitro RNA silencing system in direct competition experiments, indicating its function as an RNA silencing inhibitor in insects where RHBV replicates. In vitro mechanistic studies on the Drosophila embryo antiviral RNA silencing pathway showed that Dcr-2 activity is uncoupled from RISC assembly. Long dsRNAs are processed into siRNAs by Dcr-2, generating a pool of siRNAs. To initiate RISC assembly, siRNAs are then rebound by R2D2/Dcr-2 according to the rule of strand preference and facilitate RISC assembly (Preall et al. 2006). Thus, the antiviral RNA silencing pathway in insects can be efficiently inhibited either by masking long dsRNA with a dsRNA binding protein (van Rij et al. 2006) or sequestering the siRNA pool by a siRNA binding protein (this study).
Inhibition of RISC assembly by sequestering siRNAs was recently also observed for RNA silencing suppressor proteins of plus strand tombusviruses, closteroviruses, and potyviruses. However, these plant viruses do not replicate in their insect vectors and are therefore unlikely to encounter antiviral RNA silencing in insect cells. Since all these suppressors bind siRNAs with high affinity, competition for these molecules during RISC assembly might inhibit the assembly of functional RISC complexes in a viral infection, thus inhibiting strand separation of siRNA duplexes. Alternative modes of operation appear to be adopted by the p88 and p27 proteins of Red clover necrotic mosaic virus, which seem to recruit Dicer (like) proteins during viral RNA replication (Takeda et al. 2005). Though the various viral RNA silencing suppressors have a surprising lack of conservation in their protein sequences, a limited number of modes of action seem to be followed, each targeting a distinct part of the RNA silencing machinery. As the chemical structure of siRNAs is ubiquitous, it seems not surprising for a virus such as RHBV that has to replicate in both plants and insects to interfere with a part of the RNA silencing machinery that is identical in both organisms, i.e., siRNA.
NS3 has high affinity for miRNA/miRNA* complexes independent of RNA duplex strand mismatches
RNA silencing not only plays an important role in antiviral defense in plants and insects but is also involved in developmental processes. In the regulation of plant and animal development, host-encoded miRNAs are key players in timed targeting of mRNAs for cleavage or translational arrest. The systemic infection of plants by viruses often results in symptoms resembling developmental defects, which can be characterized by loss of leaf polarity, cell division control, and reproductive functions (Mallory and Vaucheret 2006). Occurrence of these phenotypes is regularly associated with viral suppressor proteins or virulence factors. Constitutive expression of NS3 in Arabidopsis thaliana causes defects resembling developmental defects, such as loss of leaf polarity similar but not identical to those reported previously (data not shown; Chapman et al. 2004; Dunoyer et al. 2004). These developmental phenotypes are most likely the result of miRNA/miRNA* duplex binding by NS3 independent of mismatches present in the miRNA/miRNA* duplexes, resulting in inhibition of duplex unwinding and preventing miRNA function. Naturally, interference of NS3 with the siRNA part of RNA silencing serves to inhibit this antiviral defense mechanism but it can be speculated that interference of the NS3 protein with plant and insect gene expression regulation may also serve to establish an environment in plants and insects that might be advantageous for the infecting virus.
Binding of NS3 to siRNAs and different miRNAs with comparably high affinities gives rise to the question of whether physical interactions of these molecules reflect two distinct interference strategies or are variations on a single theme. With the discovery that several sequences of 20–25 nt located within Arabidopsis intergenic regions share perfect or near-perfect complementarity with a variety of plant virus genomes (Llave 2004), it is tempting to speculate that plant viruses, besides inducing antiviral RNA silencing and the subsequent production of viral siRNAs, are also targeted by host-encoded miRNAs. However, there is no direct evidence that plant host or insect vector of RHBV encode RHBV-specific miRNAs able to target RHBV viral sequences during replication. So next to the obvious need to suppress antiviral RNA silencing in both plants and insects, the NS3 protein may perform a similar role as the Primate foamy virus type 1 (PFV-1) RNA silencing suppressor protein Tas. The latter was shown to suppress human miRNA-32-mediated translational inhibition and probably functions to inhibit restriction of PFV-1 accumulation by miRNA-32 (Lecellier et al. 2005).
Dual roles for NS3 in RNA silencing in plants and insects?
It was shown that a plant virus, which is propagatively transmitted by an insect vector, is able to counteract antiviral RNA silencing in both insect and plant hosts. Binding siRNAs offer this possibility as the host organisms generate identical 21 nt siRNAs in antiviral RNA silencing. Targeting such conserved components of RNA silencing, both hosts cannot evade the action of proteins such as NS3. The downside for the virus however is that large quantities of suppressor proteins need to be produced to quench the bulk of the antiviral siRNAs. Alternatively, interfering with specific protein components of the RNA silencing machinery could result in a stronger suppression because a smaller number of targets need to be incapacitated. This approach, however, presents the risk that these proteins may alter their primary sequence and become unsuitable as targets, or alternatively (partially) redundant gene copies can take over the targeted function. By sequestering siRNAs the NS3 protein prevents these kinds of host adaptation strategies. Moreover, by targeting siRNAs the effect of the suppressor may not be perfectly efficient, allowing modulation of virulence.
MATERIALS AND METHODS
Plasmid constructs
The coding sequence of RHBV NS3 was PCR amplified introducing a unique BamHI site at the 5′ end and a unique KpnI site at the 3′ end. The PCR product was originally ligated into the bacterial expression vector pQE30 (Qiagen). For recombinant protein expression the NS3 coding sequence was removed from the pQE vector as a BamHI–PstI fragment and cloned in frame with the MBP coding sequence into the pMAL-c2x vector (New England Biolabs). For expression of NS3 in Drosophila S2 cells (Invitrogen) the NS3 open reading frame (ORF) from the pQE30–NS3 was ligated as a BamHI–SmaI fragment into the pMK33/pMtHy vector, which was digested with SpeI, end-filled with Klenow, and subsequently cut with BamHI. pMK33/pMtHy (kindly provided by Lee Fradkin, Leiden University Medical Center, Leiden, The Netherlands, and originally constructed by Michael Koelle) was used to clone the NS3 ORF immediately downstream of the CuSO4-inducible metallothionein promoter, giving rise to pMK33–NS3.
dsRNA preparation
Double-stranded RNA was generated using T7 RNA polymerase (Promega) according to protocol using a gel purified (High Pure PCR purification kit; Roche) PCR template. The primers used introduced T7 RNA polymerase promoters at both ends of the PCR product. Primers used were T7 ds_eGFP F: 5′-GTAATACGACTCACTATAGGGGGCGTGCAGTGCTTCAGCCGC-3′ and T7 ds_eGFP R: 5′-GTAATACGACTCACTATAGGGGTGGTTGTCGGGCAGCAGCAC-3′ for eGFP-specific dsRNA (400 nt). Following transcription the reaction mixture was incubated at 70°C for 10 min and cooled down to RT. Template and single-stranded RNA molecules were removed by treatment with DNase I and RNase A and dsRNA precipitated using 0.1 V 3 M NaAc (pH 5.2) and 1 V iso-propanol.
Cell culture, transfection, and RNA silencing assay
Drosophila S2 cells (Invitrogen) were cultured at 27°C in Schneider's Drosophila medium (Invitrogen) supplemented with 20% heat-inactivated fetal bovine serum (FBS). For the RNA silencing assay, 2 mL aliquots of a S2 cell suspension (1 × 106 cells/mL) were seeded in a 35 mm tissue culture dish the day before transfection. The cells were then transfected with 1 μg/well pAc-eGFP and 2 μg/well pMK33–NS3 vector. Transfections were performed using Cellfectin (Invitrogen) in serum-free medium as described by the manufacturer. Expression of NS3 protein was induced by adding CuSO4 to 0.5 mM final concentration directly after transfection. RNA silencing was induced by adding 5 μg/well dsRNA 3 h after the transfection. GFP fluorescence was monitored 28–72 h after transfection.
S2 cell expression analysis
Expression of NS3 was analyzed by Western blotting. S2 cells were harvested by centrifugation for 5 min at 1500 rpm (Heraeus Labofuge I). Cells were resuspended in 1 pellet volume PBS and disrupted by sonification on ice with 30-sec intervals for three times at 30 sec. Proteins were TCA precipitated and separated by SDS-PAGE. Proteins were semi-dry transferred to Immobilon-P (Millipore) and detected using suppressor-specific rat primary and goat alkaline phosphatase conjugated secondary antibodies and visualized with NBT-BCIP as substrate (Roche) according to the manufacturer's recommendations.
Agrobacterium tumefaciens infiltration
A. tumefaciens infiltration was performed according to Bucher et al. (2003). For coinfiltration of N. benthamiana leaves, a mixture of a strain carrying the GFP construct (OD600 = 0.5) and a strain carrying the suppressor construct (OD600 = 0.5) was used. The GFP imaging photographs were taken 5 d after infiltration using a yellow 022 Proline B&W filter.
Recombinant protein expression
The RHBV NS3 protein was expressed from BL21 DE3 cells according to the manufacturer's recommendations. After induction for 3 h at 37°C with 0.3 mM IPTG, cells were harvested by centrifugation for 15 min at 4000 rpm (Sorvall GSA rotor) at 4°C. Cells were lysed by sonification on ice with 30-s intervals for three times at 30 sec in buffer A (100 mM NaCl, 20 mM Tris 7.4, 1 mM DTT, 2 mM MgCl2, 10% [v/v] glycerol, 0.5% Triton X-100). The soluble fraction was recovered by centrifugation at 9000g for 30 min at 4°C. Recombinant protein was purified using amylose resin (New England Biolabs) and eluted with 2.5 packed bed volumes (PBV) buffer C (buffer A lacking Triton X-100 and containing 10 mM maltose) after washing with 15 PBV buffer B (buffer A lacking Triton X-100). Protein fractions were flash frozen in aliquots in liquid nitrogen and stored at −80°C until use. Protein concentrations of elution fractions were determined using the Micro BCA protein assay kit (PIERCE) according to the manufacturer's recommendations and the purification process analyzed by SDS-PAGE and subsequent staining with Coomassie.
Electrophoretic mobility shift assay
Annealing of siRNAs and miRNAs was as described by Lakatos et al. (2004). Bacterial expressed MBP–NS3 was incubated for 20 min at RT with 100 pM 32P-labeled siRNAs or miRNAs in 1 × binding buffer (100 mM NaCl, 20 mM Tris 7.4, 1 mM DTT, 2.5 mM MgCl2, 10% glycerol) in a 10 μL reaction volume. Separation of NS3 siRNA/miRNA complexes was performed at 4°C at 150 V on a 1 mm thick, large format, 5% (38:2 acrylamide:bisacrylamide), 0.5 × TBE native gel. After running, gels were dried, exposed to a phosphor screen, and scanned (Molecular Dynamics Typhoon PhosphorImager, Amersham Biosciences), and bands were quantified using Genius Image Analyser software (Syngene).
Gel filtration
A binding reaction of 250 μL was assembled as for the electrophoretic mobility shift assay using 100 pM 32P-labeled siRNAs and 2.45 nM MBP–NS3 and chromatographed at 4°C on a Superdex-200 HR 10/30 column (Pharmacia) at 0.4 mL/min in column buffer containing 100 mM NaCl, 20 mM Tris 7.4, 1 mM DTT, 2.5 mM MgCl2, and 10% glycerol. After a void volume of 7.5 mL, fifty 200 μL fractions were collected and used for RNA isolation. Per fraction, 80 μL NaCl, 2 μL 10 mg/mL glycogen, and 3 vol 96% ethanol were added, and RNA was precipitated by incubation at −80°C. From 19 out of 50 collected fractions RNA molecules were separated on a 12% polyacrylamide and 8 M urea containing sequencing gel.
RISC assembly assays
Drosophila embryo extract preparation, target RNA labeling, and siRNA annealing were as described previously by Haley et al. (2003). In direct competition assays, embryo extracts were incubated for 30 min at 25°C with 5 nM 32P-labeled siRNA duplexes and suppressor protein, diluted with 10 μL of loading buffer (1× lysis buffer, 6% ficoll 400) and analyzed on a 4.1% (40:1 acrylamide:bisacrylamide) native acrylamide gel. In preassembled RISC assays, 32P-labeled siRNA duplexes and embryo extracts were preincubated for 30 min at 25°C to allow RISC assembly prior to addition of suppressor protein. Native gel electrophoresis for separation of silencing complexes was essentially as described previously by Lakatos et al. (2006). Gels were dried and exposed to a phosphor screen, and bands were quantified using Genius Image Analyser software (Syngene).
RISC-mediated target cleavage assays
Reaction conditions were as described by Lakatos et al. (2006). In direct competition assays, reactions were incubated for 1 h at 25°C. In indirect competition assays, siRNA and embryo extracts were preincubated for 30 min at 25°C to allow RISC assembly prior to addition of target RNA and suppressor protein. Samples were deproteinized and RNA was analyzed on an 8% denaturing gel.
Statistical analysis
All in vitro target cleavage, RISC formation, and RNA binding experiments were performed in triplicate. The percentage of RISC complex formed relative to the control experiment without MBP–NS3 (Fig. 5A,B, lanes 2), was determined as well as the percentage of cleaved target relative to controls in lanes 1 in Figure 6, A and B. The curves were best fitted to the indicated sets of data with the computer program Microcal Origin 5.00. The average with standard error is shown in all graphs.
ACKNOWLEDGMENTS
The authors thank Anikó Szigeti for excellent technical assistance and Dr. Dániel Silhavy for providing the 26 nt species RNA. This research was financially sponsored by grants from the European Union (QLG2-CT-2002-01673), SIROCCO FP6-IP (Contract no. 037900), and the Hungarian Scientific Research Fund (OTKA, T048852; OTKA, NK60352). H.H. is a recipient of an EMBO short-term fellowship (ASTF 241.00-05). L.L. is a recipient of a Bolyai János fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.444007.
REFERENCES
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bucher, E., Sijen, T., De Haan, P., Goldbach, R., Prins, M. Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J. Virol. 2003;77:1329–1336. doi: 10.1128/JVI.77.2.1329-1336.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher, E., Hemmes, H., de Haan, P., Goldbach, R., Prins, M. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J. Gen. Virol. 2004;85:983–991. doi: 10.1099/vir.0.19734-0. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., Ambros, V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chao, J.A., Lee, J.H., Chapados, B.R., Debler, E.W., Schneemann, A., Williamson, J.R. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat. Struct. Mol. Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chapman, E.J., Prokhnevsky, A.I., Gopinath, K., Dolja, V.V., Carrington, J.C. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Dev. 2004;18:1179–1186. doi: 10.1101/gad.1201204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgadillo, M.O., Saenz, P., Salvador, B., Garcia, J.A., Simon-Mateo, C. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J. Gen. Virol. 2004;85:993–999. doi: 10.1099/vir.0.19735-0. [DOI] [PubMed] [Google Scholar]
- Dunoyer, P., Lecellier, C.H., Parizotto, E.A., Himber, C., Voinnet, O. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell. 2004;16:1235–1250. doi: 10.1105/tpc.020719. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., Vaucheret, H. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. 2000;97:11650–11654. doi: 10.1073/pnas.200217597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk, B.W., Tsai, J.H. Biology and molecular biology of viruses in the genus Tenuivirus . Annu. Rev. Phytopathol. 1998;36:139–163. doi: 10.1146/annurev.phyto.36.1.139. [DOI] [PubMed] [Google Scholar]
- Galiana-Arnoux, D., Dostert, C., Schneemann, A., Hoffmann, J.A., Imler, J.L. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila . Nat. Immunol. 2006;7:590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- Haley, B., Tang, G., Zamore, P.D. In vitro analysis of RNA interference in Drosophila melanogaster . Methods. 2003;30:330–336. doi: 10.1016/s1046-2023(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., Baulcombe, D.C. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., Hannon, G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Herr, A.J. Pathways through the small RNA world of plants. FEBS Lett. 2005;579:5879–5888. doi: 10.1016/j.febslet.2005.08.040. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Lakatos, L., Szittya, G., Silhavy, D., Burgyan, J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E.J., Carrington, J.C., Liu, Y.P., Dolja, V.V., Calvino, L.F., Lopez-Moya, J.J., Burgyan, J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecellier, C.H., Dunoyer, P., Arar, K., Lehmann-Che, J., Eyquem, S., Himber, C., Saib, A., Voinnet, O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Li, H., Li, W.X., Ding, S.W. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- Li, W.X., Li, H., Lu, R., Li, F., Dus, M., Atkinson, P., Brydon, E.W., Johnson, K.L., Garcia-Sastre, A., Ball, L.A., et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc. Natl. Acad. Sci. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel, A., Simon, B., Izaurralde, E., Sattler, M. The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition. EMBO Rep. 2005;6:1149–1155. doi: 10.1038/sj.embor.7400583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C. MicroRNAs: More than a role in plant development. Mol. Plant Pathol. 2004;5:361–366. doi: 10.1111/j.1364-3703.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006;38(Suppl):1079–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Merai, Z., Kerenyi, Z., Molnar, A., Barta, E., Valoczi, A., Bisztray, G., Havelda, Z., Burgyan, J., Silhavy, D. Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J. Virol. 2005;79:7217–7226. doi: 10.1128/JVI.79.11.7217-7226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merai, Z., Kerenyi, Z., Kertesz, S., Magna, M., Lakatos, L., Silhavy, D. Double-stranded RNA binding may be a general plant RNA viral strategy to suppress RNA silencing. J. Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H., Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar, A., Csorba, T., Lakatos, L., Varallyay, E., Lacomme, C., Burgyan, J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, J.W., Pellino, J.L., Lee, Y.S., Carthew, R.W., Sontheimer, E.J. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila . Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Preall, J.B., He, Z., Gorra, J.M., Sontheimer, E.J. Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila . Curr. Biol. 2006;16:530–535. doi: 10.1016/j.cub.2006.01.061. [DOI] [PubMed] [Google Scholar]
- Ramirez, B.C., Macaya, G., Calvert, L.A., Haenni, A.L. Rice hoja blanca virus genome characterization and expression in vitro. J. Gen. Virol. 1992;73:1457–1464. doi: 10.1099/0022-1317-73-6-1457. [DOI] [PubMed] [Google Scholar]
- Ramirez, B.C., Lozano, I., Constantino, L.M., Haenni, A.L., Calvert, L.A. Complete nucleotide sequence and coding strategy of Rice hoja blanca virus RNA4. J. Gen. Virol. 1993;74:2463–2468. doi: 10.1099/0022-1317-74-11-2463. [DOI] [PubMed] [Google Scholar]
- Rand, T.A., Petersen, S., Du, F., Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Reavy, B., Dawson, S., Canto, T., MacFarlane, S.A. Heterologous expression of plant virus genes that suppress post-transcriptional gene silencing results in suppression of RNA interference in Drosophila cells. BMC Biotechnol. 2004;4:18. doi: 10.1186/1472-6750-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Silhavy, D., Burgyan, J. Effects and side effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 2004;9:76–83. doi: 10.1016/j.tplants.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Soldan, S.S., Plassmeyer, M.L., Matukonis, M.K., Gonzalez-Scarano, F. La Crosse virus nonstructural protein NSs counteracts the effects of short interfering RNA. J. Virol. 2005;79:234–244. doi: 10.1128/JVI.79.1.234-244.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer, E.J. Assembly and function of RNA silencing complexes. Nat. Rev. Mol. Cell Biol. 2005;6:127–138. doi: 10.1038/nrm1568. [DOI] [PubMed] [Google Scholar]
- Takeda, A., Sugiyama, K., Nagano, H., Mori, M., Kaido, M., Mise, K., Tsuda, S., Okuno, T. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002;532:75–79. doi: 10.1016/s0014-5793(02)03632-3. [DOI] [PubMed] [Google Scholar]
- Takeda, A., Tsukuda, M., Mizumoto, H., Okamoto, K., Kaido, M., Mise, K., Okuno, T. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., Zamore, P.D. A biochemical framework for RNA silencing in plants. Genes & Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari, Y., Zamore, P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., Zamore, P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004a;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., Zamore, P.D. A protein sensor for siRNA asymmetry. Science. 2004b;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., Sharp, P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rij, R.P., Saleh, M.C., Berry, B., Foo, C., Houk, A., Antoniewski, C., Andino, R. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes & Dev. 2006;20:2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargason, J.M., Szittya, G., Burgyan, J., Tanaka Hall, T.M. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- Voinnet, O. Induction and suppression of RNA silencing: Insights from viral infections. Nat. Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- Wang, X.H., Aliyari, R., Li, W.X., Li, H.W., Kim, K., Carthew, R., Atkinson, P., Ding, S.W. RNA interference directs innate immunity against viruses in adult Drosophila . Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijkamp, I., van Lent, J., Kormelink, R., Goldbach, R., Peters, D. Multiplication of tomato spotted wilt virus in its insect vector, Frankliniella occidentalis . J. Gen. Virol. 1993;74:341–349. doi: 10.1099/0022-1317-74-3-341. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Yuan, Y.R., Pei, Y., Lin, S.S., Tuschl, T., Patel, D.J., Chua, N.H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]