Abstract
Poly(C)-binding proteins (PCBPs) are KH (hnRNP K homology) domain-containing proteins that recognize poly(C) DNA and RNA sequences in mammalian cells. Binding poly(C) sequences via the KH domains is critical for PCBP functions. To reveal the mechanisms of KH domain-D/RNA recognition and its functional importance, we have determined the crystal structures of PCBP2 KH1 domain in complex with a 12-nucleotide DNA corresponding to two repeats of the human C-rich strand telomeric DNA and its RNA equivalent. The crystal structures reveal molecular details for not only KH1-DNA/RNA interaction but also protein–protein interaction between two KH1 domains. NMR studies on a protein construct containing two KH domains (KH1 + KH2) of PCBP2 indicate that KH1 interacts with KH2 in a way similar to the KH1–KH1 interaction. The crystal structures and NMR data suggest possible ways by which binding certain nucleic acid targets containing tandem poly(C) motifs may induce structural rearrangement of the KH domains in PCBPs; such structural rearrangement may be crucial for some PCBP functions.
Keywords: X-ray crystallography, NMR, KH domain–nucleic acid interaction
INTRODUCTION
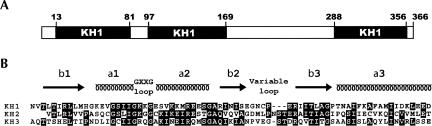
Poly(C)-binding proteins (PCBPs) are hnRNP K homology (KH) domain-containing proteins capable of recognizing poly(C) nucleic acid sequences with high affinity and specificity (for review, see Gamarnik and Andino 2000; Makeyev and Liebhaber 2002). In mammalian cells, five evolutionarily related PCBPs have been identified, namely, PCBP1–4 and hnRNP K. Typically, the PCBPs contain three KH domains, two consecutive KH domains at the N terminus, and a third KH domain at the C terminus, separated by an intervening sequence of variable length (Fig. 1).
FIGURE 1.
(A) Schematic diagram of the domain structure of human poly(C) binding protein-2 (PCBP2). Similar domain structures are observed in other members of the PCBP family. (B) Sequence alignment of the three KH domains from PCBP2. Alignments were carried out using the program ClustalX. The sequence shown for PCBP2-KH1 corresponds to residues 11–82 in the full-length protein. Secondary structures indicated for the KH1 domain were based on the crystal structures. The protein construct containing KH1 + KH2 for NMR studies has an intervening sequence of ISSSMTNSTAASRPP between the shown KH1 and KH2 sequences.
The established biological functions of PCBPs indicate that these proteins are critical players in a number of important regulation mechanisms. Unusual stability of a number of mRNAs, including α-globin, collagen-α1, erythropoietin, hydroxylase, and tyrosine, are dependent on specific binding of PCBP1 or PCBP2 to C-rich sequences within the 3′-UTRs of these mRNAs (Makeyev and Liebhaber 2002). In the most studied case of α-globin, it was established that the stoichiometry of the RNA–protein complex (the so-called α-complex) is 1:1, and a minimum RNA sequence of 20 nucleotides (nt) (5′-CCCAACGGGCCCUCCUCCCC-3′) was able to form the α-complex (Waggoner and Liebhaber 2003b). Translational silencing of 15-lipoxygenase (LOX) mRNA is mediated by the specific binding of two PCBPs (hnRNP K and PCBP1 or -2) to a CU-rich repetitive sequence motif known as the differentiation-control element (DICE) within the 3′-UTR (Ostareck-Lederer et al. 1994; Ostareck et al. 1997, 2001).
PCBPs are also involved in the regulation of viral RNA functions. Cap-independent translation of Poliovirus mRNA (which is also the genomic RNA) requires binding of PCBP2 to a structural element known as loop B RNA within IRES (internal ribosomal entry site) at the 5′-UTR. Switching from translation to RNA replication is controlled by the formation of a ternary complex involving PCBP1 or PCBP2, viral protein 3CD (precursor of the viral protease 3C and the viral polymerase 3D), and a so-called cloverleaf-like RNA structure located at the very 5′-end of the 5′-UTR (Blyn et al. 1996, 1997; Gamarnik and Andino 1997, 1998; Parsley et al. 1997).
The functional roles of PCBPs in RNA regulation certainly will expand. It was shown that PCBP2 associated with 160 mRNA species in vivo in a human hematopoietic cell line (Waggoner and Liebhaber 2003a), suggesting a general significance of PCBPs in post-translational regulation. PCBPs may also target other kinds of RNA species besides mRNA. Interaction between PCBP4b (an isoform of PCBP4) and the C-rich RNA template sequence of human telomerase was postulated to be involved in the functional role of PCBP4b as a p53-induced regulator in apoptosis and cell cycle arrest at G2-M (Zhu and Chen 2000).
PCBPs can also play a role in mechanisms dependent on DNA binding. Specific binding of hnRNP K to the single-stranded pyrimidine-rich sequence in the promoter of human c-myc gene activates transcription (Tomonaga and Levens 1996). Binding of hnRNP K and PCBP1 to the C-rich strand of human telomeric DNA was established in vitro (Lacroix et al. 2000) and in the K562 human cell line (Bandiera et al. 2003); assignment for possible functional roles of such interactions awaits further studies.
Although PCBP functions are diverse, they are all dependent on recognition of C-rich RNA or DNA sequences by the KH domains. It is therefore important to reveal the molecular details defining specificity and affinity of the nucleic acid–protein interactions and the mechanisms correlating binding of C-rich nucleic acid sequences to various functions. As a result of our continuing structural study of KH domain–nucleic acid interactions, we report here the crystal structures of PCBP2 KH1 in complex with a 12- nt DNA molecule (5′-AACCCTAACCCT-3′, harboring two C-rich recognition motifs corresponding to two repeats of the human C-rich strand telomeric DNA) and its RNA equivalent (5′-AACCCUAACCCU-3′) at 2.1 Å and 2.6 Å resolution, respectively. The KH1–RNA complex structure represents the first structure of poly(C) RNA sequence recognition by a KH domain from the PCBPs. The two structures reveal that recognition of RNA and DNA sequences by the PCBP2 KH1 domain is very similar. A network of regular hydrogen bonds achieves specific nucleic acid recognition. Protein–protein interaction between two KH1 domains using a hydrophobic molecular surface opposite to the nucleic acid binding surface is observed in both crystal structures, which is virtually identical to the KH1 homodimer in our previous crystal structure of KH1 in complex with a 7-nt DNA (Du et al. 2005). We also carried out NMR studies on a protein construct containing the first and second KH domains (KH1 + KH2) of PCBP2. Chemical shift data indicated that both KH domains assume the secondary structure of the classical type-I KH motif folding. Chemical shift difference mapping and NOEs further suggest that the KH1 domain interacts with the KH2 domain in a way similar to the KH1–KH1 interaction observed in the crystal structures.
RESULTS
Overall structure of the KH1-D/RNA complexes
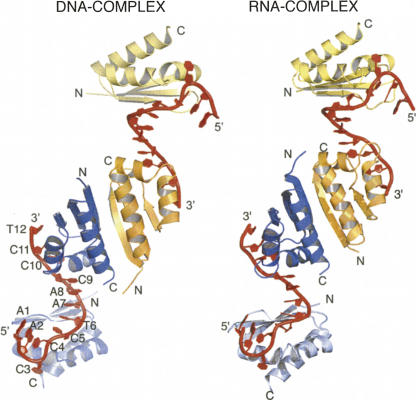
The crystal structures of PCBP2 KH1 in complex with a 12-nt DNA molecule (5′-AACCCTAACCCT-3′, harboring two C-rich recognition motifs corresponding to two repeats of the human C-rich strand telomeric DNA) and its RNA equivalent (5′-AACCCUAACCCU-3′) were determined at 2.1 Å and 2.6 Å resolution, respectively. The two structures are very similar (Fig. 2; Table 1). RMSD between the two structures (including all common heavy atoms of the protein and RNA/DNA) is merely 1.12 Å. There is no indication that the hydroxyl groups of the RNA bases play any role in specific interaction with the protein.
FIGURE 2.
Overall structures of the PCBP2 KH1–DNA (left) and –RNA (right) complexes in one asymmetric unit. There are four KH1 domains (colored in gold, orange, deep blue, and light blue, respectively) and two DNA/RNA strands (colored in red) in one asymmetric unit. Each DNA/RNA strand is bound to two KH1 domains. Every KH1 domain in the crystal forms a homodimer with another KH1 domain, as illustrated by the dimer formed by the orange and deep blue KH1 domains. Each of the KH domains in gold/light blue forms a dimer with another symmetry-related KH1 domain (not shown). Residues in one of the DNA strands are labeled by residue type and number.
TABLE 1.
Data collection and refinement statistics
There are four PCBP2 KH1 domains and two DNA (or RNA) molecules in one asymmetric unit; each DNA (or RNA) strand is recognized by two KH1 domains at the two C-rich motifs. Every KH1 domain in the crystal lattice is involved in homodimerization with another KH1 domain, as illustrated by the dimer formed by the orange and deep blue KH1 domains (Fig. 2). Each of the other two KH domains (in gold/light blue) forms a dimer with another symmetry-related KH1 domain (not shown in Fig. 2).
The structure of the PCBP2 KH1 domain is virtually identical to what we have previously determined in our 1.7 Å crystal structure of the domain in complex with a 7-nt DNA sequence 5′-AACCCTA-3′ (Du et al. 2005). The structure consists of three α helices and three β strands arranged in the order β1-α1-α2-β2-β3-α3. The evolutionarily conserved invariable G30-K31-K32-G33 loop is located between α1 and α2; the variable loop (S50 to P55) is between β2 and β3. The three β strands form an antiparallel β-sheet, with a spatial order of β1-β3-β2; the three α helices are packed against one side of the β-sheet. The D/RNA binding groove is defined by the juxtaposition of helices α1 and α2, strands β2 and β3, and both loops (the GKKG loop and the variable loop).
The crystallization D/RNA, 5′-AACCCT/UAACCCT/U-3′ contains two C-rich motifs. Each 5′-CCCT/U-3′ tetranucleotide motif constitutes the core recognition sequence directly interacting with the KH1 domain (Fig. 3A). Continuous base stacking is seen from the last base of the 5′-end recognition sequence through the two intervening adenosines to the first base of the 3′-end recognition sequence (Fig. 2).
FIGURE 3.
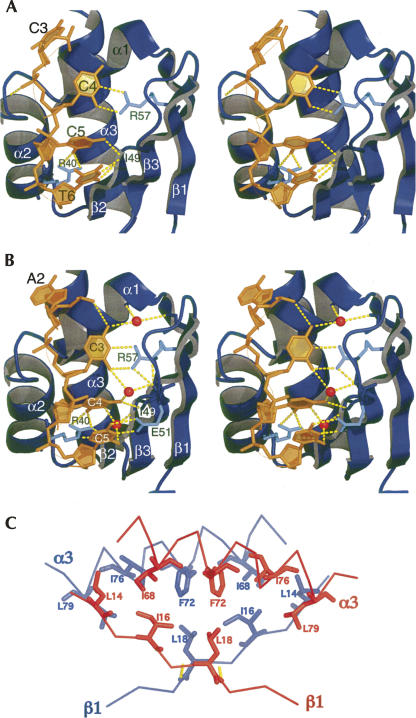
Structure of the PCBP2 KH1-DNA complex. (A) Recognition of the tetranucleotide core sequence 5′-CCCT-3′ (in orange). The KH1 domain is rendered as a ribbon representation in deep blue. Secondary structure elements of the KH1 domain are labeled. Direct hydrogen bonds (depicted as yellow dashed lines) are observed from the DNA residues to the backbone/side chain (shown as sticks) functional groups of K32, R40, I49, and R57. (B) As a comparison, our previous structure for the recognition of the tetranucleotide core sequence 5′-ACCC-3′ is shown. (C) Dimerization interface of the KH1 domains. The two monomers are colored deep blue and red in the ribbon representation; residues located at the dimerization interface are shown as sticks.
Specific recognition of the core sequence
In our previous PCBP2 KH1–DNA cocrystal structure, the crystallization DNA is 5′-AACCCTA-3′, corresponding to one repeat of the human C-rich telomeric DNA sequence. The tetranucleotide core recognition sequence was 5′-ACCC-3′ in that structure. In the present structures, the crystallization D/RNA corresponds to two repeats of the human C-rich telomeric sequence. Each repeat is recognized by one KH1 domain. Interestingly, the tetranucleotide core recognition sequence for both domains is 5′-CCCT/U-3′. The registration of the recognition sequence is therefore shifted one base from the previous cocrystal structure. We believe the registration of nucleic acid bases observed in the current two structures is more biologically relevant because the crystallization D/RNA are longer in length and exhibit less intermolecular base stacking driven by crystal packing. In spite of the difference in registration, the detailed molecular interactions for recognition, especially for the central two cytosines, are quite similar (C4 and C5 in Fig. 3A for the current structure, C3 and C4 in Fig. 3B for the previous structure). Several forces, including hydrogen bonding, electrostatic interactions, van der Waals contacts, and shape complementarities, combine to achieve specific molecular recognition.
Recognition of each of the two tetranucleotide core sequences is virtually identical. For the first position cytosine, no specific interactions are observed: van der Waals contacts are mainly provided by side chains of Gly-26, Ser-27, and Lys-31. For the second position cytosine, van der Waals contacts are provided by side chains of Val-25, Gly-26, and Ile-29. Specificity for a cytosine at this position is dictated by the side chain of Arg-57, which provides two hydrogen bond donors to the O2 and N3 acceptors of the cytosine (Fig. 3A). The extended side-chain conformation of Arg-57 is further stabilized by two hydrogen bonds to the backbone carbonyl oxygen of Ser-50 (not shown). In our previous 1.7 Å crystal structure of KH1 in complex with a 7-nt DNA shown in Figure 3B (Du et al. 2005), the N4 amino group of the second cytosine forms a hydrogen bond to a water molecule that bridges to the backbone carbonyl oxygen atoms of Gly-22 and Cys-54. Recognition of the N3 group of the cytosine at the third position also involves a water-mediated hydrogen bond to the amide of Ile-49. The structured waters in the present structures are not as well defined due to lower resolution. However, it is very likely that similar water molecules are involved in the specific recognition of the cytosines in the present structures because of the high degree of structural similarities (RMSD ∼ 0.9 Å). The phosphate group of the second position cytosine also forms an intermolecular hydrogen bond to the backbone amide of Lys-32 within the conserved GKKG loop (Fig. 3A).
For recognition of the cytosine at the third position, the O2 group of the cytosine forms hydrogen bonds with the side chain of Arg-40 from helix α2 (Fig. 3A). The extended conformation of the Arg-40 side chain is stabilized by hydrogen bonds to the backbone carbonyl of Ile-47 from strand β2 (not shown). The N4 amino group of the cytosine forms a hydrogen bond with the backbone carbonyl oxygen of Ile-49. Besides hydrogen bonding interactions, van der Waals interactions with the hydrophobic side chains from Ile-29, Val-36, and Ile-49 on one face of the third position cytosine, and intrastrand base stacking interaction with the fourth position nucleotide on the other face also contribute to the definition of the binding environment.
The last residue in the tetranucleotide core recognition motif is a thymine/uridine. The O2 and N3 group of this residue forms hydrogen bonds with the backbone amide and carbonyl group of Ile-49, respectively (Fig. 3A). The base of this uridine engages in stacking interactions with the base of the 5′-side cytosine on one face and the base of the 3′-side adenosine on the other face (Fig. 3A). In our previous structure, the last residue in the core recognition motif is a cytosine (C5 in Fig. 3B); N3 and N4 functional groups of this cytosine are recognized by hydrogen bonds to the side chain of E51 (Fig. 3B). Comparing all of the available PCBP KH domain structures in complex with DNA/RNA, the mode of recognition for the residue at this position shows a great deal of variation.
Conformation of the bound DNA and RNA
The crystallization D/RNA consists of two 5′-AACCCT/U-3′ repeats. In each repeat, the CCCT/U tetranucleotide motif constitutes the core recognition sequence directly contacting the KH1 domain. Conformation of this motif is virtually the same in both repeats. The first position of the core recognition motif does not involve base-specific interactions. The base of this nucleotide is positioned on top of helix α1; the backbone of the nucleic acids winds around the helix downward, placing the base of the second position nucleotide close to the bottom side of the helix. The bases of the first and second nucleotides act like a pair of molecular tongs grasping the helix (Fig. 3A). Two glycines at this helical segment are absolutely conserved (G26 and G30; see Fig. 1B for sequence alignments). Other amino acids with bigger side chains at these positions presumably would create steric hindrance for binding. Base stacking is observed between the third and fourth position nucleotides of the core recognition sequence (Fig. 3A).
Although conformations of the two core tetranucleotide motifs are virtually identical, the two preceding adenosine nucleotides show different structures. In the first 5′-AACCCT/U-3′ repeat, the bases of the two adenosines (A1 and A2 in Fig. 2) are stacked with each other and on top of the base of the cytosine at the second position in the core recognition motif (C4 in Fig. 2). This stacking interaction results in an almost 180° reversal of the direction of the polynucleotide chain (Fig. 2). In the second 5′-AACCCT-3′ repeat, the bases of the two adenosines (A7 and A8 in Fig. 2) are also stacked with each other, but instead of stacking on top of the second position base, they stack with the first position base in the core recognition motif (C9 in Fig. 2). Continuous base stacking is observed among the last 2 nt of the first repeat (C5 and T6 in Fig. 2) and the first 3 nt of the second repeat (A7, A8, and C9 in Fig. 2).
Dimerization of the PCBP2-KH1 domain
The PCBP2 KH1 homodimer we observe in the D/RNA complex structures is virtually identical to the one in our previous crystal structure of the KH1 domain in complex with a 7-nt DNA (Du et al. 2005), although the packing of the homodimers in the crystal lattices is different. The reoccurrence of this form of dimerization suggests that it is realized by strong intermolecular interactions. A detailed inspection of the dimerization interface reveals this is indeed the case.
The dimerization interface is defined by the anti-parallel positioning of the longest α helix (α3) and β strand (β1) in the protein domain (Fig. 3C). Dimerization orients the two three-stranded anti-parallel β sheets of the monomers in such a way that a six-stranded anti-parallel β sheet is formed. Two generic (backbones only) hydrogen bonds (Leu-19 amide to Arg-17 carbonyl oxygen, represented as yellow dashed lines in Fig. 3B) stabilize the anti-parallel arrangement of the two β1 strands. Hydrophobic interactions seem to be pivotal for dimerization. The hydrophobic side chains of Leu-14, Ile-16, and Leu-18 from the two β strands (β1s) and Ile-68, Phe-72, Ile-76, and Leu-79 from the two α3 helices define a hydrophobic interior core. Stacking of the aromatic rings of the two Phe-72s is also observed. The molecular surface forming the dimerization interface is rather hydrophobic in nature (Fig. 4, left). Formation of the homodimer buries ∼ 1932 Å2 of surface area of the proteins. Such a large area of hydrophobic surface should provide a significant driving force for the protein domain to engage in a protein–protein interaction in order to keep this hydrophobic surface from being exposed to solvent.
FIGURE 4.
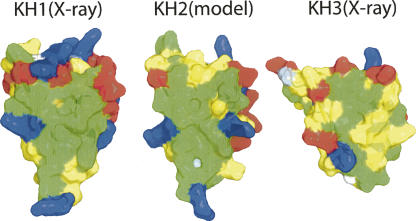
Surface representations of the three KH domains in the PCBP2 protein. (Left) Crystal structure of the PCBP2 KH1 domain. (Middle) A homologous model (built by the program Modeller based on the crystal structure of PCBP2 KH1) of the PCBP2 KH2 domain. (Right) Crystal structure of the PCBP2 KH3 domain (Fenn et al. 2007). Positively charged, negatively charged, and uncharged hydrophilic and hydrophobic residues are colored in blue, red, yellow, and green, respectively; glycines are in white. Note the large, continuous hydrophobic surface area (green) of the KH1 and KH2 domains.
NMR evidence for KH1–KH2 interaction
Since the major driving force for KH1 homodimerization is hydrophobic interaction, it is quite possible that KH1 can interact with other KH domains or other protein partners that have a compatible hydrophobic molecular surface. One such protein domain could be the second KH domain (KH2) of PCBP2, which is separated from the KH1 domain by a linker of 14 amino acids. So far there is no atomic resolution structure of any KH2 domain from PCBPs available, but a homologous model of PCBP2 KH2 displays a large hydrophobic surface area comparable to that of PCBP2 KH1 (Fig. 4).
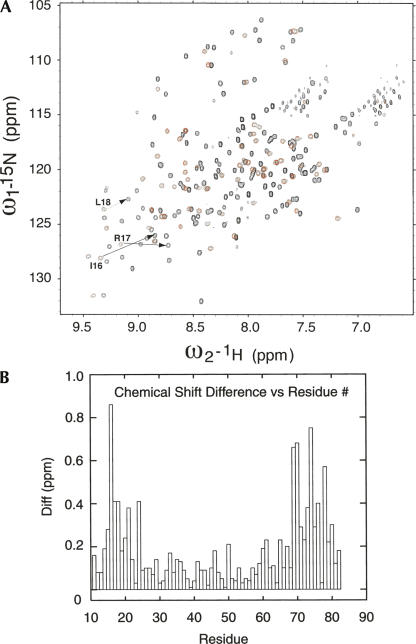
We were not able to express PCBP2 KH2 as an individual domain. But a protein construct containing both KH1 and KH2, as well as the linker between them, could be successfully expressed, refolded, purified, and concentrated to a high concentration for NMR studies. The KH1 + KH2 construct yielded excellent quality NMR spectra (Fig. 5A). Very substantial 13C/15N/1H chemical shift assignments have been obtained (BMRB accession 15,049). The chemical shift data unambiguously indicate that both KH1 and KH2 domains adopt the classical type-I KH fold with β1-α1-α2-β2-β3-α3 as the secondary structure arrangement in solution. No secondary structure was predicted for the linker region (residues 84–97 inclusive, SSSMTNSTAASRPP). Resonance cross-peaks from these residues are much stronger than those from the two KH domains, indicating different dynamic properties between the nonstructured and structured parts of the protein construct.
FIGURE 5.
KH1 chemical shift differences between KH1 in the KH1 + KH2 construct and in the individual KH1 construct. (A) Overlay of two 15N-TROSY-HSQC spectra of PCBP2 KH1 + KH2 (in black) and KH1 (in red). The spectrum was recorded at 25°C in a 90% H2O/10% D2O buffer containing 50 mM deuterated sodium acetate (pH 5.4), 2 mM DTT. Examples of significant chemical shift difference for residues of the KH1 domain are indicated by arrows. (B) The graphs show the composite chemical shift differences (in parts per million) plotted against the residue numbers. The composite difference is calculated as: ([ΔH]2 + [0.154*ΔN]2 + [0.256*ΔCA]2 + [0.256*ΔCB]2)1/2, where Δ is the chemical shift difference value in parts per million (Mulder et al. 1999). The amide proton, amide nitrogen, Cα carbon, and Cβ carbon chemical shift differences are calculated as Δ = value in KH1 + KH2 construct − value in individual KH1 construct.
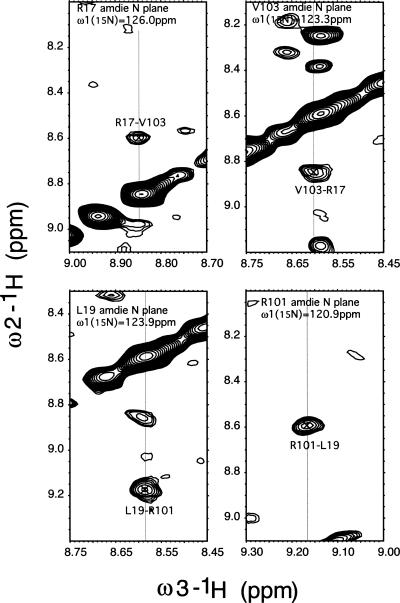
When the chemical shifts of the KH1 domain in the KH1 + KH2 construct were compared to those we obtained previously for the KH1 domain alone (Du et al. 2004), some significant chemical shift changes were noticed (Fig. 5B). The residues that exhibit these chemical shifts all belong to residues from or near the first β strand (β1) and the last α helix (α3), which are the structural elements defining the protein interaction interface of KH1. Since the KH1 homodimer is observed in various crystal structures, the isolated KH1 construct also probably exists in solution as a homodimer. Regardless of the solution state of the isolated KH1 construct, chemical shift changes of residues on the KH1 dimerization surface indicate these residues experience a different chemical environment in the context of the KH1 + KH2 construct. We observed inter-KH domain NOEs between amide protons from the first β strands of KH1 and KH2 (Fig. 6), indicating that KH1 is interacting with KH2 via the dimerization interface. The NOEs also indicate that the two β1 strands of KH1 and KH2 are arranged in an anti-parallel manner, similar to that in the KH1–KH1 homodimer crystal structures.
FIGURE 6.
Amide-to-amide NOEs between KH1 and KH2 in the KH1 + KH2 construct. The 3D-NOESY-15N-HSQC spectrum was acquired using a 13C/15N/2H/(Ile/Leu/Val)-methyl-protonated sample at 25°C in a 90% H2O/10% D2O buffer containing 50 mM deuterated sodium acetate (pH 5.4), 2 mM DTT.
DISCUSSION
Although most of the established biological functions of PCBPs involve specific recognition of poly(C) RNA sequences, there was no atomic resolution structure available for this critical protein–RNA interaction prior to our present study. We reveal that recognition of poly(C) RNA sequences is quite similar to recognition of poly(C) DNA sequences; the 2′-hydroxyl group of the ribose does not contribute to specificity and affinity of the interaction. This is in contrast to the structure of NOVA2 KH3–RNA interaction, the only other published KH domain–RNA complex crystal structure, in which the 2′-hydroxyl groups of the four core recognition residues did form hydrogen bonds with the KH domain (Lewis et al. 2000). It was believed that NOVA2 KH3 should not be able to recognize the corresponding DNA sequence in a similar manner. The ability of PCBP2 KH1 to interact with both RNA and DNA sequences enables PCBP2 to assume dual functional roles, i.e., not only in processes dependent on RNA binding but also in those entailing DNA binding.
The interactions between the PCBP2 KH1 domain and human telomere/telomerase DNA/RNA suggest that PCBPs may participate in the regulation of telomere and telomerase activities through specific binding to the C-rich telomeric DNA and telomerase RNA sequences.
We observed the same PCBP2 KH1 homodimer three times in the three nucleic acid–protein cocrystal structures we have solved (present study; Du et al. 2005). The reoccurrence of this dimer, together with the strong driving forces for dimerization as revealed by the crystal structures, strongly suggests that the dimer is stable. Each of the PCBPs contains three KH domains. Crystal structures of the KH3 domain from hnRNP K in its free and DNA-bound form and the KH3 domain from PCBP1 in its free form have also been published (Backe et al.2005; Sidiqi et al. 2005). We have also determined the crystal structure of PCBP2 KH3 in complex with a 7-nt human telomeric DNA (Fenn et al. 2007). Interestingly, the kind of dimerization we observed in PCBP2 KH1 structures was not observed in the KH3 domain structures. In the PCBP2 KH1 domain, a large hydrophobic surface area provides a critical driving force for the dimerization. Such a large hydrophobic surface area is not present in the KH3 domains of PCBP1, PCBP2, or hnRNP-K (Fig. 4, right; only the PCBP2 KH3 domain is shown). Although no structure of the KH2 domain from any PCBP protein is available at this point, the very high degree of sequence and structural homology among the KH domains should permit a reasonably realistic structural model to be built based on the available crystal structures of PCBP2 KH1, hnRNP-K KH3, and PCBP1/2 KH3 domains. Such a MODELLER-built structure of PCBP2 KH2 readily displays a large hydrophobic surface area comparable to that of PCBP2 KH1 (Fig. 4).
Similarity and difference in the properties of the molecular surfaces of the KH domains may play a pivotal role in the assembly of the KH domains in the structure of the full-length PCBP protein; hence they are functionally important. Using NMR, we showed that KH1 could interact with KH2 via the hydrophobic molecular interface. It therefore seems like both KH1 and KH2 domains are able to engage in protein–protein interaction using a large hydrophoblic molecular surface located opposite to the established or putative nucleic acid binding groove. While in the present case protein–protein interaction takes place intramolecularly between KH1 and KH2, it is not difficult to envisage that KH1 and KH2 can use the protein interaction interface to interact with other proteins. In order to do that, however, the intramolecular KH1–KH2 association has to be disrupted and the relative orientation of the two KH domains be adjusted. Whether such a conformational change plays a role in the regulation of PCBP function and how it might correlate to nucleic acid binding of the KH domains await further studies.
MATERIALS AND METHODS
Preparation of protein samples
Samples of unlabeled or Se-Met labeled PCBP2 KH1 (residues 11–82) used for crystallization were prepared as previously described (Du et al. 2005). For NMR studies, a protein construct containing the first and second KH domains (KH1 + KH2) of PCBP2 (residues 11–169) was expressed with N-terminal His-tag (MKH6K, all but the last K could be removed by TAGzyme from QIAGEN). The proteins were overexpressed in a BL21(DE3) strain of Escherichia coli (Strategene). The bacteria cultures were grown in Luria-Bertani medium for unlabeled samples, in minimal medium (with either H2O or D2O or a mixture of H2O/D2O, depending on the isotope-labeling scheme) supplemented with 15NH4Cl, [13C6]glucose, [2H7,13C6]glucose, [13C5,3-2H1]α-ketoisovalerate, and/or [13C4,3,3-2H2]α-ketobutyrate, to produce various isotopically labeled samples, including fully deuterated at all but the Ile/Leu/Val methyl groups, 13C/15N-labeled proteins. Protein expression was induced with 0.4 mM IPTG at OD600 = 0.6–0.8 at 37°C for 3 h and cells were harvested. The His-tagged protein present in the inclusion bodies was washed by 2 M urea three times and dissolved in 8 M urea (1 g of wet protein pellet in 100 mL of urea) plus 5 mM DTT. The denatured protein in 8 M urea was drop-wise added to a 4 L buffer containing 100 mM Na/K phosphate (pH 7.0) under vigorous stirring. The refolded protein was further purified by Ni-NTA resin. The purified protein was concentrate/buffer exchanged by centrifugation. The final NMR buffer contains 50 mM deuterated sodium acetate (pH 5.4), 2 mM DTT, 0.1 M EDTA, in either 90% H2O/10% D2O or 100% D2O.
Crystallization
The 12-nt crystallization DNA and RNA oligomers (5′-AACCCTAACCCT-3′ and 5′-AACCCUAACCCU-3′) were purchased from IDT. Crystals of the PCBP2 KH1-DNA/RNA complexes were obtained by hanging-drop vapor diffusion against 25% PEG 8000, 100 mM sodium acetate, 100 mM sodium cacodylate (pH 6.1) at 22°C. The protein concentration was ∼ 250 μM with a 1:1.2 protein:DNA/RNA ratio. Orthorhombic crystals grew to useful size within 1 d with diffraction to 2.1 Å for the DNA complex and 2.6 Å for the RNA complex. The crystals are in space group P21212, with four KH1 domains and two DNA/RNA strands in one asymmetric unit.
Crystallographic data collection, structure determination, and refinement
Diffraction data were collected on frozen crystals using Beamline 8.3.1 of the Advanced Light Source (ALS) at Berkeley National Laboratory. Diffraction intensities were integrated and reduced using the programs DENZO and SCALEPACK (Otwinowski and Minor 1997). The structures were determined by SAD phasing for the DNA complex and by the molecular replacement method for the RNA complex using the PCBP2-KH1 crystal structure (Du et al. 2005) as the search model. MR solutions for all four KH1 domains in the asymmetric unit were obtained with Phaser (Storoni et al. 2004) and were used to calculate the initial electron density maps. The DNA/RNA molecules were built into the density during iterative refinements. Model building of the DNA/RNA and rebuilding of the protein were carried out by either MOLOC (Gerber and Muller 1995) or Coot (Emsley and Cowtan 2004). Refinement was carried out using REFMAC5 (CCP4 1994; Murshudov et al. 1997). The four protein domains and DNA/RNA molecules were each treated as separate TLS groups, and NCS restraints were not applied during the refinement. The quality of the final structure was verified by PROCHECK (Laskowski et al. 1996). A structural model of the PCBP2 KH2 domain was built by the program MODELLER (Marti-Renom et al. 2000) using the crystal structure of PCBP1 KH1 as the homologous structure. Structure figures were prepared using PyMOL (DeLano Scientific).
NMR spectroscopy
NMR data were collected on three samples: 13C/15N, 13C/15N with 60% random fractional deuteration, and 13C/15N/2H/(Ile/Leu/Val)-methyl-protonated KH1 + KH2. The samples were in a 90% H2O/10% D2O buffer containing 50 mM deuterated sodium acetate (pH 5.4), 2 mM DTT. The typical protein concentration for the NMR experiments was ∼ 1 mM. For most of the NMR experiments, the MKH6K-tag was not cleaved.
All NMR experiments were performed on Varian Inova spectrometers operating at 600 MHz for protons. Spectra were processed with NMRPipe/NMRDraw (Delaglio et al. 1995) and analyzed with SPARKY (Goddard and Kneller 1998). All NMR experiments were carried out at 25°C. 1H, 15N, and 13C resonance assignments were achieved by using the following heteronuclear NMR experiments: 15N-HSQC, 13C-HSQC, HNCO, HNCA, HN(CO)CA (Bax and Ikura 1991), CBCANH (Grzesiek and Bax 1992a), CBCA(CO)NH (Grzesiek and Bax 1992b), HBHA(CO)NH, C(CO)NH, H(CCO)NH, and 15N TOCSY-HSQC.
ACKNOWLEDGMENTS
Partial support for this work was provided by National Institutes of Health grants AI46967 (T.L.J.) and GM51232 (R.M.S.). Thanks are also due to Chris Waddling for managing the UCSF X-ray Crystallization Laboratory. The coordinates for the PCBP2 KH1 complexes with 12-nt DNA and 12-nt RNA have been deposited in the Protein Data Bank (PDB codes 2PQU and 2PYQ, respectively).
Footnotes
Abbreviations: PCBP, poly(C)-binding protein; KH domain, hnRNP-K homology domain; KH1, the first KH domain of PCBP2; 5′-UTR, 5′-untranslated region; 3′-UTR, 3′-untranslated region; RMSD, root-mean-square deviation.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.410107.
REFERENCES
- Backe, P.H., Messias, A.C., Ravelli, R.B., Sattler, M., Cusack, S. X-ray crystallographic and NMR studies of the third KH domain of hnRNP K in complex with single-stranded nucleic acids. Structure. 2005;13:1055–1067. doi: 10.1016/j.str.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Bandiera, A., Tell, G., Marsich, E., Scaloni, A., Pocsfalvi, G., Akindahunsi, A.A., Cesaratto, L., Manzini, G. Cytosine-block telomeric type DNA-binding activity of hnRNP proteins from human cell lines. Arch. Biochem. Biophys. 2003;409:305–314. doi: 10.1016/s0003-9861(02)00413-7. [DOI] [PubMed] [Google Scholar]
- Bax, A., Ikura, M. An efficient 3D NMR technique for correlating the proton and 15N backbone amide resonances with the α-carbon of the preceding residue in uniformly 15N/13C enriched proteins. J. Biomol. NMR. 1991;1:99–104. doi: 10.1007/BF01874573. [DOI] [PubMed] [Google Scholar]
- Blyn, L.B., Swiderek, K.M., Richards, O., Stahl, D.C., Semler, B.L., Ehrenfeld, E. Poly(rC) binding protein 2 binds to stem–loop IV of the poliovirus RNA 5′ noncoding region—Identification by automated liquid chromatography tandem mass spectrometry. Proc. Natl. Acad. Sci. 1996;93:11115–11120. doi: 10.1073/pnas.93.20.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blyn, L.B., Towner, J.S., Semler, B.L., Ehrenfeld, E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Du, Z., Yu, J., Chen, Y., Andino, R., James, T.L. Specific recognition of the C-rich strand of human telomeric DNA and the RNA template of human telomerase by the first KH domain of human poly(C)-binding Protein-2. J. Biol. Chem. 2004;279:48126–48134. doi: 10.1074/jbc.M405371200. [DOI] [PubMed] [Google Scholar]
- Du, Z., Lee, J.K., Tjhen, R., Li, S., Pan, H., Stroud, R.M., James, T.L. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 Å. J. Biol. Chem. 2005;280:38823–38830. doi: 10.1074/jbc.M508183200. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fenn, S., Du, Z., Lee, J.K., Tjhen, R., Stroud, R.M., James, T.L. Crystal structure of the third KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.6 Å resolution. Nucleic Acids Res. 2007;35:2651–2660. doi: 10.1093/nar/gkm139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Two functional complexes formed by KH domain containing proteins with the 5′-noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes & Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′-untranslated region of the poliovirus genome. J. Virol. 2000;74:2219–2226. doi: 10.1128/jvi.74.5.2219-2226.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, P.R., Muller, K. MAB: A generally applicable molecular force field for structural modeling in medicinal chemistry. J. Comput. Aided Mol. Des. 1995;9:251–268. doi: 10.1007/BF00124456. [DOI] [PubMed] [Google Scholar]
- Goddard, T.D., Kneller, D.G. University of California; San Francisco: 1998. SPARKY. [Google Scholar]
- Grzesiek, S., Bax, A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 1992a;114:6291–6293. [Google Scholar]
- Grzesiek, S., Bax, A. Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J. Magn. Reson. 1992b;96:432–440. [Google Scholar]
- Lacroix, L., Lienard, H., Labourier, E., Djavaheri-Mergny, M., Lacoste, J., Leffers, H., Tazi, J., Helene, C., Mergny, J.-L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmann, J.A.C., MacArthur, M.W., Kaptein, R., Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lewis, H.A., Musunuru, K., Jensen, K.B., Edo, C., Chen, H., Darnell, R.B., Burley, S.K. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Makeyev, A.V., Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A., Fiser, A., Sánchez, R., Melo, F.A.S. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Mulder, F.A., Schipper, D., Bott, R., Boelens, R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ostareck, D.H., Ostareck-Lederer, A., Wilm, M., Thiele, B.J., Mann, M., Hentze, M.W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′-end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Ostareck, D.H., Ostareck-Lederer, A., Shatsky, I.N., Hentze, M.W. Lipoxygenase mRNA silencing in erythroid differentiation: The 3′-UTR regulatory complex controls 60S ribosomal subunit joining. Cell. 2001;104:281–290. doi: 10.1016/s0092-8674(01)00212-4. [DOI] [PubMed] [Google Scholar]
- Ostareck-Lederer, A., Ostareck, D.H., Standart, N., Thiele, B.J. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′-untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parsley, T.B., Towner, J.S., Blyn, L.B., Ehrenfeld, E., Semler, B.L. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Sidiqi, M., Wilce, J.A., Vivian, J.P., Porter, C.J., Barker, A., Leedman, P.J., Wilce, M.C. Structure and RNA binding of the third KH domain of poly(C)-binding protein 1. Nucleic Acids Res. 2005;33:1213–1221. doi: 10.1093/nar/gki265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storoni, L.C., McCoy, A.J., Read, R.J. Likelihood enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;59:1145–1153. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Tomonaga, T., Levens, D. Activating transcription from single-stranded DNA. Proc. Natl. Acad. Sci. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner, S.A., Liebhaber, S.A. Identification of mRNAs associated with αCP2-containing RNP complexes. Mol. Cell. Biol. 2003a;23:7055–7067. doi: 10.1128/MCB.23.19.7055-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner, S.A., Liebhaber, S.A. Regulation of {α}-globin mRNA stability. Exp. Biol. Med. 2003b;228:387–395. doi: 10.1177/153537020322800409. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Chen, X. MCG10, a novel p53 target gene that encodes a KH domain RNA-binding protein, is capable of inducing apoptosis and cell cycle arrest in G2-M. Mol. Cell. Biol. 2000;20:5602–5618. doi: 10.1128/mcb.20.15.5602-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]