Abstract
The potential for water to participate in RNA catalyzed reactions has been the topic of several recent studies. Here, we report crystals of a minimal, hinged hairpin ribozyme in complex with the transition-state analog vanadate at 2.05 Å resolution. Waters are present in the active site and are discussed in light of existing views of catalytic strategies employed by the hairpin ribozyme. A second structure harboring a 2′,5′-phosphodiester linkage at the site of cleavage was also solved at 2.35 Å resolution and corroborates the assignment of active site waters in the structure containing vanadate. A comparison of the two structures reveals that the 2′,5′ structure adopts a conformation that resembles the reaction intermediate in terms of (1) the positioning of its nonbridging oxygens and (2) the covalent attachment of the 2′-O nucleophile with the scissile G+1 phosphorus. The 2′,5′-linked structure was then overlaid with scissile bonds of other small ribozymes including the glmS metabolite-sensing riboswitch and the hammerhead ribozyme, and suggests the potential of the 2′,5′ linkage to elicit a reaction-intermediate conformation without the need to form metalloenzyme complexes. The hairpin ribozyme structures presented here also suggest how water molecules bound at each of the nonbridging oxygens of G+1 may electrostatically stabilize the transition state in a manner that supplements nucleobase functional groups. Such coordination has not been reported for small ribozymes, but is consistent with the structures of protein enzymes. Overall, this work establishes significant parallels between the RNA and protein enzyme worlds.
Keywords: hairpin ribozyme; 2′,5′-phosphodiester; vanadate; reaction intermediate; transition-state stabilization; active-site waters
INTRODUCTION
Protein enzymes have evolved numerous strategies to lower the energetic barrier required to convert substrates into products (Knowles 1991). Polanyi and Pauling perceptively envisioned that this could be accomplished by distorting the precatalytic geometry of the reactants into that of the transition state, which entails the expenditure of binding energy derived from the substrate–protein interaction (Lienhard 1973; Borman and Wolfenden 2004). As such, knowledge of the stereochemical interactions employed by an enzyme during the transition state can provide great insight into the chemical strategies utilized by the catalyst to accelerate a reaction rate (Lienhard 1973). Although envisioning the transition state of a phosphoryl-transfer reaction would appear somewhat trivial (Dennis and Westheimer 1966; Knowles 1980), understanding the factors leading to formation and stabilization of such intermediates for a given enzyme is not. This statement is especially true in the case of RNA enzymes, whose rate-enhancing features are only beginning to be understood (Doherty and Doudna 2001; Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006), despite the conserved and essential nature of ribocatalysts in biology.
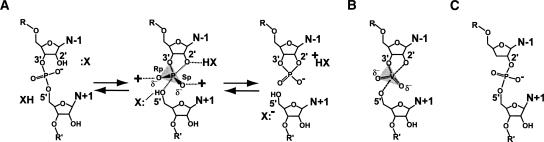
Although it is difficult and rare to capture bona fide intermediates of enzyme reactions in structural studies, high-resolution methods have provided insight into such species by use of analogs that mimic the geometric and/or electronic properties of the transition state, thereby providing inert complexes for analysis (Hiratake 2005). In the study of phosphoryl-transfer reactions, the use of vanadium oxide has proven particularly effective to trap conformations representative of the pentacoordinate, oxyphosphorane transition state (for review, see Davies and Hol 2004). However, this strategy has been employed for only a single RNA enzyme, the hairpin ribozyme (Rupert et al. 2002), a small catalytic motif derived from the negative polarity strand of the satellite RNA of tobacco ringspot virus. The hairpin ribozyme catalyzes a reversible, site-specific phosphodiester bond cleavage reaction producing a cyclic 2′,3′-phosphodiester and a 5′-OH terminus as its products (Fig. 1A; van Tol et al. 1990). In their seminal study employing the vanadium oxide mimic of the oxyphosphorane intermediate, Ferré-D'Amaré and colleagues revealed key interactions with nucleobase functional groups that appeared to stabilize the electronic structure and geometry of the transition state (Rupert et al. 2002). A role for nucleobase involvement in transition-state stabilization was supported by several ensuing studies involving the effects of abasic ribozymes (Lebruska et al. 2002; Kuzmin et al. 2004, 2005). Other studies suggested key residues at A38 or G8 may be involved as general acid/base catalysts (Pinard et al. 2001; Bevilacqua 2003; Kuzmin et al. 2005; Thomas and Perrin 2006; Wilson et al. 2006), which has been proposed as a general scheme to achieve rate acceleration by small ribozymes (Fig. 1A; Das and Piccirilli 2005; Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006; Perrotta et al. 2006).
FIGURE 1.
The chemical reaction of small ribozymes and reaction-intermediate analogs. (A) The cleavage reaction utilizes the 2′-OH of the N-1 base, which is activated by a base catalyst (:X). The pentacoordinate, trigonal–bipyramidal transition state is formed by in-line nucleophilic attack of the 2′-nucleophile on the electrophilic phosphorus. The 5′-oxygen leaving group of the N+1 base is protonated by an acid (XH). A free 5′-OH and a cyclic 2′, 3′-phosphodiester are products. (B) Schematic representation of vanadate bound between two RNA strands flanking the cleavage site. (C) Schematic representation of a 3′-deoxy, 2′,5′-phosphodiester linkage at the site of cleavage in a small ribozyme.
Recently, we reported the “precatalytic” structure of a minimal junctionless hairpin ribozyme construct with several waters bound in the active site (Salter et al. 2006). In light of hypothesized roles for water in the hairpin ribozyme mechanism of action, we sought to identify water molecules in the active site that might be implicated in proton transfer networks, specific acid/base catalysis, or transition-state stabilization (Seyhan and Burke 2000; Pinard et al. 2001; Rupert et al. 2002; Kuzmin et al. 2005; Park and Lee 2006; Rhodes et al. 2006). Herein, we report the use of vanadate and a 3′-deoxy, 2′,5′-phosphodiester linkage as reaction-intermediate analogs (Fig. 1B,C, respectively). We describe the active sites of the hairpin ribozyme constructs that accommodate these analogs, as well as ordered water molecules that are present only in “reaction-intermediate” states. The observation that both ordered water molecules and nucleobase interactions may participate in geometric and electrostatic stabilization of the transition state corroborates existing views of catalytic strategies employed by the hairpin ribozyme. Finally, we propose the use of the 2′,5′-phosphodiester linkage to induce reaction-intermediate conformations of other ribozymes that are not amenable to cocrystallization with vanadate. Employing this analog in structural studies has the potential to reveal catalytic strategies applicable to larger, less tractable RNA molecules.
RESULTS
Global fold and quality of hinged hairpin ribozyme structures
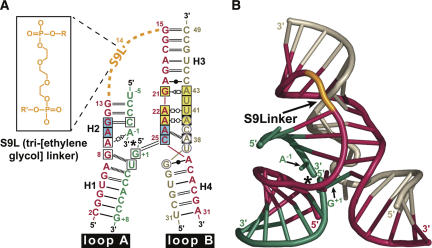
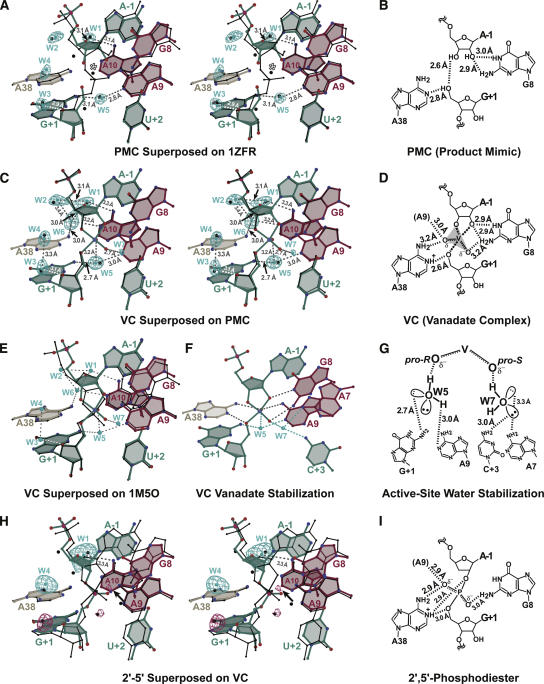
To examine the effects of reaction-intermediate analogs on the active site structure of a minimal hairpin ribozyme construct, it was necessary to create a break in the substrate backbone between A-1 and G+1 capable of coordinating vanadium oxide (Fig. 1B). Previous crystal structures of a minimal, all-RNA hairpin ribozyme employed a junctionless variant in which loop A and B domains were untethered by removal of the nucleotide at position 14 (Grum-Tokars et al. 2003; Alam et al. 2005; Salter et al. 2006). The individual loop A and B domains for this construct were formed separately in solution and then docked to yield the active enzyme. However, this construct failed to produce crystals of a metavanadate–ribozyme complex, which necessitated the development of a minimal hinged construct. Previously, Walter and coworkers described the use of a synthetic linker between positions 14 and 15 of a minimal hairpin ribozyme that led to greater catalytic efficiency compared with the native sequence (Rueda et al. 2004). Using this strategy, we incorporated a 10-atom “S9” linker at position 14, thus reattaching the loop A and B domains (Fig. 2A) in the two constructs of this investigation in which the scissile phosphate was removed. These hinged constructs crystallized readily from solutions of poly(ethylene) glycol 2000 monomethyl ether and Li2(SO4), as described for the junctionless variant (Alam et al. 2005). A more detailed description of this linker will be presented elsewhere (C.M. MacElrevey, R.C. Spitale, J. Krucinska, and J.E. Wedekind, unpubl.). Significantly, the global fold of hinged hairpin ribozyme variants (Fig. 2B) closely resembles those of both junctionless (Alam et al. 2005; Salter et al. 2006) and four-way helical junction forms (Rupert and Ferré-D'Amaré 2001). This similarity is indicated by a modest 0.48 Å RMSD resulting from an all-atom least-squares superposition between a precatalytic junctionless variant (PDB ID: 1ZFR [Salter et al. 2006]) and the hinged vanadate complex ([VC], a structure exhibiting vanadate bound between A-1 and G+1; of this study solved to 2.05 Å (atoms comprising alternate conformations for U-5 and G8 were excluded from this comparison). Such structural similarity was observed for both hinged structures whose representative fold is depicted in Figure 2B.
FIGURE 2.
Schematic representations of the minimal hinged hairpin ribozyme construct used in this study. (A) RNA strands and secondary structure of the minimal form of the hairpin ribozyme. (Blue-green) Nucleotides of the substrate strand fragments, (asterisk) the site of cleavage for an intact substrate strand. The scissile phosphate was removed for the PMC and VC structures. Alternatively, in the case of the 2′–5′ structure, the substrate strand fragments are covalently joined by a 2′,5′-phosphodiester bond (Fig. 1C). (Tan, red) Other ribozyme strands. This color scheme is preserved throughout the figures. The red 29-mer strand also contains the synthetic S9 linker (S9L, orange inset) that covalently tethers the loop A and B domains in the VC and PMC constructs. (Black labels) Helical regions. H-bond pairings: (open square) Hoogsteen; (open triangle) trans-sugar; (open circle) W-C face; (closed circle) wobble pair; (double and single solid black lines) W–C H-bond pairs; (dashed black lines) single H-bonds. (Boxes) conserved nucleotides of the loop A and B domains, (blue boxes) residues participating in a ribose zipper, (yellow boxes) residues of the E-loop, (gray circles) nucleotides within the S-turn. (B) Ribbon diagram of the hinged hairpin ribozyme representative of those reported in this study. (Asterisk) The site of cleavage. Positions A-1 and G+1 in the active site are labeled. A was adapted from the output of RNAview (Yang et al. 2003). B was generated using Nuccyl (Jovine 2003; Yang et al. 2003) and PyMOL (DeLano 2004).
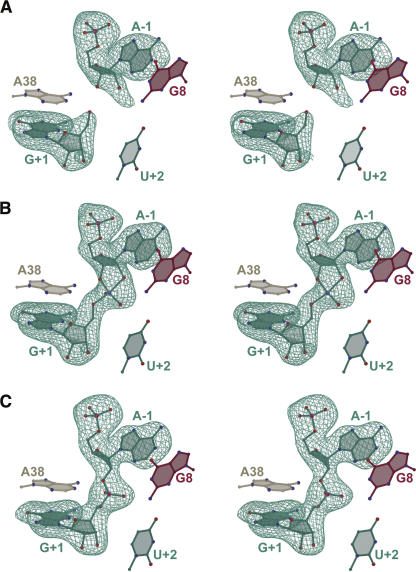
Reasonable overall refinement statistics were observed for each of the three structures of this study, including R cryst values between 22% and 26%, R free within 3% of R cryst, and highly complete diffraction data extending to resolutions of 2.05 Å, 2.25 Å, and 2.35 Å, respectively (Table 1). The quality of each structure is indicated by the agreement of the atomic coordinates to the electron density (Fig. 3) with modest distortion of the model from ideal geometry (Table 1).
TABLE 1.
X-ray diffraction and refinement statistics
FIGURE 3.
Stereographic simulated annealing omit electron density maps for the active sites. (A) PMC product mimic strands. (B) The VC complex with bound vanadate. (C) The 2′-5′ structure with a 3′-deoxy, 2′,5′-phosphodiester linkage at the site of cleavage. Local active site nucleotides are depicted for reference; the G8 syn conformation (PMC and VC structures only) was omitted for clarity. Atoms are colored: (red) oxygen, (blue) nitrogen, (pink) phosphorus, (violet) vanadate; carbon atoms and bonds are colored as in Fig. 2. Maps have reduced bias (Pannu and Read 1996) coefficients of the form m|F o| − D|F c|, are contoured at 3.0σ, and are colored as in Fig. 2. All ball-and-stick figures were created with Bobscript (Kraulis 1991; Esnouf 1999) and rendered with Raster3d (Merritt and Bacon 1997).
The product mimic complex (PMC) structure
The product mimic complex ([PMC], a structure solved in complex with 5′ and 3′ product strand fragments), was solved and refined to 2.25 Å as a basis for comparison to the 2′,5′-linked and vanadate structures of this study. Despite the fact that the PMC coordinates were derived from the precatalytic 1ZFR hairpin ribozyme model, the new structure adopts distinctly different features at the scissile bond (Fig. 4A). The PMC structure represents an inactive complex in which two synthetic RNA strands were bound to the ribozyme in lieu of genuine substrate or product strands (Fig. 2, green strands; Fig. 3A). The structure of the 5′-most strand is distinguished from a true product by the absence of a cyclic 2′,3′-phosphoanhydride linkage connecting the A-1 cis-diols, which causes the strand to be inert for ligation (Fig. 1A). Features of the PMC electron density map indicated a 2′-endo ribose pucker for A-1, supported by putative H-bonds between its 2′-OH and both the imino and exocyclic amine groups of G8 (Fig. 4B). The 3′-product fragment exhibited a well-defined electron density for the 5′-OH group of G+1 (Fig. 3A), and implies a 2.8 Å H-bond with the N1 atom of A38 (Fig. 4B). Given that this structure was buffered at pH 6.0, position N1 of A38 would be expected to be deprotonated (adenine nucleoside solution pKa = 3.5 [Saenger 1984]) and therefore act as an H-bond acceptor, although at present the true pKa of A38 is uncertain. The 5′-OH of G+1 is further stabilized by an apparent 2.6 Å H-bond with the 3′-OH of A-1.
FIGURE 4.
Overlays of various hairpin ribozyme structures with accompanying active site schematic representations. (A) Stereoview of the PMC structure (colored ball-and-stick) superimposed on the precatalytic structure of the minimal, junctionless hairpin ribozyme (PDB ID: 1ZFR [Salter et al. 2006]) (black bonds). Local active-site residues are included for reference. (Cyan spheres) Water molecules observed in the PMC structure with blue electron density resulting from simulated annealing omit maps calculated with coefficients m|F o| − D|F c| and contoured at 6.5σ; (black spheres) waters observed in the previously reported “precatalytic” structure (1ZFR); (open asterisk) water 52 from the latter structure; (dashed gray or white lines) selected putative H-bonds involving water molecules observed in the PMC structure with labeled distances. (B) Schematic diagram of the PMC active site. (Black dashed lines) Putative hydrogen bonds between nucleotide atoms. (C) Stereographic superposition of the VC structure (colored) with the PMC structure (black). A 2.8 Å putative hydrogen bond between W1 and the O4′ of A-1 was omitted for clarity. (D) Schematic diagram of the VC structure with bound vanadate from panel B. (E) Overlay of the VC structure (colored) with the 4WJ crystal structure of the hairpin ribozyme–vanadate complex (PDB ID: 1M5O [Rupert et al. 2002]) (black bonds). No waters were observed in the active site of the 1M5O structure. (F) Interactions in the VC structure predicted to promote transition state stabilization. (Gray lines) Interactions supported by nucleobase groups, (cyan lines) interactions supported by active-site waters W5 and W7. (G) Predicted orientation of W5 and W7 hydrogens and electron pairs based on the tetrahedral coordination geometry and identity of the H-bonding partners in VC. (H) Stereographic overlay of the 2′–5′ structure (colored) on the VC structure (black). Cyan electron density covering waters W1 and W4 in the 2′–5′ structure resulted from a simulated annealing omit map calculated with coefficients m|F o| − D|F c| and contoured as in panel A; violet electron density is contoured at 3.0σ to emphasize electron density peaks that were consistent with waters located in the accompanying structures, but did not fully meet the criteria used to define waters in the current study. (Black arrow) An electron density peak observed near the location of the new water W7 in the VC structure. (I) Schematic diagram of the 2′–5′ structure depicted as in B.
Differences exist between the PMC structure of this study and the genuine product complex solved in the context of the 4WJ hairpin ribozyme (PDB ID: 1M5V [Rupert et al. 2002]). While an H-bond between N1 of A38 and the 5′-OH of G+1 is predicted by both structures, the interatomic distance in 1M5V is 0.7 Å longer and exhibits an average H-bond angle of 74° (i.e., between N1 of A38 and the C5′–O5′ bond of G+1); the equivalent angle for PMC is 93°, which is also more acute than the ideal sp3 angle of ∼109°. One possible explanation for these differences arises from the presence of the cyclic phosphate group in 1M5V. At ∼2.5 Å distance from the 5′-OH of G+1, the pro-R oxygen equivalent of the cyclic anhydride sterically restricts the H-bond geometry achievable between N1 of A38 and the 5′-OH of G+1, which functions as a nucleophile in the reverse ligation reaction (Fig. 1A). These observations have implications for the role ascribed to A38 as a putative general base in ligation (Bevilacqua 2003; Doudna and Lorsch 2005; Fedor and Williamson 2005) and/or the ability to reliably trap structural conformations representative of precatalytic product states as cautioned by Ferré-D'Amaré (2004).
The vanadate complex structure
To induce an active site conformation representative of the transition state, vanadium oxide was included in the crystallization medium of the PMC complex. The resulting structure was solved and refined to 2.05 Å resolution and is referred to as VC throughout this study. A σA-weighted simulated annealing omit electron density map confirmed the presence of vanadate between the free 2′, 3′, and 5′ oxygens of A-1 and G+1 by comparison to the PMC (Fig. 3, cf. A and B). The model of vanadate fit well into the electron density while closely resembling the slightly distorted trigonal–bipyramidal geometries observed for related small molecule structures in the Cambridge Structural Database (CSD) (Allen 2002) (CSD ID: IZITEQ [Ouellette et al. 2004]; CSD ID: NONLEH [Salta and Zubieta 1997]). Refinement of the VC structure with reduced energy constraints on the τ angle (the in-line attack angle of SN2 reactions defined in the case of the hairpin ribozyme by the 2′-OH of A-1, the scissile phosphorus atom, and the 5′-OH of G+1) resulted in convergence upon an in-line geometry of 154°, compared with 163.8° or 165.5° observed for the analogous small molecule coordinates. The vanadate moiety also exhibited B factors that were noticeably higher than nearby RNA atoms during early refinement. Slightly elevated vanadate B factors exist in other enzyme–vanadate crystal structures (PDB ID: 1B8J [Holtz et al. 1999]; PDB ID: 1BO6 [Kakuta et al. 1998]), including the previous, independently determined 4WJ hairpin–vanadate complex (PDB ID: 1M5O previously reported structure of a four-way junction hairpin ribozyme construct in complex with vanadate as a transition-state analog at the active site [Rupert et al. 2002]). The presence of negative electron density peaks in σA-weighted m|F o| − D|F c| electron density maps suggested the vanadate in the current structure was bound with incomplete atomic occupancy. Collectively, these factors were used in conjunction with the R factor and R free as metrics to refine the occupancy, which converged to a final value of 50%.
An overlay of the VC and PMC structures revealed a high degree of similarity in their active sites (Fig. 4C). For example, the VC structure exhibited a putative H-bond between the N1 position of A38 and O5′ of G+1 (Fig. 4D). Unlike PMC, however, there is some ambiguity over whether the 5′ oxygen of G+1 is protonated in the structure due to its coordination with vanadium (discussed below). Notably, a comparably short N1-to-O5′ distance was observed in the 4WJ hairpin–vanadate complex (PDB ID: 1M5O [Rupert et al. 2002]), which lends confidence to both studies. At other positions, the imino group of G8 appeared to form a 2.9 Å H-bond with the 2′-oxygen of A-1, similar to that of PMC, and is consistent with the expectation that the former group would be protonated at a pH of 6.0, as used in the current study (guanine solution pKa = 9.4 [Saenger 1984]). The arrangement of the active-site residues in VC exhibited close agreement with the 4WJ (1M5O) coordinates (Fig. 4E). Aside from small variations in interatomic RNA distances, the most significant differences were observed in the relative positions of the vanadate and the G8 base as well as the presence of ordered water molecules in the current study. A likely source of the differences in the vanadate/G8 positions is the assignment of a C2′-endo ribose pucker at A-1 in the VC structure versus an O4′-endo pucker reported previously (Rupert et al. 2002). The distinctly different ribose conformation of the present study led to an “upward” shift of the A-1 diols coordinating the vanadate moiety. The spatial locations of these atoms correlate well with the positions of the imino and exocyclic amines of the G8 base, which maintain cross-strand H-bond interactions with the 2′-oxygen of A-1 and the pro-S oxygen of vanadate, respectively (Fig. 4D). Such interactions are also present in the 1M5O structure, but the G8 base in the current study is positioned noticeably higher, whereas neighboring nucleotide bases are virtually superimposed throughout the local environment (Fig. 4E). One effect from these differences is that the in-line geometry, described by the τ angle, is slightly more obtuse in the present study, 154° compared with 144° and 153° for the two molecules in the asymmetric unit of the 4WJ structure (Rupert et al. 2002). From a practical vantage point, these differences do not seem significant, and the underlying causes cannot be pinpointed at this time; contributing factors may include differences in vanadate occupancy between structures or resolution of electron density maps.
In both the VC and 1M5O structures, the nonbridging oxygens of the vanadate are stabilized by multiple, favorable interactions involving the exocyclic amines of A9, A38, and G8 (Fig. 4F, gray lines). An additional distinguishing feature of the VC structure is the presence of two active site waters, W5 and W7, which also coordinate the vanadate nonbridging oxygen positions (Fig. 4F). Each water is itself positioned by two additional hydrogen bonds with proximal nucleobase groups (Fig. 4F, cyan lines; Fig. 4G). This observation is intriguing in light of the requirement for electrostatic transition state stabilization of the phosphoryl transfer intermediate and the paucity of charged functional groups available in the nucleobase repertoire.
Overall, the similarities between the active sites of the hinged and 4WJ hairpin ribozyme structures are evidence of a robust architecture that appears preserved despite contextual differences arising from crystallization constructs or crystal packing environments. The quality of the electron density reported herein, combined with the higher-resolution X-ray diffraction data used in refinement, establish with even greater confidence the “induced-fit” interactions resulting from placement of a geometric mimic of the transition state into the active site of the hairpin ribozyme as first described by Ferré-D'Amaré (2004).
2′–5′ Reaction-intermediate structure
Due to the reduced atomic occupancy of vanadate and the goal of observing well-ordered water molecules in the hairpin ribozyme active site, alternative analogs were considered that might also lead to reaction-intermediate-like conformations. As such, a 3′-deoxy, 2′,5′-phosphodiester substitution was introduced at the site of cleavage (Fig. 1C) in the structure referred to as 2′–5′, which was solved and refined to 2.35 Å resolution. The electron density for this 2′,5′-phosphodiester linkage was well defined (Fig. 3C), and the active site organization resembled the VC structure (Fig. 4H). Significantly, the distances between atom N1 of A38 and the 5′-O of G+1, as well as the 2′-O of A-1, were consistent with a bifurcated H-bond in the 2′–5′ structure (Fig. 4I) and exhibited a pattern of nucleobase interactions similar to PMC and VC. This finding provides further evidence that position N1 of A38 serves as an H-bond donor in this interaction because the covalent ester linkage of the O2′ and O5′ oxygens require that they be deprotonated. At the buffered pH 7.0 in the crystal, this observation suggests a > 3 pH unit elevation in the imino pKa of A38, which has been speculated to be a prerequisite for this base if it is in fact serving as a general acid catalyst in cleavage (Rupert et al. 2002; Bevilacqua 2003; Fedor and Williamson 2005; Kuzmin et al. 2005).
There are other similarities between the VC and the 2′–5′ structures as well. The N2 exocyclic amine of G8 is positioned 3.0 Å from the pro-S oxygen of the scissile phosphorus (Fig. 4I), consistent with a role in electrostatic stabilization as observed for the VC complex (Fig. 4D). Another remarkable feature of the 2′–5′ structure is that the orientation of the nonbridging oxygens at the scissile phosphate closely resembles that of oxo-vanadium in the VC complex (Fig. 4H). This similarity persists despite the markedly different geometry of each phosphorus (or equivalent) center. The τ angle of the tetrahedral G+1 phosphate in the 2′–5′ structure must maintain sp3 hybridization, and thus is far from in-line (∼103°). However, the nonbridging oxygens appear to be spatially oriented by the same key interactions observed in the VC structure (Fig. 4, cf. D and I). This observation supports the notion that the 2′,5′-phosphodiester linkage is representative of a reaction-intermediate state, exhibiting an orientation of the nonbridging oxygens that is relevant to catalysis by analogy to vanadate. In light of the “catalytic” orientation of the nonbridging oxygens, only small spatial movements would be required to reach the requisite in-line geometry for cleavage. The latter orientation clearly distinguishes the VC and 2′–5′ structures from the 1ZFR precatalytic structure, a point that is discussed below in light of preferential stabilization of the transition state. As such, the 2′–5′ structure shares features with both transition-state analog structures prepared in the presence of vanadate (VC, this study; 1M5O [Rupert et al. 2002]) as well as prior “precatalytic” structures in which the 2′-O-nucleophile of A-1 is blocked by a methyl group (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002; Salter et al. 2006), and must experience an induced fit of the nonbridging oxygens as a prelude to phosphoryl transfer.
Conformational heterogeneity at nucleobase G8
Contrary to prior structural studies of minimal and 4WJ hairpin ribozymes, the base of G8 was observed in mixed anti and syn conformations in both structures missing the scissile phosphate (75% anti in VC and 50% anti in PMC). This heterogeneity manifested as distinctive positive electron density peaks in σA-weighted m|F o| − D|F c| maps corresponding to positions of the syn conformation that did not overlap with the modeled anti conformation. Modeling of syn–anti alternate conformations resulted in improved R factor and R free values, corrected the distinctive m|F o| − D|F c| electron density features, and improved the fit of the models to 2m|F o| − D|F c| electron density maps. Notably, the ribose of G8 in all three structures adopted a 2′-endo conformation, consistent with an NMR study of the isolated loop A domain in solution (Cai and Tinoco 1996). Stabilization was conferred to this pucker by a putative H-bond with position N1 of A-1 ranging in length from 2.9–3.4 Å, similar to what was observed in a previous crystal structure of the hairpin ribozyme incorporating a G8I (inosine) modification (Salter et al. 2006). This ribozyme–substrate cross-strand hydrogen bond is further corroborated by a study in which removal of the 2′-OH of G8 resulted in a ΔΔG of 0.7 kcal/mol (approximately equivalent to one H-bond) affecting primarily the K M of the cleavage reaction (Chowrira et al. 1993). The stability afforded by the A-1 purine may also explain why selection experiments demonstrated that A-1 confers greater catalytic activity than pyrimidines (Perez-Ruiz et al. 1999) and is preferred in other naturally occurring hairpin ribozyme sequences (DeYoung et al. 1995).
Waters in the active site interact with reaction-intermediate mimics
The active sites of the VC and 2′–5′ structures were observed to contain ordered water molecules bound near key nucleobases, providing important structural evidence for the potential role of water in the mechanism of action. Most of these waters were present in the PMC structure, as well as a “precatalytic” structure determined previously (Salter et al. 2006) (PDB ID: 1ZFR; Fig. 4A). An interesting exception is “water 52,” identified only in the precatalytic 1ZFR structure (Fig. 4A, asterisk), which appears to mimic the pro-R oxygen of the transition state (Salter et al. 2006). Interestingly, a new water molecule was observed in the VC complex located 2.7 Å from the pro-S nonbridging oxygen of the vanadate (W7; Fig. 4C), and is stabilized through contacts with the exocyclic amines of C+3 and A7 (Fig. 4F,G). A weak electron density peak near this position also exists in the 2′–5′ structure, suggesting an equivalent water molecule; however, insufficient resolution (or occupancy) prevented formal assignment as a water (Fig. 4H, arrow). Another new water molecule was identified in all three structures (W4; Fig. 4A,C,H) within hydrogen bonding distance of the pro-S oxygen of A38 (not shown) and 3.3 Å from the previously observed water W3 (W65 in 1ZFR), as measured in VC. In light of the consideration that water might serve as a specific acid in cleavage while coordinating to the imino position of A38 (Kuzmin et al. 2005), this possibility is unlikely for water W4 considering its distance is 4.4 Å and 4.0 Å from positions N1 and N3 of A38, respectively. Likewise, there is insufficient space to accommodate a water between the O5′ leaving group of G+1 and the imino position of A38 (Fig. 4D,I).
Hydroxyl radical footprinting suggested the active site core of the hairpin ribozyme is solvent-protected (Hampel et al. 1998), leading to the expectation that the active site waters would be unable to exchange readily with bulk solvent. A search for solvent cavities using the program VOIDOO (Kleywegt and Jones 1994; Kleywegt et al. 2001) confirmed that waters W1, W2, and W6 are located in a pocket near the Watson–Crick faces of A9 and A10 (Fig. 4C). Ferré-D'Amaré and colleagues recognized this region as having the potential to accommodate bound waters (Rupert et al. 2002). In the current structures, this pocket appears nominally sequestered from bulk solvent as a result of the van der Waals surface of the surrounding bases. A second discrete pocket was also identified containing W5 near the exocyclic amine of G+1 and is likewise solvent inaccessible. If exchange with bulk solvent occurs, the most likely pathway would be formation of a narrow channel between the ribose of A40 and the base of A38 that could form from relatively small movements at these positions. This possibility is in agreement with molecular dynamics simulations that find explicitly modeled waters occupying various positions within the active site (Park and Lee 2006; Rhodes et al. 2006) and suggests that pathways leading to entry and egress of such waters occur most likely through transient molecular deformations relative to known crystallographic structures. Such a phenomenon was originally proposed as molecular “breathing” (Rupert & Ferré-D'Amaré 2001). An alternative explanation would be that the waters in the two pockets described here were trapped during interdomain docking and could exchange during substrate turnover. In contrast, waters W3, W4, and W7 are predicted to easily exchange with bulk solvent due to their positions lining a channel formed by the substrate strand at the site of cleavage and the flanking major groove of helix 1.
DISCUSSION
This study provides the first structural evidence for ordered water molecules in the active site of a small ribozyme crystallized in the presence of a reaction-intermediate analog, and thus provides a starting point to address the significance of specific waters in the hairpin ribozyme mechanism of action. Importantly, the locations of a subset of these water molecules are consistent with a role in electrostatic stabilization of the transition state as previously attributed to RNA nucleobase functional groups (Ferré-D'Amaré 2004). Hence, the current observations are significant in that they not only complement, but also extend existing hypotheses regarding the catalytic strategies employed by ribozymes to enhance their functional group repertoire (Fedor 2002a; Rupert et al. 2002; Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006). Secondly, in light of the caveats associated with the use of vanadate as a mimic of the transition state, the 2′,5′-phosphodiester analog provides compelling new evidence that the N1 position of A38 exists in a protonated state at the buffered pH of 7.0 in the crystal. This observation and the absence of a suitably poised water located between the A38 imino group and the O5′-leaving group at G+1 are most consistent with a role for A38 as a general acid catalyst in cleavage. Finally, the novel and successful use of the 2′,5′-phosphodiester linkage as a reaction-intermediate analog has led to a proposal for its use to study the reaction coordinate of other ribozymes that are not amenable to cocrystallization with vanadate. Collectively, the chemical and structural precedents described here hold great potential to advance our understanding of the catalytic strategies employed by RNA enzymes.
Rationale for determining multiple reaction-intermediate structures
The use of molecular structure determinations has revolutionized the ability to locate and identify key residues in ribozyme active sites, and to relate specific functional groups to rate enhancement (Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006). However, a first look at a structure rarely reveals all there is to know about the mechanism of action as well as relevant conformational stabilization and dynamics (McKay 1996; Nelson and Uhlenbeck 2006). The earliest structure of the hairpin ribozyme accounted for much of the solution data regarding essential functional groups (Ryder and Strobel 2002) and revealed the spatial details of active site residues juxtaposed against the scissile bond (Rupert and Ferré-D'Amaré 2001). Most notably, the phosphoryl-transfer angle, τ, of the initial structure was nearly in-line at 158° or 169° (for each of two molecules in the asymmetric unit), suggestive of an active site poised for transesterification, despite the presence of a CH3 group blocking the 2′-nucleophile at A-1 (Rupert and Ferré-D'Amaré 2001). However, a subsequent structure in complex with vanadate revealed a markedly different orientation of the scissile bond featuring an ∼45° rotation of the nonbridging oxygens about the A-1 2′-nucleophile to G+1 5′-leaving-group axis that was accompanied by significant H-bonding to these groups in a manner unforeseen in the prior “precatalytic” structure (Rupert et al. 2002). Yet despite the more favorable and extensive interactions to the nonbridging oxygens of the vanadate moiety, the τ angle deviates significantly from ideal 180° in-line geometry, and was observed to be 153° or 144° for the two ribozyme molecules in the asymmetric unit (Rupert et al. 2002) as well as 154° in this study (Fig. 4E). Therefore, if such vanadate structures were truly representative of the transition state, one might conclude that in-line geometry plays only a minor role in the formation of a productive reaction coordinate or possibly that the reaction proceeds from a side-on attack (Brown et al. 1983). Indeed, the problem of non-in-line geometry in vanadate coordination is not unique to RNA enzymes, but is also observed in the protein world: PDB ID 1RUV, τ = 149.5° (Ladner et al. 1997); PDB ID 6RSA, τ = 165.7° (Wlodawer et al. 1983), to name a few. It follows, therefore, that a safe course of action would be the analysis of multiple reaction-intermediate analogs to discern those commonalities observed at high resolution that are sensible with respect to the stereochemical requirements of the reaction (Dennis and Westheimer 1966). Precedents for this strategy exist in the protein field. Nucleoside diphosphate kinase (Xu et al. 1997), myosin (Fisher et al. 1995; Smith and Rayment 1996), acid phosphatase (Lindqvist et al. 1994), and alkaline phosphatase (Holtz et al. 1999; Le Du et al. 2002) have each been solved with no fewer than two reaction-intermediate analogs. The commonalities of such structures have undoubtedly been useful to correlate structure with functional and biochemical data, while helping to identify the enzymatic attributes that promote phosphoryl transfer. By analogy to the protein field, the study of ribozymes will experience the same multistructure benefits.
Active-site waters and stabilization of the reaction-intermediate
An important benefit from the use of reaction-intermediate analogs for structural studies is that they often “reorganize” the active site by forming H-bonds analogous to those that stabilize the transition state. This effect was observed in the previous hairpin ribozyme–vanadate complex and provided convincing evidence that geometric and electrostatic stabilization of the transition state by active site nucleobases is a catalytic strategy employed by this ribozyme (Rupert et al. 2002). It is not surprising that the same interactions are observed in the VC structure, refined to 2.05 Å in the current study, in light of the close agreement between the two active sites (Fig. 4E). However, the use of vanadate in the present study in conjunction with a minimal hinged hairpin ribozyme variant exhibiting favorable diffraction properties, described previously (Salter et al. 2006), undoubtedly contributed to the ability to also observe active site waters. Such waters provide compelling evidence for the notion that water is an important component in the activity of the hairpin ribozyme (Pinard et al. 2001; Fedor and Williamson 2005; Kuzmin et al. 2005; Bevilacqua and Yajima 2006; Park and Lee 2006; Rhodes et al. 2006; Salter et al. 2006). Remarkably, two tightly bound waters were observed coordinating to the nonbridging oxygens of the vanadate moiety in the VC structure. W7 and W5 coordinate to the pro-S and pro-R oxygens, respectively (Fig. 4C), and contribute either a second or third stabilizing interaction to each oxygen in addition to contacts with conserved nucleobase groups (Fig. 4F). It is notable that the presence of a water–hydrogen bonding to the pro-S oxygen was predicted in a molecular dynamics simulation employing the previous coordinates of the 4WJ hairpin ribozyme–vanadate complex assuming an explicit solvent model (Park and Lee 2006), although details of any additional RNA groups serving to position these waters were not described. Collectively, these observations (Fig. 4F) complement the previous view of transition-state stabilization (Rupert et al. 2002) by providing important structural evidence that water molecules can assist in transition-state binding, which may be a universal phenomenon for members of the small ribozyme family.
Achieving correct geometry is critical for catalysis; however, it is also important to ameliorate the negative charge that accumulates during the transition state of an associative phosphoryl-transfer reaction (Knowles 1980), such as that performed by the hairpin ribozyme. For example, the structures of three well-characterized phosphoryl-transfer protein enzymes in complex with transition-state analogs revealed two or three interactions with the nonbridging oxygens, commonly by backbone amide hydrogens of the polypeptide chain, but also with charged amino acid side chains and divalent metal ions (Smith and Rayment 1996; Ladner et al. 1997; Baxter et al. 2006). While the utility of formally charged metal ions and amino acid side chains is obvious, backbone amide hydrogens have an estimated partial positive charge of +0.31e (MacKerell et al. 1998), suggesting they contribute significant electrostatic stabilization to the transition state. In contrast, without a complement of charged functional groups or bound active site ions, the question arose as to how the hairpin ribozyme could achieve a comparable degree of charge stabilization. The partial positive charges of exocyclic amino group hydrogens have been estimated for adenine, cytosine, and guanine as + 0.40e, ∼ + 0.29e, and ∼ + 0.32e, respectively, while the partial positive charges for the imino hydrogens of uracil and guanine are + 0.36e and + 0.28e (MacKerell et al. 1995). This observation suggests that neutral nucleobase groups can confer a level of electrostatic stabilization comparable to the amide hydrogens of the peptide backbone, although they are incapable of the level of charge stabilization conferred by a lysine or arginine on a group-for-group basis. Intriguingly, the partial positive charge on water hydrogens has been estimated to be between + 0.41e and + 0.54e (Jorgensen et al. 1983; Berendsen et al. 1987). Therefore, in the absence of active site metals and positively charged amino acid groups, it is plausible that the coordination of water molecules to stabilize negative charge buildup may represent an important, but heretofore unappreciated, strategy for RNA enzymes to confer charge stabilization, so long as such molecules are incapable of serving as phosphoryl group acceptors (e.g., the ATPase activity of hexokinase). In light of the evidence that the imino group of A38 exists as a positively charged H-bond donor to the leaving group at pH 7.0, additional electrostatic contributions at the nonbridging oxygens of G+1 by water appear to underscore the catalytic strategy of providing an active site that complements the negative charge of the transition state, while geometrically orienting the reacting groups. Indeed, the diminutive size of water molecules allows their use in crowded, compact active sites that cannot otherwise accommodate hydrated metal ions or additional nucleobases with positively polarized functional groups. Albeit, it is likely that coordinating water molecules in an intermolecular interaction is not as desirable or effective as positioning constituent functional groups (e.g., charged/polar amino acids in protein enzymes) in an intramolecular manner. However, for ribozymes that otherwise lack sufficiently charged groups, selective coordination of water and proper positioning of nucleobases may provide an alternative means to produce dense, local regions of charge complementarity that promote formation of the transition state.
The idea that water may stabilize negative charge buildup has been suggested for the tetrahedral intermediate associated with peptide bond formation in the ribosome (Schmeing et al. 2005). It is notable that a water molecule observed in the ribosome was oriented geometrically for electrostatic stabilization by two additional RNA groups, as observed for the two waters presented herein (Fig. 4G). Additionally, the 2′–5′ structure, refined to 2.35 Å, exhibited evidence of a very similar distribution of active site water molecules supporting their catalytic relevance (Fig. 4H). Examination of the H-bond donor/acceptor properties of the groups responsible for the tetrahedral coordination to W5 and W7 led to the proposed scheme depicted in Figure 4, F and G. In each case a hydrogen atom from water is oriented toward a nonbridging oxygen of the vanadate transition-state analog to enhance the charge neutralization of this electron-rich group. As such, each nonbridging oxygen interacts with two or three H-bond donors in a tetrahedral configuration. This is consistent with the expectation that each oxygen accumulates negative charge during the transition state to the extent that it exists transiently as an oxyanion with three lone pairs of electrons. By analogy to vanadate, W5 and W7 would geometrically and electrostatically stabilize the nonbridging oxygens of the scissile phosphate in the bona fide transition state; of course, these interactions would occur exclusively under conditions in which the nonbridging oxygens have adopted a “catalytic” orientation coincident with greater negative charge character. As a result of their small size, versatile H-bonding properties, and the comparatively large O–H bond polarity, the structural and electrostatic role for waters observed here may prove to be a common strategy employed by other members of this or other ribozyme families.
Significance of G8 conformational heterogeneity
The base of G8 was observed to occupy a mixture of syn (25%) and anti (75%) conformations in the VC structure. The functional significance of the syn conformation of G8, if any, has not been established. Intriguingly, a 7-deaza-guanosine substitution for G8 results in a ∼14-fold decrease in k cat and an approximately fivefold increase in KM in cleavage assays employing a similar three-stranded hairpin ribozyme (Grasby et al. 1995). This effect is difficult to rationalize considering the position of N7 in the anti conformation; however, assuming the vanadate moiety in the VC structure is representative of the transition state, the N7 position of the syn conformation could ostensibly support solvent-mediated contact(s) to the 2′-nucleophile and/or pro-S oxygen of the scissile phosphate. On the other hand, the anti conformation is strongly supported by compelling evidence that position N1 of G8 is involved in ribozyme chemistry (Thomas and Perrin 2006) and that mutating the exocyclic moiety at position 2 has significant effects on activity (Pinard et al. 2001). Furthermore, in the context of the anti conformation, the detrimental effects of a G8 7-deaza modification could arise from changes in the base-stacking properties or pKa of the base (Grasby et al. 1995), as evidenced by the observation that this mutation altered the reactivity of position N1 (Thomas and Perrin 2006). Considering that exchange between the syn and anti conformations of G8 in situ would require significant movements of the nearby bases, such conformational heterogeneity may arise during folding.
Importantly, no conformational heterogeneity at nucleobase G8 was observed in the 2′–5′ structure. A likely reason is that the 2′–5′ structure maintains a covalent phosphodiester bond between A-1 and G+1, unlike the PMC and VC structures. Therefore, removal of the scissile phosphate may lead to undesirable flexibility, as evidenced by the presence of syn conformation at G8 in both the VC and PMC structures. It is also interesting that the VC structure exhibited a higher prevalence of the anti conformation than the PMC structure, which may be due to the stabilizing presence of vanadate in place of the scissile phosphate. The persistence of even a small degree of syn conformation may in turn contribute to the incomplete occupancy of vanadate. Overall, the lack of conformational heterogeneity in the 2′–5′ structure represents another potential advantage of this linkage as a reaction-intermediate analog.
Despite the base heterogeneity at G8 observed in the VC and PMC structures, the ribose conformation at this position maintained a favorable 2′-endo pucker in all three structures. Furthermore, the syn conformation appears to have little effect on the local architecture of the active site, as indicted by the modest 0.48 Å rmsd between all atoms of the VC and precatalytic structures. It is likely that as the resolution of the minimal hairpin ribozyme continues to improve, evidence for additional conformational heterogeneity will become apparent, which was observed in prior studies of catalytic RNA molecules (Wedekind and McKay 2003; Alam et al. 2005).
Caveats for the use of vanadate
Vanadate naturally adopts a trigonal–bipyramidal conformation and inhibits the activity of a variety of enzymes. This propensity has been exploited in numerous studies of phosphoryl-transfer enzymes (for review, see Davies and Hol 2004) in which vanadate has been used to trap conformations pertinent to the transition state. The chemical properties of vanadate, along with its biological and inhibitory roles, have been studied extensively (for review, see Crans et al. 2004). Despite the general acceptance of vanadate as a mimic of the transition state of phosphoryl transfer reactions, subtle differences in its geometric and electronic character relative to phosphate warrant careful interpretation of enzyme–vanadate complexes. The most obvious concern is that vanadate coordination is often distorted from an ideal trigonal–bipyramid compared with the true phosphorane transition state of the SN2 catalyzed reaction (van Tol et al. 1990). In fact, the τ angle was determined to be either 144° or 153° in the previous study of a hairpin ribozyme–vanadate complex (Rupert et al. 2002) and 154° in the VC structure reported herein. The origin of these deviations may reside in the inability of vanadate to perfectly bind in an active site evolutionarily tuned to engage the phosphoryl transition state. Furthermore, the 0.46 Å ionic radius of vanadium(V) (Shannon and Prewitt 1969) is significantly smaller than the 1.1 Å covalent radius of phosphorus (Sanderson 1971). Collectively, the geometric discrepancies can be downplayed somewhat, but only in light of the corroborating biochemical information supporting the observed interactions between vanadate and key nucleobases, such as G8 and A38, that are likely to support transition state stabilization (Pinard et al. 2001; Lebruska et al. 2002; Rupert et al. 2002; Kuzmin et al. 2004, 2005; Thomas and Perrin 2006).
A second fundamental concern is whether vanadium can be considered an accurate electronic mimic of phosphorus. Supporting evidence for electronic similarities includes the similarity between vanadate and phosphate pKa values (Crans et al. 2004 and references therein), as well as their categorization as group V elements in the periodic table, which makes them likely to adopt Noble electronic configurations. Despite these similarities, theoretical calculations have determined that V–O bonds are significantly less polar than P–O bonds (Krauss and Basch 1992), which can affect the propensity of the axial, nonbridging oxygens to engage in hydrogen bonding or Coulombic interactions. For example, a difference in the electronic character of vanadate has been invoked to account for the disparity between interactions observed in an early structural study of RNase A in complex with vanadate versus those expected based on knowledge of the reaction mechanism. In this case study, unexpected interactions were observed between vanadate and the key active site residues Lys41 and His12 (Wlodawer et al. 1983). Specifically, these amino acids interacted with vanadate in a manner that was incongruous with mechanistic evidence supporting a concerted, imidazole-mediated acid/base mechanism in which Lys41 ameliorates negative charge buildup on the nonbridging oxygens. Later studies offered explanations citing the apparent plasticity of the “oxyvanadate framework” (Ladner et al. 1997) and fundamental differences in the vanadate electronics compared with phosphate (Messmore and Raines 2000). The salient and admonitory conclusion is that the bound vanadate was stabilized by the active site of RNase A, but through interactions irrelevant to a widely accepted view of the transition state. Distinguishing between artifacts and chemically relevant interactions may be difficult and underscores the need for supporting biochemical and functional evidence, as well as a strategy that includes the use of multiple reaction-intermediate analogs.
Aside from caveats regarding the structural interpretation of enzyme–vanadate complexes, there are technical considerations as well. The first is that removal of the scissile phosphate may lead to conformational heterogeneity/alterations of local active site groups, such as that observed for position G8 in the PMC and VC structures reported here. Similarly, vanadate may not bind with 100% occupancy, as documented here for VC and for some protein enzymes (PDB ID: 1M7G [Lansdon et al. 2002]) including RNase A (PDB ID: 1RUV [Ladner et al. 1997]). This may be due in part to the tendency of vanadate to polymerize in solution across the acidic and neutral pH range (Crans et al. 2004), thereby diminishing the molecular species that can bind in an enzyme active site. Second, vanadate may be susceptible as a transition metal to oxidation/reduction during X-ray diffraction experiments resulting from ionizing radiation. Although crystals are cryoprotected in modern X-ray diffraction studies, the deleterious effects of radiation have been noted in several recent studies that possess metal centers, even before the other more obvious effects of radiation damage are manifest (Grabolle et al. 2006; Ravelli and Garman 2006, and references therein). With respect to the hairpin ribozyme, this effect may have important implications as well. Small molecule structures indicate that reducing vanadium(V) to vanadium(IV) does not necessarily change the trigonal–bipyramidal geometry. However, coordination can be filled by alcohol rather than alkoxide moieties (CSD ID: PASCAN [Barr-David et al. 1992]), even at an apical position (CSD ID: ELEBOM [Nunes et al. 2003]). Therefore, the presumption that N1 of A38 is a protonated H-bond donor due to its close proximity to the 5′-O of G+1 (VC, this study; Rupert et al. 2002) assumes such reduction has not taken place. To provide insight into the likelihood of this reduction, a comparison of the one-electron reduction potential for the vanadium(V) ion with those of RNA nucleotides is warranted. The reduction potential for vanadium(V) to vanadium(IV) versus the normal hydrogen electrode is + 0.99 V (Slebodnick et al. 1997). While we are not aware of the purine one-electron reduction potentials measured at near-neutral pH, those for uridine and cytidine are ∼ − 1.2 V versus the normal hydrogen electrode under slightly alkaline conditions (Steenken et al. 1992). In a study employing polar, aprotic solvent conditions (i.e., a dielectric more representative of a solvent inaccessible active site), the one-electron reduction potentials were measured for guanine and adenine, and these were observed to be even more negative than those measured for pyrimidine nucleobases (Seidel et al. 1996). It is therefore plausible that reduction of vanadium(V) would be favored relative to RNA bases; however, the influence of the local environment complicates this aspect of radiation damage studies and precludes a conclusive determination at present. Nevertheless, this possibility has important implications in the case of the hairpin ribozyme regarding the protonation state of position N1 of A38. Therefore, in addition to corroborating biochemical evidence, it is desirable to employ a reaction-intermediate that is not as susceptible to such reduction effects in structural studies.
Benefits of a 2′,5′ covalent linkage
The 2′,5′-phosphodiester bond is not susceptible to the same transition-metal radiation damage that could present an ambiguity in the assigned protonation state of the G+1 O5′ as noted for vanadate. Significantly, the 2′–5′ structure also demonstrates that the 5′-O of G+1 is within 3.0 Å of the A38 N1 position (Fig. 4H,I). Since the covalent phosphodiester linkage precludes the existence of a 5′-OH at G+1, the results are consistent with the imino group of A38 serving as an H-bond donor to O5′ of G+1 under neutral pH conditions utilized in crystallization. Such a shift in the pKa of A38 toward neutral could be consistent with a general acid, as noted by Bevilacqua (2003) and Kuzmin et al. (2005). The use of the 2′–5′ substitution also circumvents another prohibitive technical aspect of vanadate for ribozyme studies, i.e., the need to remove the scissile phosphate between N-1 and N+1. In the hairpin ribozyme, extensive nucleobase interactions from the ribozyme stabilize the substrate/product strands 5′ and 3′ of the cleavage site. Fortunately, in this stabilizing context the generation of a PMC complex (Fig. 3A) as a preface to vanadate binding has little or no effect on the global hairpin ribozyme fold (Fig. 4A). Conversely, structures of the hepatitis δ virus (Ke et al. 2004) and metabolite-sensing glmS (Klein and Ferré-D'Amaré 2006) ribozymes lack analogous interactions that stabilize the 5′ portion of the substrate/product strand, thus making them more susceptible to dissociation from the ribozyme complex. Hence, without the benefit of three oxygen ligands donated by both 5′ and 3′ substrate/product fragments, vanadate is unlikely to coordinate productively in these active sites, thereby confounding high-resolution structural studies. Importantly, the cis diols of terminating nucleotides in the hairpin ribozyme were not observed to coordinate vanadate in the 4WJ complex (Rupert et al. 2002; Supplemental Data) or the VC reported here. Therefore, employing a covalent linkage as a reaction-intermediate analog represents a valuable strategy for studying ribozymes recalcitrant to the use of vanadate.
Although the 2′,5′-phosphodiester linkage elicits a stereochemical configuration of neighboring RNA functional groups similar to vanadate, the former structure cannot be considered a transition-state mimic because the scissile phosphorus equivalent exhibits tetrahedral geometry (Fig. 4H). As such, the most similar feature between the 2′–5′ and VC structures is the orientation of the nonbridging oxygens at the scissile phosphate (Fig. 4H). This similarity appears to be the result of reduced conformational freedom afforded to the 2′,5′-phosphodiester bond linkage relative to the normal 3′,5′-phosphodiester bond, which exhibits ∼160° of rotational freedom in the hairpin ribozyme active site (Salter et al. 2006). Thus, the 2′,5′ linkage is restricted in its ability to sample conformational space (by design) and adopts an orientation resembling the transition state. As stated, the favorability of this conformation is substantiated by the evidence of nearly the same ordered water molecules in both the VC and 2′–5′ structures (Fig. 4H). Taken together, we propose the 2′–5′ structure represents a more “precatalytic” state than prior 2′-O-methyl structures (e.g., PDB ID: 1ZFR [Salter et al. 2006]).
A supporting line of evidence for the “catalytic” conformation of the scissile bond in the present study relates to the location of W52 in the precatalytic structure (Salter et al. 2006). This position (Fig. 4A, open asterisk) is occupied by the pro-R oxygen of the scissile phosphate, as observed for both the VC (by analogy to vanadate) and 2′–5′ structures (Fig. 4H). It has been noted that protein enzymes often bind water molecules in their active sites at positions poised to receive electrostatic stabilization during phosphoryl-transfer (Pujadas and Palau 2001; Gil-Ortiz et al. 2003). As expected, W52 is stabilized in the precatalytic structure by W86 (equivalent to W5 in the current study), as well as the exocyclic amines of both A38 and A9, analogous to the stabilization of the pro-R oxygen in the VC and 2′–5′ structures. Taken together, these observations affirm that the hairpin ribozyme active site is predisposed to stabilize a “catalytic” orientation of the scissile bond through coordination of its nonbridging oxygens (Rupert et al. 2002). Conversely, when the scissile phosphate is not oriented properly, water molecules or the 2′-OH moiety of position −1 can mimic the nonbridging oxygens to engage in nonproductive ground-state stabilization (Salter et al. 2006).
The conclusion that the 2′,5′ linkage induces a bona fide precatalytic state is tempered by the observation that the hairpin ribozyme, unlike the hepatitis δ virus (Shih and Been 1999), cannot cleave a substrate comprising a 3′-OH with a 2′,5′ linkage (Feldstein et al. 1990). At this time, it is unclear whether the presence of the 3′ group would affect the conformation of the 2′–5′ structure described here. Nevertheless, it is significant that the 2′,5′-phosphodiester analog predicts similar stabilizing interactions observed in the vanadate complex while also avoiding the complications associated with removal of a covalent bond in the substrate strand.
Application of a 2′,5′-phosphodiester linkage to other small ribozymes
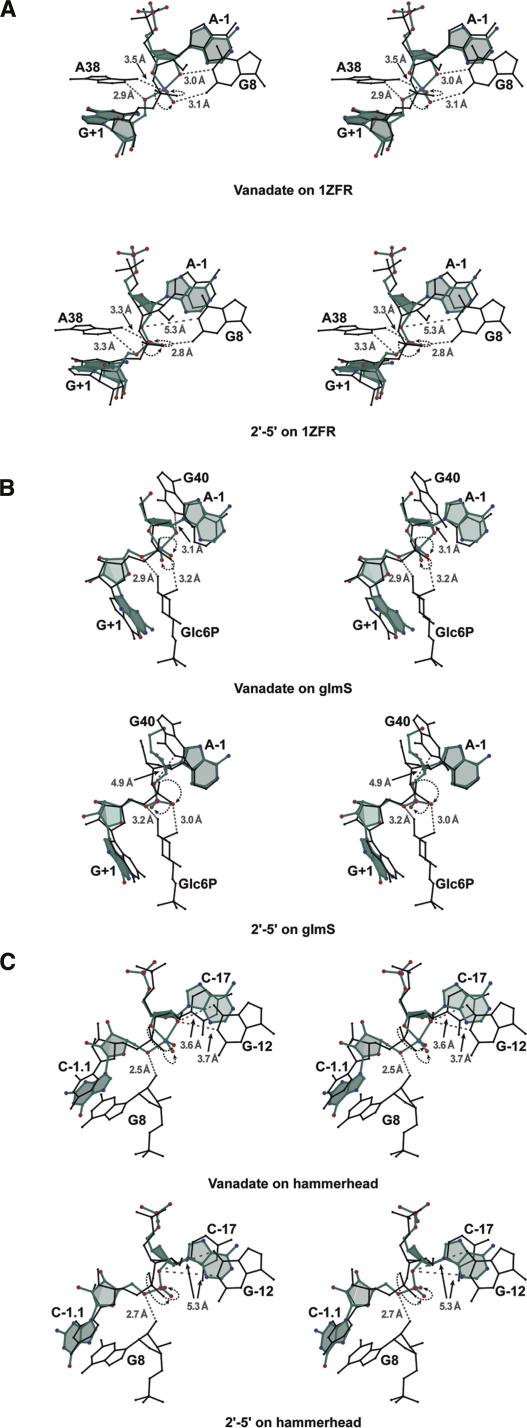
Members of the small ribozyme family catalyze the same chemical reaction as the hairpin ribozyme (Fig. 1A), albeit utilizing distinct strategies (Doudna and Lorsch 2005; Fedor and Williamson 2005; Bevilacqua and Yajima 2006). By analogy with the hairpin ribozyme, it may be concluded that the transition-state geometry cannot be identified solely on the basis of the in-line τ angle (discussed above). For small ribozymes that are refractory to the use of vanadate due to the absence of 5′-substrate stabilizing interactions, the 2′,5′-phosphodiester linkage offers an alternative means to achieve an “induced fit” of the active site to the reaction-intermediate analog, which may be observable by crystallographic analysis. To demonstrate this concept, the vanadate and 2′,5′ linkages were superimposed on a local level with precatalytic structures of the hairpin ribozyme (Salter et al. 2006) as well as the glmS (Klein and Ferré-D'Amaré 2006) and hammerhead ribozyme structures (Martick and Scott 2006) (Fig. 5). Overlays of the hairpin ribozyme (Fig. 5A) illustrate that the interactions predicted by this local superposition method corroborate the bond rotations and interactions necessary to achieve reaction-intermediate structures (Rupert et al. 2002; VC and 2′–5′ structures). For the metabolite-sensing glmS ribozyme (Fig. 5B), the overlays predict favorable interactions shared between the metabolite and nonbridging oxygen as well as 5′-O leaving groups, respectively. The amine group equivalent of the metabolite, which was mutated to oxygen to prevent cleavage in the crystal lattice, is situated notably close to the leaving group, while a vicinal hydroxyl serves to stabilize the pro-S nonbridging oxygen. The vanadate overlay also predicts an H-bond between the 2′-O attacking group of A-1 and G40, a nucleobase identified as potentially serving as a general base catalyst (Klein and Ferré-D'Amaré 2006). The equivalent interaction is somewhat more distant in the 2′,5′-phosphodiester overlay. For the hammerhead ribozyme, the 2′-OH of nucleobase G8 has been posited as a candidate general acid that protonates the 5′-O leaving group of C-1.1 (Martick and Scott 2006). In the vanadate overlay, this interaction is predicted to occur, but in the 2′,5′-phosphodiester overlay, this distance is slightly longer owing to the tetrahedral geometry of the electrophilic phosphorus. However, the nonbridging oxygens are positioned similarly compared with vanadate, and it is plausible that both analogs would form similar induced-fit interactions to stabilize their orientations. In both overlays, some of the distances predicted for the interactions are not ideal, which is undoubtedly due in part to a limitation of the superposition technique. Notably, nucleobase G12 has been identified as important for interacting with the 2′-OH attacking group (Martick and Scott 2006). However unlike the vanadate overlay, the 2′,5′ overlay does not predict this interaction.
FIGURE 5.
Reaction-intermediate analogs of this study overlaid on small ribozyme coordinates. (A) Precatalytic hairpin ribozyme (Salter et al. 2006), (B) glmS ribozyme in complex with glucose-6-phosphate inhibitor (Klein and Ferré-D'Amaré 2006), (C) tertiary-contact-stabilized hammerhead ribozyme (Martick and Scott 2006). Models of the vanadate (upper panel images) and 2′,5′-phosphodiester linkage (lower panel images) from this study are depicted as ball-and-stick models (colored), including the respective flanking A-1 and G+1 bases. (Dashed lines) Selected H-bond interactions predicted by the investigators in each study to support the transition state are shown in the context of the two reaction-intermediate linkages of this study; no phosphate was present at position A-1 in the glmS crystal structure; (arrows) direction of rotation about the scissile bond necessary for the nonbridging oxygens of the respective precatalytic structures to reach the “catalytic” positions predicted by the vanadate and 2′–5′ overlays.
Interestingly, the phosphate groups of the glmS and hammerhead ribozymes are predicted to undergo a rotation similar in magnitude and direction to achieve a “catalytic” orientation by analogy to the hairpin ribozyme. In all three structures, the pro-R oxygen is predicted to rotate back into the plane of the page while its former position is nearly accommodated by the pro-S oxygen (Fig. 5, dashed black arrows). It remains to be seen whether or not such stabilizing interactions, potentially the result of induced-fit distortion of the precatalytic conformation, could be identified in the context of the reaction-intermediate analogs as proposed here. Future high-resolution structures of these ribozymes in the presence of vanadate or the 3′-deoxy 2′–5′ linkage may also reveal significant interactions with solvent molecules and provide insight into transition-state stabilization as well as the putative proton-transfer pathways postulated (Klein and Ferré-D'Amaré 2006; Martick and Scott 2006).
SUMMARY AND CONCLUSIONS
The use of a minimal, junctionless hairpin ribozyme construct provided the first view of ordered water molecules bound in the active site of a small RNA enzyme in a precatalytic conformation. To determine how the location and distribution of water molecules changes on the reaction coordinate, this ribozyme was cocrystallized with vanadate using a hinged construct and refined to 2.05 Å resolution (VC). Intriguingly, two waters were observed near the nonbridging positions of the oxyphosphorane mimic in a manner that suggests a role for solvent atoms in electrostatic stabilization of the transition state. To independently confirm these observations, we solved an additional structure with a 3′-deoxy, 2′,5′-phosphodiester linkage at the cleavage site in place of the canonical 3′,5′ phosphodiester scissile bond and refined it to 2.35 Å (2′–5′). Comparison of the VC and 2′–5′ structures provided strong evidence that N1 of A38 is capable of serving as a protonated H-bond donor at physiological pH, and also revealed similar orientations of the atoms involved in phosphoryl-transfer including solvent molecules. Taken together, the findings extend our knowledge of the mechanism of the hairpin ribozyme, including water molecules. Finally, we conclude that these analogs represent valuable tools for structural studies aimed at further elucidating the mechanism of action of the hairpin ribozyme, as well as small RNA enzymes in general.
MATERIALS AND METHODS
Hairpin ribozyme constructs
Previous studies of the 61-nucleotide (nt), all-RNA minimal hairpin ribozyme utilized a junctionless variant requiring loop A and B domains to be annealed separately and then docked together to form the folded active site (Grum-Tokars et al. 2003; Alam et al. 2005; Salter et al. 2006). To maximize the prevalence of complete, folded ribozyme molecules, a hinged form of the ribozyme was developed (C.M. MacElrevey, R.C. Spitale, J. Krucinska, and J.E. Wedekind, unpubl.) and employed in constructs that lack the scissile phosphate. The “hinge” refers to a synthetic S9 linker (Fig. 2A) at position 14, a site occupied by adenine in the 4WJ sequence, which covalently joins the loop A and B domains. This modification reduced the complexity of the construct by decreasing the number of prerequisite intermolecular annealing events required to form the individual loop domains and docked active site.
The active sites of the three constructs described in this study incorporate different linkages at the scissile bond as mimics of either the product fragment complex, transition-state, or reaction-intermediate states and are referred to as the “PMC,” “VC,” or “2′–5′” constructs, respectively. In all structures, position 39 was mutated to a cytidine to prevent conformational heterogeneity in the S-turn observed for the native, U39 structure (Alam et al. 2005). The 29-mer (contains S9 linker or separate 12- and 17-mer strands if S9L is absent) and 19-mer ribozyme strands, as well as the 8-mer and 5-mer substrate strand fragments (Fig. 2A) were from Dharmacon. The 2′,5′-phosphodiester linkage at the scissile bond was created by incorporating a 5′-DMT, 3′-deoxy adenosine, 2′-cyanoethyl-diisopropyl phosphoramidite (Glen Research) into the 13-mer substrate strand (synthesized by the W.M. Keck Facility, Yale University). All RNA strands were subjected to in-house deprotection, HPLC purification, and domain docking as described previously (Wedekind and McKay 2000; Grum-Tokars et al. 2003).
Crystallization and X-ray diffraction experiments
Crystals of ribozyme constructs were obtained by the hanging-drop vapor diffusion method with screening around conditions established previously for the all-RNA 61-mer (Alam et al. 2005; Salter et al. 2006). Conditions common to all three constructs included: 250 mM Li2SO4, 2.5 mM Co(NH3)6Cl3, and 2 mM spermidine-HCl. Empirically formulated additions included: 18.5% PEG 2K MME, 100 mM sodium cacodylate (pH 6.0), 20 mM NaF, and 50 mM ascorbic acid (Sigma Aldrich) for the PMC construct; 20% PEG 2K MME, 100 mM sodium cacodylate (pH 6.0), and 4.5 mM NH4VO3 (Sigma Aldrich) for the VC construct; and 20.5% PEG 2K MME, 100 mM sodium HEPES (pH 7.0), and 15 mM nicotinic acid (Fluka) for the 2′–5′ construct. The ammonium metavanadate solution was prepared as a 125 mM stock in 10 mM sodium cacodylate pH 6.5, heated to 95°C with frequent vortexing until dissolved, and immediately added to the annealing step of ribozyme preparation. The solution was colored yellow, indicative of the vanadium(V) oxidation state (Crans et al. 2004). Crystals grew at 20°C to their maximum size over ∼2 wk: 0.21 mm × 0.21 mm × 0.21 mm (PMC crystal), 0.38 mm × 0.4 mm × 0.43 mm (VC crystal), and 0.08 mm × 0.1 mm × 0.25 mm (2′–5′ crystal). Cryoprotection was achieved by serial transfers of 3 min duration into synthetic mother liquor solutions containing 5%, 10%, 15%, or 17.5% (v/v) glycerol. Crystals were captured in thin loops (Hampton Research) and flash-frozen by plunging into N2 (l). The crystals were loaded into a cassette (Crystal Positioning Systems) and shipped to the Stanford Synchrotron Radiation Laboratory for remote data collection on the Stanford Automounting System at beamline 11–1 (PMC and 2′–5′, λ = 0.97945 Å) or 9–1 (VC, λ = 0.98789 Å); collection was performed in dose mode. Low-resolution passes were collected first: 500 mm detector distance, 0.5° per frame, and 1-sec exposures (PMC and 2′–5′ crystals) or 2-sec exposures (VC crystal). The high-resolution data were collected at a 300-mm detector distance with 0.5° per frame, and 10-sec (PMC and 2′–5′ crystals) or 20-sec exposures (VC crystal). A total of 60° (PMC and VC) or 62.5° (2′–5′) of data were collected for each low- or high-resolution pass. Data collection strategies were predicted using MOSFLM (Leslie 1992) and recorded using ADSC Quantum-315 CCD detectors. Intensity and data reduction statistics are provided in Table 1.
Data processing, structure determination, and refinement
X-ray diffraction data for each construct were processed with Crystal Clear (Rigaku-MSC; Pflugrath 1999); low- and high-resolution passes were integrated separately and merged together during scaling. Intensity and data reduction statistics are shown in Table 1. Each construct crystallized in space group P6122 with a single molecule per asymmetric unit as observed previously for the minimal, junctionless all-RNA 61-mer construct (Alam et al. 2005; Salter et al. 2006). Preliminary structures were determined by difference Fourier methods followed by rigid body refinement at 3.0 Å resolution as implemented in CNS (Brünger et al. 1998). The initial coordinates were derived from the all-RNA hairpin ribozyme refined previously at 2.05 Å resolution (PDB ID: 1ZFR) (Salter et al. 2006). Rigid body refinement was first applied to the entire structure followed by independent treatment of the respective loop A and B domains. Subsequent refinements to resolutions of 2.25 Å (PMC), 2.05 Å (VC), and 2.35 Å (2′–5′) were achieved through cycles of positional minimization and individual B-factor refinement utilizing a maximum-likelihood target. Manual adjustments to the models were carried out with the interactive graphics program O (Jones et al. 1991). Occupancies of the anti and syn G8 base conformations as well as vanadate in the active site of the VC structure were adjusted to minimize the R factor and R free statistics, and differences between the refined B factors were compared with nearby atoms. Simulated annealing omit maps were employed to guide manual refinement and reduce model bias. All structures exhibited alternate conformations for position U-5 that engages in a (designed) U–U pair (C.M. MacElrevey, R.C. Spitale, J. Krucinska, and J.E. Wedekind, unpubl.). RMSD values were calculated using LSQKAB as part of the CCP4 Suite (Kabsch 1976; CCP4 1994).
Assignment and analysis methods for waters and bound ions
Oxygen atoms representing waters were added manually to each structure. Water molecules were assigned to electron density peaks that met the following criteria: (1) the peak was located between 2.6 Å and 3.4 Å of at least one suitable H-bond donor or acceptor; (2) putative H-bond partners conformed to the tetrahedral coordination expected for water molecules; and (3) electron density was exhibited in σA-weighted (reduced-bias) electron density maps with coefficients 2m|Fo| − D|Fc| and m|Fo| − D|Fc| at levels of ≥0.9σ and ≥3.0σ, respectively. Solvent molecules have atomic B factors representative of neighboring RNA and/or solvent atoms. See Table 1 for average RNA and solvent B factors.
Ions were ruled out as occupying the sites assigned to water molecules. While Li+ was present in the crystallization medium at a concentration of 500 mM with a SO4 2− counterion, it was deemed an unlikely alternative due to its electron deficiency. Sulfate (250 mM) and cobalt hexamine (2.5 mM) ions were distinguished from waters by their greater electron density and larger van der Waals radii. Co(NH3)6Cl3 is also relatively inert to ligand exchange, thus preventing direct inner sphere Co3+ coordination with RNA atoms (Fedor 2002b). The most likely ionic candidate to occupy sites assigned as waters is Na+ because it is isoelectronic with water and present in the crystallization medium at concentrations of 100 mM (VC and 2′–5′) or 140 mM (PMC). However, Na+ ions prefer negatively charged ligands such as oxygen, octahedral coordination geometry (although other coordination numbers are tolerated), and more commonly interacts at distances shorter than that expected for water molecules (Harding 2002). The interactions observed for the assigned waters of the active site are consistent with tetrahedral coordination to both H-bond donors and acceptors. Furthermore, the coordination and location of several waters in the active site match those identified previously under conditions of very low sodium concentration (Salter et al. 2006). A single Na+ ion was assigned in the PMC structure in apparent coordination to the cis-diols of A-1. The interaction angles, negative character of the 2′ and 3′ hydroxyls, and interatomic distances were more consistent with those expected for Na+ ions versus a water molecule. A similar interaction between sodium ions and cis-diols was observed in a small molecule structure deposited in the Cambridge Crystallographic Database (CSD ID: WOVLUO [Park and Lee 2001]).
In the three structures reported in this study, 2 Co(NH3)6 3+ ions and a single SO4 2− ion occupied sites identified previously (Alam et al. 2005; Salter et al. 2006). The coordination geometry of these ions is distinct from water molecules, and the previous observations corroborated their respective assignments. Attempts to model these positions as waters resulted in local positive electron density peaks in m|Fo| − D|Fc| electron density maps, suggesting their van der Waals radii were insufficient to accommodate the electron density.
Modeling vanadate in the active site
The bond lengths and angles defining the vanadate moiety were obtained from a well-refined small-molecule structure deposited in the Cambridge Crystallographic Database (CSD ID: IZITEQ [Ouellette et al. 2004]); a supporting structure was also found (CSD ID: NONLEH [Salta and Zubieta 1997]). The energy restraint imposed upon the τ angle (165.5° in the IZITEQ structural template) of the VC complex was reduced to reflect the uncertainty of this parameter in the context of the active site of the hairpin ribozyme. The bond distances between vanadate and the nonbridging oxygens were both set to 1.62 Å, the average of the two distances observed in the Cambridge template, despite evidence from neutron diffraction experiments that one oxygen may exist with a hydrogen bound in the context of vanadate present in the active site of RNase A (Wlodawer et al. 1983). The vanadate occupancy (q) was adjusted from 100% to minimize the values of R work and R free, respectively. The final q-value of 50% also eliminated the negative electron density peak for the vanadate moiety observed in σA-weighted m|F o| − D|F c| electron density maps.
Modeling C2′-endo conformations
Modeling C2′-endo conformations for select nucleotide riboses was performed to account for residual electron density features that persisted after refinement with C3′-endo (A-form) puckers. Features that were considered to support modeling a 2′-endo conformation include: (1) accounting for positive and negative electron density peaks situated adjacent to the 2′-OH; (2) counterbalancing electron density peaks situated along the C4-C3 bond; (3) achieving favorable interactions accessible only to the 2′-endo conformation; and (4) removal of clashes/unfavorable interactions arising from the 3′-endo conformation. Coordinates for the refined structures were deposited in the RCSB Protein Data Bank with accession codes 2P7D, 2P7E, and 2P7F.
Modeling vanadate and 2′,5′-phosphodiester overlays
The A-1 and G+1 nucleotide coordinates, along with either the vanadate (Fig. 5, upper panels) or 2′,5′-phosphodiester linkage (Fig. 5, lower panels), were overlaid on the active sites of the hairpin (PDB ID: 1ZFR [Salter et al. 2006]), glmS (PDB ID: 2H0Z [Klein and Ferré-D'Amaré 2006]), and hammerhead (PDB ID: 2GOZ [Martick and Scott 2006]) ribozyme “precatalytic” structures. The overlay of the modeled −1 and +1 nucleotides was optimized primarily through manual adjustment of the dihedral angles of the vanadate or 2′,5′ linkage. The N9–C1′ dihedral angle was also changed for G+1 of the vanadate and 2′,5′-phosphodiester overlays to optimize the fit with the equivalent bases of the glmS and hammerhead crystal structures; a slight adjustment of the 2′–5′ ribose was performed as well to fit the respective crystal structures.
SUPPLEMENTAL DATA
The Cambridge Crystallographic Data Center contains the supplementary crystallographic data for the small molecule structures referenced in this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk/data_request/cif.
ACKNOWLEDGMENTS
We thank Celeste MacElrevey, who contributed valuable comments during refinement and the preparation of this manuscript. We thank Professor Edward Snell for generous assistance in the remote collection of diffraction data at the Stanford Synchrotron Radiation Laboratory (SSRL) and for insightful discussions. Professor William Bernhard and Dr. Elspeth Garman added significantly to the discussion regarding the effects of ionizing radiation. We also thank Hao Zhang and Weiye Wang for purifying the RNA strand containing the 2′,5′-phosphodiester linkage, as well as Rob Spitale, Jason Salter, and Krystle Williams for providing feedback on the manuscript. We acknowledge the staff at SSRL for technical support. This work was support by grants to J.E.W. from the NIH/NIGMS R01 GM63162 and the Petroleum Research Fund 45534-AC 4. A.T.T. was supported by an Elon Huntington-Hooker Predoctoral Fellowship in Chemistry and through support from the Biophysics and Structural Biology graduate program. Portions of this research were carried out at SSRL, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Footnotes
Abbreviations: PMC, product mimic complex; VC, vanadate complex; H-bond, hydrogen bond.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.510807.
REFERENCES
- Alam, S., Grum-Tokars, V., Krucinska, J., Kundracik, M.L., Wedekind, J.E. Conformational heterogeneity at position U37 of an all-RNA hairpin ribozyme with implications for metal binding and the catalytic structure of the S-turn. Biochemistry. 2005;44:14396–14408. doi: 10.1021/bi051550i. [DOI] [PubMed] [Google Scholar]
- Allen, F.H. The Cambridge Structural Database: A quarter of a million crystal structures and rising. Acta Crystallogr. B. 2002;58:380–388. doi: 10.1107/s0108768102003890. [DOI] [PubMed] [Google Scholar]
- Barr-David, G., Hambley, T.W., Irwin, J.A., Judd, R.J., Lay, P.A., Martin, B.D., Bramley, R., Dixon, N.E., Hendry, P. Supression by vanadium(IV) of chromium(V)-mediated DNA cleavage and chromium(VI/V)-induced mutagenesis. Synthesis and crystal structure of the vanadium(IV) complex (NH4) [V(O){HOC(Et)2COO}{OC(Et)2COO}] Inorg. Chem. 1992;31:4906–4908. [Google Scholar]
- Baxter, N.J., Olguin, L.F., Golicnik, M., Feng, G., Hounslow, A.M., Bermel, W., Blackburn, G.M., Hollfelder, F., Waltho, J.P., Williams, N.H. A Trojan horse transition state analogue generated by MgF3 formation in an enzyme active site. Proc. Natl. Acad. Sci. 2006;103:14732–14737. doi: 10.1073/pnas.0604448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H.J.C., Grigera, J.R., Straatsma, T.P. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- Bevilacqua, P.C. Mechanistic considerations for general acid-base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Yajima, R. Nucleobase catalysis in ribozyme mechanism. Curr. Opin. Chem. Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Borman, S., Wolfenden, R. Enzyme catalysis: Transition-state theory has venerable history. Chem. Eng. News. 2004;82:35–39. [Google Scholar]
- Brown, R.S., Hingerty, B.E., Dewan, J.C., Klug, A. Pb(II)-catalysed cleavage of the sugar–phosphate backbone of yeast tRNAPhe—Implications for lead toxicity and self-splicing RNA. Nature. 1983;303:543–546. doi: 10.1038/303543a0. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cai, Z., Tinoco I., Jr Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (CCP4) The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chowrira, B.M., Berzal-Herranz, A., Keller, C.F., Burke, J.M. Four ribose 2′-hydroxyl groups essential for catalytic function of the hairpin ribozyme. J. Biol. Chem. 1993;268:19458–19462. [PubMed] [Google Scholar]
- Crans, D.C., Smee, J.J., Gaidamauskas, E., Yang, L. The chemistry and biochemistry of vanadium and the biological activities exerted by vanadium compounds. Chem. Rev. 2004;104:849–902. doi: 10.1021/cr020607t. [DOI] [PubMed] [Google Scholar]
- Das, S.R., Piccirilli, J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Davies, D.R., Hol, W.G. The power of vanadate in crystallographic investigations of phosphoryl transfer enzymes. FEBS Lett. 2004;577:315–321. doi: 10.1016/j.febslet.2004.10.022. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific LLC; 2004. [Google Scholar]
- Dennis, E.A., Westheimer, F.H. The geometry of the transition state in the hydrolysis of phosphate esters. J. Am. Chem. Soc. 1966;88:3432–3433. doi: 10.1021/ja00966a046. [DOI] [PubMed] [Google Scholar]
- DeYoung, M., Siwkowski, A.M., Lian, Y., Hampel, A. Catalytic properties of hairpin ribozymes derived from Chicory yellow mottle virus and arabis mosaic virus satellite RNAs. Biochemistry. 1995;34:15785–15791. doi: 10.1021/bi00048a024. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Doudna, J.A. Ribozyme structures and mechanisms. Annu. Rev. Biophys. Biomol. Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A., Lorsch, J.R. Ribozyme catalysis: Not different, just worse. Nat. Struct. Mol. Biol. 2005;12:395–402. doi: 10.1038/nsmb932. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. The catalytic mechanism of the hairpin ribozyme. Biochem. Soc. Trans. 2002a;30:1109–1115. doi: 10.1042/bst0301109. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J. The role of metal ions in RNA catalysis. Curr. Opin. Struct. Biol. 2002b;12:289–295. doi: 10.1016/s0959-440x(02)00324-x. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J., Williamson, J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Feldstein, P.A., Buzayan, J.M., van Tol, H., deBear, J., Gough, G.R., Gilham, P.T., Bruening, G. Specific association between an endoribonucleolytic sequence from a satellite RNA and a substrate analogue containing a 2′-5′ phosphodiester. Proc. Natl. Acad. Sci. 1990;87:2623–2627. doi: 10.1073/pnas.87.7.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré, A.R. The hairpin ribozyme. Biopolymers. 2004;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- Fisher, A.J., Smith, C.A., Thoden, J.B., Smith, R., Sutoh, K., Holden, H.M., Rayment, I. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- Gil-Ortiz, F., Ramon-Maiques, S., Fita, I., Rubio, V. The course of phosphorus in the reaction of N-acetyl-L-glutamate kinase, determined from the structures of crystalline complexes, including a complex with an AlF(4)(−) transition state mimic. J. Mol. Biol. 2003;331:231–244. doi: 10.1016/s0022-2836(03)00716-2. [DOI] [PubMed] [Google Scholar]
- Grabolle, M., Haumann, M., Muller, C., Liebisch, P., Dau, H. Rapid loss of structural motifs in the manganese complex of oxygenic photosynthesis by X-ray irradiation at 10–300 K. J. Biol. Chem. 2006;281:4580–4588. doi: 10.1074/jbc.M509724200. [DOI] [PubMed] [Google Scholar]
- Grasby, J.A., Mersmann, K., Singh, M., Gait, M.J. Purine functional groups in essential residues of the hairpin ribozyme required for catalytic cleavage of RNA. Biochemistry. 1995;34:4068–4076. doi: 10.1021/bi00012a025. [DOI] [PubMed] [Google Scholar]
- Grum-Tokars, V., Milovanovic, M., Wedekind, J.E. Crystallization and X-ray diffraction analysis of an all-RNA U39C mutant of the minimal hairpin ribozyme. Acta Crystallogr. D Biol. Crystallogr. 2003;59:142–145. doi: 10.1107/s0907444902019066. [DOI] [PubMed] [Google Scholar]
- Hampel, K.J., Walter, N.G., Burke, J.M. The solvent-protected core of the hairpin ribozyme–substrate complex. Biochemistry. 1998;37:14672–14682. doi: 10.1021/bi981083n. [DOI] [PubMed] [Google Scholar]
- Harding, M.M. Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Crystallogr. D Biol. Crystallogr. 2002;58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- Hiratake, J. Enzyme inhibitors as chemical tools to study enzyme catalysis: Rational design, synthesis, and applications. Chem. Rec. 2005;5:209–228. doi: 10.1002/tcr.20045. [DOI] [PubMed] [Google Scholar]
- Holtz, K.M., Stec, B., Kantrowitz, E.R. A model of the transition state in the alkaline phosphatase reaction. J. Biol. Chem. 1999;274:8351–8354. doi: 10.1074/jbc.274.13.8351. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar, C., Madura, J.D., Impey, R.W., Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Jovine, L. Nuccyl: Nucleic acid add-on to Pymol: 2003 www.biosci.ki.se/groups/ljo/software/nuccyl.html
- Kabsch, W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. 1976;A32:922–923. [Google Scholar]
- Kakuta, Y., Petrotchenko, E.V., Pedersen, L.C., Negishi, M. The sulfuryl transfer mechanism. Crystal structure of a vanadate complex of estrogen sulfotransferase and mutational analysis. J. Biol. Chem. 1998;273:27325–27330. doi: 10.1074/jbc.273.42.27325. [DOI] [PubMed] [Google Scholar]
- Ke, A., Zhou, K., Ding, F., Cate, J.H., Doudna, J.A. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Ferré-D'Amaré, A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Jones, T.A. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J., Zou, J.Y., Kjeldgaard, M., Jones, T.A. Around O. In: Rossman M.G., Arnold E., editors. International tables for crystallography, vol. F. Springer and International Union of Crystallography; Berlin: 2001. pp. 353–356.pp. 366–367. [Google Scholar]
- Knowles, J.R. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- Knowles, J.R. Enzyme catalysis: Not different, just better. Nature. 1991;350:121–124. doi: 10.1038/350121a0. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Krauss, M., Basch, H. Is the vanadate anion an analogue of the transition state of RNAse A? J. Am. Chem. Soc. 1992;114:3630–3634. [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Fedor, M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Cottrell, J.W., Fedor, M.J. Role of an active site adenine in hairpin ribozyme catalysis. J. Mol. Biol. 2005;349:989–1010. doi: 10.1016/j.jmb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Ladner, J.E., Wladkowski, B.D., Svensson, L.A., Sjolin, L., Gilliland, G.L. X-ray structure of a ribonuclease A-uridine vanadate complex at 1.3 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 1997;53:290–301. doi: 10.1107/S090744499601582X. [DOI] [PubMed] [Google Scholar]
- Lansdon, E.B., Segel, I.H., Fisher, A.J. Ligand-induced structural changes in adenosine 5′-phosphosulfate kinase from Penicillium chrysogenum . Biochemistry. 2002;41:13672–13680. doi: 10.1021/bi026556b. [DOI] [PubMed] [Google Scholar]
- Le Du, M.H., Lamoure, C., Muller, B.H., Bulgakov, O.V., Lajeunesse, E., Menez, A., Boulain, J.C. Artificial evolution of an enzyme active site: Structural studies of three highly active mutants of Escherichia coli alkaline phosphatase. J. Mol. Biol. 2002;316:941–953. doi: 10.1006/jmbi.2001.5384. [DOI] [PubMed] [Google Scholar]
- Lebruska, L.L., Kuzmine, I.I., Fedor, M.J. Rescue of an abasic hairpin ribozyme by cationic nucleobases: Evidence for a novel mechanism of RNA catalysis. Chem. Biol. 2002;9:465–473. doi: 10.1016/s1074-5521(02)00130-8. [DOI] [PubMed] [Google Scholar]
- Leslie, A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB newsletter on protein crystallography 26 1992 [Google Scholar]
- Lienhard, G.E. Enzymatic catalysis and transition-state theory. Science. 1973;180:149–154. doi: 10.1126/science.180.4082.149. [DOI] [PubMed] [Google Scholar]
- Lindqvist, Y., Schneider, G., Vihko, P. Crystal structures of rat acid phosphatase complexed with the transition-state analogs vanadate and molybdate. Implications for the reaction mechanism. Eur. J. Biochem. 1994;221:139–142. doi: 10.1111/j.1432-1033.1994.tb18722.x. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D., Jr, Wiórkiewicz-Kuczera, J., Karplus, M. An all-atom empirical energy function for the simulation of nucleic acids. J. Am. Chem. Soc. 1995;117:11946–11975. [Google Scholar]
- MacKerell A.D., Jr, Bashford, D., Bellott, M., Dunbrack R.L., Jr, Evanseck, J.D., Field, M.J., Fischer, S., Gao, J., Guo, H., Ha, S., et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay, D.B. Structure and function of the hammerhead ribozyme: An unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- Merritt, E.A., Bacon, D.J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Messmore, J.M., Raines, R.T. Pentavalent organo-vanadates as transition state analogues for phosphoryl transfer reactions. J. Am. Chem. Soc. 2000;122:9911–9916. doi: 10.1021/ja0021058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.A., Uhlenbeck, O.C. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Nunes, G.G., Friedermann, G.R., Herbst, M.H., Barthem, R.B., Vugman, N.V., Barclay, J.E., Evans, D.J., Hitchcock, P.B., Leigh, G.J., Sa, E.L., et al. The first hetero-binuclear alkoxide of iron and vanadium: Structural and spectroscopic features. Inorg. Chem. Commun. 2003;6:1278–1281. [Google Scholar]
- Ouellette, W., Koo, B.K., Burkholder, E., Golub, V., O'Connor, C.J., Zubieta, J. Solid state coordination chemistry: Structural consequences of variations in tether length in the oxovanadium-copper-bisterpy-[O3P(CH2)nPO3]4- system, n = 1-6 (bisterpy = 2,2′:4′,4′′:2′′,2′′′-quarterpyridyl-6′,6′′-di-2-pyridine) Dalton Trans. 2004;10:1527–1538. doi: 10.1039/b401669f. [DOI] [PubMed] [Google Scholar]
- Pannu, N.S., Read, R.J. Improved structure refinement through maximum liklihood. Acta Crystallogr. A. 1996;A52:659–668. [Google Scholar]
- Park, Y.J., Lee, B.H. Sodium α-glucoheptonate dihydrate. Acta Crystallogr. C. 2001;57:12–13. doi: 10.1107/s010827010001355x. [DOI] [PubMed] [Google Scholar]
- Park, H., Lee, S. Role of solvent dynamics in stabilizing the transition state of RNA hydrolysis by hairpin ribozyme. J. Chem. Theory Comput. 2006;2:858–862. doi: 10.1021/ct0503015. [DOI] [PubMed] [Google Scholar]
- Perez-Ruiz, M., Barroso-DelJesus, A., Berzal-Herranz, A. Specificity of the hairpin ribozyme. Sequence requirements surrounding the cleavage site. J. Biol. Chem. 1999;274:29376–29380. doi: 10.1074/jbc.274.41.29376. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Wadkins, T.S., Been, M.D. Chemical rescue, multiple ionizable groups, and general acid–base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath, J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Pinard, R., Hampel, K.J., Heckman, J.E., Lambert, D., Chan, P.A., Major, F., Burke, J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas, G., Palau, J. Molecular mimicry of substrate oxygen atoms by water molecules in the β-amylase active site. Protein Sci. 2001;10:1645–1657. doi: 10.1110/ps.8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravelli, R.B., Garman, E.F. Radiation damage in macromolecular cryocrystallography. Curr. Opin. Struct. Biol. 2006;16:624–629. doi: 10.1016/j.sbi.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Rhodes, M.M., Reblova, K., Sponer, J., Walter, N.G. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc. Natl. Acad. Sci. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda, D., Bokinsky, G., Rhodes, M.M., Rust, M.J., Zhuang, X., Walter, N.G. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert, P.B., Ferré-D'Amaré, A.R. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B., Massey, A.P., Sigurdsson, S.T., Ferré-D'Amaré, A.R. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Strobel, S.A. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer; New York: 1984. [Google Scholar]
- Salta, J., Zubieta, J. Studies of the oxovanadium–organophosphonate system: Hydrothermal synthesis and crystal structure of the mixed valence cluster [V5O9(PhPO3)3(PhPO3H)2]2- and a comparison to the structure of the fully oxidized parent cluster [V5O7(OCH3)2(PhPO3)5]1- . J. Cluster Sci. 1997;8:361. [Google Scholar]
- Salter, J., Krucinska, J., Alam, S., Grum-Tokars, V., Wedekind, J.E. Water in the active site of an all-RNA hairpin ribozyme and effects of Gua8 base variants on the geometry of phosphoryl transfer. Biochemistry. 2006;45:686–700. doi: 10.1021/bi051887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson, R.T. Chemical bonds and bond energy. Academic Press; New York: 1971. [Google Scholar]
- Schmeing, T.M., Huang, K.S., Kitchen, D.E., Strobel, S.A., Steitz, T.A. Structural insights into the roles of water and the 2′ hydroxyl of the P-site tRNA in the peptidyl transferase reaction. Mol. Cell. 2005;20:437–448. doi: 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Seidel, C.A.M., Schulz, A., Sauer, M.H.M. Nucleobase-specific quenching of fluorescent dyes. 1. Nucleobase one-electron redox potentials and their correlation with static and dynamic quenching efficiencies. J. Phys. Chem. 1996;100:5541–5553. [Google Scholar]
- Seyhan, A.A., Burke, J.M. Mg2+-independent hairpin ribozyme catalysis in hydrated RNA films. RNA. 2000;6:189–198. doi: 10.1017/s1355838200991441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, R.D., Prewitt, C.T. Effective ionic radii in oxides and fluorides. Acta Crystallogr. B. 1969;25:925–946. [Google Scholar]
- Shih, I.H., Been, M.D. Ribozyme cleavage of a 2,5-phosphodiester linkage: Mechanism and a restricted divalent metal-ion requirement. RNA. 1999;5:1140–1148. doi: 10.1017/s1355838299990763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebodnick, C., Hamstra, B.J., Pecoraro, V.L. Modeling the biological chemistry of vanadium: Structural and reactivity studies elucidating biological function. In: Allen H., et al., editors. Structure and bonding. Springer; Berlin: 1997. pp. 51–108. [Google Scholar]
- Smith, C.A., Rayment, I. X-ray structure of the magnesium(II).ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- Steenken, S., Telo, J.P., Novais, H.M., Candeias, L.P. One-electron-reduction potentials of pyrimidine bases, nucleosides, and nucleotides in aqueous solution. Consequences for DNA redox chemistry. J. Am. Chem. Soc. 1992;114:4701–4708. [Google Scholar]
- Thomas, J.M., Perrin, D.M. Active site labeling of G8 in the hairpin ribozyme: Implications for structure and mechanism. J. Am. Chem. Soc. 2006;128:16540–16545. doi: 10.1021/ja063942y. [DOI] [PubMed] [Google Scholar]
- van Tol, H., Buzayan, J.M., Feldstein, P.A., Eckstein, F., Bruening, G. Two autolytic processing reactions of a satellite RNA proceed with inversion of configuration. Nucleic Acids Res. 1990;18:1971–1975. doi: 10.1093/nar/18.8.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind, J.E., McKay, D.B. Purification, crystallization, and X-ray diffraction analysis of small ribozymes. Methods Enzymol. 2000;317:149–168. doi: 10.1016/s0076-6879(00)17013-2. [DOI] [PubMed] [Google Scholar]
- Wedekind, J.E., McKay, D.B. Crystal structure of the leadzyme at 1.8 Å resolution: Metal ion binding and the implications for catalytic mechanism and allo site ion regulation. Biochemistry. 2003;42:9554–9563. doi: 10.1021/bi0300783. [DOI] [PubMed] [Google Scholar]
- Wilson, T.J., Ouellet, J., Zhao, Z.Y., Harusawa, S., Araki, L., Kurihara, T., Lilley, D.M. Nucleobase catalysis in the hairpin ribozyme. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodawer, A., Miller, M., Sjolin, L. Active site of RNase: Neutron diffraction study of a complex with uridine vanadate, a transition-state analog. Proc. Natl. Acad. Sci. 1983;80:3628–3631. doi: 10.1073/pnas.80.12.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.W., Morera, S., Janin, J., Cherfils, J. AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP. Proc. Natl. Acad. Sci. 1997;94:3579–3583. doi: 10.1073/pnas.94.8.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Jossinet, F., Leontis, N., Chen, L., Westbrook, J., Berman, H., Westhof, E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]