Abstract
Tumors must adapt to the hypoxic environment in order to grow beyond a benign microscopic mass. In addition to transcriptional activation mediated by HIF-1α, hypoxia has also been reported to inhibit translation. The degree of translational inhibition is dependent on the duration as well as the severity of the hypoxic insult. Anoxia (<0.02% O2) seems to have a more rapid and dramatic effect on translation as compared to hypoxia. We show here that prolonged hypoxia dramatically and reversibly inhibits translation in PC-3 cells. We also found that mTOR is inactivated and eIF-2α is phosphorylated during hypoxic treatment but only the eIF-2α phosphorylation correlates with the translational repression. We further used polysome analysis and microarray technology to analyze the impact of this translational repression on gene expression. We found that 33 mRNAs were refractory to this translational repression and that there was no correlation between mRNA induction and the ability to recruit ribosomes during hypoxia. We also found that ribosomal protein encoding mRNAs are more sensitive to this translational repression as compared to the majority of mRNAs. Although other reports have analyzed the effect of translation inhibition on gene expression under anoxic conditions, we believe that this is the first report in hypoxic cells. Our results show that the translational repression that occurs during hypoxia does impact gene expression in the highly transformed prostate cancer cell line, PC-3.
Keywords: hypoxia, polysomes, translation, gene expression, HIF-1α
INTRODUCTION
Hypoxia is a term used to describe cellular environments in which oxygen levels are below that which is normally found in healthy tissues. Cellular oxygen environments above 0.02% oxygen but below 3% are considered hypoxic, while environments devoid of oxygen (<0.02% O2) are considered anoxic. Due to the aberrant nature of the tumor vasculature, established tumors normally have areas of varying degrees of hypoxia and often the center of a tumor is anoxic (Semenza 2002; Dvorak 2003; Kizaka-Kondoh et al. 2003; Brown and Wilson 2004). Although hypoxia is often used to describe anoxia, these are two distinct conditions that can elicit different cellular responses.
Tumors must adapt to the hypoxic environment, for example, by inducing angiogenesis and/or blocking apoptosis, in order to develop beyond a microscopic benign mass (Gimbrone et al. 1972; Vaupel et al. 1989; Parangi et al. 1996; Brown and Giaccia 1998; Bergers and Benjamin 2003). To adapt to the hypoxic environment, the cell induces the Hypoxia Inducible Factor-1, HIF-1, which is the primary transcription factor involved with the transcriptional activation that occurs during hypoxia (Huang et al. 1998; Semenza 2001, 2002, 2003; Sonna et al. 2003). Although not as extensively studied as the transcriptional effects, hypoxia has also been reported to have a significant impact on translation.
Metabolic labeling experiments have shown that oxygen deprivation inhibits translation in a wide variety of cell types under both hypoxic and anoxic conditions (Kraggerud et al. 1995; Stein et al. 1998; Tinton and Buc-Calderon 1999; Koumenis et al. 2002; Lang et al. 2002; Liu and Simon 2004; Wouters et al. 2005; Koritzinsky et al. 2006; Liu et al. 2006). Interestingly, the kinetics of this translation inhibition in tissue culture cells differs between these two conditions. Anoxia has been shown to have an immediate effect on translation, inhibiting it by 40%–50% within 4 h of exposure (Koumenis et al. 2002; Bi et al. 2005; Blais et al. 2006; Koritzinsky et al. 2006). In contrast, hypoxia appears to require prolonged exposure (>16 h) and inhibits translation by 30%–50% in most cell lines tested (Connolly et al. 2006; Liu et al. 2006).
The molecular mechanism responsible for the shutdown in translation during hypoxia and anoxia is not completely understood but appears to involve the repression of cap-dependent translation (Pain 1996; Kozak 1999; Arsham et al. 2003; Liu and Simon 2004; Merrick 2004). Hypoxia and anoxia appear to inhibit cap-dependent translation initiation, in part, through modulating the activity of two kinases, mammalian target of rapamycin (mTOR) and PERK (Koumenis et al. 2002; Arsham et al. 2003; Blais et al. 2006; Koumenis and Wouters 2006; van den Beucken et al. 2006).
The mTOR kinase is a major regulator of translation in response to stress and nutrient deprivation and affects both global translation and the translation of mRNAs containing 5′- terminal oligopyrimidine tracts (5′-TOPs) (Gingras et al. 2001, 2004; Hay and Sonenberg 2004). When mTOR is inhibited, the 4E-binding proteins (4E-BPs) become hypophosphorylated, which increases their affinity for eIF-4E and inhibits cap-dependent translation by sequestering eIF-4E (Richter and Sonenberg 2005). Inactivation of mTOR also results in the specific translational repression of 5′-TOP-containing mRNAs (Gingras et al. 2004; Hay and Sonenberg 2004; Wouters et al. 2005) through phosphorylation of p70S6K (Gingras et al. 2004; Hay and Sonenberg 2004; Wouters et al. 2005). A major class of 5′-TOP-containing mRNAs is the mRNAs that encode ribosomal proteins (rpmRNAs).
It has recently been reported that mTOR inactivation is required for translational repression in MCF10A cells during prolonged hypoxic treatment (Connolly et al. 2006). However, the precise role of mTOR in mediating the translational repression during hypoxia remains unclear because previous reports have shown that mTOR is rapidly inactivated by hypoxia but translational repression requires prolonged exposure (Arsham et al. 2003; Liu and Simon 2004; Liu et al. 2006).
In addition to inactivating mTOR, anoxia and under certain conditions hypoxia activate PERK (Koumenis et al. 2002). PERK is responsible for the phosphorylation of the translation initiation factor eIF-2α. Phosphorylated eIF-2α cannot exchange GDP for GTP, which prevents the conversion of the inactive eIF2-GDP into the active eIF2-GTP form. In the absence of eIF2-GTP, the ternary complex does not form and global translation is inhibited (Pain 1996). The activation of PERK by hypoxia seems to be mediated through the unfolded protein response and occurs rapidly after exposure to acute anoxia and prolonged hypoxia under starvation conditions (Koumenis et al. 2002; Blais et al. 2006; Koumenis and Wouters 2006). There are significant differences in the kinetics of eIF-2α phosphorylation under hypoxic and anoxic conditions (Koumenis and Wouters 2006). Anoxia results in the rapid phosphorylation of eIF-2α, which corresponds well with the inhibition of translation during acute anoxia. Conversely, short-term exposure to hypoxia has little or no effect on eIF-2α phosphorylation, while long-term exposure has been reported to increase eIF-2α phosphorylation. However, these changes in phosphorylation during hypoxia do not correlate with the translational repression in HEK293 cells (Liu et al. 2006). Additionally, long-term exposure to hypoxia had no effect on eIF-2α phosphorylation in MCF10A cells even though translation was repressed (Connolly et al. 2006). These data strongly suggest that anoxia and hypoxia utilize different molecular mechanisms to inhibit translation. Additionally, this may also imply that the mechanisms used for translational repression during hypoxia may be cell line dependent.
This inhibition of cap-dependent translation during hypoxia does not affect all mRNAs. For example, the HIF-1α and VEGFA mRNAs are refractory to this translational repression (Stein et al. 1998; Gorlach et al. 2000; Lang et al. 2002). It has been reported that these mRNAs contain an internal ribosome entry site (IRES) in their 5′ UTRs (Huez et al. 1998; Stein et al. 1998; Lang et al. 2002). IRESs are cis-acting elements that promote internal initiation of translation by directly recruiting ribosomes to the mRNA (Pelletier and Sonenberg 1988; Macejak and Sarnow 1991; Vagner et al. 2001; Sarnow et al. 2005). Internal initiation of translation is a cap-independent process and has less of a requirement for translation initiation factors than normal cap-dependent translation. It has been hypothesized that the IRES elements in the VEGFA and HIF-1α mRNAs allow these mRNAs to be translated during hypoxia when global cap-dependent translation is inhibited (Stein et al. 1998; Lang et al. 2002). The fact that certain mRNAs are not translationally repressed during hypoxia can have dramatic implications on gene expression, because proteins encoded by these mRNAs will continue to be synthesized when cap-dependent protein synthesis is impaired.
There are three reports that analyzed the impact of this translational repression on global gene expression and they all focused on anoxic conditions (Blais et al. 2004, 2006; Koritzinsky et al. 2005). Since there appear to be differences in the molecular mechanisms involved with the translational repression that occurs under anoxic and hypoxic conditions, it is likely that the impact of the translational repression on gene expression will be different. To address this point we analyzed the effect of prolonged hypoxia (1.0% O2) on translation and gene expression in PC-3 cells. We show that translation in PC-3 cells is extremely sensitive to prolonged hypoxia and that this effect correlates well with the phosphorylation of eIF-2α. Additionally, we analyzed the impact of this repression on gene expression and identified several novel genes that are refractory to, and others that are extremely sensitive to, the translational repression that occurs during hypoxia.
RESULTS
Hypoxia inhibits translation in PC-3 cells
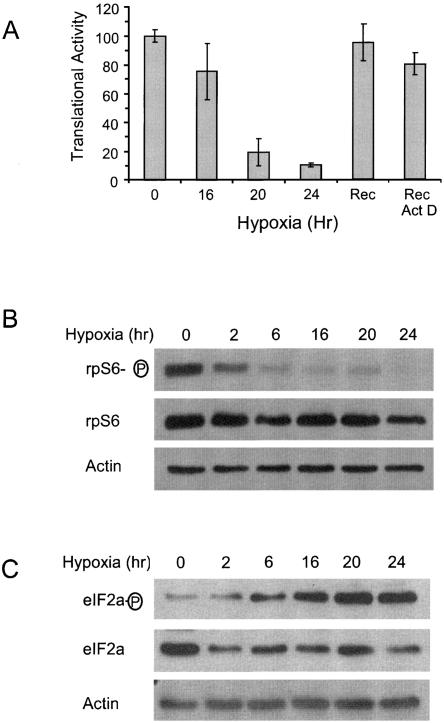
PC-3 cells were grown under normoxic conditions (20.9% O2, 5.0% CO2) and placed into a hypoxic chamber (1.0% O2, 5.0% CO2) for 0, 16, 20, and 24 h. The cells were then labeled with 35S-met/cys for 30 min and the amount of label incorporated into protein, as measured by TCA precipitable (TCA-ppt) counts, was determined. To control for altered label uptake and differences in cell number after treatment, the amount of label in the total extract was also determined. The relative translational activity of each sample was determined by measuring the amount of label incorporated into TCA-ppt counts normalized to the total amount of label in the extract. This ratio was then compared to that found in normoxic cells (Fig. 1, 0 h). As shown in Figure 1A, 16, 20, and 24 h of hypoxic treatment inhibited translation by 25%, 80%, and 90%, respectively. The dramatic inhibition of translation that occurs during prolonged hypoxia was completely eliminated when hypoxic cells (1.0% O2 24 h) were returned to normoxic conditions for 1 h (Fig. 1A, Rec). This recovery occurred even in the presence of the transcriptional inhibitor actinomycin D (Fig. 1A, Rec Act D), indicating that continued transcription was not needed for this recovery. Transcriptional inhibition by Act D was monitored by measuring the decay of the c-myc mRNA. Act D treatment resulted in the rapid loss of the c-Myc mRNA (apparent t 1/2 = 1.3 h), indicating that Act D is inhibiting transcription in hypoxic PC-3 cells (data not shown). The fact that the loss of label into TCA-ppt counts is completely reversible and is not dependent on continued transcription is consistent with an inhibition of translation. This requirement for prolonged exposure to hypoxia is similar to what others have reported in other tissue culture cells (Tinton and Buc-Calderon 1999; Koumenis et al. 2002; Lang et al. 2002; Arsham et al. 2003; Blais et al. 2004; Koritzinsky et al. 2006; Liu et al. 2006).
FIGURE 1.
Hypoxia reversibly inhibits translation in PC-3 cells. (A) PC-3 cells were exposed to hypoxia (1.0% O2) for 0, 16, 20, and 24 h. After 24 h of hypoxic treatment additional plates were returned to normoxic conditions for 1 h in the absence (Rec) or presence of Actinomycin D (5 μg/mL) (Rec + Act D). After treatment cells were labeled with 35S met/cys for 30 min. The translational activity was determined by measuring the amount of label incorporated into TCA-ppt material relative to total label uptake. This ratio was set to 100% for normoxic cells (0 h). The mean of triplicate samples are shown (± SD). (B,C) Cells were exposed to hypoxia for various times and the amount of phosphorylated and total RPS6 (B) and phosphorylated and total eIF2α (C) were determined by immuno-blot analysis. Actin was used as a loading control.
Hypoxia results in the inactivation of mTOR and phosphorylation of eIF-2α
mTOR inactivation and eIF-2α phosphorylation have both been implicated in the translational repression that occurs under hypoxic conditions (Gingras et al. 2001; Hay and Sonenberg 2004; Wouters et al. 2005; Connolly et al. 2006; Koritzinsky et al. 2006; Liu et al. 2006). In order to investigate the role of mTOR on the translational inhibition that we observe during prolonged hypoxia, we analyzed the phosphorylation status of an indirect target of mTOR, ribosomal protein S6 (RPS6). Immunoblot analysis revealed that hypoxic treatment resulted in the rapid and dramatic loss of RPS6 phosphorylation (Fig. 1B), indicating that mTOR is rapidly inactivated during hypoxia. Since hypoxia does not effect global translation at these early time points (Fig. 1A), it is unlikely that mTOR inactivation is responsible for this global repression.
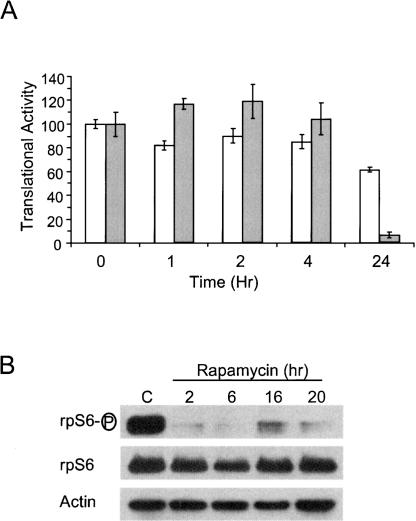
To further confirm this result we analyzed the effect of rapamycin (a potent inhibitor of mTOR) on translation and compared it to the effect of hypoxia. Figure 2A shows that rapamycin treatment for 1, 2, and 4 h had only a modest effect on translation, inhibiting it by no more than 20%. Longer exposure to rapamycin (24 h) inhibited translation by 40%. As expected, immunoblot analysis of the phosphorylation status of RPS6 confirmed that rapamycin treatment resulted in a rapid inhibition of mTOR activity (Fig. 2B). We conclude from these data that hypoxic treatment has a rapid and dramatic effect on mTOR activity. While this inactivation likely has an effect on the translation of specific mRNAs, i.e., 5′-TOP-containing mRNAs, it is unlikely to be solely responsible for the 90% reduction in global translation that occurs after 20 h of hypoxic treatment (Figs. 1, 2; J.D. Thomas and G.J. Johannes, unpubl. observations). However, we cannot completely rule out the possibility that prolonged inactivation of mTOR has a negative impact on translation.
FIGURE 2.
Rapamycin treatment does not mimic the translational repression that occurs during prolonged hypoxic treatment. PC-3 cells were exposed to rapamycin (open bars) or hypoxia (gray bars) for 0, 1, 2, 4 and 24 h. (A) Translational activity was determined as in Figure 1. The averages of triplicate samples are shown (± SD). (B) Immuno-blot analysis was used to analyze the amount of phosphorylated and total rpS6 in control C or rapamycin treated PC-3 cells. Actin was used as a loading control.
In contrast to the mTOR data, phosphorylation of eIF-2α does correlate with the translational repression that occurs during hypoxia. As shown in Figure 1C, hypoxia results in the slow but dramatic increase in the phosphorylation of eIF-2α that reaches a maximum by 20 h of treatment. This correlates well with the translational repression and is in direct contrast to what is seen when MCF10A cells and HEK293 cells are exposed to moderate hypoxia (Connolly et al. 2006; Liu et al. 2006). Thus these data suggest that eIF-2α phosphorylation is involved with the translational repression that occurs when PC-3 cells are exposed to prolonged hypoxia.
The global translational repression that occurs during hypoxia does not affect the polysomal distribution of the HIF-1α mRNA
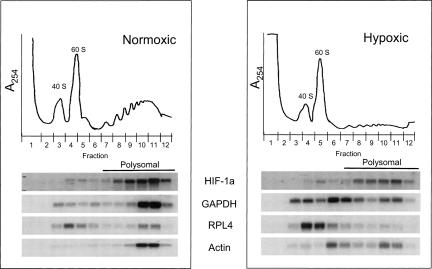
It has been reported that the HIF-1α mRNA polysome distribution is unaffected by hypoxic treatment in both NIH3T3 cells and HeLa cells when global translation is inhibited (Gorlach et al. 2000; Lang et al. 2002). We wanted to determine if this occurs in PC-3 cells and also evaluate the effect of hypoxia on the polysomal distribution of the GAPDH, actin, and the ribosomal protein L4 (RPL4) mRNAs. To this end we grew PC-3 cells under normoxic and hypoxic conditions and fractionated the ribosome/mRNA complexes by sucrose gradient centrifugation. The A254 traces in Figure 3 show the loss of polysomal material in cells grown under hypoxic conditions (compare fractions 7–12 in the A254 traces). This reflects the redistribution of translating ribosomes (polysomes) into their individual subunits resulting from a global shutdown in translation and is consistent with the metabolic labeling experiments (Figs. 1A, 2A). In order to accommodate the larger 40S and 60S peaks due to the disruption of ribosomes, the hypoxic gradient was graphed on a 2× scale. Additionally, to ensure good separation between polysomal and nonpolysomal material we used high salt gradients, which results in the dissociation of 80S ribosomal monomers that are not associated with mRNA into their individual subunits (Martin and Hartwell 1970).
FIGURE 3.
Polysome distribution of the HIF-1α, GAPDH, RPL4, and the actin mRNAs are differentially affected by hypoxic treatment. PC-3 cells were grown under normoxic and hypoxic conditions (1.0% O2 20 h) and subjected to polysome analysis. Polysome lysates were fractionated by sucrose gradient centrifugation and collected into twelve 1 mL fractions. Absorbance profiles (A254) are shown at the top of each panel. The top of the gradient is on the left and peaks representing the 40 S and 60 S ribosomal subunits are denoted. The nonpolysomal region of the gradient included fractions 1–6 and the polysomal portion is fractions 7–12. RNA was isolated from each fraction and subjected to Northern Analysis using 32P-labeled probes to detect the HIF-1α, GAPDH, RPL4, and actin mRNAs.
The distribution of an mRNA within the polysome gradient is reflective of its translational efficiency. The greater the percentage of an mRNA that is associated with polysomes the greater its translational efficiency. As shown in Figure 3, during normoxia the vast majority of the HIF-1α, GAPDH, and actin mRNAs were found in the polysomal portion of the gradient with 83%, 81%, and 91% of the mRNA associated with high molecular weight (HMW) polysomes (fractions 9–12), respectively. Moreover, the majority of the GAPDH and actin mRNAs was found predominately in fractions 10 and 11, while the HIF-1α mRNA was more broadly distributed throughout the polysomal portion of the gradient (fractions 8–12). This more even distribution of the HIF-1α mRNA within the polysomal portion of the gradient indicates that this mRNA is translated less efficiently than the actin and GAPDH mRNAs, under normoxic conditions. The RPL4 mRNA had a bimodal distribution with 40% of the RPL4 mRNA associated with HMW polysomes (fractions 10 and 11), while the other 60% was found in the nonpolysomal portion of the gradient (fractions 4 and 5). This suggests that the RPL4 mRNA exists as two distinct pools in the cell, one that is efficiently translated and one that is not. This bimodal distribution is consistent with the known translational regulation of 5′-TOP-containing mRNAs (see the Introduction) (Avni et al. 1997; Meyuhas 2000).
Under hypoxic conditions the GAPDH, RPL4, and actin mRNAs all redistributed to the nonpolysomal portion of the gradient with only 31%, 31%, and 49% of the polysomal mRNA remaining associated with HMW polysomes (fractions 9–12), respectively. It is important to note that although the bulk of the actin and GAPDH mRNAs redistributed to the nonpolysomal/monosomal portion of the gradient (fractions 6 and 7), a small portion of each mRNA remained associated with HMW polysomes (fractions 10 and 11). This may indicate that a small portion of mRNA within the cell can still efficiently recruit ribosomes or that a small number of cells may be refractory to the translational repression. Unlike the GADPH and actin mRNAs, the RPL4 mRNA completely redistributed to the top of the gradient (fractions 4 and 5), indicating the inability to recruit ribosomes. Since the RPL4 mRNA is a 5′-TOP mRNA, this redistribution most likely reflects the active translational repression of this mRNA by the inactivation of mTOR (Fig. 1B). In contrast to these three mRNAs that were sensitive to the translational repression, the polysomal distribution of the HIF-1α mRNA was relatively unaffected by hypoxia. Specifically, 92% of the HIF-1α polysomal mRNA remained associated with HMW polysomes during hypoxia. Together these data demonstrate that the HIF-1α mRNA is refractory to the translational repression that occurs during hypoxia, which is similar to what others have reported (Gorlach et al. 2000; Lang et al. 2002). Thus it is likely that there are other mRNAs that are also refractory to this hypoxia-mediated translational repression.
The HIF-1α mRNA is reduced in abundance in response to hypoxia
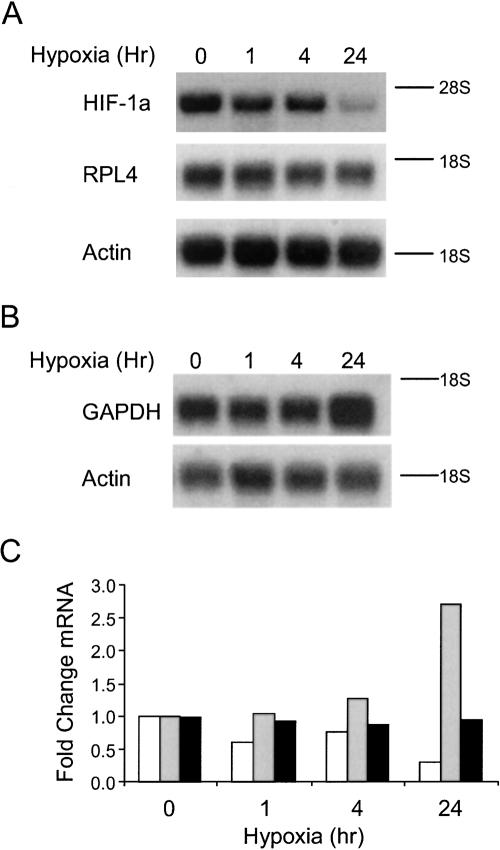
We next analyzed the effect of hypoxia on the abundance of the HIF-1α, RPL4, GAPDH, and actin mRNAs to evaluate the relationship between mRNA induction and polysome association during hypoxia. Figure 4, A and B, shows the Northern blots for the selected mRNAs after hypoxic treatment for 0, 1, 4, and 24 h. Quantitation of the Northern blots (Fig. 4C) revealed that the HIF-1α mRNA was reduced approximately threefold, the RPL4 mRNA was unaffected, and the GAPDH mRNA was induced ∼2.5-fold by 24 h of hypoxic treatment. These data show that the effect of hypoxia on the abundance of the mRNA does not necessarily reflect the sensitivity of the mRNA to the translational repression. For example, the GAPDH mRNA increases in abundance in response to hypoxia and is sensitive to the translational repression, whereas the HIF-1α mRNA is reduced in abundance and is refractory to the translational repression that occurs during hypoxia.
FIGURE 4.
The effect of hypoxic treatment on the abundance of the GAPDH, HIF-1α, RPL4, Actin, and GAPDH mRNAs. PC-3 cells were grown under normoxic (0 h) and hypoxic conditions for 1, 4, and 24 h. RNA was isolated and subjected to Northern analysis using 32P-labeled probes to detect the HIF-1α (open bars), RPL4 (black bars), actin, and GAPDH (gray bars) mRNAs. The 18 S and 28 S ribosomal RNAs are denoted by lines to the right of the image (when appropriate). Actin mRNA was used as a loading control. (A) Northern blot showing the abundance of the HIF-1α and RPL4 mRNAs. (B) Northern blot showing the abundance of the GPADH mRNA during hypoxia. (C) Results from A and B were quantitated and normalized to the actin mRNA using phosphorimage analysis and ImageQuant software. The value for the 0 time point was set to 100%.
Identification of other mRNAs that remain associated with polysomes during hypoxia
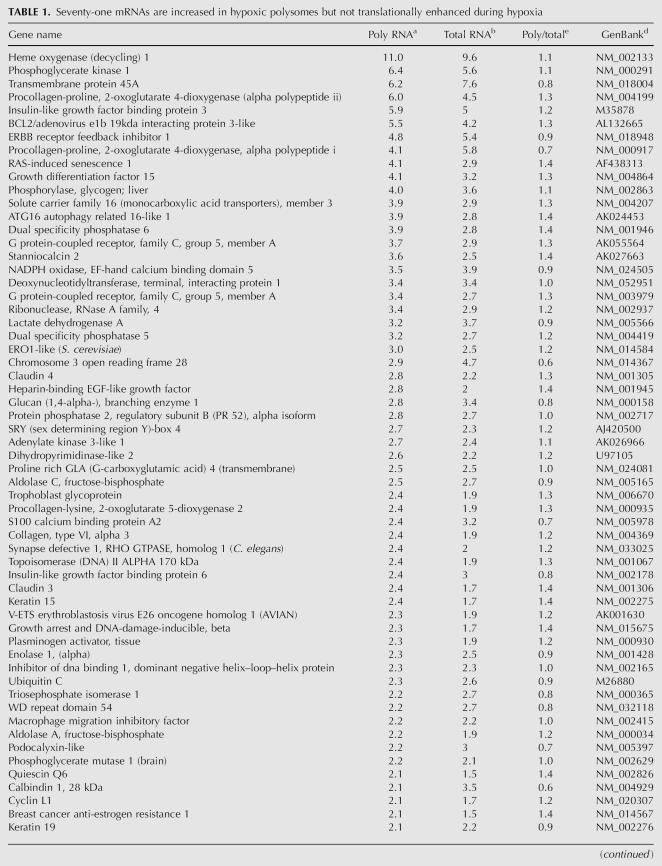
In order to analyze the impact of this translational repression on gene expression during hypoxia, we used global gene expression analysis to identify other mRNAs that had the ability to be maintained on polysomes during hypoxia. To this end we carried out microarray analysis on polysomal RNA from hypoxic and normoxic PC-3 cells. The data presented in Tables 1, 2, 3, and 4 were obtained from three independent biological samples that were analyzed using the GenePix Auto-Processing (GPAP3, Oklahoma State University) program and annotated with DAVID 2006 (NIAID/NIH) (Dennis et al. 2003). All of the values reported in Tables 1, 2, 3, and 4 represent fold changes observed during hypoxia relative to normoxia. Each gene is annotated in the table with the gene name, the fold change in abundance in polysomal RNA (Poly RNA), the fold change in mRNA abundance (Total RNA), the relative translatability (Poly/ Total) and the GenBank accession number.
TABLE 1.
Seventy-one mRNAs are increased in hypoxic polysomes but not translationally enhanced during hypoxia
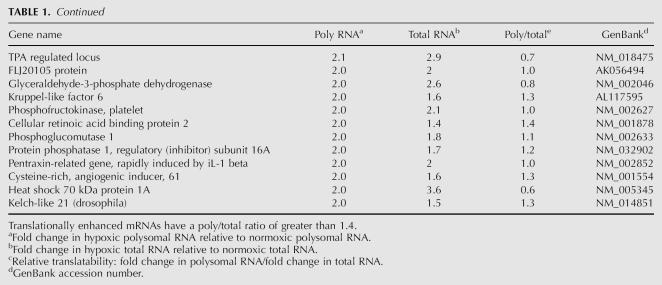
TABLE 2.
Thirty-three mRNAs are predicted to be translationally enriched on hypoxic polysomes
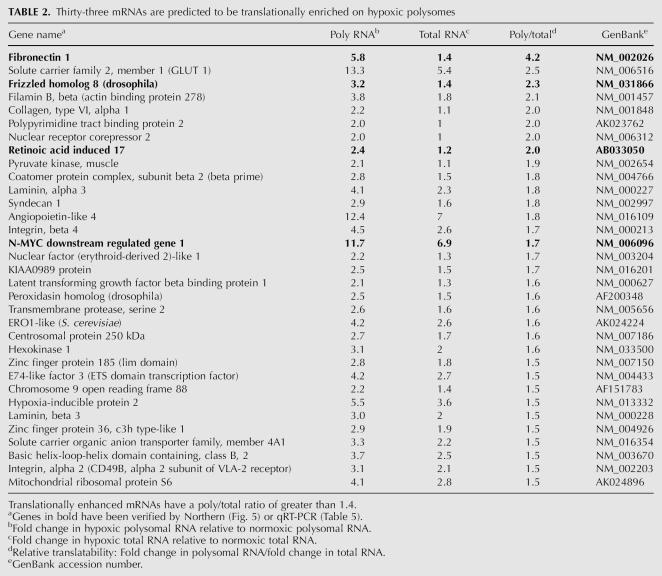
TABLE 3.
Twenty-five mRNAs are reduced in abundance in hypoxic polysomes
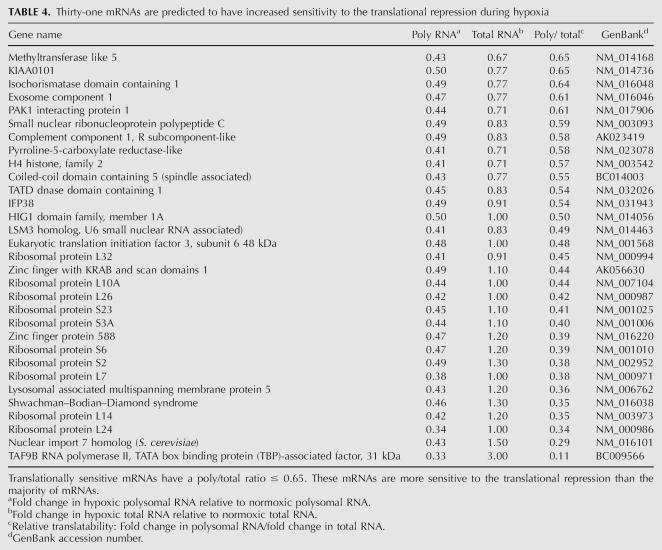
TABLE 4.
Thirty-one mRNAs are predicted to have increased sensitivity to the translational repression during hypoxia
Tables 1 and 2 show 104 mRNAs that were increased in abundance in hypoxic polysomes by 2.0-fold or greater (Tables 1 and 2 combined). Many of the genes that were increased in hypoxic polysomes are involved with glucose metabolism and glycolysis, such as heme oxygenase 1, phosphoglycerate kinase, and lactate dehydrogenase A.
An increase in polysomal RNA can reflect changes in mRNA abundance and/or changes in translatability of an mRNA. Since we were interested in identifying the mRNAs that were refractory to the translational repression, it was important to separate the contributions of these two phenomena. To this end, we determined the relative translatability of each mRNA. The relative translatability of an mRNA was determined by normalizing the change in abundance in polysomal RNA to the change in abundance in total RNA for each mRNA (poly/total).
Table 1 shows that 71 of these polysome-enriched mRNAs were induced in abundance greater than or equal to the amount they were increased in polysomal RNA, such as GAPDH, heme oxygenase 1, and phosphoglycerate kinase 1. This indicates that these mRNAs, although increased in abundance, do not efficiently associate with polysomes and thus are sensitive to the translational repression that occurs during hypoxia. This prediction is confirmed by the polysomal redistribution of the GAPDH mRNA (Fig. 3 and below) and the lack of GAPDH protein induction (Fig. 5) in response to hypoxic treatment.
FIGURE 5.
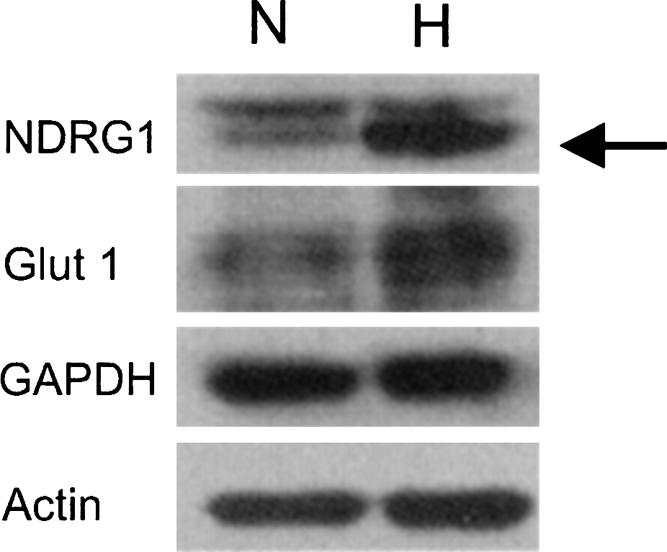
Proteins encoded by two translationally enhanced mRNAs, NDRG1 and Glut1 are induced during hypoxia. PC-3 cells were grown under normoxic (N) or hypoxic conditions (H) for 20 h. Twenty micrograms of protein were separated by 10% SDS-PAGE and the amount of NDRG1, GLUT1, GAPDH, and actin proteins were analyzed by immuno-blot analysis. Actin was used as a loading control. Arrow denotes the NDRG1 band.
Identification of mRNAs that are translationally enhanced during hypoxia
In contrast, Table 2 shows that 33 of these 104 mRNAs that were enriched in hypoxic polysomes were translationally enhanced during hypoxia. We have defined translationally enhanced mRNAs as those that are increased in hypoxic polysomes by 50% or greater relative to changes in mRNA abundance (poly/total ≥ 1.5). Therefore, these mRNAs are present on hypoxic polysomes to a greater extent than can be explained by increases in mRNA abundance alone. These mRNAs are predicted to be maintained on polysomes and thus be refractory to the translational repression that occurs during hypoxia. Fourteen of these 33 translationally enhanced genes were both induced in abundance and maintained on polysomes, such as solute carrier family 2, angiopoietin-like 4 (ANGPTL4), and N-Myc downstream regulated gene 1 (NDRG1), while the rest did not increase in abundance more than twofold in response to hypoxia but were maintained on polysomes.
To verify that the proteins encoded by the translationally enhanced mRNAs were induced during hypoxia, we examined the expression of NDRG1 and solute carrier family 2, member 1 (GLUT1) by immuno-blot analysis. As shown in Figure 5 both the NDRG1 and GLUT1 proteins are induced by hypoxic treatment, while actin and GAPDH are unaffected. It is important to note that even though the GAPDH mRNA is induced 2.5-fold in response to hypoxia, the protein remains unchanged, reflecting the reduced translational efficiency of its mRNA. Additionally, protein yields from hypoxic cells are ∼30% (29% ± 4.6%) lower, compared to an equivalent number of normoxic cells (data not shown). This reduction in protein yield most likely reflects the reduction in global protein synthesis during hypoxia. Since the immuno-blot analysis compares equal micrograms of protein, the specific proteins in the hypoxic samples are overestimated.
Direct measurement of the rate of synthesis of these proteins during hypoxia was not possible because pulse labeling of the hypoxic cells resulted in increased cell death and low protein yield. We attribute this to the combined stresses of hypoxia (20 h) and the addition of met/cys-depleted media during the pulse-labeling step.
Ribosomal protein mRNAs are most sensitive to the translational repression that occurs during hypoxia
Our analysis also identified 56 genes (Tables 3 and 4 combined) that were reduced by twofold or greater in polysomal RNA from hypoxic cells. Since these mRNAs are reduced in abundance, the fold change is expressed as a factor of less than one. The reduced presence of these mRNAs in polysomal RNA can result from a reduced abundance of the mRNA, a reduced translatability of the mRNA, or a combination of both. To evaluate the contribution of changes in mRNA levels and changes in translatability we also evaluated the relative translatability (poly/total) of these mRNAs (see above). For our analysis, mRNAs that had a poly/total ratio of < 0.65 were considered translationally sensitive. This analysis identified 31 mRNAs that were predicted to be translationally sensitive to the translational repression during hypoxia (Table 4). Interestingly, 11 out of these 31 genes encode ribosomal proteins. This increased sensitivity to the translational repression of these 5′-TOP-containing mRNAs is consistent with the redistribution of the RPL4 mRNA (Fig. 3) and most likely reflects the inhibition of mTOR during hypoxia (Fig. 1B).
Since we knew that HIF-1α mRNA remains associated with polysomes during hypoxia (Fig. 3) even though it is reduced threefold in abundance (Fig. 4), we looked for other down-regulated genes that were enriched in polysomes. We found one gene, matrix metallopeptidase 1 (MMP1), which was reduced in abundance more than it was lost on polysomes (Table 3, bold type). This indicates that even though this mRNA is reduced in abundance it is still maintained on polysomes during hypoxia.
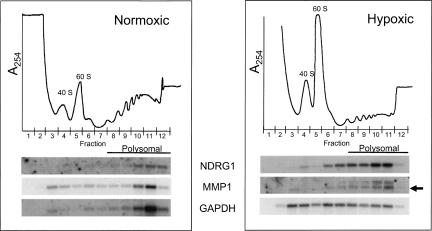
Confirmation that the NDRG1 and MMP1 mRNAs remain associated with polysomes during hypoxia
We used Northern analysis to determine the polysomal distribution of two mRNAs, MMP1 and NDRG1, which we identified from our screen that remained associated with polysomes and were predicted to be translationally enhanced during hypoxia. As predicted by the microarray analysis, both of these mRNAs remained associated with polysomes during hypoxia. Figure 6 shows the loss of polysomal material (compare A254 profiles for normoxic and hypoxic cells) and the redistribution of the GAPDH mRNA into the nonpolysomal portion of the gradient. Both of these findings reflect the global shutdown in translation during hypoxia and are consistent with the data presented in Figures 1–3. In contrast, 92% of the NDRG1 and 85% of the MMP1 mRNA remained associated with HMW polysomes during hypoxia. The band seen above the MMP1 band, indicated by the asterisk, is residual NDRG1 that was incompletely stripped off the blot. This is only seen in the hypoxic gradient because the NDRG1 mRNA is significantly induced in abundance. These results confirm that the mRNAs encoding MMP-1 and NDRG1 continue to associate with polysomes during hypoxia, thus validating our microarray analysis.
FIGURE 6.
The NDRG1 and MMP1 mRNAs remain associated with polysomes during hypoxic treatment. PC-3 cells were subjected to polysome analysis as in Figure 3. 32P-labeled probes were used to detect the polysomal distribution of the NDRG1, MMP1, and GAPDH mRNAs in normoxic and hypoxic conditions.After probing with NDRG1 the blot was stripped and probed for MMP1. In the hypoxic blot for MMP1 the arrow head denotes the MMP1 band. The upper band, denoted by an asterisk, is incompletely stripped NDRG1 mRNA from the previous probing.
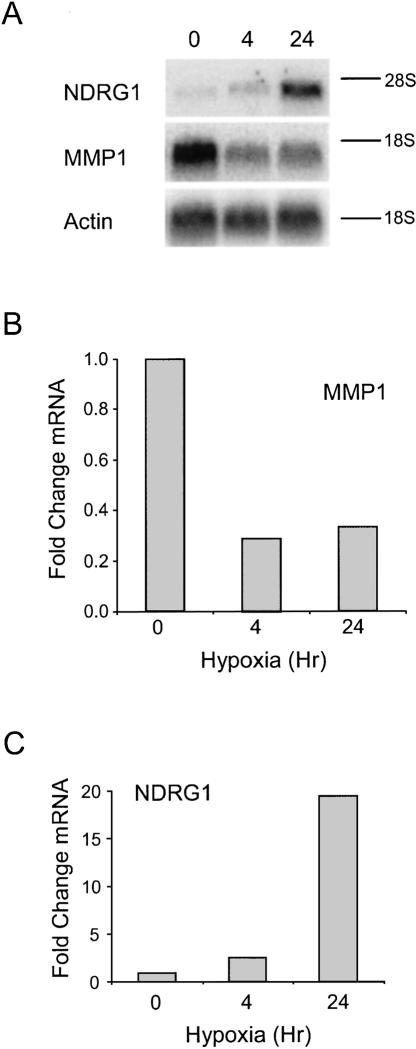
We also analyzed the changes in total mRNA abundance of these mRNAs after 0, 4, and 24 h of hypoxic exposure (Fig. 7A). Quantitation of the Northern blots revealed that the MMP1 mRNA decreased 3.3-fold (Fig. 7B) and the NDRG1 mRNA increased 19-fold (Fig. 7C) in response to 24 h of hypoxic treatment. These data are in general agreement with the microarray study, which showed that the NDRG1 mRNA was induced by 6.9-fold while the MMP-1 was reduced by 2.6-fold in response to hypoxia. Taken together, the data in Figures 6 and 7 show that continued translation during hypoxia does not require a corresponding increase in mRNA abundance. For example, the MMP1 mRNA is translationally enhanced but the abundance of its mRNA is reduced during hypoxia. Additionally, these data show that induction of an mRNA does not ensure translational enhancement during hypoxia as shown by the finding that the GAPDH mRNA is translationally repressed but the abundance of this mRNA is increased during hypoxia. Thus there does not appear to be a direct correlation between mRNA induction and translational enhancement during hypoxia.
FIGURE 7.
The effect of hypoxic treatment of the steady-state levels of the NDRG1, MMP-1, and actin mRNAs. PC-3 cells were grown under normoxic (0 h) and hypoxic conditions for 4 and 24 h. (A) Equal micrograms (5 μg) of RNA were subjected to Northern analysis using 32P-labeled probes to detect the NDRG1, MMP1, and actin mRNAs. The 18 S or 28 S ribosomal RNAs are denoted by a line to the right of the image (when appropriate). Actin mRNA was used as a loading control. Results were quantitated for NDRG1 (B) and MMP1 (C) after normalization to the actin mRNA using phosphorimage analysis and ImageQuant software. The value for the 0 time point was set to 1.
Confirmation of four additional mRNAs that remain associated with polysomes during hypoxia
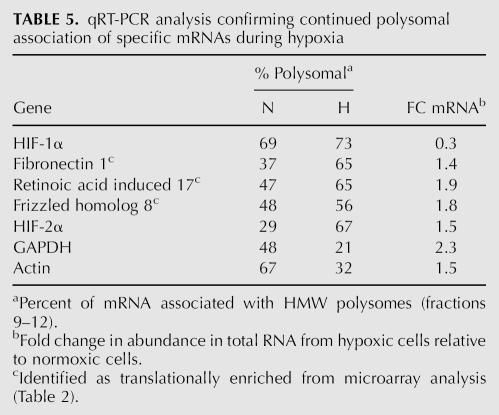
We utilized qRT-PCR to analyze the polysomal distribution of three other mRNAs, fibronectin, retinoic acid induced 17, and frizzled homolog 8, that were predicted by microarray analysis to be maintained on polysomes during hypoxia (bold type, Table 2). As shown in Table 5, the percentage of polysomal mRNA (% polysomal) for all three of these mRNAs was greater in hypoxic cells (H) than in normoxic cells (N). In addition, we tested the polysome association of the hypoxia inducible factor 2α (HIF-2α) mRNA and found that this mRNA was also significantly enriched in polysomes from hypoxic cells. We believe that this is the first report that shows that the HIF-2α mRNA is refractory to the translational repression that occurs during hypoxia. All four of these mRNAs were maintained on polysomes to a greater extent than HIF-1α (which is our positive control for mRNAs that remain associated with polysomes during hypoxia) as indicated by the net percent polysomal increase from normoxic to hypoxic polysomes. This indicates that these mRNAs are translated efficiently under hypoxic conditions and are thus refractory to the translational repression. In contrast, the percentage of polysomal mRNA of the GAPDH and actin mRNAs, both of which are sensitive to the translational repression (see Figs. 3, 5), are reduced by more than twofold under hypoxic conditions.
TABLE 5.
qRT-PCR analysis confirming continued polysomal association of specific mRNAs during hypoxia
We also used qRT-PCR to determine the change in the abundance of these mRNAs during hypoxia, relative to normoxic conditions. This is represented as the fold change in mRNA (FC mRNA) during hypoxia. As shown in Table 5, we observed a threefold loss of the HIF-1α mRNA (expressed as a 0.3-fold change) and a 2.3-fold increase in the GAPDH mRNA similar to what was determined by Northern analysis (Fig. 4).
Thus the qRT-PCR data confirm the microarray data and show that the fibronectin 1, frizzled homolog 8, and retinoic acid induced 17, HIF-1α, and HIF-2α mRNAs are maintained on polysomes in hypoxic cells when global translation is inhibited. We are currently investigating the molecular mechanism(s) that allow these mRNAs to continue to be translated during hypoxia.
DISCUSSION
We have analyzed the effect of hypoxia on translation and gene expression in the prostate cancer cell line PC-3. We found that translation in PC-3 cells is inhibited by > 80% in response to prolonged exposure to hypoxia and is much more dramatic than what has been reported for other cell lines (Tinton and Buc-Calderon 1999; Koumenis et al. 2002; Connolly et al. 2006; Liu et al. 2006). Several lines of evidence indicate that this reduction in translation is not due to a reduction in mRNA. First, we find that this inhibition is completely reversed within 1 h after cells are returned to normoxia even in the presence of the transcriptional inhibitor Act D (Fig. 1A). This strongly indicates that new transcription is not needed for restoration of translational activity. Second, the microarray analysis only identified 14 mRNAs that were reduced in abundance more than twofold in response to hypoxic treatment (Table 3; data not shown), indicating a negligible effect on the global mRNA population. Third, the majority of mRNAs associate with fewer ribosomes during hypoxia, as shown by the loss of polysomes and redistribution of mRNAs (Figs. 3, 6; Table 4), indicating a block in translation initiation. Thus it is unlikely that the reduction in translational activity observed during hypoxia is due to a loss of the global mRNA population.
This strong translational repression that we observe in the highly transformed prostate cancer cell line, PC-3, is in contrast to what has been reported for breast cancer cell lines. Connolly et al. (2006) reported that as breast cancer cells become more transformed they become resistant to this translational repression. This increased sensitivity of PC-3 cells may reflect fundamental differences between the transformation of breast cancer cell lines versus prostate cancer cell lines or may reflect a unique characteristic of the PC-3 cell line. They also show that this translational repression is mediated through the inactivation of mTOR and that this inactivation is lost in the more transformed breast cancer cell lines. In contrast, we show that mTOR is rapidly inactivated in PC-3 cells during hypoxia but this inactivation does not correlate with the global translational repression (Figs. 1A,B, 2). However, our data are consistent with what has been reported for serum-starved HEK293 cells in which mTOR inactivation does not correlate with translational repression (Arsham et al. 2003).
Unlike the data for mTOR, we find that eIF-2α phosphorylation correlates well with the hypoxia-induced translational repression in PC-3 cells. This is contradictory to what has been reported for the weakly transformed MCF10A breast cancer cell line and is similar to what has been reported for serum-starved HEK293 cells (Connolly et al. 2006; Liu et al. 2006) Although the PC-3 cells were not serum starved in our study, they are much more sensitive to the translational repression than the HEK293 cells. Together, these data suggest that different cell lines can use different mechanisms to inhibit translation during hypoxia. It is plausible to hypothesize that eIF-2α phosphorylation is required for translational repression in highly transformed cell lines, while mTOR inactivation has a much more dominant effect in less transformed cells.
This correlation of eIF-2α phosphorylation and translational repression under hypoxic conditions is similar to what has been reported to occur under anoxic conditions. However the kinetics of eIF-2α phosphorylation is different under the two conditions. Anoxic treatment results in the rapid and transient phosphorylation of eIF-2α (Koritzinsky et al. 2006) that correlates with the translational repression. During this initial phase, mRNAs that have upstream open reading frames such as ATF4 are translated with more efficiency (Blais et al. 2004). Longer exposure to anoxic conditions results in the loss of eIF-2α phosphorylation even though translation remains repressed. This is the opposite of what we observe during hypoxia in PC-3 cells. Initial exposure of PC-3 cells to hypoxia results in the inactivation of mTOR (Fig. 1B) while prolonged exposure increases eIF-2α phosphorylation (Fig. 1C).
Although the mechanism leading to eIF-2α phosphorylation by prolonged hypoxia is not known, it is possible that this process may also involve the PERK kinase as has been shown under anoxic conditions (Koumenis et al. 2002; Blais et al. 2006; Koumenis and Wouters 2006). Prolonged exposure to hypoxia may result in the slow accumulation of misfolded protein in the ER. Over time, this misfolded protein may reach a threshold level that activates the unfolded protein response (UPR), resulting in the activation of PERK, which in turn phosphorylates eIF-2α. Thus chronic hypoxia may utilize a mechanism similar to that of acute anoxia, but unlike anoxia, prolonged exposure to hypoxia may be required for activation of the UPR. However, it is also likely that there are other mechanisms involved with the translational repression, which occurs during hypoxia, that are distinct from the mechanisms used under anoxic conditions. In fact it has recently been reported that blocking the expression of PERK does not completely prevent the translational repression that occurs when transformed mouse embryonic fibroblasts are exposed to acute anoxia (Blais et al. 2006). Thus other mechanisms must be involved with the translational repression that occurs during oxygen deprivation and it may be these other mechanisms that are responsible for the differences in genes that are translationally enhanced during hypoxia and anoxia. We are currently investigating the molecular mechanism(s) involved with the translational repression that occurs during hypoxia to address this issue.
In order to analyze the effect of hypoxia on gene expression we carried out microarray analysis of polysomal RNA (translational profiling) from hypoxic cells. We found that out of the 20,000 genes on the array, only 104 mRNAs were shown to be increased in hypoxic polysomes when compared to normoxic polysomes (Tables 1 and 2 combined). Many of these mRNAs are not efficiently translated during hypoxia, such as GAPDH (see Table 1). This mRNA is induced in abundance 2.6-fold in response to hypoxia but is translationally repressed during hypoxia. The GAPDH mRNA is enriched in hypoxic polysomes only because it is increased in abundance more than it is reduced in polysome association. Since the GAPDH mRNA is induced in abundance during hypoxia we cannot say if the newly synthesized mRNA is unable to recruit ribosomes while the older mRNA continues to be translated. However, since the actin mRNA, which is not induced, also redistributes to the top half of the gradient, it is likely that both previously synthesized and newly synthesized mRNAs are unable to efficiently recruit ribosomes during hypoxia.
Comparison of these genes to those found enriched in polysomes from anoxic DU145 cells identified only five mRNAs, NDRG1, ANGPTL4, hypoxia inducible protein 2, BHLHB2, and Stanniocalcin 2, which were enriched in polysomes during hypoxic and anoxic conditions (Koritzinsky et al. 2005). All of these mRNAs are significantly induced in abundance during hypoxia (>3.0-fold) and anoxia, and this may contribute to their enrichment on polysomes in both systems. The other 99 mRNAs that we identified are unique to our system and may reflect mRNAs that are regulated by hypoxia in PC-3 cells and not DU145 cells or may be due to differences in the microarrays used in the different experiments. Alternatively, these differences may reflect fundamental differences in gene expression at both the translational and transcriptional levels between anoxic and hypoxic cells. To further evaluate the differences between anoxia and hypoxia we examined the translational enrichment of three mRNAs that have been reported to be translationally enriched during anoxic treatment (Blais et al. 2004). In PC-3 cells we found that ATF4 (NM_001675), eIF5 (NM_001969), and TXBP151 (NM_006024) were not translationally enriched in hypoxic polysomes with fold changes in poly RNA/change in total RNA of 1.4/1.9, −1.2/−1.2, and 1.4/1.3, respectively. Thus it is likely that anoxia and hypoxia impact the translation of specific mRNAs differently, resulting in certain mRNAs being translationally enhanced during hypoxia while others are translationally enhanced during anoxia. It would be interesting to directly address this question by identifying translationally enhanced mRNAs under hypoxic and anoxic conditions in the same cell line using the same methodology.
Of the 104 mRNAs enriched in hypoxic polysomes (Tables 1 and 2 combined), we further classified a subset of these as translationally enhanced during hypoxia (Table 2). These 33 mRNAs showed the most robust polysome association during hypoxia and were increased in hypoxic polysomes to a greater extent than could be explained by increases in mRNA abundance. We tested four mRNAs from this subset, fibronectin 1, frizzled homolog 8, retinoic acid induced 17, and NDRG1, and found that all continued to associate with polysomes during hypoxia (Fig. 5; Table 5). This indicates that these mRNAs have the ability to be translated during hypoxia when global translation was inhibited. Evaluation of the 5′ UTRs of the translationally enriched mRNAs showed that 16 out of the 33 translationally enriched mRNAs have a GC content > 60% but no other distinguishing features.
It has been hypothesized that IRES-mediated translation may be responsible for the continued efficient recruitment of ribosomes during hypoxia. Recently, it has been shown that the VCIP mRNA is translationally enhanced during acute anoxia and that this mRNA contains an IRES element that is active during acute anoxia (Blais et al. 2006). It has also been reported that certain IRES elements continue to function when eIF2α is phosphorylated (Fernandez et al. 2002; Komar and Hatzoglou 2005) and our data indicate that mRNAs that have the ability to recruit ribosomes during hypoxia must do so in the presence of phosphorylated eIF-2α. Together these data strongly support the hypothesis that IRES elements allow mRNAs to be translated during hypoxia. In order to test this hypothesis we are currently cloning the 5′ UTRs of several of the translationally enhanced genes into reporter constructs to test for IRES activity.
Since some of the mRNAs, such as actin and GAPDH, which are translationally inhibited during hypoxia appear to still associate, albeit inefficiently, with the translational machinery, it is possible that the proteins encoded by these mRNAs continue to be synthesized during hypoxia but at an extremely low rate. Thus an mRNA that is translationally suppressed during hypoxia may be able to overcome this suppression by inducing the mRNAs to a sufficient level to maintain protein production. This is consistent with our gene expression data that show many induced mRNAs are enriched on hypoxic polysomes but not translationally enhanced (Table 1). In contrast to this mechanism of maintaining protein production during hypoxia, there appears to be a unique class of mRNAs that are unaffected by this translational repression (Table 2). These latter mRNAs likely use an alternative molecular mechanism of translation initiation, such as internal initiation of translation. The proteins encoded by these mRNAs will continue to be efficiently synthesized when global translation is repressed and thus are likely to be induced in the proteome of hypoxic cells to an extent not predicted by total mRNA analysis.
The impact of the translation repression that occurs during hypoxia on actual tumor growth is still unknown; however, the fact that two critical factors in the cellular response to hypoxia, HIF-1α and HIF-2α, are encoded by mRNAs that are refractory to this translation repression indicates that overcoming this inhibition is important in the cellular adaptation to hypoxia and tumor growth (Gorlach et al. 2000; Lang et al. 2002). Consistent with this hypothesis, it has been recently reported that mRNAs that are translationally enhanced during acute anoxia, in a PERK dependent manner, are involved with the cellular adaptation to hypoxia and induction of angiogenesis (Blais et al. 2006). Interestingly, this anoxic study identified the MMP13 mRNA as being translationally activated during acute anoxia. This protein is very similar to MMP1, which we identified to be translationally enhanced during chronic hypoxia. MMP13 and MMP1 are both interstitial collagenases involved with remodeling of the extracellular matrix for angiogenesis and cancer cell migration (Goldberg et al. 1986; Freije et al. 1994; Deryugina and Quigley 2006). It is possible that MMP13 is preferentially translated during acute anoxia while MMP1 is preferentially expressed during chronic hypoxia. Since tumors exhibit varying degrees of oxygenation and often contain areas of chronic hypoxia as well as areas of acute anoxia, it is interesting to speculate that MMP13 is preferentially translated and expressed in areas of acute anoxia while MMP1 is preferentially translated and expressed in areas of chronic hypoxia (Vaupel et al. 1989; Kizaka-Kondoh et al. 2003) . Thus mRNAs that are translationally enhanced during hypoxia may help the cells adapt to the chronic hypoxic condition, which can occur in the center of a tumor, while mRNAs that are translationally enhanced during acute anoxia may help the cells to adapt to acute anoxia, which occurs when tumor blood vessels are occluded. This blockage is usually transient, and the genes translationally enhanced under acute anoxia may help the cell adapt to this rapid loss of oxygen. Therefore, understanding the molecular mechanism(s), whether it be internal ribosome binding, low cap requirement, or some other mechanism, that allows mRNAs to be translated during hypoxia may lead to the discovery of novel targets for attacking cancer by blocking its ability to adapt to the hypoxic environment.
MATERIALS AND METHODS
Tissue culture and hypoxic treatment
PC-3 (ATCC # CRL-1435) cells were grown in Ham's F12-K media (MediaTech) supplemented with 10% FBS (Mediatech) and Pen/Strep at 37°C in 5% CO2. For hypoxic treatment, cells were plated under normoxic conditions and grown for 16–20 h and then placed into a hypoxic chamber (Coy Laboratory) pre-equilibrated to 1.0% O2, 5% CO2 at 37°C. Oxygen levels were verified using an independent oxygen sensor (Drager, Inc.). Actinomycin D (Act D; MP Biomedical, Inc.) was used at 5 μg/mL and rapamycin (Alexis Biochemicals) was used at 2 μg/mL.
Metabolic labeling
Cells were labeled for 30 min by adding 70 μCi/mL of Pro-mix (Pro-mix L-35S, Amersham Bioscience) to the media (1 mL) in 12-well plates. After labeling, cycloheximide was added to 100 μg/mL and incubated for 5 min (to block further protein synthesis). Cells were then washed twice with PBS, lysed in 1 mL of RIPA buffer, transferred to Eppendorf tubes, and placed at −20°C. For the TCA precipitation (ppt), 75 μL of extract were added to 1 mL of ice cold 10% trichloroacetic acid (TCA; Fisher Scientific) and incubated on ice for 30 min. The mixture was then passed through a glass filter (GF/C Millipore) and washed using a vacuum manifold, according to the manufacturer's instructions (Millipore). The dried filters were placed into 5 mL of scintillation fluid and read in a scintillation counter. For total counts per minute, 20 μL of total extract were placed onto a glass filter, dried, and read as above. Counts were normalized to microliters of extract and percent of TCA incorporation was determined by dividing the TCA ppt cpm/μL extract by total cpm/μL extract. All experiments were performed a minimum of three times with duplicate wells.
Immuno-blot analysis
Whole-cell extracts were prepared by lysis in RIPA buffer containing Halts protease inhibitor cocktail (Pierce) using standard procedures. Protein concentration was determined using the Biorad DC Kit. Thirty-five micrograms of protein were separated by 10% SDS-PAGE and transferred to Immobilion-P membrane (Millipore). Transfer efficiency was evaluated by Ponceu S staining of the membrane. Antibodies for RPS6 (5G10), phosphorylated RPS6 (ser 235/236), eIF2α, and phosphorylated eIF-2α(Ser 51) were purchased from Cell Signaling Technology. Anti-GAPDH (Ambion, Inc.), anti-NDRG1 (N-19, Santa Cruz Biotech), and GLUT 1 (ab652, Abcam) were also purchased. The blot was developed using an ECL Plus Detection Kit (Amersham Biosciences). After detection the blot was striped and probed using anti-actin (Sigma).
Polysome analysis
PC-3 cells were untreated or placed in a hypoxic chamber for 20 h and subjected to polysome analysis as described (Johannes and Sarnow 1998). Briefly, after treatment, cycloheximide was added to 100 μg/mL, 3 min prior to harvesting. Polysome lysates were prepared in polysome extraction buffer (15 mM Tris-CL at pH 7.6, 15 mM MgCl2, 0.3 M NaCL, 1% Trition X-100, 0.1 mg/mL cycloheximide, and 1 mg/mL heparin), and subjected to sucrose gradient (10%–50%) centrifugation. After centrifugation, gradients were collected into twelve 1 mL fractions while the Abs254 was continuously monitored using an ISCO fractionator (Brandel, Inc.). The RNA in the fractions was extracted and equal volumes of RNA from each sample were analyzed by Northern analysis as described below. Semiquantitative analysis of the polysome gradients (A254) was performed by analyzing the area under the polysomal (fractions 7–12) and nonpolysomal portion (fractions 2–6). All polysomal analysis was done a minimum of three times.
Northern analysis
Polysomal RNA (see above) or 5 μg total RNA, isolated using the RNAeasy kit (Qiagen), was fractionated through a 1.2% formaldehyde gel, transferred to a Genescreen Plus membrane (Perkin Elmer), cross-linked, and probed with body labeled cDNA probes generated using the RadPrime Kit (Invitrogen) following standard protocols (Ausubel 1987). Transfer and RNA integrity were evaluated by methylene blue staining of the membrane. Hybridization was performed using standard techniques in aqueous buffer (Ausubel 1987). After washing, blots were visualized and quantitated using PhosphorImager analysis and Imagequant software (Amersham). All Northerns, total and polysomal, were done a minimum of three times.
Quantitative real-time PCR (qRT-PCR)
RNA templates for qRT-PCR consisted of total RNA or pooled sucrose gradient fractionated RNA. RNA isolated from the nonpolysomal (fractions 2–5), monosomal/low molecular weight polysomal (fractions 6–8), and HMW polysomal (fractions 9–12) fractions was pooled and 0.2 μg of the RNA were then subjected to qRT-PCR. The percentage of each mRNA associated with HMW polysomes (% polysomal) is shown. Validated primer/probe sets were purchased from ABI and the gene and catalog numbers are as follows: actin: Hs99999903_m1; GAPDH: Hs99999905-m1; HIF-1α: Hs00153153-m1; HIF-2α: Hs01026149_m1; NDRG1: Hs00608389-m1; fibronectin 1: Hs00415006_m1; frizzled homolog 8: Hs00259040-s1; and retinoic acid induced 17: Hs00277476-m1. For total RNA analysis, cyclophilin A (Hs99999904_m1) was used as a loading control and normalized to actin mRNA. Analysis was done using the MxPro Software (Stratagene) using the MX 3000 and the Brilliant qRT-PCR system according to the manufacturer's instructions. All probes were tested in no RT control reactions, and all reactions were performed in duplicate or triplicate and had percent errors of less than 10%.
Microarray analysis
Polysomal RNA from normoxic and hypoxic cells was obtained by pooling equal volumes of RNA from fractions 9–12 of the gradients. Polysomal or total RNA was used to synthesize Cy3- and Cy5-labeled aRNA using the Amino Allyl MessageAmpII aRNA Amplification Kit (Ambion). Equal micrograms of each labeled aRNA were combined and hybridized to the DNA microarray (Human Genome Oligo Set V.2.0-; Qiagen) using SlideHyb buffer #1 (Ambion) at 65°C for 16 h. The slide was then washed, dried, and scanned using an Axon dual laser GenePix 4000 scanner. The data were collected and initially analyzed with the GenePix Pro 5.1 software. The data from three biological replicates were then analyzed by the GenePix Auto-Processor Software (GPAP3; Oklahoma State University) using local lowess normalization-pin by pin intensity dependent normalization. The correlation among biological replicates was > 0.85. The resulting quantitation is shown in Tables 1, 2, 3, and 4. The cutoff value for increase in abundance in polysomal or total RNA was twofold. After analysis the genes were annotated using the DAVID 2006 software (NIH). The microarrays were purchased from the Genomics Facility at the Drexel University College of Medicine.
ACKNOWLEDGMENTS
We thank Dr. Shelly Waggoner for helpful discussions and critical reading and editing of the manuscript and Dr. Peter Sarnow for insightful comments. We also thank the OSU Microarray Core Facility, which was supported by grants from NSF (EOS-0132534) and NIH (2P20RR016478-04, 1P20RR16478-02, and 5P20RR15564-03) for the GPAP program and the Genomics Facility at the Drexel University College of Medicine for providing the DNA microarrays. This work is supported by Public Health Service grant CA112086 from the National Cancer Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.534807.
REFERENCES
- Arsham, A.M., Howell, J.J., Simon, M.C. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M. Current protocols in molecular biology. Wiley; New York: 1987. [Google Scholar]
- Avni, D., Biberman, Y., Meyuhas, O. The 5′-terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type- and sequence context-dependent manner. Nucleic Acids Res. 1997;25:995–1001. doi: 10.1093/nar/25.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers, G., Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- Bi, M., Naczki, C., Koritzinsky, M., Fels, D., Blais, J., Hu, N., Harding, H., Novoa, I., Varia, M., Raleigh, J., et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais, J.D., Filipenko, V., Bi, M., Harding, H.P., Ron, D., Koumenis, C., Wouters, B.G., Bell, J.C. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 2004;24:7469–7482. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais, J.D., Addison, C.L., Edge, R., Falls, T., Zhao, H., Wary, K., Koumenis, C., Harding, H.P., Ron, D., Holcik, M., et al. PERK-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol. Cell. Biol. 2006;26:9517–9532. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.M., Giaccia, A.J. The unique physiology of solid tumors: Opportunities (and problems) for cancer therapy. Cancer Res. 1998;58:1408–1416. [PubMed] [Google Scholar]
- Brown, J.M., Wilson, W.R. Exploiting tumor hypoxia in cancer treatment. Nat. Rev. Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- Connolly, E., Braunstein, S., Formenti, S., Schneider, R.J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman, B.T., Hosack, D.A., Yang, J., Gao, W., Lane, H.C., Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- Deryugina, E.I., Quigley, J.P. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- Dvorak, H.F. Rous–Whipple award lecture. How tumors make bad blood vessels and stroma. Am. J. Pathol. 2003;162:1747–1757. doi: 10.1016/s0002-9440(10)64309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez, J., Yaman, I., Sarnow, P., Snider, M.D., Hatzoglou, M. Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 2002;277:19198–19205. doi: 10.1074/jbc.M201052200. [DOI] [PubMed] [Google Scholar]
- Freije, J.M., Diez-Itza, I., Balbin, M., Sanchez, L.M., Blasco, R., Tolivia, J., Lopez-Otin, C. Molecular cloning and expression of collagenase-3, a novel human matrix metalloproteinase produced by breast carcinomas. J. Biol. Chem. 1994;269:16766–16773. [PubMed] [Google Scholar]
- Gimbrone M.A., Jr, Leapman, S.B., Cotran, R.S., Folkman, J. Tumor dormancy in vivo by prevention of neovascularization. J. Exp. Med. 1972;136:261–276. doi: 10.1084/jem.136.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 2004;279:169–197. doi: 10.1007/978-3-642-18930-2_11. [DOI] [PubMed] [Google Scholar]
- Goldberg, G.I., Wilhelm, S.M., Kronberger, A., Bauer, E.A., Grant, G.A., Eisen, A.Z. Human fibroblast collagenase. Complete primary structure and homology to an oncogene transformation-induced rat protein. J. Biol. Chem. 1986;261:6600–6605. [PubMed] [Google Scholar]
- Gorlach, A., Camenisch, G., Kvietikova, I., Vogt, L., Wenger, R.H., Gassmann, M. Efficient translation of mouse hypoxia-inducible factor-1α under normoxic and hypoxic conditions. Biochim. Biophys. Acta. 2000;1493:125–134. doi: 10.1016/s0167-4781(00)00172-x. [DOI] [PubMed] [Google Scholar]
- Hay, N., Sonenberg, N. Upstream and downstream of mTOR. Genes & Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- Huang, L.E., Gu, J., Schau, M., Bunn, H.F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huez, I., Creancier, L., Audigier, S., Gensac, M.C., Prats, A.C., Prats, H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol. Cell. Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G., Sarnow, P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizaka-Kondoh, S., Inoue, M., Harada, H., Hiraoka, M. Tumor hypoxia: A target for selective cancer therapy. Cancer Sci. 2003;94:1021–1028. doi: 10.1111/j.1349-7006.2003.tb01395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar, A.A., Hatzoglou, M. Internal ribosome entry sites in cellular mRNAs: Mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- Koritzinsky, M., Seigneuric, R., Magagnin, M.G., van den Beucken, T., Lambin, P., Wouters, B.G. The hypoxic proteome is influenced by gene-specific changes in mRNA translation. Radiother. Oncol. 2005;76:177–186. doi: 10.1016/j.radonc.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Koritzinsky, M., Magagnin, M.G., van den Beucken, T., Seigneuric, R., Savelkouls, K., Dostie, J., Pyronnet, S., Kaufman, R.J., Weppler, S.A., Voncken, J.W., et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis, C., Naczki, C., Koritzinsky, M., Rastani, S., Diehl, A., Sonenberg, N., Koromilas, A., Wouters, B.G. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2α. Mol. Cell. Biol. 2002;22:7405–7416. doi: 10.1128/MCB.22.21.7405-7416.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis, C., Wouters, B.G. “Translating” tumor hypoxia: Unfolded protein response (UPR)-dependent and UPR-independent pathways. Mol. Cancer Res. 2006;4:423–426. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- Kozak, M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Kraggerud, S.M., Sandvik, J.A., Pettersen, E.O. Regulation of protein synthesis in human cells exposed to extreme hypoxia. Anticancer Res. 1995;15:683–686. [PubMed] [Google Scholar]
- Lang, K.J., Kappel, A., Goodall, G.J. Hypoxia-inducible factor-1α mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol. Biol. Cell. 2002;13:1792–1801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, L., Simon, M.C. Regulation of transcription and translation by hypoxia. Cancer Biol. Ther. 2004;3:492–497. doi: 10.4161/cbt.3.6.1010. [DOI] [PubMed] [Google Scholar]
- Liu, L., Cash, T.P., Jones, R.G., Keith, B., Thompson, C.B., Simon, M.C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak, D.G., Sarnow, P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Martin, T.E., Hartwell, L.H. Resistance of active yeast ribosomes to dissociation by KCl. J. Biol. Chem. 1970;245:1504–1506. [PubMed] [Google Scholar]
- Merrick, W.C. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Meyuhas, O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Pain, V.M. Initiation of protein synthesis in eukaryotic cells. Eur. J. Biochem. 1996;236:747–771. doi: 10.1111/j.1432-1033.1996.00747.x. [DOI] [PubMed] [Google Scholar]
- Parangi, S., O'Reilly, M., Christofori, G., Holmgren, L., Grosfeld, J., Folkman, J., Hanahan, D. Antiangiogenic therapy of transgenic mice impairs de novo tumor growth. Proc. Natl. Acad. Sci. 1996;93:2002–2007. doi: 10.1073/pnas.93.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Richter, J.D., Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- Sarnow, P., Cevallos, R.C., Jan, E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005;33:1479–1482. doi: 10.1042/BST0331479. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. Hypoxia-inducible factor 1: Oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 2001;7:345–350. doi: 10.1016/s1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. HIF-1 and tumor progression: Pathophysiology and therapeutics. Trends Mol. Med. 2002;8:S62–S67. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. Angiogenesis in ischemic and neoplastic disorders. Annu. Rev. Med. 2003;54:17–28. doi: 10.1146/annurev.med.54.101601.152418. [DOI] [PubMed] [Google Scholar]
- Sonna, L.A., Cullivan, M.L., Sheldon, H.K., Pratt, R.E., Lilly, C.M. Effect of hypoxia on gene expression by human hepatocytes (HepG2) Physiol. Genomics. 2003;12:195–207. doi: 10.1152/physiolgenomics.00104.2002. [DOI] [PubMed] [Google Scholar]
- Stein, I., Itin, A., Einat, P., Skaliter, R., Grossman, Z., Keshet, E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinton, S.A., Buc-Calderon, P.M. Hypoxia increases the association of 4E-binding protein 1 with the initiation factor 4E in isolated rat hepatocytes. FEBS Lett. 1999;446:55–59. doi: 10.1016/s0014-5793(99)00185-4. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Galy, B., Pyronnet, S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Beucken, T., Koritzinsky, M., Wouters, B.G. Translational control of gene expression during hypoxia. Cancer Biol. Ther. 2006;5:749–755. doi: 10.4161/cbt.5.7.2972. [DOI] [PubMed] [Google Scholar]
- Vaupel, P., Kallinowski, F., Okunieff, P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: A review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- Wouters, B.G., van den Beucken, T., Magagnin, M.G., Koritzinsky, M., Fels, D., Koumenis, C. Control of the hypoxic response through regulation of mRNA translation. Semin. Cell Dev. Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]