Abstract
Sequence comparison of several RNA m5C methyltransferases identifies two conserved cysteine residues that belong to signature motifs IV and VI of RNA and DNA methyltransferases. While the cysteine of motif IV is used as the nucleophilic catalyst by DNA m5C methyltransferases, this role is fulfilled by the cysteine of motif VI in Escherichia coli 16S rRNA m5C967 methyltransferase, but whether this conclusion applies to other RNA m5C methyltransferases remains to be verified. Yeast tRNA m5C methyltransferase Trm4p is a multisite-specific S-adenosyl-L-methionine-dependent enzyme that catalyzes the methylation of cytosine at C5 in several positions of tRNA. Here, we confirm that Cys310 of motif VI in Trm4p is essential for nucleophilic catalysis, presumably by forming a covalent link with carbon 6 of cytosine. Indeed, the enzyme is able to form a stable covalent adduct with the 5-fluorocytosine-containing RNA substrate analog, whereas the C310A mutant protein is inactive and unable to form the covalent complex.
Keywords: 5-fluorocytosine, RNA methyltransferase, Trm4, cysteine nucleophile, m5C
INTRODUCTION
All types of RNA molecules contain modified nucleosides that are formed post-transcriptionally by specific RNA modification enzymes. The role of these modifications is to ensure translation fidelity as well as correct RNA folding, three-dimensional RNA structure stabilization, and proper recognition of RNA by its translation partners (Agris 2004; Grosjean 2005). Transfer RNAs are, by far, the RNA species that contain the most diverse types of modification, with >90 chemically different modified nucleosides (Rozenski et al. 1999; Dunin-Horkawicz et al. 2006). One of the most frequently encountered modifications, the methylation of cytosine at carbon 5 (m5C, 5-methylcytosine), occurs at different positions in most eukaryotic and archaeal tRNAs, but also in DNAs, rRNAs, and possibly other cellular RNAs of all three domains of life. From sequence alignments and phylogenetic analysis of the enzymes catalyzing this modification in RNAs, the RNA m5C methyltransferases (MTases), 12 subfamilies have been identified (Reid et al. 1999; Bujnicki et al. 2004). They all contain signature motifs that are highly homologous to those found in DNA m5C MTases, including different S-adenosyl-L-methionine (SAM)-binding motifs and a ProCys sequence in motif IV containing the essential cysteine nucleophile catalyst of DNA m5C MTases (Chen et al. 1991).
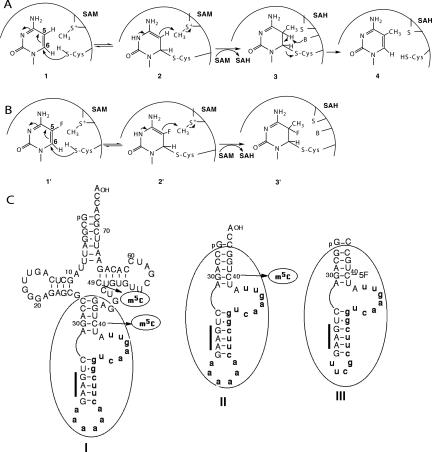
The mechanism of m5C formation in DNA and RNA involves first the attack of the thiol group of a cysteine nucleophile on C6 of cytosine 1, forming an enolate (or enol) intermediate 2 with a covalent bond between the thiol group and C6 of the pyrimidine ring (Fig. 1A). This step allows activation of C5 for the subsequent electrophilic substitution by the methyl group of SAM, generating a methylated nucleic acid • protein intermediate 3. Finally, proton abstraction from C5 by a general base and β elimination of the enzyme lead to m5C-containing product 4 and regeneration of the free enzyme. The same mechanism is used by enzymes that modify uridine or dihydrouridine at C5 of the pyrimidine ring, like Escherichia coli tRNA m5U54 MTase (Kealey and Santi 1991; Gu and Santi 1992; Kealey et al. 1994), pseudouridine synthases (Huang et al. 1998; Gu et al. 1999), and thymidylate synthase (Carreras and Santi 1995). In the cases of E. coli 23S rRNA m5U1939 MTase and tRNA pseudouridine synthase TruB, a 5,6-dihydropyrimidine intermediate (Ivanetich and Santi 1992) has been directly visualized in crystal structures of covalent complexes between enzymes and 5-fluoropyrimidine-containing RNAs that act as mechanism-based inhibitors (Hoang and Ferre-D'Amare 2001; Pan et al. 2003; Lee et al. 2005). Indeed, when a fluorinated substrate analog 1′ is used, the stable carbon–fluorine bond prevents β elimination of the enzyme, which may allow trapping a covalent methylated RNA • enzyme complex 3′ (Fig. 1B; Lee et al. 2005).
FIGURE 1.
Schemes of the mechanism for methylation of cytosine nucleosides and different substrates of Trm4p. (A) Attack by the nucleophilic thiol group of a catalytic cysteine on C6 of cytosine 1 creates a covalent intermediate 2 that serves to activate C5 as a carbanion that is stabilized by resonance. The carbanion attacks the methyl group of SAM nucleophilically, which causes the formation of a covalent methylated enzyme • nucleoside complex 3, containing 5-methyl-5,6 dihydrocytosine. Then, deprotonation of C5 leads to the m5C-containing product 4, S-adenosyl-homocysteine (SAH), and free enzyme. (B) Upon incubation with 5-fluoro-containing nucleoside 1′, the covalent intermediate 2′ is formed. Methylation of 2′ by SAM leads to trapping of the covalent 5-methyl-5-fluorohydrocytosine complex 3′ since fluorine at C5 cannot be abstracted. (C) Sequences and secondary structures of yeast tRNAPhe precursor (panel I), mini-tRNAPhe (panel II), and 36-mer FC-mini-RNA substrate analog (panel III). The intron is indicated in bold small letters and the anticodon sequence by a thick bar.
Unlike DNA m5C MTases, in the case of E. coli 16S rRNA m5C967 MTase (also called Fmu or RsmB), the nucleophile in the first reaction step is not provided by the cysteine of the ProCys sequence of motif IV. Instead, this role appears to be fulfilled by an additional cysteine residue belonging to motif VI that is conserved in RNA m5C MTases. This cysteine is not found in DNA m5C MTases but aligns with the nucleophilic cysteine of E. coli tRNA m5U54 MTase (Kealey and Santi 1991; Liu and Santi 2000; Bujnicki et al. 2004). The nucleophilic role for this cysteine in E. coli 16S rRNA m5C967 MTase has been shown by testing the activity and the ability to form a covalent complex with 5-fluorocytosine-containing RNA of site-directed mutants of the two conserved cysteine residues present in motifs IV and VI (Liu and Santi 2000).
In the case of yeast Nop2p, a putative rRNA m5C MTase essential for ribosome assembly (Hong et al. 1997; Wu et al. 2001), the other conserved cysteine (ProCys cysteine of motif IV) has been proposed to be important for the breakdown of the covalent enzyme • RNA catalytic intermediate. Indeed, the mutation of this cysteine into either serine or alanine leads to cell lethality (King et al. 1999) together with the accumulation of covalent complexes between the mutant protein and uncharacterized cellular RNA. Although the catalytic activity of yeast Nop2p has not yet been characterized, this observation is consistent with a mechanism involving a nucleophilic attack by the cysteine of motif VI to form a covalent Michael adduct, followed by the release of methylated RNA from the enzyme assisted by the cysteine of motif IV (Fig. 1A). The lethality of the ProCys cysteine mutation in Nop2p could then be explained by an abnormal stability of these covalent complexes sequestering rRNA. Indeed, when the putative nucleophilic cysteine of motif VI is also mutated, the ability of the enzyme to form covalent complexes is lost, and the cell is viable (King and Redman 2002).
Yeast tRNA m5C MTase (Trm4p) catalyzes the SAM-dependent formation of 5-methylcytosine at four different positions depending on the tRNAs (Motorin and Grosjean 1999). In Trm4p, the conserved cysteines in motifs IV and VI are, respectively, Cys260 and Cys310. The importance of these two cysteines has previously been investigated by studying the C260A, C260S, C310S, and C260S:C310S mutants of Trm4p (King and Redman 2002; Redman 2006). The C310S and C260S:C310S mutants of Trm4p had no detectable methyl transfer activity, showing that Cys310 of motif VI is important for catalysis. A nucleophilic role of Cys310 is suggested by the sequence homology of Trm4p with E. coli 16S rRNA m5C967 MTase for which the corresponding cysteine was shown to be essential for nucleophilic catalysis (Liu and Santi 2000). In addition, covalent complexes of C260A and C260S Trm4p mutants with endogenous RNA of the protein expression cell were shown to accumulate, and such complexes were not observed in the cases of the native enzyme or Cys310 mutants (King and Redman 2002; Redman 2006). These experiments support the predictions of a nucleophilic role for Cys310 and the assistance of Cys260 of motif IV for the breakdown of the covalent adduct in catalysis (Fig. 1A). Here, we show that Trm4p is able to form a covalent complex with a 5-fluorocytosine-containing mini-RNA substrate analog (Fig. 1B), in contrast to the C310A mutant, which has also lost the capacity to catalyze m5C formation. These results confirm that Cys310 of motif VI is essential for nucleophilic catalysis by Trm4p and agree with the postulated mechanism for Trm4p.
RESULTS AND DISCUSSION
The three-dimensional structure of tRNA is not required for recognition by Trm4p
In yeast tRNAs, m5C is found at positions 48 and/or 49 in most tRNAs and at positions 34 and 40 only in two yeast tRNAs, tRNALeu and tRNAPhe, respectively (Sprinzl and Vassilenko 2005). The enzymatic formation of m5C at positions 34 and 40 catalyzed by Trm4p is strictly dependent on the presence of the intron but not the formation of m5C in positions 48 or 49. A mini-RNA called mini-tRNAPhe, composed of the anticodon-stem–loop extended by the intron of the yeast tRNAPhe precursor (Fig. 1C, panels I and II), was shown to be an efficient in vitro substrate for Trm4p (Motorin and Grosjean 1999). We designed a new 36-mer mini-RNA that could be used for further NMR and crystallographic studies. This mini-RNA binds Trm4p with similar affinity as mini-tRNAPhe (data not shown) and is also modified by Trm4p (Fig. 2A).
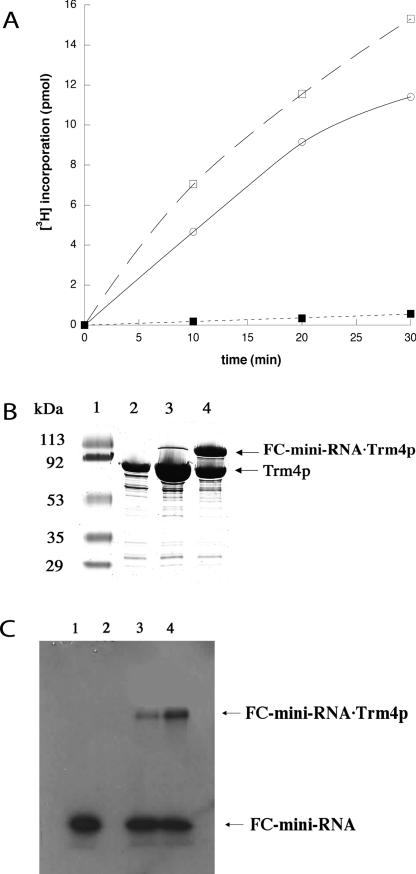
FIGURE 2.
Formation of a SAM-dependent covalent complex between Trm4p and FC-mini-RNA substrate analog. (A) Trm4p was incubated in the presence of [3H]SAM with mini-RNA (open squares), FC-mini-RNA (closed squares), or yeast tRNAPhe (open circles). The transfer of the labeled methyl group of [3H]SAM to RNA was monitored by filtering RNA, precipitated under acidic conditions after reaction, and quantifying its radioactivity. (B) SDS-PAGE analysis testing the formation of a covalent complex of Trm4p with 36-mer FC-mini-RNA substrate analog after Coomassie blue staining. (Lane 1) Molecular weight markers; (lane 2) 3 μg of purified Trm4p; incubation mixture of Trm4p and FC-mini-RNA after 3 h reaction in the (lane 3) absence or (lane 4) presence of SAM. (C) SDS-PAGE analysis of the covalent complex of Trm4p with [32P]FC-mini-RNA substrate analog (lane 4) in the presence of SAM after autoradiography. The controls are as follows: (lane 1) no Trm4p; (lane 2) no RNA; (lane 3) no SAM.
Trm4p is able to form a covalent stable complex with 5-fluorocytosine-containing RNA
An analog of the 36-mer mini-RNA substrate containing 5-fluorocytosine (FC) at the target position (FC-mini-RNA) (Fig. 1C, panel III), was used to test the ability of Trm4p to form a covalent dihydrocytosine intermediate in catalysis (Fig. 1A,B). Trm4p binds the FC-mini-RNA substrate analog and the mini-tRNAPhe substrate with similar affinities (data not shown). The ability of Trm4p to form a covalent adduct with FC-mini-RNA was tested in the presence or absence of SAM (Fig. 2B,C). As visualized on SDS-PAGE gel after protein staining with Coomassie blue, Trm4p forms a SAM dependent complex with FC-mini-RNA that migrates more slowly than the free enzyme (Fig. 2B). The adduct appears to be covalent since it is stable on heating in SDS and during SDS-PAGE. The presence of RNA in the complex was confirmed by carrying out the same experiment again with 3′-end-labeled [32P]FC-mini-RNA. Indeed, the complex was then visualized on SDS-PAGE by autoradiography (Fig. 2C). The FC-mini-RNA can be methylated (Fig. 2A) since the incorporation of tritium from [3H]SAM into the RNA product is detectable. The slight observed incorporation corresponds to less than one enzyme cycle since only 0.06 mol of product per mole of enzyme is formed in 30 min. The covalent trapping of the enzyme by the suicide FC-mini-RNA substrate, inducing no regeneration of free enzyme, is likely responsible for the slow methylation rate. Moreover, the electronegativity of the fluor that makes complex 2′ (Fig. 1B) less reactive than complex 2 (Fig. 1A) for the nucleophilic attack of the SAM methyl group could also slow down the pre-steady-state reaction. The covalent C5-fluoro-methyl cytosine complex 3′ (Fig. 1B) can also be purified by chromatography on an anion exchange monoQ column (Fig. 3A, peak B; Fig. 3B, lane B). Altogether, these results are consistent with the formation of a stable covalent adduct between a thiol group of the enzyme and C6 of 5-fluorocytosine-containing RNA, which is subsequently methylated at C5 (Fig. 1B).
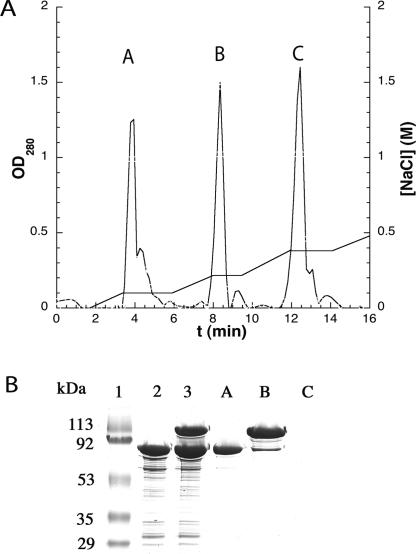
FIGURE 3.
Isolation of a covalent Trm4p • FC-mini-RNA complex. (A) MonoQ chromatography of the incubation mixture of Trm4p with FC-mini-RNA substrate analog in the presence of SAM using a NaCl gradient. (B) SDS-PAGE analysis of the incubation mixture of Trm4p and FC-mini-RNA in the presence of SAM before and after monoQ chromatography, after Coomassie blue staining. (Lane 1) Molecular weight markers; (lane 2) 3 μg of purified Trm4p; incubation mixture of Trm4p and FC-mini-RNA after 3 h reaction (lane 3) in the presence of SAM and after monoQ chromatography: (lane A) peak A; (lane B) peak B; (lane C) peak C. Peak C does not contain protein since it does not give a band that can be stained by Coomassie blue. Yet, peak C could react again with Trm4p to form additionnal Trm4p • FC-mini-RNA complex (data not shown) and therefore contains RNA.
The C310A-Trm4p mutant is correctly folded, binds RNA substrates with similar affinity as the native enzyme, but is not able to form a covalent complex
To confirm the function of Cys310 of motif VI in the covalent complex formation, the mutant C310A-Trm4p was produced. As a control, the far-UV-CD spectra of the native and mutated proteins were compared (Fig. 4A), in order to verify that the mutation does not affect the protein folding. The data show that the C310A mutant is correctly folded since there is no significant difference between the two proteins in the secondary structure. In addition, the apparent dissociation constants of the native and mutated proteins both for a full-length yeast tRNAPhe and mini-tRNAPhe were determined by a gel shift assay and compared (Fig. 4B). The apparent K d value of Trm4p for tRNAPhe of 45 ± 5 nM is ∼22-fold lower than that for mini-tRNAPhe (K d = 1 ± 0.1 μM). The C310A mutant shows only a slightly lower affinity for RNA compared to Trm4p with K d values of 62.5 ± 7 nM and 1.2 ± 0.1 μM, for tRNAPhe and mini-tRNAPhe, respectively. These affinities ensure that the mutant protein interacts with RNA substrates similarly to the native enzyme. Finally, the catalytic properties of the mutant enzyme were examined by testing its ability to form a covalent complex with the FC-mini-RNA substrate analog (Fig. 4C) and to catalyze the transfer of the labeled methyl group of [3H]SAM to the yeast tRNAPhe transcript that is modified at position 49 by Trm4p (Motorin and Grosjean 1999) or the 36-mer mini-RNA substrate (data not shown). Both these tests indicate that the C310A mutant of Trm4p is unable to bind RNA covalently as well as to transfer the methyl group from SAM to the RNA substrate since no activity was detectable. Therefore, Cys310 of motif VI is likely the nucleophilic catalyst of Trm4p.
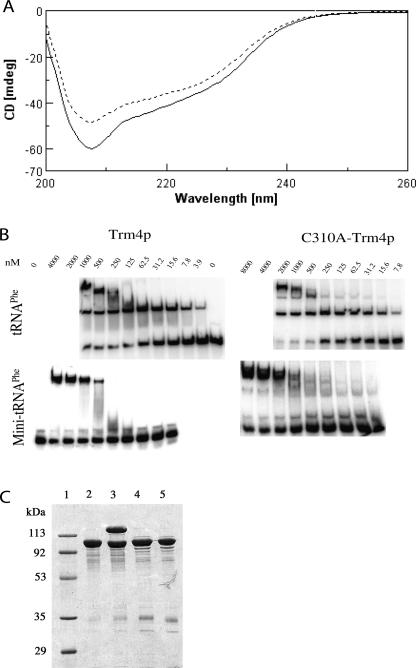
FIGURE 4.
Characteristics of the C310A mutant protein. (A) Far-UV CD spectra in 0.02 M Tris-HCl buffer (pH 8), 50 mM NaF at a protein concentration of 10 μM or 8 μM for Trm4p (continuous line) and C310A-Trm4p (dashed line), respectively. (B) Comparison of the affinity constants of Trm4p and C310A-Trm4p for yeast tRNAPhe and mini-tRNAPhe using a gel retardation assay with labeled RNA and different concentrations of proteins. (C) SDS-PAGE analysis testing the formation of a covalent complex of C310A-Trm4p with 36-mer FC-mini-RNA substrate analog after Coomassie blue staining. (Lane 1) Molecular weight markers; 3 μg of purified Trm4p (lane 2) alone or (lane 3) in the presence of FC-mini-RNA and SAM; 3 μg of purified C310A-Trm4p (lane 4) alone or (lane 5) in the presence of FC-mini-RNA and SAM.
Conclusion
Enzymatic methylation on C5 of cytosine in DNA and RNA implies an initial nucleophilic attack at C6 by a cysteine of the catalytic center, leading to a covalent link between the target nucleotide and the MTase. Direct evidence for covalent catalysis by the m5C MTases is usually obtained by trapping the enzyme as a denaturant-resistant complex with a 5-fluoro-containing-nucleotic acid substrate analog.
DNA and RNA m5C MTases contain homologous sequence motifs, but whereas DNA m5C MTases use the cysteine of motif IV as the nucleophilic catalyst, the cysteine nucleophile of E. coli 16S rRNA m5C967 MTase belongs to motif VI and is not conserved in DNA m5C MTases. Although it was anticipated that other RNA m5C MTases would use the conserved cysteine of motif VI as the nucleophilic catalyst (Liu and Santi 2000), it remained to establish that it also holds true for other RNA species like tRNA.
Here, we show the formation of a stable covalent complex between yeast tRNA m5C MTase Trm4p and a mini-RNA substrate that was modified at the C5 position of target cytosine by a fluorine (Fig. 1B). Since previous attempts to isolate the C260A mutant of Trm4p have shown that the protein is covalently linked with endogenous RNA of the protein expression cell (King and Redman 2002), a residue different from Cys260 of motif IV appears to be the nucleophile. Indeed, we demonstrate that Cys310 of motif VI is essential for nucleophilic catalysis of Trm4p by showing that the C310A mutant protein, which is correctly folded and binds RNA with similar affinity as the native protein, does not catalyze m5C formation and is not able to form a covalent complex with 5-fluoro-cytosine-containing RNA substrate analog. In other respects, the stable covalent FC-mini-RNA • Trm4p complex is easily purified, and is therefore amenable to crystallization studies. This strategy has already been proved to be efficient to lead to the structures of RNA modification enzymes in complex with RNA (Hoang and Ferre-D'Amare 2001; Pan et al. 2003; Lee et al. 2005). Solving the crystal structure of Trm4p in complex with 5-fluoro-containing RNA should illuminate further details about the reaction mechanism and the mode of RNA recognition by the enzyme.
The mutation of the other conserved cysteine, Cys260, in Trm4p, belonging to the ProCys sequence of motif IV, leads to the accumulation of stable protein • RNA complexes, which supports the idea that the cysteine of motif IV is important for release of methylated RNA (King and Redman 2002; Redman 2006). This cysteine likely functions as a base, deprotoning C5 (Fig. 1A) in the final step of catalysis. This proposition is reinforced by the crystal structures of E. coli 16S rRNA m5C967 MTase (Foster et al. 2003), of E. coli 16S rRNA m5C1407 MTase (Hallberg et al. 2006), and of a putative m5C RNA MTase from Pyrococcus horikoshii (Ishikawa et al. 2004), in which both conserved cysteines of motifs IV and VI are located adjacent to each other.
In summary, our experiments are consistent with the hypothesis that Cys310 is the catalytic nucleophile in Trm4p. This is the second example, after 16S rRNA m5C967 MTase (Liu and Santi 2000), indicating that the majority if not all RNA m5C MTases use the conserved cysteine of motif VI as a nucleophile, in contrast to DNA m5C MTases.
MATERIALS AND METHODS
Preparation of RNA substrates
Nonlabeled RNA, [α-32P]CTP yeast tRNAPhe, and 40-mer [α-32P]CTP mini-tRNAPhe transcripts were obtained by in vitro transcription with T7 RNA polymerase of linearized plasmids and purification of the resulting transcripts on urea gels as described (Grosjean et al. 1990). 36-mer FC-mini-RNA was bought from Dharmacon and was labeled at the 3′ end with [5′-32P]pCp using T4 RNA ligase (England et al. 1980), then purified by electrophoresis on 7 M urea–10% PAGE.
Mutagenesis
pET28b-C310ATrm4 was prepared by mutagenesis of pET28b/YBL024w (Motorin and Grosjean 1999) with the QuickChange site-directed mutagenesis kit (Stratagene) by using two complementary primers 5′-CGGTAGATTGGTTTACTCAACCGCTTCTTAAAATCC-3′. An Ala codon GCT (underlined) replaced the wild-type Cys codon TGT. Clones carrying the mutant plasmid were identified by restriction enzyme digestion with BamHI and NheI and confirmed by DNA sequencing of the whole inserted gene. The single-point mutant (C310A) of Trm4p encoded by the recombinant plasmid was expressed in the E. coli BL21(DE3) host strain (Novagen). To improve the expression level of the recombinant protein, the strain was cotransformed by the plasmid pDC952 bearing the gene argU coding for minor E. coli tRNAArg UCU and plated out on LB agar containing kanamycin (25 μg/mL) and chloramphenicol (30 μg/mL). The mutant protein was shown by SDS-PAGE to be present in the soluble fraction of the total cell lysate.
Trm4p and C310A-Trm4p proteins purification
The bacteria were grown in MM9 minimal medium supplemented with 1 mM MgSO4, 0.3 mM CaCl2, 0.2% glucose, 0.5% casamino acids, 30 μg/mL chloramphenicol, and 50 μg/mL kanamycin. Cultures (1 L) were grown at 37°C to an OD600 of 0.8, and expression was induced at 20°C by addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 1 mM. After 3 h of induction, the cells were collected by centrifugation. All purification steps were conducted at 4°C. E. coli cells expressing the recombinant enzymes were resuspended in five volumes of buffer A (0.3 M NaCl, 10 mM β-mercaptoethanol, 1% [v/v] protease inhibitor cocktail for use in purification of Histidine-tagged proteins [Sigma], 50 mM sodium phosphate at pH 8) supplemented with 20 mM imidazole. Cells were disrupted by sonication, and the lysate was centrifuged at 30,000g for 30 min. The supernatant was loaded on Ni2+-NTA affinity column (QIAGEN) equilibrated in buffer A supplemented with 30 mM imidazole and eluted with buffer A supplemented with 150 mM imidazole. EDTA at a final concentration of 5 mM was added to the eluted protein fractions, which were then dialyzed against 20 mM Tris-HCl (pH 8), 50 mM NaCl, 2 mM DTT, and 5 mM EDTA.
Measurement of RNA binding affinity
[32P]-labeled yeast tRNAPhe or mini-tRNAPhe (10 fmol) was incubated for 20 min at 25°C in 20 μL of 20 mM Tris-HCl (pH 8), 50 mM NaCl, 0.1 mg/mL BSA, 2 mM DTT, and 10% glycerol at different concentrations of Trm4p. After electrophoresis at 4°C on a 6% native polyacrylamide gel, the mobility shift of RNA was visualized by autoradiography.
In vitro enzymatic assay for tRNA m5C methyltransferase activity
The m5C methyltransferase activity was tested by monitoring the [3H]methyl group incorporation from [3H]SAM (15 Ci/mmol; Amersham) into nonradioactive RNA. Enzyme (200 mM) was incubated at 30°C with 660 nM [3H]SAM, 1 μM RNA, in 50 μL of 20 mM Tris-HCl (pH 8), 50 mM NaCl, and 2 mM DTT. The reaction was stopped by addition of cold 5% trichloroacetic acid, and the precipitate containing protein and RNA was collected by filtration through a GF/C filter (Whatman). The filter was washed with cold 5% trichloroacetic acid and dried, and the radioactivity was measured by liquid scintillation counting.
Formation and purification of the FC-mini-RNA • Trm4p complex
The formation of the FC-mini-RNA • enzyme complex was analyzed on a 12% acrylamide gel. Enzyme (35 μM) was incubated for 2 h at 25°C with FC-mini-RNA (35 μM) and SAM (0.115 mM) in 25 mM Tris-HCl (pH 7.5), 50 mM KCl, and 1 mM DTT. For the quantitative formation of the FC-mini-RNA • Trm4p complex, 88 μM Trm4p was mixed with 11.5 mM SAM and 74 μM FC-mini-RNA in the previous buffer, then loaded onto a Mono Q HR 5/5 column (Pharmacia) and eluted at 1 mL/min with a step gradient of NaCl in 20 mM Tris-HCl (pH 8).
CD spectra
CD spectra were obtained with a Jasco JB10 spectrophotometer.
ACKNOWLEDGMENTS
We thank Bernard Guibert for help in CD spectrophotometry, Magali Carbonneaux for purifying the mutant protein, Dominique Fourmy for help in the design and synthesis of the 36-mer mini-RNA substrate, Florence Lederer for helpful discussions, and Henri Grosjean for critical reading of the manuscript and advice throughout this work.
Footnotes
Abbreviations: MTase, methyltransferase; SAM, S-adenosyl-L-methionine; m5C, 5-methylcytosine; FC, 5-fluorocytosine; Trm4p, yeast tRNA m5C methyltransferase.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.515707.
REFERENCES
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki, J.M., Feder, M., Ayres, C.L., Redman, K.L. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res. 2004;32:2453–2463. doi: 10.1093/nar/gkh564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras, C.W., Santi, D.V. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- Chen, L., MacMillan, A.M., Chang, W., Ezaz-Nikpay, K., Lane, W.S., Verdine, G.L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Dunin-Horkawicz, S., Czerwoniec, A., Gajda, M.J., Feder, M., Grosjean, H., Bujnicki, J.M. MODOMICS: A database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England, T.E., Bruce, A.G., Uhlenbeck, O.C. Specific labeling of 3′ termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65:65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Foster, P.G., Nunes, C.R., Greene, P., Moustakas, D., Stroud, R.M. The first structure of an RNA m5C methyltransferase, Fmu, provides insight into catalytic mechanism and specific binding of RNA substrate. Structure. 2003;11:1609–1620. doi: 10.1016/j.str.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Grosjean, H. Fine-tuning of RNA functions by modification and editing. Springer; Berlin: 2005. [Google Scholar]
- Grosjean, H., Droogmans, L., Giege, R., Uhlenbeck, O.C. Guanosine modifications in runoff transcripts of synthetic transfer RNA-Phe genes microinjected into Xenopus oocytes. Biochim. Biophys. Acta. 1990;1050:267–273. doi: 10.1016/0167-4781(90)90179-6. [DOI] [PubMed] [Google Scholar]
- Gu, X., Santi, D.V. Covalent adducts between tRNA (m5U54)-methyltransferase and RNA substrates. Biochemistry. 1992;31:10295–10302. doi: 10.1021/bi00157a017. [DOI] [PubMed] [Google Scholar]
- Gu, X., Liu, Y., Santi, D.V. The mechanism of pseudouridine synthase I as deduced from its interaction with 5-fluorouracil-tRNA. Proc. Natl. Acad. Sci. 1999;96:14270–14275. doi: 10.1073/pnas.96.25.14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, B.M., Ericsson, U.B., Johnson, K.A., Andersen, N.M., Douthwaite, S., Nordlund, P., Beuscher A.E., IV, Erlandsen, H. The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain. J. Mol. Biol. 2006;360:774–787. doi: 10.1016/j.jmb.2006.05.047. [DOI] [PubMed] [Google Scholar]
- Hoang, C., Ferre-D'Amare, A.R. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- Hong, B., Brockenbrough, J.S., Wu, P., Aris, J.P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997;17:378–388. doi: 10.1128/mcb.17.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Pookanjanatavip, M., Gu, X., Santi, D.V. A conserved aspartate of tRNA pseudouridine synthase is essential for activity and a probable nucleophilic catalyst. Biochemistry. 1998;37:344–351. doi: 10.1021/bi971874+. [DOI] [PubMed] [Google Scholar]
- Ishikawa, I., Sakai, N., Tamura, T., Yao, M., Watanabe, N., Tanaka, I. Crystal structure of human p120 homologue protein PH1374 from Pyrococcus horikoshii . Proteins. 2004;54:814–816. doi: 10.1002/prot.10645. [DOI] [PubMed] [Google Scholar]
- Ivanetich, K.M., Santi, D.V. 5,6-Dihydropyrimidine adducts in the reactions and interactions of pyrimidines with proteins. Prog. Nucleic Acid Res. Mol. Biol. 1992;42:127–156. doi: 10.1016/s0079-6603(08)60575-9. [DOI] [PubMed] [Google Scholar]
- Kealey, J.T., Santi, D.V. Identification of the catalytic nucleophile of tRNA (m5U54)methyltransferase. Biochemistry. 1991;30:9724–9728. doi: 10.1021/bi00104a022. [DOI] [PubMed] [Google Scholar]
- Kealey, J.T., Gu, X., Santi, D.V. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie. 1994;76:1133–1142. doi: 10.1016/0300-9084(94)90042-6. [DOI] [PubMed] [Google Scholar]
- King, M.Y., Redman, K.L. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry. 2002;41:11218–11225. doi: 10.1021/bi026055q. [DOI] [PubMed] [Google Scholar]
- King, M., Ton, D., Redman, K.L. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J. 1999;337:29–35. [PMC free article] [PubMed] [Google Scholar]
- Lee, T.T., Agarwalla, S., Stroud, R.M. A unique RNA Fold in the RumA-RNA-cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell. 2005;120:599–611. doi: 10.1016/j.cell.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Santi, D.V. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. 2000;97:8263–8265. doi: 10.1073/pnas.97.15.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin, Y., Grosjean, H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, H., Agarwalla, S., Moustakas, D.T., Finer-Moore, J., Stroud, R.M. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl. Acad. Sci. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redman, K.L. Assembly of protein–RNA complexes using natural RNA and mutant forms of an RNA cytosine methyltransferase. Biomacromolecules. 2006;7:3321–3326. doi: 10.1021/bm051012l. [DOI] [PubMed] [Google Scholar]
- Reid, R., Greene, P.J., Santi, D.V. Exposition of a family of RNA m5C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999;27:3138–3145. doi: 10.1093/nar/27.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., McCloskey, J.A. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Vassilenko, K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–D140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Wu, P., Aris, J.P. Nucleolar protein Nop12p participates in synthesis of 25S rRNA in Saccharomyces cerevisiae . Nucleic Acids Res. 2001;29:2938–2949. doi: 10.1093/nar/29.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]