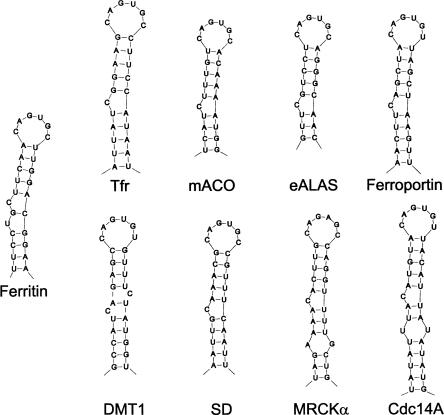
Abstract
An RNA hairpin structure referred to as the iron-responsive element (IRE) and iron regulatory proteins (IRPs) are key players in the control of iron metabolism in animal cells. They regulate translation initiation or mRNA stability, and the IRE is found in a variety of mRNAs, such as those encoding ferritin, transferrin receptor (Tfr), erythroid aminolevulinic acid synthase (eALAS), mitochondrial aconitase (mACO), ferroportin, and divalent metal transporter 1 (DMT1). We have studied the evolution of the IRE by considering all mRNAs previously known to be associated with this structure and by computationally examining its occurrence in a large variety of eukaryotic organisms. More than 100 novel sequences together with ∼50 IREs that were previously reported resulted in a comprehensive view of the phylogenetic distribution of this element. A comparison of the different mRNAs shows that the IREs of eALAS and mACO are found in chordates, those of ferroportin and Tfr1 are found in vertebrates, and the IRE of DMT1 is confined to mammals. In contrast, the IRE of ferritin occurs in a majority of metazoa including lower metazoa such as sponges and Nematostella (sea anemone). These findings suggest that the ferritin IRE represents the ancestral version of this type of translational control and that during the evolution of higher animals the IRE structure was adopted by other genes. On the basis of primary sequence comparison between different organisms, we suggest that some of these IREs developed by “convergent evolution” through stepwise changes in sequence, rather than by recombination events.
Keywords: iron-responsive element, RNA, evolution, IRP, bioinformatics, RNA secondary structure
INTRODUCTION
Local RNA structures in untranslated regions of mRNAs play an important role in translational control. Examples include riboswitches, which are frequent in bacteria (Nudler and Mironov 2004; Winkler 2005), and the selenocysteine insertion sequence (SECIS), which is present in both bacteria and eukaryotes and is responsible for incorporation of selenocysteine in proteins (Hubert et al. 1996; Walczak et al. 1997). Another RNA structural element is the iron-responsive element (IRE), which is part of a well-characterized system of translational control in eukaryotes. In the transferrin receptor (Tfr) mRNA there are five copies of the IRE located in the 3′-UTR, and in ferritin mRNA a single IRE is located in the 5′-UTR. In iron-starved cells the IREs are targets of two cytoplasmic iron regulatory proteins, IRP1 and IRP2. The binding of these proteins results in stabilization of Tfr mRNA and inhibition of ferritin translation. Conversely, when iron is abundant, IRP proteins tend to have lower affinity to IREs, and, as a result, Tfr mRNA degradation as well as ferritin translation is stimulated. IREs are also present in other mRNAs whose products are related to iron metabolism. One example is the erythroid aminolevulinic acid synthase (eALAS or ALAS2) (Duncan et al. 1999), which is responsible for the first step in tetrapyrrole synthesis. Other proteins whose mRNA contain IREs are the iron transporters ferroportin (Gunshin et al. 2001) and DMT1 (Abboud and Haile 2000). IREs have also been identified in mRNAs encoding mitochondrial aconitase and succinate dehydrogenase (Kohler et al. 1995; Gray et al. 1996), myotonic dystrophy kinase-related Cdc42-binding kinase α (MRCKα) (Cmejla et al. 2006), cdc14A (Sanchez et al. 2006), glycolate oxidase (Kohler et al. 1999), the Complex I 75-kDa subunit (Lin et al. 2001), and Alzheimer's amyloid precursor protein (Rogers et al. 2002).
IREs are 26–30-nucleotide (nt)-long hairpin-forming sequences with a CAGUGN apical loop sequence, which is conserved in all IREs (Thomson et al. 1999). N at position 6 can be A, C, or U but never G (Addess et al. 1997). The C in position 1 forms a base pair with G in position 5 (Addess et al. 1997; McCallum and Pardi 2003). This base pair is presumably required for efficient IRP binding (Henderson et al. 1994; Addess et al. 1997). Most IREs have a conserved C residue five bases upstream of the CAGUGN sequence, creating a bulge in the hairpin, whereas others, present in ferritin mRNAs, instead have a conserved bulge/loop UGC/C (Fig. 1). In the UGC/C-type IRE the G and C are paired (Gdaniec et al. 1998), and the results of in vitro selection experiments indicated that this base pair favors high-affinity binding by IRPs (Butt et al. 1996). IRE stems form an A-helix with a small distortion due to the conserved, unpaired C or the internal loop bulge (Gdaniec et al. 1998). The helix between the UGC/C distortion and the apical loop seems significant since it contributes to protein binding (Leibold et al. 1990).
FIGURE 1.
Classes of iron-responsive elements based on occurrence in mRNAs. The IREs are those of ferritin (Hentze et al. 1987), transferrin receptor (Tfr) (Casey et al. 1988), mitochondrial aconitase (mACO) (Butt et al. 1996), erythroid aminolevulinic synthase (eALAS) (Dandekar et al. 1991), ferroportin (Gunshin et al. 2001), divalent metal ion transporter (DMT1) (Abboud and Haile 2000), succinate dehydrogenase (SD) (Melefors 1996), myotonic dystrophy kinase-related Cdc42-binding kinase α (MRCKα) (Cmejla et al. 2006), and cell division cycle 14a (Cdc14a) (Sanchez et al. 2006). Human sequences are shown, except in the case of succinate dehydrogenase, where the Drosophila melanogaster sequence is shown.
The mechanism involving IRE and IRP proteins is an important mechanism from a physiological point of view as illustrated by diseases in which the molecular basis of the disease can be traced to these regulatory components. Thus, in the human hereditary disease hyperferritinemia cataract syndrome, ferritin L-chain IRE mutations interfere with IRP binding and translational repression (Girelli et al. 1995; Roetto et al. 2002). Autosomal dominant iron overload is caused by a mutation in the apical loop sequence of the H-ferritin IRE (Kato et al. 2001). Furthermore, a syndrome of progressive neurodegenerative disease and anemia develops in adult mice that are lacking IRP2 (LaVaute et al. 2001).
The iron-responsive element has previously been identified in a number of different mRNAs and in many different organisms. In order to better understand the evolution of IREs, we have here systematically examined the occurrence of these elements using computational screening of metazoan genome and mRNA sequences. Like other ncRNAs, the computational identification of IREs requires methods that make use of a combination of primary sequence as well as secondary structure motifs. We have previously identified and characterized different ncRNA families, mainly SRP RNA and RNase P and MRP RNA (Regalia et al. 2002; Rosenblad et al. 2004, 2006; Piccinelli et al. 2005). We have used methods based on primary sequence matching such as HMM profiles, pattern matching methods that exploit both primary sequence and secondary structure, and methods based on covariance models as used in Rfam (Griffiths-Jones et al. 2003). Here we have used similar methods to inventory IRE sequences in metazoa and present a number of elements that were not previously reported. Using this novel information, we are able to compare the evolution of IRE in different mRNAs.
RESULTS
We performed computational searches of IREs in available mRNAs and genomic sequences corresponding to all proteins related to iron metabolism. We first identified nucleotide sequences matching the protein under study. Sequences flanking the coding sequence and that were expected to contain the entire UTR region were then extracted and analyzed with respect to IRE. We also carried out a more unbiased search of all untranslated regions of human mRNAs with the Infernal software (Griffiths-Jones et al. 2003). This search did not identify IRE-containing mRNAs in addition to those encoding proteins previously known to be associated with IRE. The probability of finding a hairpin with the IRE consensus properties in UTR sequences in a random sequence is very low (<1e-4 in a 3000-nt sequence). Therefore, we believe that a vast majority of the IRE predictions reported here are reliable.
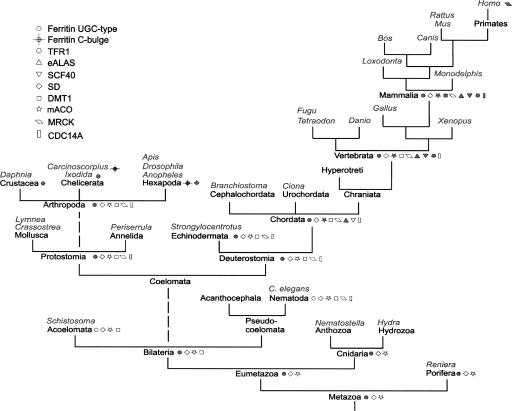
All IREs identified in this work (>100) as well as a majority of IREs that were previously reported (49) are listed in Tables 1–3. Secondary structure models of representative IREs are shown in Figure 1. In Tables 1–3 the distances to the AUG codon or to the termination codon of the coding sequence are indicated, as well as the distances for the 5′-UTR IREs to the 5′-end of the mRNA. The position of IRE relative to the 5′-end has been shown to be important for its function as a translational regulator (Goossen and Hentze 1992). Based on the numbers in Tables 1 and 3, it would seem that the ferritin (with the exception of insects) (Nichol and Winzerling 2002) and eALAS IREs typically are close to the 5′-end of the mRNA, whereas the ferroportin and mitochondrial aconitase (mACO) IREs are more distantly located. In the mACO mRNA the AUG codon is part of the IRE hairpin structure (Gray et al. 1996), and this position of AUG is highly conserved in mACO (Table 3). The sequences, structure, and phylogenetic distribution of the different IRE families (Fig. 1; Tables 1–3) are discussed in more detail below. The phylogenetic distribution of IREs and the corresponding proteins is summarized in Figure 2. In addition to the proteins listed in Table 1 and Figure 2, we also examined IRE structures of glycolate oxidase (Kohler et al. 1999), the Complex I 75-kDa subunit (Lin et al. 2001), and Alzheimer's amyloid precursor protein (Rogers et al. 2002). However, these IREs offer a problem from a bioinformatics perspective as they deviate strongly from the consensus structure of the IRE. Furthermore, these IRE sequences do not seem to be conserved among mammals.
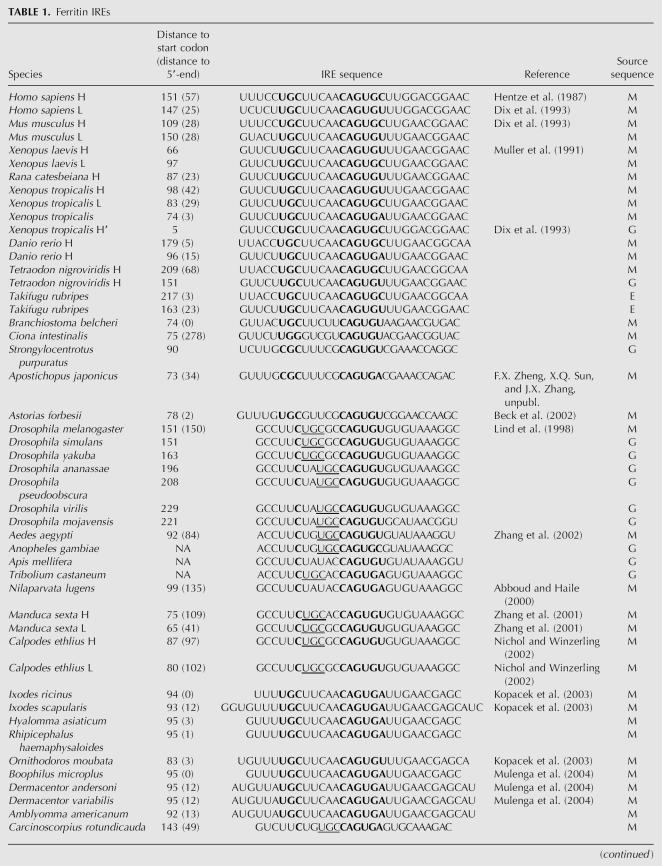
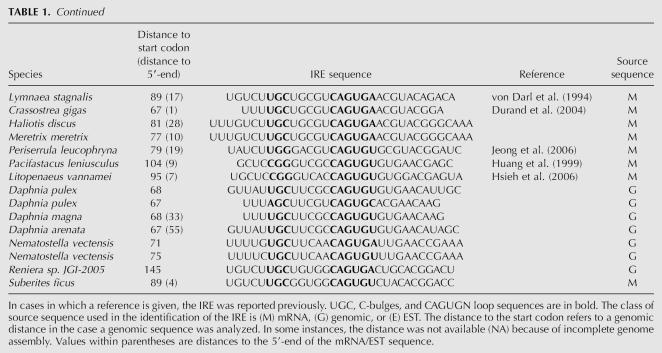
TABLE 1.
Ferritin IREs
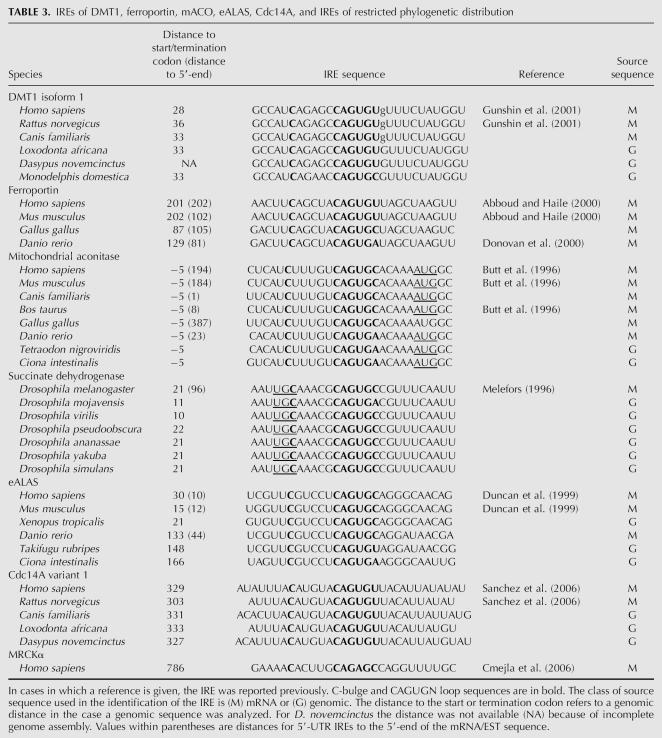
TABLE 3.
IREs of DMT1, ferroportin, mACO, eALAS, Cdc14A, and IREs of restricted phylogenetic distribution
FIGURE 2.
Phylogenetic distribution of iron-responsive elements and corresponding proteins. Open symbols refer to a situation in which a protein ortholog is found but not an IRE in the UTR. A filled symbol means that the protein as well as an IRE have been identified. Example species or genera analyzed in this work are shown in italics.
Ferritin
Ferritin is a highly conserved protein that is found in metazoa, plants, eubacteria, and archaea. In metazoa two ferritin chains are identified, H and L. Xenopus has an additional chain referred to as the H′ or M form (Dickey et al. 1987). H and H′ chains are characterized by catalytic activity associated with specific amino acid residues (Liu and Theil 2004), and these properties in combination with phylogenetic analysis were exploited to classify protein sequences as H/H′ or L. In Xenopus tropicalis we discovered yet another ferritin chain, but were not able to classify this on the basis of protein sequence (Table 1). Danio rerio (zebrafish) is unusual in the respect that one of the ferritin chain genes is present in many copies (on chromosome 3).
The IREs identified in ferritin mRNAs are shown in Table 1. In addition, we identified IREs in a number of mammals that are not shown in Table 1. For a majority of organisms where we found more than one ferritin chain, each of the corresponding mRNAs has an IRE. This includes human and mouse L and H chain mRNAs (Dix et al. 1993) as well as the X. tropicalis L, H, and H′ chain mRNAs (Table 1). As a rule, this IRE is of the UGC/C type as will be discussed further below. Human and mouse ferritin IREs have base-paired regions that flank the IRE (Dix et al. 1993). However, a comparison of all available ferritin mRNA sequences shows that such regions occur in very few metazoan species and are not evolutionarily conserved.
The only metazoan species where we failed to identify an IRE were Caenorhabditis elegans and Schistosoma mansoni, consistent with previous observations (Schussler et al. 1996; Gourley et al. 2003). We were not able to find IREs in organisms outside the metazoan group. IRE-like sequences have been identified in Bacillus subtilis (Alen and Sonenshein 1999) and in Escherichia coli (Dandekar et al. 1998). However, these IREs have structures that are different from the consensus IRE considered here, and it was not possible to find phylogenetic support for the bacterial structures as homologs in closely related bacteria could not be identified.
It is particularly interesting to note that 5′-UTR UGC/C-type IREs are identified even in lower metazoa such as Porifera (sponges) and Cnidaria (Fig. 3), showing that the ferritin IRE is remarkably conserved. Thus, the IREs in the lower metazoa are in most cases very similar to those in mammals (Fig. 3). The presence of IRE in lower metazoan ferritin mRNAs strongly suggests that IRE is a very early metazoan invention.
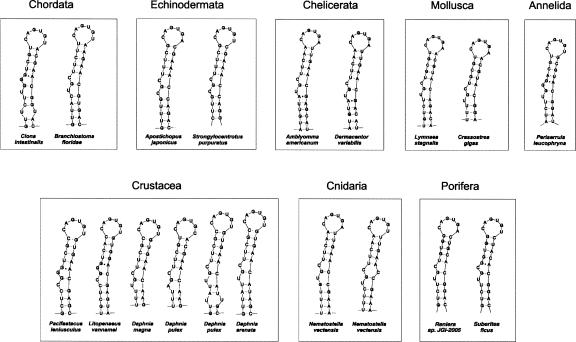
FIGURE 3.
Structure of ferritin IREs. Selected IREs of nonvertebrate metazoa are shown. All the IREs shown were identified in this work except for those of L. stagnalis (von Darl et al. 1994), C. gigas (Durand et al. 2004), P. leucophryna (Jeong et al. 2006), P. leniusculus (Huang et al. 1999), D. variabilis (Mulenga et al. 2004), and L. vannamei (Hsieh et al. 2006). Structural features of this class of IRE include (1) the UGC bulge where the G is able to pair with the C in the opposite strand; (2) a variability of the UGC consensus sequence; and (3) a G-A pair in some of the structures.
UGC/C-type IRE
As a rule, the ferritin IRE is of the UGC/C type. Exceptions are found in the group of insects and in the horseshoe crab as will be discussed further below. In this investigation we have collected a large number of novel sequences that are of the UGC/C type. They all strongly support a secondary structure with a UGC loop and C bulge. Variants of the UGC sequence occur in Strongylocentrotus purpuratus (sea urchin) and Apostichopus japonicus (CGC) (F.X. Zheng, X.Q. Sun, and J.X. Zhang, unpubl.), Daphnia pulex (water flea; AGC), Ciona intestinalis, and Periserrula leucophryna (UGG) (Jeong et al. 2006), Pacifastacus leniusculus (signal crayfish) (Huang et al. 1999), and Litopenaeus vannamei (Pacific white shrimp; CGG) (Hsieh et al. 2006). All the UGC/C-type sequences consistently support a G-C pairing (Fig. 3) as previously shown by structural studies of selected IREs (Addess et al. 1997; Gdaniec et al. 1998; Walden et al. 2006).
In some instances, the same sequence can be folded into either the UGC/C type or the C type. Examples are IREs from vertebrates, D. pulex, Lymnaea stagnalis (great pond snail), Crassostrea gigas (Pacific oyster), and Reniera. However, in these cases, the C-type structure seems thermodynamically less favorable.
C-bulge-type IRE
Whereas most IREs that we identified seem to be of the UGC/C-bulge type, a restricted number of phylogenetic groups presents the C-type element. Thus, in the group of insects including six species of Drosophila, Anopheles gambiae, Apis mellifera, and Tribolium castaneum as well as in the Carcinoscorpius rotundicauda (Southeast Asian horseshoe crab), IREs were found to be of the C type as previously shown for Drosophila (Georgieva et al. 1999). In Drosophila the IRE is located in an intron part of the gene, although this intron is sometimes included in the spliced product (Lind et al. 1998). A comparison of this element from the different insect species shows that the IRE is highly conserved with multiple compensatory mutations that preserve the hairpin structure, providing further evidence that this sequence element is functionally important.
The only sequence outside of the insect group that seems to be of the C-bulge-type IRE is that from C. rotundicauda, a member of Chelicerata. It is an intriguing observation that it is much more similar in sequence and secondary structure to insect IREs than to other members of Chelicerata like the ticks (Table 1).
Comparison of the base-pairing schemes of ferritin IREs shows that the non-Watson–Crick pair G-A is rather frequent in the invertebrates (it occurs in Amblyomma americanum, Dermacentor variabilis, P. leniusculus, L. vannamei, P. leucophryna, and Suberites ficus and is commonly located next to the bulge region) (Fig. 3). The G-A pair is, however, not found in vertebrate ferritin IREs, or in any nonferritin IREs.
In summary, it would seem that the UGC/C type of IRE was the ancestral version of the element that has been conserved in evolution, although it was converted into the C type in insects and horseshoe crab.
Transferrin receptor
In mammals there are two types of transferrin receptor, Tfr1 and Tfr2. In the fishes there are two isoforms of Tfr1, called Tfr1a and Tfr1b. Tfr1 and Tfr2 are related to the proteins NLDL, NLD2, and PSMA (Mahadevan and Saldanha 1999; Ghosh and Heston 2004), and these proteins share a domain structure with the three Pfam domains PA, Peptidase M28, and the Tfr dimerization domain. In an attempt to identify possible homologs to Tfr1 and Tfr2 in lower animals such C. intestinalis and Branchiostoma, we searched available protein sequence databases. A number of Ciona proteins were identified with domains characteristic of the Tfr/NLDL/NLD2/PSMA family. However, in a phylogenetic analysis these Ciona proteins do not clearly associate with Tfr (data not shown). Although we cannot formally exclude that there is a Tfr homolog in Ciona, we were not able to identify an iron-responsive element in any of the mRNAs in Ciona related to the Tfr/NLDL/NLD2/PSMA family.
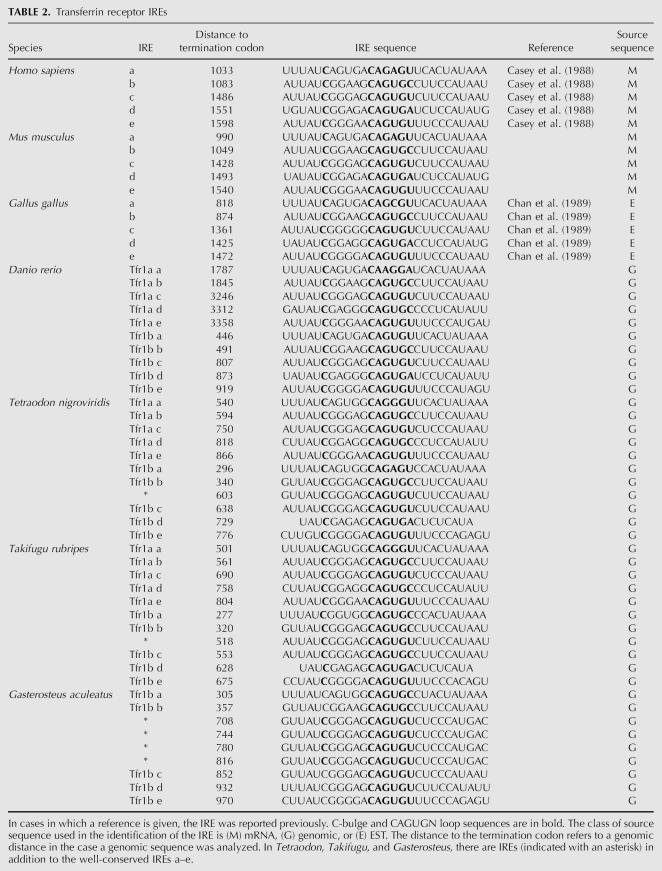
In mammals the Tfr1 mRNA has five iron-responsive elements (IRE-a–IRE-e) (Fig. 4), whereas Tfr2 mRNA is lacking IREs. Here we identified IREs also in other vertebrates, such as the fishes D. rerio, Fugu rubripes, Tetraodon nigroviridis, and Gasterosteus aculeatus (stickleback). In these fishes IREs were present in both Tfr1a and Tfr1b mRNAs (Fig. 4; Table 2). Surprisingly, we identified additional IREs in the Tfr1b mRNAs of F. rubripes, T. nigroviridis, and G. aculeatus in the region between IRE-b and IRE-c (Fig. 4; Table 2). For instance, in G. aculeatus there are four such IREs, all identical.
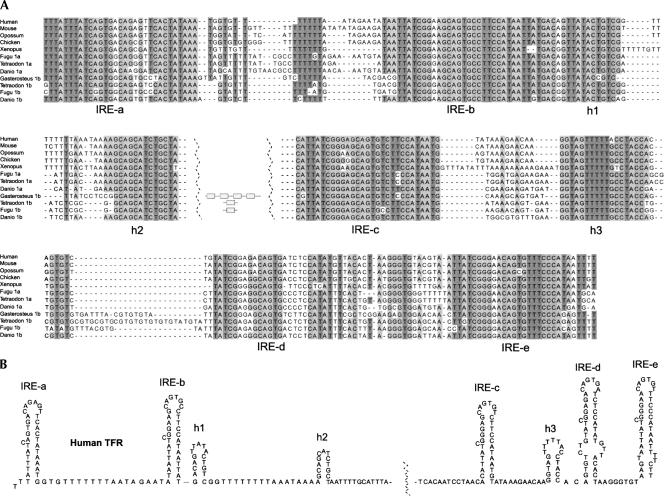
FIGURE 4.
Transferrin receptor IREs. (A) A multiple alignment of selected vertebrate sequences of the Tfr 3′-UTR region containing five IREs. In the fish D. rerio, T. nigroviridis, and F. rubripes, both Tfr1a and Tfr1b mRNAs contain IREs. (In the case of Gasterosteus we have no information whether the sequence is 1a or 1b.) Three postulated hairpins (h1, h2, and h3) are also shown (Casey et al. 1989; Chan et al. 1989; Koeller et al. 1989; Schlegl et al. 1997). Additional IREs in the fishes between the IRE-b and -c elements are represented as shaded boxes. (B) Proposed secondary structure of the region using the human Tfr sequence.
TABLE 2.
Transferrin receptor IREs
The canonical sequence of the IRE apical loop is CAGUGN. In the human Tfr1 mRNA the first IRE (IRE-a) has a noncanonical CAGAGU loop sequence. Also in other species this IRE is characterized by having a noncanonical loop sequence (chicken CAGCGU, D. rerio Tfr1a CAAGGA, and T. rubripes Tfr1a CAGGGU) (Fig. 4). An exception is the corresponding fish Tfr1b IRE, which is more similar to the canonical sequence (Fig. 4; Table 2).
A comparison of the mRNA region containing the five IREs from different species shows a remarkable degree of conservation. A phylogenetic analysis strongly suggests that the order of IREs has been preserved during evolution. Not only are the IREs highly conserved, but also are three additional regions as indicated in Figure 4. All of these regions are able to form short hairpin structures, as previously noted for the human Tfr1 mRNA (Casey et al. 1989; Chan et al. 1989; Koeller et al. 1989; Schlegl et al. 1997).
Mitochondrial aconitase
Human, porcine, murine, and bovine mitochondrial aconitase mRNAs have previously been shown to contain an IRE (Butt et al. 1996). Here we identified such an element also in chicken Gallus gallus, in X. tropicalis, the fishes D. rerio and T. nigroviridis, and in the sea squirt C. intestinalis. The IRE is extremely well conserved in sequence from mammals to Ciona as indicated in Table 3. Consistently, the AUG codon is part of the IRE hairpin. We have not been able to find an IRE in any nonvertebrate species among the metazoa, including S. purpuratus (sea urchin), Branchiostoma (lancelet), nematodes, insects, and lower metazoa, although all these organisms have a mitochondrial aconitase. It would therefore seem that the IRE was acquired at a stage of evolution close to the development of Chordata (Fig. 2).
Erythroid aminolevulinic acid synthase
In vertebrates there are two forms of eALAS, referred to as H (housekeeping form) and E (erythroid form). The phylogeny of this enzyme as well as its IRE has been studied by Duncan et al. (1999). The enzyme is widely distributed and is found in metazoa, fungi, protists, and eubacteria. Both H and E forms are present in vertebrates. Previous analysis of the distribution of IREs showed that it is present in the E-form of vertebrate eALAS genes (Duncan et al. 1999). Thus, an IRE was identified in mRNA of human, whale, rat, mouse, chicken, and toadfish, whereas it seems to be absent in hagfish. In addition, we here found IREs in the E-chain mRNAs of T. rubripes, D. rerio, X. tropicalis, and C. intestinalis. Consistent with previous observations (Duncan et al. 1999), we were not able to identify an IRE in the sea urchin or in the lower metazoa like Nematostella. Therefore, the distribution of the eALAS IRE seems to be very similar to that of mitochondrial aconitase (Fig. 2).
Ferroportin
Ferroportin is a protein found in vertebrates, many invertebrates, plants, and the fungal groups Basidiomycota and Pezizomycotina, as judged by PSI-BLAST searches as well as searches using the Pfam model of the ferroportin domain (data not shown; Finn et al. 2006). However, a search of IREs in ferroportin mRNAs revealed such elements only in vertebrates. In more deeply branching organisms like C. intestinalis, Ciona savignyi, Branchiostoma, and Strongylocentrotus, homologs to ferroportin were identified, but no IREs could be identified in their mRNAs. Table 3 lists IREs identified in man, mouse, chicken, and zebrafish. The ferroportin IRE sequence, like that of mitochondrial aconitase, is remarkably conserved.
DMT1
The DMT1 protein is found in all vertebrates and in sea urchin. We identified IREs in DMT1 mRNAs of mammals, including the marsupial opossum. A closely related sequence is identified in X. tropicalis, although with a nucleotide change in the CAGUGN consensus motif (Fig. 5). This is most likely not a sequencing error because the sequence is consistently present in all ESTs corresponding to this mRNA as well as in all primary reads of genomic sequence. Fishes also seem to have 3′-UTRs with reasonable primary sequence similarity to the corresponding mammalian sequences (Fig. 5). However, neither is a CAGUGN motif present, nor a hairpin structure characteristic of IRE (Fig. 5). These observations would suggest that the mammalian DMT1 IRE evolved from a non-IRE sequence by gradual changes involving nucleotide substitutions of the ancestral sequences.
FIGURE 5.
Alignment of 3′-UTR region of DMT1 mRNA. Sequences immediately downstream from the termination codon of DMT1 mRNAs were subjected to a multiple alignment using T-Coffee (Notredame et al. 2000) with default parameters. The sequences are from six fish species (on top) in addition to frog, opossum, mouse, and human. The IRE consensus loop sequence CAGUGN is shown, as well as the location of the IRE hairpin.
IREs with a restricted phylogenetic distribution
Succinate dehydrogenase is a ubiquitous Krebs cycle enzyme. The Drosophila mRNA of succinate dehydrogenase has been reported to contain a physiologically significant IRE (Gray et al. 1996). We found the sequence of this IRE to be perfectly conserved in all available Drosophila species (Table 3). However, we have failed to identify such an IRE in other insects, including the closely related A. gambiae. Neither was an IRE identified in other metazoan succinate dehydrogenase mRNAs. It would therefore seem that the recruitment of the IRE to the succinate dehydrogenase mRNA is an event highly specific to Drosophila.
A number of IREs has been reported that seem to be restricted to primates or to primates, rodents, and dog. Such an example is Cdc14a (Sanchez et al. 2006), where the IRE is found only in human, rat, and dog, but the protein is also present in frog and chicken. The human MRCKα has homologs in chimpanzee, mouse, and dog. However, only the human sequence can adopt the structure postulated by Cmejla et al. (2006). The chimpanzee sequence has a nucleotide change that disrupts a base pair present in the proposed human IRE hairpin structure. In mouse, rat, and dog there is weak sequence similarity to the human sequence, but none of these sequences has the consensus features of the IRE.
DISCUSSION
We have assembled 49 previously known IREs and have identified >100 additional sequences (Tables 1–3). These novel sequences were not previously reported in the literature or annotated as such elements in sequence databases. Out of the 107 novel IREs listed in Tables 1–3, only 20 are reported also in Rfam (Griffiths-Jones et al. 2003). This is partly because not all possible genomic sequences have been analyzed in the context of that database. We also noted that some of the IRE sequences presented in Rfam are false positives.
The compilation of IREs reported here allows us to examine structural properties and consensus features of the IRE, and it also allows interesting conclusions as to the evolution of this regulatory element.
Structure and evolution of ferritin IREs
The ferritin IRE is widely distributed among metazoa. Thus, we identified it in all metazoa examined, except C. elegans and S. mansoni. We found an IRE also in the deeply branching metazoa Nematostella and the sponges Reniera and S. ficus.
Most of the ferritin IREs fold into a UGC/C-type structure. It has been shown that a base pair is formed between the G in the UGC bulge and the C in the other strand (Addess et al. 1997; Gdaniec et al. 1998; Walden et al. 2006). This type of pairing is strongly supported by the IREs that we have identified here (Fig. 3). A certain variation in the UGC sequence is apparently allowed, with C or A in the first position and with G in the third position. Another common feature of the ferritin IRE is an A-G pair adjacent to the UGC/C bulges (Table 1; Fig. 3).
Whereas most ferritin IREs are of the UGC/C type, the insect species Drosophila, Apis, Aedes, and Tribolium as well as the horseshoe crab C. rotundicauda are unusual in that they have an IRE of the C-bulge type. It is interesting that the chelicerate C. rotundicauda has a C-type IRE, whereas other chelicerates like D. variabilis and A. americanum have a UGC/C-type IRE. Assuming that the UGC/C-type IRE is the ancestral form, this would suggest that evolution to a C-type IRE occurred more than once.
We noted that some of the ferritin IRE sequences can fold into either a UGC/C-type or C-type structure. This suggests a possible mechanism of evolution from UGC/C-type to C-type structure. First, one or more nucleotide substitutions allowed a C-type structure. Then further changes stabilized this structure. As an alternative mechanism, the C-type structure could be obtained more directly by deletion of the U being part of the UGC loop.
It is interesting to note that a vast majority of insect ferritin IREs that are of the C type also have a UGC triplet as part of the stem on top of the C-bulge (underlined in Table 1). This might be a coincidence but could also be of functional significance, relating this type of IRE more closely to the UGC/C type.
Why are most ferritin IREs of the UGC/C type, whereas others are of the C type? A C-type IRE is known to bind IRP2 much less efficiently than a UGC/C-type of IRE (Ke et al. 1998), whereas the binding of IRP1 is less influenced by the IRE structure. Therefore, a conversion during evolution from UGC to C type may be a way to fine tune the IRE/IRP regulatory mechanism.
Transferrin receptor IREs
We identified a number of vertebrate Tfr IREs that were not previously reported. The presence of IREs in Tfr1a as well as Tfr1b mRNAs in fishes is also a novel finding. The transferrin receptor, as well as the IRE contained within its mRNA, occurs in vertebrates. The only fish species in which we failed to identify Tfr IREs is Petromyzon marinus, a more deeply branching vertebrate. However, only single-pass DNA sequencing reads are available for this organism, and we cannot exclude the possibility that such IREs will be identified once we have access to sequences from a more advanced stage of the genome sequencing project. On the other hand, if such IREs are, indeed, missing in Petromyzon, this is an interesting observation, as it would suggest that Tfr IREs have appeared early in the evolution of vertebrates.
Multiple alignments of vertebrate Tfr 3′-UTR regions show a high degree of conservation in the parts that contain the IRE structures. All IRE sequences as well as their order are remarkably conserved. Neighbor-joining analysis of all Tfr IRE sequences showed that all the elements are grouped according to their position in the cluster (data not shown). Not only are the IREs highly conserved but also three additional potential hairpin regions as indicated in Figure 4. For the first of these hairpins (Fig. 4, h1), there are compensatory mutations that give support to the proposed hairpin structure. For the two other helices (Fig. 4, h2,h3), the sequences are all identical. Two additional helices were suggested in Schlegl et al. (1997). However, we could not find comparative genomics support for any of these. The Tfr 3′-UTR regions are known to contain AU-rich elements, for instance, between IRE-a and IRE-b and between h1 and h2 (Koeller et al. 1989; Theil 1993). These elements are identified also in fishes, although some of them are less pronounced (Fig. 4).
Notable differences between Tfr 3′-UTRs of fishes and other vertebrates are that some fishes have more than five IREs and that the IRE-a of many of the fish Tfr1b mRNAs have a canonical CAGUGN apical loop sequence. Thus, the T. rubripes, D. rerio, and G. aculeatus Tfr1b have the sequence CAGUGY (Fig. 4). Based on these observations, a model of IRE evolution could be envisaged in which the Tfr1b 3′-UTR represents a more ancestral form of the IRE/IRP regulatory region. Originally a single IRE was duplicated, giving rise to five or more IREs as in Tfr1b. Evidence of such a duplication process is clearly seen in the case of G. aculeatus, where four identical copies of the IRE have been produced. The next step is a gene duplication event giving rise to the Tfr1a gene, where the IRE-a is eventually changed to confer a specific advantage. Finally the Tfr1b gene was lost during vertebrate evolution. The fact that a consensus sequence has been avoided during the evolution of the IRE-a suggests that it has an important function although it is not binding the IRP proteins effectively.
IREs of restricted phylogenetic distribution
The MRCKα IRE is identified only in primates, and only the human sequence can adopt the structure proposed in Cmejla et al. (2006). Therefore, we cannot find any phylogenetic support at all for this IRE. Further experiments are necessary to clarify its role in IRP-mediated control. In case it is a bona fide IRE, then it has apparently appeared at a late stage in evolution and possibly by a mechanism of evolution as described for DMT1 above.
A special case is the Drosophila succinate dehydrogenase mRNA that seems to use an IRE sequence (Gray et al. 1996). It is interesting to note that this IRE is restricted to Drosophila and not found in other insects examined, including the closely related A. gambiae. In Drosophila the only other known IRE is that of ferritin. It is an interesting observation that, as in ferritin mRNA, the succinate dehydrogenase IRE has a UGC sequence (Tables 1, 3). This might reflect an evolutionary process in which the succinate dehydrogenase IRE evolved from the ferritin IRE.
The ferritin IRE, an ancestral system of iron regulation
We suggest that the IRE/IRP system first operated in ferritin mRNA, as we have identified IREs in the ferritin mRNAs of deeply branching metazoa like sea anemone and sponges, phylogenetic groups where there is no evidence of IREs in other mRNAs. Therefore, the IRE/IRP system in ferritin mRNA is most likely the ancestral version of this type of translational control. Later in evolution IREs appeared in other mRNAs, presumably using in part the same repertoire of IRP proteins as the ferritin IRE. Thus, mACO and eALAS IREs appear at the level of Chordata; Tfr and ferroportin IREs appear with the development of vertebrates; the DMT1 IRE appear at the level of mammals (or possible earlier); and the remaining IREs appear at a later stage.
How does a gene acquire an IRE during evolution? Such an element may be recruited from another gene by DNA recombination, or the IRE can arise independently through changes to a UTR sequence by “convergent” evolution. In both cases, a positive selection may occur for effective binding to an already existing iron regulatory protein. For most of the IRE families, the available sequence data cannot be used to discriminate between these two mechanisms of IRE evolution. However, there seems to be evidence of a convergent evolution in the case of DMT1 mRNA and the IRE present in the 3′-UTR as shown in Figure 5. We cannot exclude that the alignment is fortuitous, and it would be more reliable if we had access to more sequences reflecting the evolution from teleosts to mammals. Nevertheless, the alignment would suggest a progression from a non-IRE-like sequence in the teleosts to a “consensus” IRE like the one observed in mammals.
MATERIALS AND METHODS
Genome and protein sequences
The majority of protein sequences were retrieved from NCBI (ftp.ncbi.nih.gov/blast/db/), SWISS-PROT (http://www.expasy.ch/sprot/), and UniProt (http://www.expasy.uniprot.org/). Genomic sequences were from NCBI (ftp.ncbi.nih.gov/genomes/), EMBL (http://www.ebi.ac.uk), ENSEMBL (http://www.ensembl.org), and TraceDB (ftp.ncbi.nlm.nih.gov/pub/TraceDB). Multiple alignments available at the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu) were used to identify evolutionarily conserved sequences.
Identification of iron-responsive elements
To identify as many potential IREs as possible, we searched all available mRNA and genomic sequences. We used pattern matching as well as the cmsearch of the Infernal software (Griffiths-Jones et al. 2003). Patterns were constructed on the basis of known properties of IREs, and cmsearch used covariance models from Rfam or more specific models, such as models based on C- or UGC/C-type IRE alignments. FASTA34 (Pearson 2000) or BLAST (Altschul et al. 1997; Altschul and Koonin 1998) searches were carried out in order to identify closely related homologs. Sequences were checked for conserved primary sequence motifs and the ability to fold into a secondary structure typical for the iron-responsive element. Secondary structure predictions were carried out with MFOLD (Zuker 1989).
Identification of protein homologs
In order to identify as many homologs as possible to previously known proteins whose mRNAs are known to contain IREs, we made use of BLAST searches (Altschul et al. 1997; Altschul and Koonin 1998). The database searched was the NCBI GenBank set of proteins (Benson et al. 2006), but some proteins were absent in this set and were retrieved from individual genome projects. Alternatively, they were identified from TBLASTN searches of genome sequences or predicted by Genewise alignments (Birney et al. 2004). Additional homologs were in some instances identified with Pfam models (http://hmmer.wustl.edu/; Eddy 1998; Bateman et al. 2004).
Phylogenetic analysis of proteins
All the proteins considered to be significant homologs based on E-values were retrieved, and multiple alignments were created using ClustalW 1.83 (Thompson et al. 1994) or T-Coffee (Notredame et al. 2000). Neighbor-joining trees were obtained with ClustalW using the bootstrap option.
Phylogenetic analysis was also carried out using programs of the PHYLIP package (Felsenstein 1996). For each ClustalW or T-Coffee alignment, we used parsimony (PROTPARS), maximum likelihood (PROML), and neighbor joining (NEIGHBOR). The neighbor joining was based on a matrix created by PROTDIST. SEQBOOT was used to generate for each of the three methods' 500 bootstrapped data sets. CONSENSE was then used to compute a consensus tree by the majority-rule consensus tree method for the three different procedures.
ACKNOWLEDGMENTS
P.P. was supported by the Swedish Research School of Genomics and Bioinformatics.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.464807.
REFERENCES
- Abboud, S., Haile, D.J. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J. Biol. Chem. 2000;275:19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- Addess, K.J., Basilion, J.P., Klausner, R.D., Rouault, T.A., Pardi, A. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 1997;274:72–83. doi: 10.1006/jmbi.1997.1377. [DOI] [PubMed] [Google Scholar]
- Alen, C., Sonenshein, A.L. Bacillus subtilis aconitase is an RNA-binding protein. Proc. Natl. Acad. Sci. 1999;96:10412–10417. doi: 10.1073/pnas.96.18.10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Koonin, E.V. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Coin, L., Durbin, R., Finn, R.D., Hollich, V., Griffiths-Jones, S., Khanna, A., Marshall, M., Moxon, S., Sonnhammer, E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, G., Ellis, T.W., Habicht, G.S., Schluter, S.F., Marchalonis, J.J. Evolution of the acute phase response: Iron release by echinoderm (Asterias forbesi) coelomocytes, and cloning of an echinoderm ferritin molecule. Dev. Comp. Immunol. 2002;26:11–26. doi: 10.1016/s0145-305x(01)00051-9. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Wheeler, D.L. GenBank. Nucleic Acids Res. 2006;34:D16–D20. doi: 10.1093/nar/gkj157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney, E., Clamp, M., Durbin, R. GeneWise and Genomewise. Genome Res. 2004;14:988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, J., Kim, H.Y., Basilion, J.P., Cohen, S., Iwai, K., Philpott, C.C., Altschul, S., Klausner, R.D., Rouault, T.A. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc. Natl. Acad. Sci. 1996;93:4345–4349. doi: 10.1073/pnas.93.9.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey, J.L., Hentze, M.W., Koeller, D.M., Caughman, S.W., Rouault, T.A., Klausner, R.D., Harford, J.B. Iron-responsive elements: Regulatory RNA sequences that control mRNA levels and translation. Science. 1988;240:924–928. doi: 10.1126/science.2452485. [DOI] [PubMed] [Google Scholar]
- Casey, J.L., Koeller, D.M., Ramin, V.C., Klausner, R.D., Harford, J.B. Iron regulation of transferrin receptor mRNA levels requires iron-responsive elements and a rapid turnover determinant in the 3′-untranslated region of the mRNA. EMBO J. 1989;8:3693–3699. doi: 10.1002/j.1460-2075.1989.tb08544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, L.N., Grammatikakis, N., Banks, J.M., Gerhardt, E.M. Chicken transferrin receptor gene: Conservation 3′ noncoding sequences and expression in erythroid cells. Nucleic Acids Res. 1989;17:3763–3771. doi: 10.1093/nar/17.10.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmejla, R., Petrak, J., Cmejlova, J. A novel iron responsive element in the 3′-UTR of human MRCKα. Biochem. Biophys. Res. Commun. 2006;341:158–166. doi: 10.1016/j.bbrc.2005.12.155. [DOI] [PubMed] [Google Scholar]
- Dandekar, T., Stripecke, R., Gray, N.K., Goossen, B., Constable, A., Johansson, H.E., Hentze, M.W. Identification of a novel iron-responsive element in murine and human erythroid δ-aminolevulinic acid synthase mRNA. EMBO J. 1991;10:1903–1909. doi: 10.1002/j.1460-2075.1991.tb07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar, T., Beyer, K., Bork, P., Kenealy, M.R., Pantopoulos, K., Hentze, M., Sonntag-Buck, V., Flouriot, G., Gannon, F., Schreiber, S. Systematic genomic screening and analysis of mRNA in untranslated regions and mRNA precursors: Combining experimental and computational approaches. Bioinformatics. 1998;14:271–278. doi: 10.1093/bioinformatics/14.3.271. [DOI] [PubMed] [Google Scholar]
- Dickey, L.F., Sreedharan, S., Theil, E.C., Didsbury, J.R., Wang, Y.H., Kaufman, R.E. Differences in the regulation of messenger RNA for housekeeping and specialized-cell ferritin. A comparison of three distinct ferritin complementary DNAs, the corresponding subunits, and identification of the first processed in amphibia. J. Biol. Chem. 1987;262:7901–7907. [PubMed] [Google Scholar]
- Dix, D.J., Lin, P.N., McKenzie, A.R., Walden, W.E., Theil, E.C. The influence of the base-paired flanking region on structure and function of the ferritin mRNA iron regulatory element. J. Mol. Biol. 1993;231:230–240. doi: 10.1006/jmbi.1993.1278. [DOI] [PubMed] [Google Scholar]
- Donovan, A., Brownlie, A., Zhou, Y., Shepard, J., Pratt, S.J., Moynihan, J., Paw, B.H., Drejer, A., Barut, B., Zapata, A., et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- Duncan, R., Faggart, M.A., Roger, A.J., Cornell, N.W. Phylogenetic analysis of the 5-aminolevulinate synthase gene. Mol. Biol. Evol. 1999;16:383–396. doi: 10.1093/oxfordjournals.molbev.a026119. [DOI] [PubMed] [Google Scholar]
- Durand, J.P., Goudard, F., Pieri, J., Escoubas, J.M., Schreiber, N., Cadoret, J.P. Crassostrea gigas ferritin: cDNA sequence analysis for two heavy chain type subunits and protein purification. Gene. 2004;338:187–195. doi: 10.1016/j.gene.2004.04.027. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Finn, R.D., Mistry, J., Schuster-Bockler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., Moxon, S., Marshall, M., Khanna, A., Durbin, R., et al. Pfam: Clans, web tools, and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdaniec, Z., Sierzputowska-Gracz, H., Theil, E.C. Iron regulatory element and internal loop/bulge structure for ferritin mRNA studied by cobalt(III) hexammine binding, molecular modeling, and NMR spectroscopy. Biochemistry. 1998;37:1505–1512. doi: 10.1021/bi9719814. [DOI] [PubMed] [Google Scholar]
- Georgieva, T., Dunkov, B.C., Harizanova, N., Ralchev, K., Law, J.H. Iron availability dramatically alters the distribution of ferritin subunit messages in Drosophila melanogaster . Proc. Natl. Acad. Sci. 1999;96:2716–2721. doi: 10.1073/pnas.96.6.2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A., Heston, W.D. Tumor target prostate specific membrane antigen (PSMA) and its regulation in prostate cancer. J. Cell. Biochem. 2004;91:528–539. doi: 10.1002/jcb.10661. [DOI] [PubMed] [Google Scholar]
- Girelli, D., Corrocher, R., Bisceglia, L., Olivieri, O., De Franceschi, L., Zelante, L., Gasparini, P. Molecular basis for the recently described hereditary hyperferritinemia-cataract syndrome: A mutation in the iron-responsive element of ferritin L-subunit gene (the “Verona mutation”) Blood. 1995;86:4050–4053. [PubMed] [Google Scholar]
- Goossen, B., Hentze, M.W. Position is the critical determinant for function of iron-responsive elements as translational regulators. Mol. Cell. Biol. 1992;12:1959–1966. doi: 10.1128/mcb.12.5.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourley, B.L., Parker, S.B., Jones, B.J., Zumbrennen, K.B., Leibold, E.A. Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans . J. Biol. Chem. 2003;278:3227–3234. doi: 10.1074/jbc.M210333200. [DOI] [PubMed] [Google Scholar]
- Gray, N.K., Pantopoulos, K., Dandekar, T., Ackrell, B.A., Hentze, M.W. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc. Natl. Acad. Sci. 1996;93:4925–4930. doi: 10.1073/pnas.93.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., Eddy, S.R. Rfam: An RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin, H., Allerson, C.R., Polycarpou-Schwarz, M., Rofts, A., Rogers, J.T., Kishi, F., Hentze, M.W., Rouault, T.A., Andrews, N.C., Hediger, M.A. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- Henderson, B.R., Menotti, E., Bonnard, C., Kuhn, L.C. Optimal sequence and structure of iron-responsive elements. Selection of RNA stem–loops with high affinity for iron regulatory factor. J. Biol. Chem. 1994;269:17481–17489. [PubMed] [Google Scholar]
- Hentze, M.W., Caughman, S.W., Rouault, T.A., Barriocanal, J.G., Dancis, A., Harford, J.B., Klausner, R.D. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- Hsieh, S.L., Chiu, Y.C., Kuo, C.M. Molecular cloning and tissue distribution of ferritin in Pacific white shrimp (Litopenaeus vannamei) Fish Shellfish Immunol. 2006;21:279–283. doi: 10.1016/j.fsi.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Huang, T.S., Melefors, O., Lind, M.I., Soderhall, K. An atypical iron-responsive element (IRE) within crayfish ferritin mRNA and an iron regulatory protein 1 (IRP1)-like protein from crayfish hepatopancreas. Insect Biochem. Mol. Biol. 1999;29:1–9. doi: 10.1016/s0965-1748(98)00097-6. [DOI] [PubMed] [Google Scholar]
- Hubert, N., Walczak, R., Sturchler, C., Myslinski, E., Schuster, C., Westhof, E., Carbon, P., Krol, A. RNAs mediating cotranslational insertion of selenocysteine in eukaryotic selenoproteins. Biochimie. 1996;78:590–596. doi: 10.1016/s0300-9084(96)80005-8. [DOI] [PubMed] [Google Scholar]
- Jeong, B.R., Chung, S.M., Baek, N.J., Koo, K.B., Baik, H.S., Joo, H.S., Chang, C.S., Choi, J.W. Characterization, cloning, and expression of the ferritin gene from the Korean polychaete, Periserrula leucophryna . J. Microbiol. 2006;44:54–63. [PubMed] [Google Scholar]
- Kato, J., Fujikawa, K., Kanda, M., Fukuda, N., Sasaki, K., Takayama, T., Kobune, M., Takada, K., Takimoto, R., Hamada, H., et al. A mutation, in the iron-responsive element of H ferritin mRNA, causing autosomal dominant iron overload. Am. J. Hum. Genet. 2001;69:191–197. doi: 10.1086/321261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Y., Wu, J., Leibold, E.A., Walden, W.E., Theil, E.C. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J. Biol. Chem. 1998;273:23637–23640. doi: 10.1074/jbc.273.37.23637. [DOI] [PubMed] [Google Scholar]
- Koeller, D.M., Casey, J.L., Hentze, M.W., Gerhardt, E.M., Chan, L.N., Klausner, R.D., Harford, J.B. A cytosolic protein binds to structural elements within the iron regulatory region of the transferrin receptor mRNA. Proc. Natl. Acad. Sci. 1989;86:3574–3578. doi: 10.1073/pnas.86.10.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, S.A., Henderson, B.R., Kuhn, L.C. Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J. Biol. Chem. 1995;270:30781–30786. doi: 10.1074/jbc.270.51.30781. [DOI] [PubMed] [Google Scholar]
- Kohler, S.A., Menotti, E., Kuhn, L.C. Molecular cloning of mouse glycolate oxidase. High evolutionary conservation and presence of an iron-responsive element-like sequence in the mRNA. J. Biol. Chem. 1999;274:2401–2407. doi: 10.1074/jbc.274.4.2401. [DOI] [PubMed] [Google Scholar]
- Kopacek, P., Zdychova, J., Yoshiga, T., Weise, C., Rudenko, N., Law, J.H. Molecular cloning, expression and isolation of ferritins from two tick species—Ornithodoros moubata and Ixodes ricinus . Insect Biochem. Mol. Biol. 2003;33:103–113. doi: 10.1016/s0965-1748(02)00181-9. [DOI] [PubMed] [Google Scholar]
- LaVaute, T., Smith, S., Cooperman, S., Iwai, K., Land, W., Meyron-Holtz, E., Drake, S.K., Miller, G., Abu-Asab, M., Tsokos, M., et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- Leibold, E.A., Laudano, A., Yu, Y. Structural requirements of iron-responsive elements for binding of the protein involved in both transferrin receptor and ferritin mRNA post-transcriptional regulation. Nucleic Acids Res. 1990;18:1819–1824. doi: 10.1093/nar/18.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, E., Graziano, J.H., Freyer, G.A. Regulation of the 75-kDa subunit of mitochondrial complex I by iron. J. Biol. Chem. 2001;276:27685–27692. doi: 10.1074/jbc.M100941200. [DOI] [PubMed] [Google Scholar]
- Lind, M.I., Ekengren, S., Melefors, O., Soderhall, K. Drosophila ferritin mRNA: Alternative RNA splicing regulates the presence of the iron-responsive element. FEBS Lett. 1998;436:476–482. doi: 10.1016/s0014-5793(98)01186-7. [DOI] [PubMed] [Google Scholar]
- Liu, X., Theil, E.C. Ferritin reactions: Direct identification of the site for the diferric peroxide reaction intermediate. Proc. Natl. Acad. Sci. 2004;101:8557–8562. doi: 10.1073/pnas.0401146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan, D., Saldanha, J.W. The extracellular regions of PSMA and the transferrin receptor contain an aminopeptidase domain: Implications for drug design. Protein Sci. 1999;8:2546–2549. doi: 10.1110/ps.8.11.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, S.A., Pardi, A. Refined solution structure of the iron-responsive element RNA using residual dipolar couplings. J. Mol. Biol. 2003;326:1037–1050. doi: 10.1016/s0022-2836(02)01431-6. [DOI] [PubMed] [Google Scholar]
- Melefors, O. Translational regulation in vivo of the Drosophila melanogaster mRNA encoding succinate dehydrogenase iron protein via iron responsive elements. Biochem. Biophys. Res. Commun. 1996;221:437–441. doi: 10.1006/bbrc.1996.0613. [DOI] [PubMed] [Google Scholar]
- Mulenga, A., Simser, J.A., Macaluso, K.R., Azad, A.F. Stress and transcriptional regulation of tick ferritin HC. Insect Mol. Biol. 2004;13:423–433. doi: 10.1111/j.0962-1075.2004.00502.x. [DOI] [PubMed] [Google Scholar]
- Muller, J.P., Vedel, M., Monnot, M.J., Touzet, N., Wegnez, M. Molecular cloning and expression of ferritin mRNA in heavy metal-poisoned Xenopus laevis cells. DNA Cell Biol. 1991;10:571–579. doi: 10.1089/dna.1991.10.571. [DOI] [PubMed] [Google Scholar]
- Nichol, H., Winzerling, J. Structured RNA upstream of insect cap distal iron responsive elements enhances iron regulatory protein-mediated control of translation. Insect Biochem. Mol. Biol. 2002;32:1699–1710. doi: 10.1016/s0965-1748(02)00110-8. [DOI] [PubMed] [Google Scholar]
- Notredame, C., Higgins, D.G., Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nudler, E., Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- Piccinelli, P., Rosenblad, M.A., Samuelsson, T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalia, M., Rosenblad, M.A., Samuelsson, T. Prediction of signal recognition particle RNA genes. Nucleic Acids Res. 2002;30:3368–3377. doi: 10.1093/nar/gkf468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roetto, A., Bosio, S., Gramaglia, E., Barilaro, M.R., Zecchina, G., Camaschella, C. Pathogenesis of hyperferritinemia cataract syndrome. Blood Cells Mol. Dis. 2002;29:532–535. doi: 10.1006/bcmd.2002.0590. [DOI] [PubMed] [Google Scholar]
- Rogers, J.T., Randall, J.D., Cahill, C.M., Eder, P.S., Huang, X., Gunshin, H., Leiter, L., McPhee, J., Sarang, S.S., Utsuki, T., et al. An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J. Biol. Chem. 2002;277:45518–45528. doi: 10.1074/jbc.M207435200. [DOI] [PubMed] [Google Scholar]
- Rosenblad, M.A., Zwieb, C., Samuelsson, T. Identification and comparative analysis of components from the signal recognition particle in protozoa and fungi. BMC Genomics. 2004;5:5. doi: 10.1186/1471-2164-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblad, M.A., Lopez, M.D., Piccinelli, P., Samuelsson, T. Inventory and analysis of the protein subunits of the ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez, M., Galy, B., Dandekar, T., Bengert, P., Vainshtein, Y., Stolte, J., Muckenthaler, M.U., Hentze, M.W. Iron regulation and the cell cycle: Identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J. Biol. Chem. 2006;281:22865–22874. doi: 10.1074/jbc.M603876200. [DOI] [PubMed] [Google Scholar]
- Schlegl, J., Gegout, V., Schlager, B., Hentze, M.W., Westhof, E., Ehresmann, C., Ehresmann, B., Romby, P. Probing the structure of the regulatory region of human transferrin receptor messenger RNA and its interaction with iron regulatory protein-1. RNA. 1997;3:1159–1172. [PMC free article] [PubMed] [Google Scholar]
- Schussler, P., Potters, E., Winnen, R., Michel, A., Bottke, W., Kunz, W. Ferritin mRNAs in Schistosoma mansoni do not have iron-responsive elements for post-transcriptional regulation. Eur. J. Biochem. 1996;241:64–69. doi: 10.1111/j.1432-1033.1996.0064t.x. [DOI] [PubMed] [Google Scholar]
- Theil, E.C. The IRE (iron regulatory element) family: Structures which regulate mRNA translation or stability. Biofactors. 1993;4:87–93. [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, A.M., Rogers, J.T., Leedman, P.J. Iron-regulatory proteins, iron-responsive elements and ferritin mRNA translation. Int. J. Biochem. Cell Biol. 1999;31:1139–1152. doi: 10.1016/s1357-2725(99)00080-1. [DOI] [PubMed] [Google Scholar]
- von Darl, M., Harrison, P.M., Bottke, W. cDNA cloning and deduced amino acid sequence of two ferritins: Soma ferritin and yolk ferritin, from the snail Lymnaea stagnalis L. Eur. J. Biochem. 1994;222:353–366. doi: 10.1111/j.1432-1033.1994.tb18874.x. [DOI] [PubMed] [Google Scholar]
- Walczak, R., Hubert, N., Carbon, P., Krol, A. Solution structure of SECIS, the mRNA element required for eukaryotic selenocysteine insertion—Interaction studies with the SECIS-binding protein SBP. Biomed. Environ. Sci. 1997;10:177–181. [PubMed] [Google Scholar]
- Walden, W.E., Selezneva, A.I., Dupuy, J., Volbeda, A., Fontecilla-Camps, J.C., Theil, E.C., Volz, K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005;9:594–602. doi: 10.1016/j.cbpa.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Albert, D.W., Kohlhepp, P., D-Pham, D.Q., Winzerling, J.J. Repression of Manduca sexta ferritin synthesis by IRP1/IRE interaction. Insect Mol. Biol. 2001;10:531–539. doi: 10.1046/j.0962-1075.2001.00293.x. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Dimopoulos, G., Wolf, A., Minana, B., Kafatos, F.C., Winzerling, J.J. Cloning and molecular characterization of two mosquito iron regulatory proteins. Insect Biochem. Mol. Biol. 2002;32:579–589. doi: 10.1016/s0965-1748(01)00138-2. [DOI] [PubMed] [Google Scholar]
- Zuker, M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]