Abstract
The human exosome is a 3′–5′ exoribonuclease complex that functions both in the nucleus and in the cytoplasm to either degrade or process RNA. Little is known yet about potential differences among core exosome complexes in these different cellular compartments and the roles of the individual subunits in maintaining a stable and functional complex. Glycerol gradient sedimentation analyses indicated that a significant subset of nuclear exosomes is present in much larger complexes (60–80S) than the cytoplasmic exosomes (∼10S). Interestingly, siRNA-mediated knock-down experiments indicated that the cytoplasmic exosome is down-regulated much more efficiently than the nuclear exosome. In addition, we observed that knock-down of hRrp41p or hRrp4p but not PM/Scl-100 or PM/Scl-75 leads to codepletion of other subunits. Nevertheless, PM/Scl-100 and PM/Scl-75 are required to maintain normal levels of three different mRNA reporters: a wild-type β-globin mRNA, a β-globin mRNA containing an AU-rich (ARE) instability element, and a β-globin mRNA bearing a premature termination codon (PTC). The increased levels of ARE- and the PTC-containing mRNAs upon down-regulation of the different exosome subunits, in particular PM/Scl-100, appeared to be due to decreased turnover rates. These results indicate that, although not required for exosome stability, PM/Scl-100 and PM/Scl-75 are involved in mRNA degradation, either as essential subunits of a functional exosome complex or as exosome-independent proteins.
Keywords: exosome, RNA degradation, mRNA turnover, RNA processing, exoribonuclease
INTRODUCTION
The exosome is a 3′–5′ exoribonuclease complex that was initially identified in yeast (Mitchell et al. 1997). Since then, a variety of functions have been described for the yeast exosome, including the processing of ribosomal and small nucle(ol)ar RNAs (Allmang et al. 1999a; van-Hoof et al. 2000; Schilders et al. 2006) and the degradation of mRNAs (Jacobs Anderson and Parker 1998; Bousquet-Antonelli et al. 2000; Das et al. 2003).
The exosome has been found to be highly conserved among eukaryotes, including humans (Allmang et al. 1999b; Chekanova et al. 2000; Brouwer et al. 2001; Estevez et al. 2001; Andrulis et al. 2002). Nine subunits of the human exosome are considered core components since they are shared by the nuclear and the cytoplasmic forms of the complex (Chen et al. 2001). Six of these, hRrp41p, hRrp42p, hMtr3p, OIP2, hRrp46p, and PM/Scl-75, are related to the bacterial RNase PH. Three other components, hRrp4p, hRrp40p, and hCsl4p, contain an S1 RNA binding domain. The RNase PH proteins assemble into a hexameric ring (Raijmakers et al. 2002; Estevez et al. 2003; Lorentzen et al. 2005; Hernandez et al. 2006) and the S1 subunits are positioned on top of the ring with their S1 domains facing toward the central pore (Fig. 1; Lorentzen et al. 2005; Pruijn 2005). The PM/Scl-100 protein is not considered a core exosome component, based on its association with only the higher molecular mass exosome complexes (Brouwer et al. 2001; Schilders et al. 2005) and the exclusively nuclear localization of its yeast counterpart Rrp6p (Allmang et al. 1999b).
FIGURE 1.
Model of the exosome. (Left) Top view of the exosome core complex. The RNase PH-like subunits assemble into a hexameric ring structure. The RNase PH domains of PM/Scl-75, hRrp42p, and OIP2 are more related to each other (RPD1, light gray) than to that of hRrp41p, hMtr3p, and hRrp46p, which also contain a related RNase PH domain (RPD2, dark gray). The S1 RNA-binding domain containing subunits hRrp4p, hCsl4p, and hRrp40p (white, black, and white, respectively) are positioned on top of the ring structure, with the S1 domains oriented toward the central pore. (Right) Side view of the exosome. Since it is currently unknown how PM/Scl-100 associates with the core of the exosome, this subunit has not been included.
Cytoplasmic mRNA degradation usually initiates with deadenylation, followed by either further degradation of the mRNA body by the exosome (3′–5′ pathway) or cleavage of the 5′ cap followed by 5′–3′ degradation by the Xrn1p exoribonuclease (5′–3′ pathway). For mRNAs containing premature translation termination codons (PTCs), a surveillance pathway exists termed nonsense-mediated decay (NMD) (Meyer et al. 2004; Schilders et al. 2006). mRNAs containing an AU-rich element (ARE) in the 3′-untranslated region (UTR) undergo rapid decay in the cytoplasm (Chen and Shyu 1995). Two studies provided evidence that the exosome-dependent 3′–5′ pathway plays a major role in the degradation of ARE-containing mRNAs in vitro (Chen et al. 2001; Mukherjee et al. 2002). A more recent in vivo analysis, however, indicated that the 5′–3′ pathway may be the major mode of decay for ARE-containing mRNAs (Stoecklin et al. 2005).
While it has been established that different cofactors associate with the exosome to modulate its various activities, little is known about heterogeneity of exosome complexes in human cells and the importance of individual subunits for their stability. Here we addressed this issue by down-regulation of three core exosome subunits (hRrp4p, hRrp41p, and PM/Scl-75) and one exosome-associated factor, PM/Scl-100. Our data reveal not only marked differences between the nuclear and cytoplasmic exosomes, but also indicate that some subunits are more important for exosome stability than others. We also demonstrate that PM/Scl-100 plays a prominent role in the decay of both ARE- and PTC-containing mRNAs, either as an essential subunit for a fully functional exosome or as an exosome-independent protein.
RESULTS
Glycerol gradient sedimentation of exosome subunits
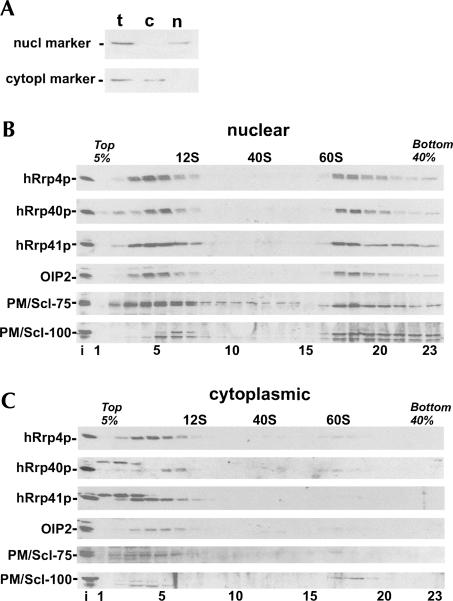
We, and others, have previously reported that human exosome core subunits are in part associated with high molecular weight complexes (Mitchell et al. 1997; Allmang et al. 1999b; Brouwer et al. 2001). In a subsequent study, we observed differential glycerol gradient sedimentation profiles of the core exosome subunit hRrp4p and the exosome-associated factor PM/Scl-100; hRrp4p accumulated partly in 10S fractions and partly in 60–80S fractions, whereas PM/Scl-100 was found almost exclusively in 60–80S fractions (Schilders et al. 2005). To extend this analysis, we investigated the sedimentation behavior of the S1 core exosome subunits hRrp4p and hRrp40p; the RNase PH core exosome subunits hRrp41p, OIP2, and PM/Scl-75; and the exosome-associated factor PM/Scl-100 using nuclear and cytoplasmic extracts of human HEp-2 cells. The relative purity of the nuclear and cytoplasmic fractions was determined by Western blotting using antibodies reactive with a nuclear and a cytoplasmic marker protein, respectively (Fig. 2A). Fractions were analyzed by Western blotting with antibodies against the different exosome subunits and by agarose gel electrophoresis to monitor the distribution of RNA. The U1 snRNA was used as a marker for 12S complexes, while the large rRNAs were used to identify 40S and 60S ribosomal subunits. In the nuclear extracts, all tested core exosome subunits distributed almost equally in ∼ 10S and 60–80S fractions. In contrast to the core subunits, however, the nuclear PM/Scl-100 protein was mainly found in 60–80S fractions (Fig. 2B). With the cytoplasmic fractions, strikingly different patterns were seen; the majority of all cytoplasmic core exosome subunits were found at ∼ 10S (Fig. 2C). The relatively small amounts of PM/Scl-100 in the cytoplasmic fraction were almost exclusively found in ∼ 60S fractions. The sedimentation profile of these cytoplasmic PM/Scl-100-containing complexes seems to be different from that observed for nuclear PM/Scl-100-containing complexes. Also small amounts of exosome core subunits were found in fractions containing the cytoplasmic ∼ 60S complexes. Taken together, these data indicate that in the nucleus a subset of exosomes exists in much larger complexes than in the cytoplasm. These complexes are likely enriched for PM/Scl-100, while the ∼ 10S subpopulation appeared to be largely devoid of PM/Scl-100. For some subunits, additional bands were observed that migrated close to the specific bands. At present, we do not know whether these bands represent isoforms of the respective exosome subunits or are due to cross-reactivity of the sera used.
FIGURE 2.
Glycerol gradient sedimentation of human exosome subunits. (A) Purity of the nuclear and cytoplasmic fractions was determined by Western blotting using antibodies to human topoisomerase I (nuclear marker) and eIF-2α (cytoplasmic marker). Lanes with total (t), cytoplasmic (c), or nuclear (n) extract are indicated. Nuclear (B) and cytoplasmic (C) HEp-2 cell extracts were fractionated in 5%–40% (v/v) glycerol gradients. The sedimentation of hRrp4p, hRrp40p, hRrp41p, OIP2, PM/Scl-75, and PM/Scl-100 was determined by Western blotting. The input (i) material is shown in the lanes on the left. The positions of 12S, 40S, and 60S marker complexes are indicated.
hRrp41p and hRrp4p, but not PM/Scl-75 and PM/Scl-100, are required for exosome stability
The differences between nuclear and cytoplasmic exosomes raised the question of whether or not heterogeneities among individual exosome subunits may also exist. To address this issue, we down-regulated different human exosome subunits by RNAi and tested the effect on the remainder of the complex. Two RNase PH subunits (PM/Scl-75 and hRrp41p), one S1 subunit (hRrp4p), and one exosome-associated factor (PM/Scl-100) were knocked down. HEp-2 cells were transfected with siRNAs and cell extracts were prepared after 24 or 48 h. Western blot analysis showed that the targeted proteins were efficiently down-regulated (Fig. 3A). In addition, the hRrp41p knock-down led to reduced levels of all other tested core exosome subunits except PM/Scl-100 (Fig. 3A, lanes 7,8). This result suggested that the absence of hRrp41p led to destabilization of the exosome core complex and, as a result, degradation of its subunits. This notion is supported by the fact that at the mRNA level reduced levels of only the hRrp41p mRNA were observed, whereas the levels of the other exosome subunit mRNAs remained unaffected (data not shown). In contrast to the hRrp41p siRNA, no codepletion of other subunits was detected with the PM/Scl-75 (Fig. 3A, lanes 5,6) and the PM/Scl-100 siRNAs (Fig. 3A, lanes 9,10). Quantification of the bands confirmed that down-regulation of PM/Scl-75 or PM/Scl-100 did not lead to reduced levels of other subunits (Fig. 3B). This was rather unexpected for PM/Scl-75, since this is also a component of the hexameric ring of RNase PH subunits at the core of the exosome complex (see Fig. 1). Possibly, stable exosome (sub)complexes can exist in the absence of PM/Scl-75. In this light it is interesting to note that exosome complexes purified from Drosophila lacked dRrp45, the Drosophila homolog of PM/scl-75 (Andrulis et al. 2002). Further experiments will be required to determine whether or not human exosome (sub)complexes lacking PM/scl-75 exist. As observed for hRrp41p, knock-down of hRrp4p led to codepletion of other core exosome subunits and also in this case PM/Scl-100 was not affected (Fig. 3A, lanes 13,14). To determine to what extend the codepletion efficiency correlated with the depletion of the siRNA targeted subunits, the protein levels were quantified. This revealed that hRrp41p or hRrp4p down-regulation led to a similar reduction in the levels of other subunits (Fig. 3B). These data suggest that hRrp41p and hRrp4p are required for exosome stability, whereas PM/Scl-75 or PM/Scl-100 depletion does not markedly affect the stability of other subunits.
FIGURE 3.
Depletion of exosome subunits affects cell growth. (A) PM/Scl-75, hRrp41p, PM/Scl-100, and hRrp4p were knocked down by siRNAs and the effect on the levels of various exosome subunits at 24 and 48 h after siRNA treatment was analyzed by Western blotting. As a control, a monoclonal antibody against γ-tubulin was used. (B) Quantification of the effect of siRNA treatment on the protein levels of PM/Scl-75, hRrp41p, and hRrp4p. The siRNAs used are indicated above the graph. The protein levels in extracts from cells treated with control siRNA were set at 100%. The mean values of three independent experiments are shown; standard deviations are indicated by error bars. (C) The effect of exosome subunit depletion on cell proliferation. Cells were harvested and counted 24, 48, and 72 h after siRNA treatment.
We also investigated the effect of hRrp41p down-regulation on glycerol gradient sedimentation profiles of other exosome subunits. In agreement with the results shown in Figure 3A, reduced levels were observed for other subunits except PM/Scl-100; no significant changes in the sedimentation profiles were seen, however (data not shown).
The different codepletion phenotypes observed prompted us to investigate effects on cell growth. HEp-2 cells were harvested and counted 24, 48, and 72 h after siRNA transfection. Knock-down of hRrp4p, hRrp41p, and PM/Scl-75 strongly inhibited cell growth, whereas only slight effects were seen during PM/Scl-100 depletion (Fig. 3C). Thus, even though the PM/Scl-75 knock-down does not lead to exosome destabilization, vital functions for cell growth are affected. The observation that PM/Scl-100 knock-down only slightly affected cell growth suggests that similar to yeast Rrp6 (Briggs et al. 1998), PM/Scl-100 may not be essential for cell proliferation. However, we cannot exclude the possibility that, even though the knock-down was rather efficient (approximately fivefold reduced protein levels), sufficient amounts of PM/Scl-100 were still present to support cell growth.
Cytoplasmic exosomes are depleted by RNAi more efficiently than nuclear exosomes
The association of exosomes with higher-order complexes in the nucleus raised the question of whether turnover rates of exosomes in the nucleus are different from those in the cytoplasm. To explore this possibility, PM/Scl-75, hRrp41p, and PM/Scl-100 were knocked down and nuclear and cytoplasmic fractions were analyzed by Western blotting. In the cytoplasmic fractions, after 24 h clearly reduced levels already were seen for all three targeted proteins (Fig. 4A, lanes 6–8). Strikingly however, at this time point no down-regulation could be detected in the nuclear fractions. After 48 h, a very efficient depletion was seen for all three proteins in the cytoplasm (Fig. 4B, lanes 6–8), whereas in the nuclear fraction, only PM/Scl-100 showed significantly reduced levels (Fig. 4B, lane 4), while hardly any down-regulation was detected for PM/Scl-75 and hRrp41p (Fig. 4B, lanes 2,3).
FIGURE 4.
Knock-down of exosome subunits is more efficient in the cytoplasm than in the nucleus. PM/Scl-75, hRrp41p, and PM/Scl-100 were down-regulated by RNAi, and subsequently HEp-2 nuclear and cytoplasmic fractions prepared after 24 (A) or 48 h (B) were analyzed by Western blotting. As controls for the fractionation, an antiserum to the RNase P/MRP subunit hRpp40p (nuclear marker) and a monoclonal antibody against eIF-2α (cytoplasmic marker) were used.
Thus, cytoplasmic exosomes are depleted much more efficiently than nuclear exosomes. These observations suggest that either nuclear exosomes are more stable than cytoplasmic exosomes or that exosomes accumulate in the nucleus due to ongoing transport from the cytoplasm.
PM/Scl-75, hRrp41p, and PM/Scl-100 are involved in mRNA turnover
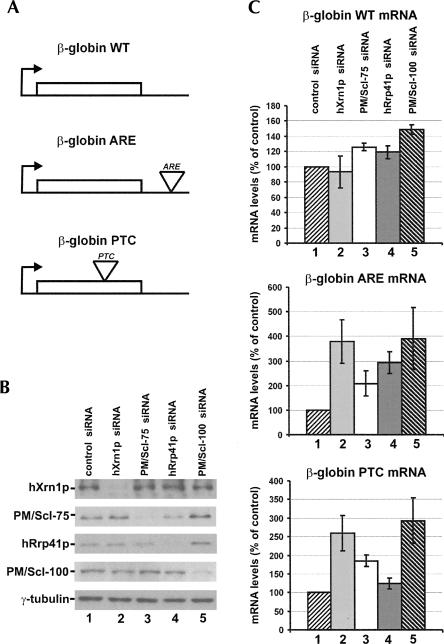
The more efficient down-regulation of the exosome in the cytoplasm allowed us to study the involvement of individual exosome components in RNA degradation in this compartment of the cell. Therefore we determined the effect of PM/Scl-75, hRrp41p, or PM/Scl-100 depletion on mRNA turnover. Since hRrp41p and hRrp4p siRNAs cause similar exosome destabilization (Fig. 3A), hRrp4p was not included in these experiments. Instead, hXrn1p, a component of the alternative 5′–3′ mRNA decay pathway, was knocked down in parallel. Human HEp-2 cells were first transfected with three different reporter constructs: (1) a construct expressing a wild-type (WT) β-globin mRNA; (2) a construct expressing a β-globin mRNA with an ARE (from the c-fos mRNA) in its 3′-UTR; and (3) a β-globin mRNA construct containing a PTC in its coding region, making this mRNA susceptible to NMD (Fig. 5A). Second, the cells were transfected with two different control siRNAs to monitor nonspecific effects on reporter mRNA levels; no differences were seen compared to untreated transfected cells (data not shown). Subsequently, reporter mRNA-expressing cells were transfected with siRNAs for PM/Scl-75, hRrp41p, and PM/Scl-100, as well as an siRNA targeting hXrn1p. Knock-down efficiencies at 48 h after siRNA transfection were verified by Western blotting (Fig. 5B). Using material from the same cells, mRNA levels of the reporter constructs were measured by real-time quantitative RT-PCR analysis; the 18S ribosomal RNA was used for normalization (see Materials and Methods). For the WT β-globin mRNA, slightly increased levels were observed upon knock-down of the different exosome subunits, PM/Scl-100 depletion having the strongest effect (1.5-fold increased mRNA levels) (Fig. 5C). No significant effects were seen upon knock-down of hXrn1p. Much stronger effects were seen with the β-globin-ARE and the β-globin-PTC mRNAs, which may be due to the faster turnover rates of these transcripts, making them more sensitive reporters for mRNA turnover. The knock-down of both hXrn1p and PM/Scl-100 causes about a fourfold increased mRNA levels of the ARE-containing transcript and 2.5- to threefold increased levels of the PTC transcript. Depletion of the core exosome subunits PM/Scl-75 and hRrp41p had less drastic effects: two- to threefold and ∼ 1.5-fold increased mRNA levels of the ARE and PTC transcripts, respectively. Together, these data suggest that both the core exosome and PM/Scl-100 are involved in the turnover of all three types of mRNA.
FIGURE 5.
Knock-down of exosome core subunits and PM/Scl-100 increases reporter mRNA steady-state levels. HEp-2 cells were transfected with three different β-globin mRNA expression contructs: a wild-type (WT) mRNA, a mRNA containing an ARE, and a mRNA bearing a PTC. Subsequently, the cells were transfected with a control siRNA (lane 1), or an siRNA for hXrn1p (lane 2), PM/Scl-75 (lane 3), hRrp41p (lane 4), or PM/Scl-100 (lane 5). (A) A schematic representation of the reporter mRNAs used. (B) Western blotting analysis of the knock-downs. Protein extracts were prepared in parallel with the RNA isolations used for quantitative RT-PCR and analyzed for the presence of the various proteins targeted by RNAi. (C) The effects of the knock-downs on the accumulation of the different reporter mRNAs were analyzed by quantitative RT-PCR. The numbering of the bars corresponds to the numbering of the lanes in A. The 18S rRNA was used for normalization. Mean values and standard deviations from three independent experiments are shown.
To discriminate between effects of exosome depletion on mRNA degradation and on other processes that influence steady-state mRNA levels, we performed mRNA decay analyses. Transcription of the reporter constructs was inhibited by the addition of tetracycline (see Materials and Methods), and mRNA turnover rates were determined. Reporter mRNA levels were determined at different time points after inhibition of transcription by real-time quantitative RT-PCR. As expected, the turnover of the wild-type β-globin mRNA was very slow, since this is known to be a rather stable mRNA species. During the first 10 h, no significant decrease in wild-type β-globin mRNA levels was seen (Fig. 6A). A much faster decay was observed for the β-globin-ARE and the β-globin-PTC mRNAs, with half-lives of ∼ 2.5 and 6 h, respectively. To investigate the effect of exosome depletion on the degradation of these mRNAs, their levels at 4 h (β-globin-ARE mRNA) or at 8 h (β-globin-PTC mRNA) after shutting off their transcription were determined. In both cases at these time points mRNA levels were reduced to 30%–40% in the cells treated with a control siRNA. Due to its very low turnover rate, the wild-type β-globin mRNA could not be included in this analysis. Both for the ARE and for the PTC transcripts, significantly reduced mRNA decay rates were observed in the hXrn1p and the PM/Scl-100 depleted cells (Fig. 6B). Less dramatic effects on mRNA decay were observed with PM/Scl-75 and hRrp41p depletion; PM/Scl-75 down-regulation moderately reduced the decay rate of the ARE transcript. These results correlate well with those of the mRNA steady-state measurements, indicating that the observed effects on the mRNA steady-state levels were largely due to effects on mRNA degradation.
FIGURE 6.
Down-regulation of hXrn1p and exosome subunits attenuates mRNA decay. (A) mRNA decay curves for the β-globin WT, the β-globin ARE, and the β-globin PTC transcripts. Transcription of the reporter constructs was inhibited by tetracycline addition. mRNA levels were determined by quantitative RT-PCR and normalized to endogenous GAPDH mRNA levels. (B) Control siRNA-treated cells or cells treated with siRNAs for hXrn1p, PM/Scl-75, hRrp41p, or PM/Scl-100 were harvested 4 or 8 h after transcription inhibition for the β-globin ARE and the β-globin PTC mRNAs, respectively. As in panel A, mRNA levels were determined by quantitative RT-PCR and normalized to endogenous GAPDH mRNA levels. Measurements were performed in triplicate; mean values and standard deviations are shown.
Taken together, these data indicate that both hXrn1p and PM/Scl-100 contribute to the turnover of the ARE- and the PTC-containing transcripts.
DISCUSSION
Previously it was reported for the Trypanosoma brucei exosome that down-regulation of the RNase PH-type subunits as well as down-regulation of TbRRP6 causes destabilization of the exosome complex, whereas the S1-type subunits could be depleted without affecting the levels of other subunits (Estevez et al. 2003). Similarly, our results indicate that depletion of the RNase PH subunit hRrp41p causes codepletion of all other human exosome subunits analyzed, except PM/Scl-100. In contrast, down-regulation of PM/Scl-75 and PM/Scl-100 did not lead to reduced levels of other subunits of the human exosome, whereas for the T. brucei exosome these components were found to be required for complex stability. Moreover, depletion of the S1-type subunit hRrp4p did lead to decreased levels of other exosome subunits. These results indicate that a subset of exosome subunits is important for complex integrity, but that these subunits may differ between different species. The observation that PM/Scl-100 is dispensable for the integrity of the human exosome is consistent with the observation that this protein shows different glycerol gradient sedimentation patterns than the other human exosome subunits (Fig. 2A,B). Also, the results of gel filtration experiments previously showed that the majority of PM/Scl-100 is associated a subset of exosome-containing complexes (Brouwer et al. 2001). The hRrp4p protein appeared to be required for maintenance of normal levels of other exosome subunits, which is consistent with a recent report indicating that the S1-type subunits are required to reconstitute stable human exosome complexes in vitro (Liu et al. 2006).
Surprisingly, down-regulation of PM/Scl-75 did not affect the levels of other exosome subunits. However, also in the T. brucei exosome, not all RNase PH-type subunits caused a complete disruption of the complex when depleted (Estevez et al. 2003). Thus, it is possible that in the absence of PM/Scl-75, exosome complexes are still able to assemble. In support of this view, exosome complexes purified from Drosophila lacked dRrp45p, the Drosophila homolog of PM/Scl-75 (Andrulis et al. 2002). An alternative possibility is that a non-exosome-associated pool of PM/Scl-75 exists, which may be preferentially affected by the siRNA treatment.
Another striking observation was that down-regulation of exosome subunits was much more efficient in the cytoplasm than in the nucleus (Fig. 4). This phenomenon can be explained in three ways: (1) nuclear and cytoplasmic exosomes are depleted with similar efficiencies but the remaining exosomes accumulate in the nucleus due to ongoing transport from the cytoplasm; (2) nuclear exosome complexes may be more stable than cytoplasmic complexes; or (3) in the cytoplasmic environment turnover of exosomes may be much faster than that in the nucleus. Additional experiments will be required to provide more insight into this issue. Regardless of the exact underlying mechanism, our data strongly suggest that there is no or hardly any export of exosomes from the nucleus.
Down-regulation of components of both the 5′–3′ and 3′–5′ RNA degradation pathways led to increased levels of reporter mRNAs. The most prominent effects were observed after depletion of PM/Scl-100 and hXrn1p. These effects were mainly due to mRNA degradation defects rather than alterations at the level of transcription, because very similar data were obtained with cells in which the transcription of the reporter mRNAs was silenced. Our results indicate that both hXrn1p and PM/Scl-100 are involved in the turnover of ARE- and PTC-containing transcripts, while the role of the core exosome subunits may be less important. Our results are in good agreement with data obtained by Lejeune et al. (2003), who reported an involvement of both 5′–3′mRNA decay factors and PM/Scl-100 in the turnover of PTC-containing transcripts. Interestingly, also in this study no effect of down-regulation of a core component (hRrp4p) on mRNA decay was observed, confirming the notion that PM/Scl-100 has a more pronounced role in nonsense-mediated decay than the core exosome. For ARE-mediated mRNA decay, a major role for the 5′–3′mRNA decay pathway was reported recently (Stoecklin et al. 2005). Our results show that also PM/Scl-75 and PM/Scl-100 are involved in ARE-mediated mRNA decay. In this regard it is important to note that Stoecklin and collaborators used a reporter containing an ARE from the GM-CSF mRNA, which is of a different class of AREs (class II) than the c-fos ARE (class I), which was used in our study (Chen and Shyu 1995). On the other hand, in cell extracts the human exosome was shown to play an important role in class II ARE-containing mRNA decay, and PM/Scl-75 was demonstrated to be directly involved (Mukherjee et al. 2002). Thus, the contribution of both pathways to the degradation of class I and II ARE-containing mRNAs may differ. An important role for 3′–5′ decay in the degradation of class I ARE-containing mRNAs was also observed in another recent study (Murray and Schoenberg 2007). It was surprising that depletion of hRrp41p, which causes the most severe codepletion effects, has the fewest effects on mRNA decay. A possible explanation for this observation is that PM/Scl-75 and PM/Scl-100 may at least in part act independently of the exosome in mRNA decay. In this respect it is interesting to note that Stoecklin et al. (2005) also observed an inhibition of ARE-mediated mRNA decay upon PM/Scl-75 knock-down, while depletion of two other exosome subunits (hRrp46p and hRrp40p) had no effect.
In conclusion, our data reveal differences among individual exosome subunits of the human exosome, both with respect to their importance for the integrity of the complex and their impact on mRNA decay. Moreover, they differentially affect cell growth. An interesting possibility is that PM/Scl-75 and/or PM/Scl-100 may act independently of the exosome.
MATERIALS AND METHODS
Preparation of cell extracts
Cytoplasmic and nuclear fractions were prepared from HEp-2 cells by resuspension of the cells in lysis buffer (25 mM Tris-HCl at pH 7.5, 150 mM KCl, 1 mM EDTA, 0.5 mM PMSF, and 1 mM DTT), permeabilization by the addition of digitonin to a final concentration of 0.025%, incubation for 10 min at room temperature, and centrifugation at 1000g at 4°C. The supernatant was transferred to a new tube and is referred to as the cytoplasmic fraction. After resuspending the pellet in lysis buffer, the nuclear fraction was prepared by sonication using a Branson microtip. Both extracts were clarified by centrifugation at 12,000g for 10 min at 4°C. Extracts were checked for proper fractionation using antisera reactive with the nuclear markers Rpp40 or topoisomerase I and the cytoplasmic marker eIF-2α, respectively. For the preparation of total cell extracts, HEp-2 cells were directly sonicated in lysis buffer followed by centrifugation.
Glycerol gradient analysis
HEp-2 cell extracts were prepared as described above and loaded on 5%–40% (v/v) glycerol gradients prepared in gradient buffer (25 mM Tris-HCl at pH 7.5, 150 mM KCl, 1 mM DTT, and 0.02% Triton X-100). Gradients were centrifuged in a TH641 rotor (Sorvall) for 16 h at 25,000 rpm at 4°C and subsequently 23 fractions per gradient were collected. The proteins in these fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes for further analysis.
siRNA treatment
siRNAs were purchased from Eurogentec. The sequences of the siRNAs for PM/Scl-100, hRrp41p, hRrp4p, PM/Scl-75, and hXrn1p are 5′-GUACAACCCAGGAUAUGUGdTdT-3′, 5′-UGUGCAGGUGCUACAGGCAdTdT-3′, 5′-GUUGAUCUGUGUGAAAGCUdTdT-3′, 5′-GCCAAGAUGCUCCCAUAAUdTdT-3′, and 5′-GUCAUGGCAAGGAGUUACCdTdT-3′, respectively. siRNA transfections were essentially done as described (Schilders et al. 2005) with the exception that 20 nM siRNA were used. As controls, cells were transfected with control siRNAs (a control siRNA from Eurogentec or an siRNA for a melanoma cell marker, which is not expressed in the cell line used in this study) or were left untreated. For the analysis of effects on cell growth, cells were harvested and counted with a Coulter AC-T series analyzer.
Immunoblotting
For the detection of proteins on Western blots, monoclonal anti-hRrp4p antibody (culture supernatant) (Modiquest) and rabbit antisera were diluted 10- and 1000-fold, respectively, in blocking buffer (5% skimmed milk, phosphate buffered saline [PBS] and 0.05% NP-40). As a secondary antibody, horseradish peroxidase-conjugated rabbit anti-mouse IgG or swine anti-rabbit IgG (Dako Immnuoglobulins) was used after 1000-fold dilution in blocking buffer. Bound antibodies were visualized by chemiluminescence. Alternatively, for quantification, goat anti-mouse IRDye 800CW or goat anti-rabbit IRDye 800CW (Li-Cor Biosciences) was used at a 1:10,000 dilution. Quantification was performed with the Odyssey Infrared Imaging System.
Transfection of HEp-2 cells with β-globin mRNA constructs
Expression plasmids pTet-BBB and pTet-BBB+ARE were kindly provided by Dr. A.-B. Shyu (Houston Medical School, University of Texas). pTet-BBB+PTC was constructed as follows: pTet-BBB was digested with AccI, treatment with Klenow DNA polymerase, and religation. This caused a frameshift resulting in a PTC. HEp-2 cells stably transfected with the plasmid pTet-tTAk (which expresses a tetracycline-inducible transcriptional activator) were cultured to 70% confluence and transfected as described (Schilders et al. 2005). After transfection, the cells were seeded in 6-well or 24-well plates for 24 h (1.2 × 105 or 3 × 104 cells per well, respectively). Subsequently, the cells were transfected with siRNAs as described above. For the steady-state mRNA measurements, RNA was isolated from the cells 48 h after siRNA treatment. For the time-course analyses, tetracycline (1 μg/mL final concentration) was added to the medium, and cells were harvested after 3, 6, 10, and 24 h (β-globin WT), 2, 3, 4, 5, 6 h (β-globin ARE), or 2, 4, 6, 8, 10 h (β-globin PTC). mRNA decay curves were obtained with control siRNA treated cells. Cells treated with control, hXrn1p, PM/Scl-75, hRrp41p, or PM/Scl-100 siRNA were harvested at 4 or 8 h after transcription arrest for the β-globin ARE and the β-globin PTC mRNAs, respectively.
Real-time quantitative RT-PCR analysis
For real-time quantitative RT-PCR analysis, total RNA was isolated from HEp-2 cells using the TRIzol reagent (Invitrogen) and treated with DNaseI (Invitrogen). cDNA was generated using the Reverse Transcription System from Promega following the manufacturer's instructions. For mRNA steady-state level measurements, the 18S rRNA was used as a control; for the mRNA turnover assays, the GAPDH mRNA was used as a control. The following primers were used: OEVD52 5′-AGCTGAGTGAACTGCACTGTGAC-3′ and OEVD31 5′-GCCTGCACCTGAGGAGTGA-3′ for the β-globin reporter mRNAs, and 18S for 5′-TACCTGGTTGATCCTGCCAGTAGC-3′ and 18Srev 5′-AACTGATTTAATGAGCCATTCGC-3′ for the 18S rRNA. For the GAPDH mRNA, primers GAPDH forward (5′-TGGAAGGACTCATGACCACA-3′) and GAPDH reverse (5′-GAGGCAGGGATGATGTTCTG-3′) were used. qPCR analysis was performed using the ABI-PRISM 7000 Sequence Detection System from Applied Biosystems.
ACKNOWLEDGMENTS
We thank Dr. Ann-Bin Shyu (Houston Medical School, University of Texas) and Dr. Jens-Lykke Andersen (University of Colorado, Boulder) for plasmids and antibodies. This work was supported in part by the Council for Earth and Life Sciences (NWO-ALW) and by the Council for Chemical Sciences (NWO-CW) of The Netherlands Organization for Scientific Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.575107.
REFERENCES
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., Tollervey, D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999a;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., Mitchell, P. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes & Dev. 1999b;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., Lis, J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila . Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Presutti, C., Tollervey, D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. doi: 10.1016/s0092-8674(00)00065-9. [DOI] [PubMed] [Google Scholar]
- Briggs, M.W., Burkard, K.T., Butler, J.S. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′-end formation. J. Biol. Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- Brouwer, R., Allmang, C., Raijmakers, R., van-Aarssen, Y., Egberts, W.V., Petfalski, E., van-Venrooij, W.J., Tollervey, D., Pruijn, G.J. Three novel components of the human exosome. J. Biol. Chem. 2001;276:6177–6184. doi: 10.1074/jbc.M007603200. [DOI] [PubMed] [Google Scholar]
- Chekanova, J.A., Shaw, R.J., Wills, M.A., Belostotsky, D.A. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem. 2000;275:33158–33166. doi: 10.1074/jbc.M005493200. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., Karin, M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Das, B., Butler, J.S., Sherman, F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae . Mol. Cell. Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, A.M., Kempf, T., Clayton, C. The exosome of Trypanosoma brucei . EMBO J. 2001;20:3831–3839. doi: 10.1093/emboj/20.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, A.M., Lehner, B., Sanderson, C.M., Ruppert, T., Clayton, C. The roles of intersubunit interactions in exosome stability. J. Biol. Chem. 2003;278:34943–34951. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
- Hernandez, H., Dziembowski, A., Taverner, T., Seraphin, B., Robinson, C.V. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson, J.S., Parker, R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., Maquat, L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Greimann, J.C., Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lorentzen, E., Walter, P., Fribourg, S., Evguenieva-Hackenberg, E., Klug, G., Conti, E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., Tollervey, D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O'Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., Wilusz, J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, E.L., Schoenberg, D.R.2007A+U-rich instability elements differentially activate 5′–3′ and 3′–5′ mRNA decay Mol. Cell Biol. 82791.– [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruijn, G.J. Doughnuts dealing with RNA. Nat. Struct. Mol. Biol. 2005;12:562–564. doi: 10.1038/nsmb0705-562. [DOI] [PubMed] [Google Scholar]
- Raijmakers, R., Egberts, W.V., van Venrooij, W.J., Pruijn, G.J. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- Schilders, G., Raijmakers, R., Raats, J.M.H., Pruijn, G.J.M. MPP6 is an exosome-associated RNA-binding protein involved in 5.8S rRNA maturation. Nucleic Acids Res. 2005;33:6795–6804. doi: 10.1093/nar/gki982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilders, G., van Dijk, E., Raijmakers, R., Pruijn, G.J. Cell and molecular biology of the exosome: How to make or break an RNA. Int. Rev. Cytol. 2006;251:159–208. doi: 10.1016/S0074-7696(06)51005-8. [DOI] [PubMed] [Google Scholar]
- Stoecklin, G., Mayo, T., Anderson, P. ARE-mRNA degradation requires the 5′–3′ decay pathway. EMBO Rep. 2005;7:72–77. doi: 10.1038/sj.embor.7400572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van-Hoof, A., Lennertz, P., Parker, R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]