Abstract
CC chemokine receptor 5 (CCR5) is the major HIV-1 coreceptor and its expression levels are a critical determinant of HIV-1 infection. However, the molecular mechanisms of CCR5 regulation in primary targets of HIV-1 remain unknown. Despite binding to conserved DNA elements, we show that the transcription factors GATA binding protein 1 (GATA-1) and GATA-3 differentially suppress the expression of CCR5 in stem-cell–derived dendritic cells and primary human T-cell subsets. In addition, GATA-1 expression was also more potent than GATA-3 in suppressing T helper 1 (Th1)–associated genes, interferon-γ (IFNγ), and CXC chemokine receptor-3 (CXCR3). GATA-1, but not GATA-3, potently suppressed CCR5 transcription, thereby rendering human T cells resistant to CCR5-tropic HIV-1 infection. However, GATA-1 could also serve as a surrogate for GATA-3 in its canonic role of programming Th2 gene expression. These findings provide insight into GATA-3–mediated gene regulation during T-cell differentiation. Importantly, decoding the mechanisms of GATA-1–mediated repression of CCR5 may offer an opportunity to develop novel approaches to inhibit CCR5 expression in T cells.
Introduction
CC chemokine receptor-5 (CCR5) is a receptor for several CC chemokines that regulate leukocyte migration and activation.1 CCR5 also serves as the primary coreceptor on CD4+ cells for the viral entry of macrophage-tropic (R5) strains of HIV-1.1,2 The clinical relevance of CCR5 expression to HIV-1 infection is highlighted by the following observations. First, individuals who are homozygous for a 32 base pair (bp) deletion within the CCR5 coding region (CCR5Δ32) have no CCR5 cell-surface expression and are highly resistant to HIV-1 infection.3-5 Second, CCR5 levels vary significantly among individuals with an intact coding region, and this, in turn, is associated with variable risks of HIV-1 acquisition and rates of disease progression to AIDS.6 Finally, CCR5 expression levels and infectivity of T cells by HIV-1 tightly correlate in vitro.7 Together, these findings emphasize the pivotal role of CCR5 in the pathogenesis of HIV-1/AIDS and underscore the importance of understanding the regulation of CCR5 expression in cells that are targets for HIV-1 infection.
In the immune system, CCR5 is expressed on CD4+ effector and memory T cells, invariant natural killer T (NKT) cells, macrophages, immature dendritic cells (DCs) and in bone marrow precursor cells during hematopoiesis.1 Although CCR5 expression in most cell types is uniform, it is highly variable in human T cells and is closely tied to the type of effector function acquired during naive T (TN) cell activation and differentiation. Specifically, CCR5 is more highly expressed on interferon-γ (IFNγ)–secreting type 1 (Th1) cells than on interleukin 4 (IL-4)–producing type 2 (Th2) cells.8 In addition to CCR5, CXC chemokine receptor-3 (CXCR3), another chemokine receptor, is also abundantly expressed on Th1 cells.8,9 By contrast, Th2 cells preferentially express CCR48 and the prostaglandin D2 receptor (CRTH2).10 These distinct chemotactic receptor expression profiles on Th1 and Th2 cells are thought to be critical for regulating their migratory propensities to different sites of inflammation and may also influence cell-mediated immune responses critical for control of HIV-1 infection.11,12
A strong link between the expression of specific transcription factors (TFs), including members of the GATA-binding protein (GATA) family, and T-cell differentiation has been well established. However, the precise repertoire of TFs that regulate CCR5 expression in human T cells is less clear, and 2 lines of evidence have implicated members of the GATA family in the regulation of CCR5 expression. First, promoter analyses of CCR5 have identified DNA binding sites for several TFs, including GATA TFs.13,14 A previous report showed that GATA TFs can bind to the CCR5 cis-regulatory region and that GATA-1 can trans-activate reporter constructs containing CCR5 promoter elements in a T-lymphoid cell line.15 However, whether CCR5 expression is regulated by GATA TFs within the context of physiologic cellular targets of HIV-1 is not known.
Structurally, GATA TFs are composed of either 1 or 2 centralized C4-type zinc-finger motifs that mediate DNA binding and protein-protein interactions.16,17 Among the GATA family members, GATA-1, GATA-2, and GATA-3 are most closely related, based on both tissue expression profiles and sequence conservation within their zinc-finger domain. The high degree of sequence similarity within the zinc-finger domain of GATA-1, -2, and -3 confer similar DNA binding preferences for the consensus DNA element (A/T)GATA(A/G),16 and these GATA members all serve as key regulators of hematopoiesis in mammals.18,19 Notably, homology in amino- and carboxy-terminal regions located outside the zinc-finger domains of GATA-1, -2, and -3 is minimal.
GATA-3 is a master regulator of TN-cell differentiation into Th2 effector cells.20,21 Indeed, expression of GATA-3 enhances Th2 cytokine production, decreases IFNγ secretion, and promotes chromatin remodeling of the IL-4 locus during T-cell differentiation.21 By contrast, GATA-1 is expressed in CD34+ hematopoietic stem cells (HSCs), erythroblasts, megakaryocytes, eosinophils, and mast cells, serving to regulate megakaryopoiesis and erythropoiesis in mice and humans.22,23 Mice lacking GATA-1 display severe hematopoietic dysfunction and do not survive gestation as a result of lethal anemia.24 In humans, mutations that interfere with GATA-1 expression or function result in severe anemic disorders or hematopoietic cancers.25-27 Interestingly, studies using GATA-1–deficient mice demonstrate that GATA-3 expression can rescue hematopoietic deficiencies and embryonic lethality, suggesting that some degree of functional redundancy exists between these 2 TFs.28,29 However, the ability of GATA-3 to reconstitute GATA-1 function is incomplete because adult mice that have GATA-3 substituted for GATA-1 generate reduced levels of improperly functioning platelets and erythrocytes.28 Nevertheless, these findings suggest that the DNA-binding zinc-finger domain of GATA-3, which is greater than 90% identical to that of GATA-1, is largely sufficient to direct hematopoiesis.
Here, we demonstrate that expression of GATA-1, but not GATA-3, markedly reduces CCR5 promoter activity and thus represses CCR5 gene expression in primary human T-cell subsets and DCs. The down-regulation of CCR5 expression by GATA-1 renders human T cells resistant to infection by CCR5-tropic HIV-1. Ectopic expression of GATA-1 also down-regulated the expression of IFNγ and CXCR3, consistent with the possibility that GATA-1 might be a global repressor of Th1 effector functions. However, similar to GATA-3, ectopic expression of GATA-1 in human naive and memory T cells programs Th2 cytokine and chemotactic receptor expression profiles, albeit with somewhat lower efficiency than GATA-3. Hence, GATA-1 and GATA-3 share similar properties with respect to their ability to influence Th2 differentiation, but they have differential effects on CCR5 expression. Decoding the molecular determinants of GATA-1–mediated repression of CCR5 could aid in the development of novel approaches to modulate its expression during HIV-1 infection. Furthermore, understanding the transcriptional control of T-cell differentiation has implications in modulating adaptive immune responses.
Materials and methods
Lentiviral vectors
Construction and use of HIV-derived vectors (HDVs) that contain bicistronic marker genes have been described.30 The HDV bicistronically expressing GATA-3 and a marker gene (murine Cd24 or GFP) has also been described.31 To generate HDVs expressing GATA-1, murine or human GATA-1 was subcloned into the SmaI site of the HDV vector and confirmed by sequencing of the insert regions. Murine and human GATA-1 share 90% and 100% sequence identity within the open reading frame and zinc-finger domains, respectively.
Purification of T-cell subsets
Peripheral blood mononuclear cells (PBMCs) were isolated from neonatal placental cord blood or from adult blood by Ficoll (Pharmacia, Piscataway, NJ) density centrifugation. Resting CD4+ T cells were purified as described using CD4-Dynabeads30,31 and were 99.5% CD4+CD3+ as determined by fluorescence-activated cell sorting (FACS) analysis. Primary T cells transduced with HDVs express the reporter protein mCD24 on the cell surface and were enriched by sorting for mCD24+ cells as described.31 In some experiments purified CD4+ T cells were further subdivided into central memory (CD45RO+CCR7+), effector memory (CD45RO+CCR7–), and naive (CD45RO–CCR7+) T cells from adult blood, using magnetic bead and flow cytometry (FACSAria) sorting as described.31 Human NKT cells were isolated from adult blood PBMCs as described.32 The blood from healthy donors was obtained in accordance with and by approval of Vanderbilt University Medical Center institutional review board for these studies. Informed consent was provided in accordance with the Declaration of Helsinki.
CD34+ hematopoietic stem-cell isolation, culture, and DC differentiation
Human CD34+ HSCs were isolated from Ficoll-separated umbilical cord blood mononuclear cells by magnetic sorting using anti-CD34 antibodies conjugated to magnetic-activated cell sorting (MACS) beads (Miltenyi Biotech, Auburn, CA). Purified stem cells were cultured in serum-free media (Stem Cell Technologies, Vancouver, BC, Canada; serum-free expansion media) supplemented with stem-cell factor (SCF; 50 ng/mL), FMS-related tyrosine kinase 3 ligand (100 ng/mL), IL-3 (20 ng/mL), thrombopoietin (200 ng/mL) (all from R&D Systems, Flanders, NJ) and penicillin/streptomycin (Cellgro, Herndon, VA) for 3 to 4 days. HSCs were subsequently expanded for an additional 3 to 4 days in stem-cell media containing SCF. To differentiate stem cells into DCs, they were washed and cultured in RPMI plus 10% fetal calf serum (FCS) supplemented with human IL-4 (50 ng/mL) and granulocyte-macrophage colony-stimulating factor (20 ng/mL; R&D systems). DC differentiation from HSCs was confirmed via FACS analysis by staining with anti-CD1a (BD Biosciences, Mountain View, CA) or anti-CD1c (Miltenyi Biotech) antibodies 4 to 6 days after differentiation.
T-cell activation and differentiation
T cells were activated through the T-cell receptor (TCR) using anti-CD3 and anti-CD28 antibodies. Briefly, 96-well plates were coated with 10 μg/mL goat-anti–mouse immunoglobulin G (IgG; Caltag, San Francisco, CA) for 1 hour at 37°C. Wells were washed twice with phosphate-buffered saline (PBS) and coated with 0.5 μg/mL anti-CD3 for an additional hour at 37°C. After washing to remove unbound CD3 antibodies, T cells were added to antibody-coated wells with soluble anti-CD28 (1 μg/mL) in the presence of Th1 or Th2 polarizing cytokine conditions as described.31 Cells were removed from activation signals after 48 hours and expanded in recombinant human IL-2 (200 U/mL; Chiron Diagnostics, Emeryville, CA) supplemented media.30 NKT cells were purified, activated, and expanded as described.32
Virus production and infections
Vesicular stomatitis virus glycoprotein (VSV-G)–pseudotyped, replication-defective HIV-derived vectors, R5-tropic replication-competent HIV-1 virions (BAL) and X4-tropic HIV-1 virions (LAI) were generated as described.30 Transduction of primary T-cell subsets and DCs with HDV or R5-tropic viruses were performed in flat-bottom 96- or 24-well plates at multiplicities of infection (MOIs) ranging from 1 to 8. For some experiments, cells inoculated with virus were centrifuged for 1 hour at 1000g to enhance infections as described.33 For single-cycle infections, HDV-transduced human T cells were infected for 2 days. The cells were then washed to remove residual virus and HIV protease inhibitors (indinavir, 10 μg/mL; ritonavir, 10 μg/mL) and azidodeoxythymidine (AZT, 10 μM), all obtained from the National Institutes of Health (NIH) AIDS Research and Reference reagent program, were added to prevent spread of the virus. All viruses were engineered to express either green fluorescent protein (GFP) or murine CD24 (mCD24) in the place of the nef gene to monitor cell transduction using flow cytometry. Viral replication was determined by quantifying the HIV-1 core protein p24 in culture supernatants by enzyme-linked immunosorbent assay (ELISA) as described.32
Antibodies and FACS analysis
Cells were stained with the relevant antibodies and analyzed using a flow cytometer (FACSCalibur) as described.31 Analysis was performed using FlowJo software (Tree Star, San Carlos, CA). The following antihuman antibodies were used for staining: CD3, CD4, CD45RO, CD45RA, CCR4, CCR5, CXCR3, CCR4, CD1a, and CD1c (all from BD Biosciences), CCR7 (R&D Sciences), anti-Vβ11, anti-Vα24 (Coulter, Miami, FL) and a murine antibody against CD24 (heat-stable antigen [HAS]; Pharmingen, San Jose, CA). The CRTH2 antibody used for these experiments has been described.10
Real-time PCR
Total RNA isolation and cDNA synthesis from primary cells was performed as described.34 The cDNA was used to perform real-time polymerase chain reaction (PCR) to determine expression levels of GATA-3, GATA-1, CCR5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH levels were used to normalize RNA content of samples. Real-time PCR was conducted using the TaqMan universal PCR Master Mix (Roche, Indianapolis, IN) in a Model MX3000P Sequence Detection System (Stratagene, La Jolla, CA). To quantify GATA-1 and GATA-3 transcript numbers, serial dilutions (50 000 copies to 2.5 copies) of respective plasmids were used to generate standard curves. Transcript numbers were quantified from the GATA-1 or GATA-3 threshold cycle (Ct) values following normalization to GAPDH values using real-time PCR analytic software (Stratagene). The sequences of the primers and probes are as follows: GATA-3 forward, GGACGAGAAAGAGTGCCTC; GATA-3 reverse, TGGGACGACTCCAGCTTCA; GATA-3 probe, FAM-AGGTGCCCCTGCCCGACAGCBHQ. Human real-time PCR primers/probe sets for CCR5 (assay ID no. Hs00152917_m1), GAPDH (assay ID no. Hs99999905_m1), and GATA-1 (assay ID no. Hs00231112_m1) were purchased from Applied Biosystems (Foster City, CA).
CCR5 promoter analysis in primary human T cells
CCR5 promoter activity was determined in GATA-1 or GATA-3 transduced primary T cells after anti-CD3 and anti-CD28 stimulation for 24 hours as described above. Cells (1-2 × 106) were washed twice with PBS and resuspended in 160 μL human T-cell nucleofection buffer (Amaxa, Cologne, Germany). Cells were transfected with 2 μg of either the promoterless pGL3-Basic vector (Promega, Madison, WI) or a vector that contains a CCR5 promoter construct that spans both promoters 1 and 2. The CCR5 promoter construct was cloned upstream of the firefly luciferase gene in this vector, is shown in Figure 4C, and corresponds to the plasmid construct pB1 as described.13 pHRL-CMV vector (0.3 μg; Promega) that contains the synthetic Renilla luciferase gene was cotransfected to normalize for transfection efficiency. Cells were transfected using a nucleofection device (Amaxa) per manufacturer's instructions and cultured 18 hours in IL-2–supplemented media. Cells were subsequently lysed, and both firefly and Renilla luciferase activities were determined using a Dual-Luciferase assay kit (Promega) and analyzed using a luminometer (Mediators PhL; Mediators Diagnostic Systems, Vienna, Austria).
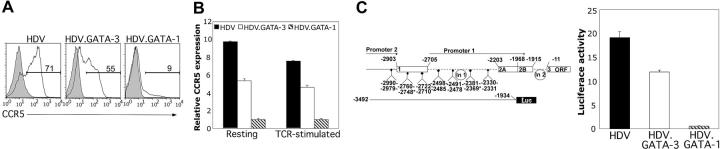
Figure 4.
GATA-1 expression in primary human T cells suppresses CCR5 promoter activity. (A) Total resting CD4+ T cells TCR activated and transduced with HDV, HDV.GATA-3, or HDV.GATA-1 were expanded, and cell-surface expression of CCR5 was assessed on unsorted cells by staining with either isotype control (gray peaks) or anti-CCR5 (white peaks) antibodies in conjunction with an anti-mCD24 antibody. The histograms are gated on the mCD24+ populations, and the percentage of CCR5-positive T cells are displayed. The histograms are representative of 5 independent stainings performed on cells isolated from different blood donors. (B) Total CD4+ human T cells expressing HDV, HDV.GATA-1, or HDV.GATA-3 as above were sorted based on mCD24 expression (see “Materials and methods”) and either left unstimulated (resting) or restimulated via anti-CD3 and anti-CD28 crosslinking for 18 hours (restimulated). Cells were lysed, cDNA was generated, and quantitative real-time PCR was performed using gene-specific primers. CCR5 mRNA expression levels are normalized to GAPDH levels and are displayed as fold differences in relative expression. These data represent 3 independent real-time PCR reactions run from cDNA generated from separate sets of cells from different donors. (C, left) Genomic organization of human CCR5 and the GATA-1 cis-binding sites. Open boxes are exons, dashed lines connecting exons are introns. Exons and introns are numbered. Nucleotide numbering and gene structure are according to Mummidi et al.39 GATA-1–binding positions are indicated by diamonds, and the GATA binding sites previously characterized14,15 are marked with asterisks. The luciferase reporter construct is schematically shown below the gene structure (see “Materials and methods”). (C, right) Transduced human T cells positively sorted for mCD24 expression were restimulated through the TCR (see “Materials and methods”) and are transfected with either the full-length CCR5 promoter driving firefly luciferase expression or a promoterless firefly luciferase plasmid. Cells were cotransfected to normalize for transfection efficiency. Transfected cells were lysed, and luciferase expression was quantified. Firefly luciferase activity was normalized to Renilla activity within samples, and the data are displayed as the relative fold induction of firefly luciferase driven by CCR5 promoter over the promoterless control vector. These data represent 3 independent sets of transfections performed on cells from separate donors. Error bars indicate standard deviation of duplicate samples.
Cytokine detection
Intracellular cytokine analysis of human T cells stimulated with PMA (phorbol 12-myristate-13-acetate) and ionomycin was performed as described.31 Cells were stained with antibodies for the following human cytokines: allophycocyanin (APC)–conjugated anti-IFNγ, phycoerythrin (PE)–conjugated anti–IL-4, PE-conjugated anti–IL-13, and APC-conjugated anti–IL-5 (all from Pharmingen). For detection of secreted cytokines, T cells were stimulated via anti-CD3/anti-CD28 stimulation for 18 hours. Supernatants were assayed using the cytometric bead array (CBA) kit (BD Biosciences) and analyzed using CBA 6-bead analysis software (BD Biosciences) using flow cytometry.
Western blot analysis of GATA-3 and GATA-1 expression
Western blot analyses on primary human T cells transduced with the control HDV, HDV.GATA-3, or HDV.GATA-1 were performed as described.31 Blots were probed with anti–GATA-3 (HG3-31; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies followed by horseradish peroxidase (HRP)–conjugated anti–mouse IgG (Jackson Laboratories, West Grove, PA), and developed using West Pico luminol/peroxide solutions (Bio-Rad Laboratories, Hercules, CA) and autoradiographed (Amersham). The membranes were stripped using a stripping solution (Pierce Chemical, Rockford, IL) for 5 minutes at 37°C and 7 minutes at room temperature. Stripped membranes were reprobed with an anti–GATA-1 (Abcam, Cambridge, United Kingdom) antibody followed by HRP-conjugated antirabbit antibody. To normalize for protein content, the membranes were stripped again and probed with anti–β-actin (I-19; Santa Cruz Biotechnology) antibody followed by anti–mouse IgG-HRP.
Results
GATA TFs have been implicated in the regulation of CCR5 expression. Indeed, CCR5 promoter regions contain as many as 7 DNA-binding sites for GATA TFs.13,14 Moreover, expression of GATA-3 during T-cell differentiation promotes Th2-effector–cell development, and these cell types have lower levels of CCR5 expression compared with Th1 cells.8,20,21 In our initial studies, however, we found that ectopic expression of GATA-3 in T cells was associated with only slight reductions in CCR5 expression (findings described below). Therefore, we sought to determine whether other GATA TFs could regulate CCR5 expression. We focused on GATA-1 because previous studies have shown that it binds to the CCR5 cis-regulatory region and trans-activates its expression in reporter assays performed in a transformed T-lymphoid cell line.15 However, GATA-1 is not expressed in T cells, but it is expressed in HSCs18 that can differentiate into DCs. In turn, these cells express robust levels of CCR535 and are targets of initial HIV-1 infection.36 Therefore, we determined whether there was a relationship between endogenous GATA-1 and CCR5 expression during HSC differentiation into DCs.
Inverse relationship between GATA-1 and CCR5 cell-surface expression in primary HSCs
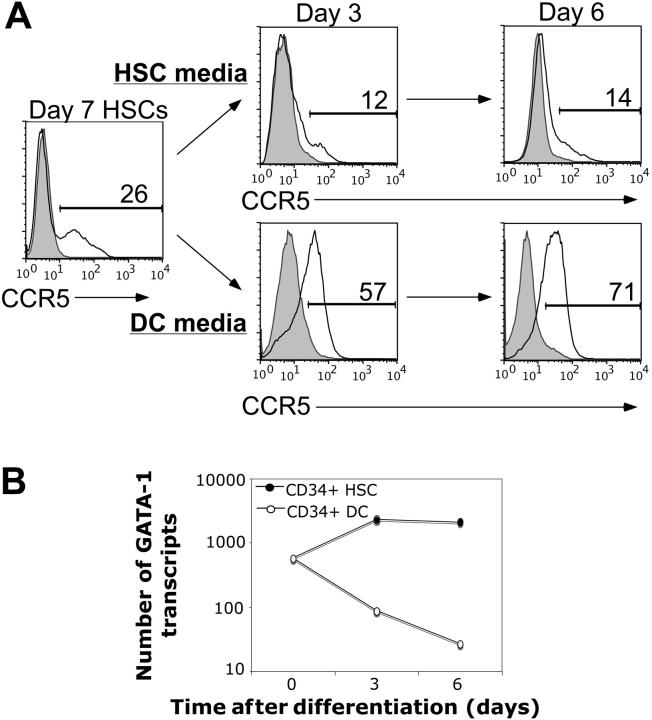
Primary human HSCs were isolated from neonatal umbilical cord blood and expanded in vitro in stem-cell media. The expanded HSCs were then either kept in stem-cell media to maintain their stem-cell pluripotency or were washed and cultured in DC-differentiating media. Cell-surface CCR5 and endogenous GATA-1 expression were determined by flow cytometric and quantitative real-time PCR analyses, respectively (Figure 1). We found that primary human HSCs express high levels of GATA-1 but only minimal amounts of CCR5 on the cell surface (Figure 1). By contrast, as HSCs differentiated into DCs, the expression levels of GATA-1 rapidly decreased, whereas CCR5 expression progressively increased (Figure 1).
Figure 1.
Endogenous GATA-1 expression is inversely related to CCR5 expression in human CD34+ hematopoietic stem cells during DC differentiation. Purified and expanded primary human HSCs were cultured in stem cell media or DC-differentiating media and 2 endpoints were analyzed: (A) cell-surface CCR5 expression was assessed at the indicated time points via flow cytometry and (B) endogenous GATA-1 mRNA levels were determined via quantitative real-time PCR (see “Materials and methods”). The findings represent 2 experiments performed using stem cells isolated from independent umbilical cord blood samples
Ectopic expression of GATA-1 potently reduces cell-surface expression of CCR5 in primary targets of HIV-1
The observed inverse relationship existing between GATA-1 and CCR5 expression levels in HSCs suggested that GATA-1 may repress CCR5 expression in these, and potentially other, primary human cell types. To directly test this possibility, we ectopically expressed either GATA-1 or GATA-3 in DCs derived from HSCs. For these experiments, stem-cell–derived DCs were generated as described above and were transduced with lentiviral vectors encoding GATA-1 (HDV.GATA-1) or GATA-3 (HDV.GATA-3). Expression of GATA-1 or GATA-3 in transduced cells was confirmed by real-time PCR analysis (data not shown). Remarkably, we found that ectopic expression of GATA-1, but not GATA-3, in DCs was associated with a marked decrease in CCR5 cell-surface levels (Figure 2A).
Figure 2.
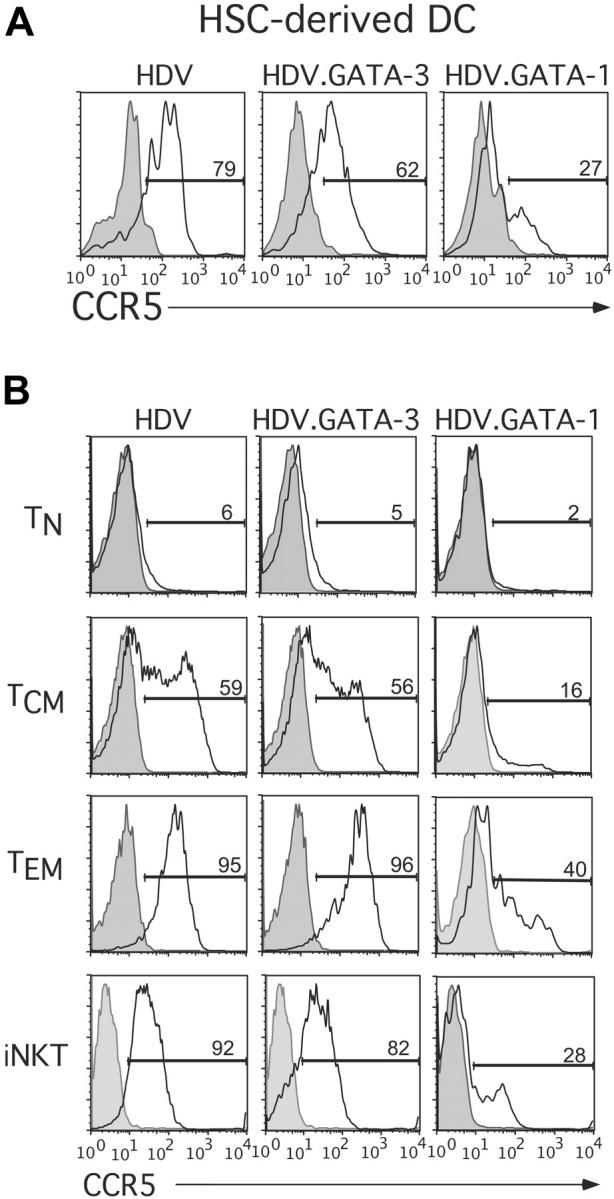
Ectopic expression of GATA-1 potently reduces cell-surface expression of CCR5 in primary human cell targets of HIV-1. (A) Primary human HSC-derived DCs were transduced with HDV, HDV.GATA-1, or HDV.GATA-3 and cultured for an additional 4 days. CCR5 expression levels on HSC-derived DCs were determined via flow cytometric analyses by costaining cells with anti-CCR5, anti-CD1c (Miltenyi Biotech) to monitor DC differentiation and anti-mCD24 to resolve DCs expressing the lentiviral transgene. (B) Primary human conventional T-cell subsets (top 3 panels) or NKT cells (bottom) were TCR activated and transduced to express GATA-1, GATA-3, or the control HDV (see “Materials and methods”). Cells were expanded in the presence of IL-2, and cell-surface CCR5 expression was determined on unsorted populations of transduced cells and costained with anti-mCD24 antibodies. Additionally, NKT cells were costained with a fluorescein isothiocyanate (FITC)–conjugated anti–human Vβ11 antibody (Coulter), to exclude any contaminating non-NKT cells. These results are representative of at least 3 experiments performed on each cell type from unique adult or umbilical cord blood preparations.
Because human effector and memory, but not naive, T cells also express high levels of CCR5 and are primary targets of HIV-1 infection, we next asked whether GATA-1 could also down-regulate CCR5 expression in human T cells. Purified CD4+ T cells were first fractionated into naive (TN; CD45RO–CCR7+), central memory (TCM; CD45RO+CCR7+), and effector memory (TEM; CD45RO+CCR7–) subsets as described.31,37 In addition, NKT cells, which also express high levels of CCR5 and are susceptible to HIV-1 infection,32,38 were prepared as described.32 Following cell purification, all T-cell subsets were activated through the TCR and concurrently transduced with HDV, HDV.GATA-3, or HDV.GATA-1. The ectopic expression of GATA TFs in transduced cells was confirmed by either real-time PCR or Western blot analyses (Supplemental Figure 1, available at the Blood website [see the Supplemental Figures link at the top of the online article]; and data not shown). Transduced T cells were expanded for 8 to 10 days in IL-2–supplemented media, and their phenotypes were analyzed to ensure all transduced cells were phenotypically similar. As expected, activation of TN and TCM cells transduced with the control HDV resulted in their conversion to a TEM phenotype, whereas expanded TEM cells remained CD45RO+CCR7– (Supplemental Figure 2). Importantly, the expression of GATA-3 or GATA-1 in TN, TCM, TEM, or NKT cells did not alter the activation and differentiation of these T-cell subsets (Supplemental Figure 2).
We next determined the expression of CCR5 on transduced T cells. Expression of GATA-1 markedly suppressed CCR5 expression in memory T-cell subsets and NKT cells (Figure 2B). By contrast, cells ectopically expressing GATA-3 displayed only a modest reduction in CCR5 levels (Figure 2B). Furthermore, GATA-1–mediated repression in CCR5 levels was stable because reduced CCR5 expression levels were evident at 7 to 12 days after transduction (data not shown). These findings indicate that GATA-1 and GATA-3 differentially regulate CCR5 expression in primary cells and demonstrate that GATA-1, but not GATA-3, potently decreases CCR5 cell-surface expression in multiple human cell types that can serve as targets for infection by HIV-1.
GATA-1 expression in human T cells inhibits R5-tropic HIV-1 infection
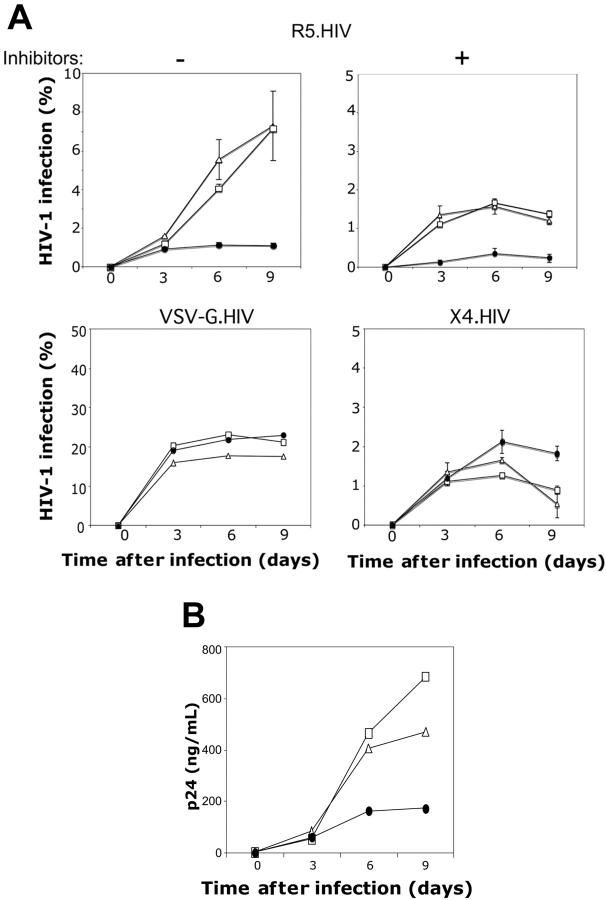
Our results indicated that GATA-1 is a potent repressor of CCR5 expression in DCs, conventional CD4+ T-cell subsets, and NKT cells. To determine the functional correlate of this finding, we asked whether GATA-1–mediated down-regulation of CCR5 expression renders human T cells less susceptible to HIV-1 infection. To test this, primary human T cells transduced with GATA-1, GATA-3, or the control HDV were purified based on mCD24 expression. These purified cells were then infected with replication-competent CCR5-tropic HIV-1 (R5.HIV), CXCR4-tropic HIV-1 (X4.HIV), or replication-defective VSV-G–pseudotyped HIV-1. HIV-1 infection and replication were assessed over the course of 9-day cultures as described.32 Human T cells expressing the control HDV and HDV.GATA-3 were readily susceptible to R5.HIV infection (Figure 3A). By contrast, T cells that ectopically expressed GATA-1 were significantly less susceptible to R5.HIV infection, and subsequent spread of viral replication in these cultures was markedly diminished (Figure 3A-B). Furthermore, the reduced infectivity of GATA-1–expressing human T cells by R5-tropic HIV-1 was still observed in the presence of HIV protease inhibitors and the reverse-transcription inhibitor AZT, which limited R5.HIV infection to a single-cycle (Figure 3A, upper right). By contrast, the susceptibility to infection with either X4.HIV or VSV-G.HIV was similar between T cells that were transduced with GATA-1, GATA-3, or the control HDV (Figure 3A, bottom). Collectively, these data suggest that GATA-1 expression specifically inhibits R5.HIV infection of T cells, and this effect is specifically due to GATA-1–mediated reduction of CCR5 expression levels.
Figure 3.
GATA-1–induced CCR5 down-regulation inhibits R5-tropic HIV infection. (A) T cells expressing HDV (▵), HDV.GATA-3 (□), or HDV.GATA-1 (•) were prepared as described in the caption of Figure 2 and superinfected with replication-competent CCR5-tropic HIV-1 (R5.HIV), at an MOI of 1 either in the absence (top left) or presence (top right) of HIV protease inhibitors and AZT, which were added at day 2 after infection. Infections were determined based on GFP expression as indicated. T cells expressing GATA-3 or GATA-1 were also infected with either VSV-G.HIV (bottom left) or X4.HIV (bottom right) at 1 MOI. All infection data are shown as the percentage of GFP+ T cells at the indicated times. (B) Culture supernatants were also collected from T cells infected with R5.HIV at the indicated times after infection, and viral replication was quantified via p24 ELISA. These data are representative of 3 independent experiments performed using T cells isolated from individual donors. Error bars indicate standard deviation of duplicate samples.
Expression of GATA-1 in primary human T cells suppresses CCR5 promoter activity
We next asked whether the inhibition of CCR5 expression by GATA-1 was due to suppression of CCR5 gene expression or posttranslational receptor internalization. To differentiate between these 2 mechanistic possibilities, we determined CCR5 mRNA levels by quantitative real-time PCR analysis in GATA TF-transduced T cells. GATA-1–expressing T cells displayed approximately 10-fold less CCR5 transcript compared with those expressing the control HDV (Figure 4B). By contrast, CCR5 mRNA levels were only 2-fold lower in GATA-3–expressing cells (Figure 4B). These findings are concordant with the GATA-1–directed reduction in CCR5 cell-surface expression levels (Figure 4A).
The findings in Figure 4A and B suggested that the effect of GATA-1 on CCR5 expression might lie at the level of influencing CCR5 gene expression rather than at a posttranslational level. We therefore determined whether the decreased CCR5 transcript levels in GATA-1–expressing T cells were due to diminished promoter activity. Transduced primary human T cells were first stimulated through the TCR for 18 to 24 hours and then transfected with a luciferase vector with or without a CCR5 promoter construct to drive its expression (Figure 4C, left). This promoter construct encompasses both CCR5 promoters 1 and 2 as previously described.39 We found that CCR5 promoter activity was significantly reduced in primary human T cells expressing GATA-1 compared with cells that were transduced with the control HDV vector (Figure 4C). Additionally, a slight but consistent decline in CCR5 promoter activity was also observed in cells expressing GATA-3 compared with cells expressing the control HDV, albeit more modest of a reduction when compared with that of GATA-1 (Figure 4C). The latter findings mirror the reductions of CCR5 message and cell-surface protein levels observed in GATA-1– and GATA-3–expressing T cells (Figure 4A-B). Collectively, these findings indicated that CCR5 promoter activity in primary human T cells is potently silenced in the presence of GATA-1, whereas it is only slightly decreased by GATA-3 expression. Additionally, our findings suggest that the reductions in CCR5 promoter activity are linked to reduced transcription and translation of CCR5.
GATA-1 expression in naive and memory T-cell subsets induces Th2 effector cytokine profiles
The aforementioned findings indicated that GATA-1 and GATA-3 have distinct regulatory effects on CCR5 expression. These findings were paradoxic because (1) these closely related GATA TFs share highly conserved zinc-finger domains and bind to very similar DNA sequences,16 and (2) previous studies suggested that a high degree of functional redundancy may exist between GATA-1 and GATA-3.28,29 Therefore, we sought to determine whether GATA-3 and GATA-1 would also exhibit differential effects with respect to their abilities to program Th2 cytokine gene expression in human naive and memory T-cell subsets.31
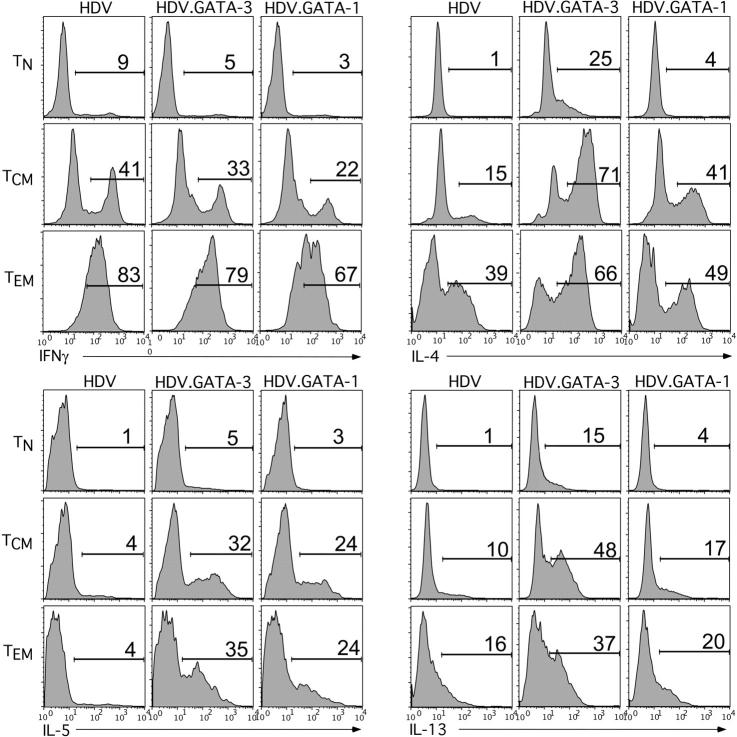
For these experiments, TN, TCM, and TEM cells were purified, TCR activated, and transduced with HDV.GATA-1, HDV.GATA-3, or the control HDV. These cells were expanded for 8 to 10 days and then probed for the intracellular production of Th1 and Th2 cytokines. Indeed, ectopic expression of either GATA-3 or GATA-1 in all T-cell subsets was associated with a marked up-regulation of IL-4 and IL-5, and to a lesser extent IL-13, compared with T cells that expressed only HDV (Figure 5). However, TCM and TEM cells ectopically expressing GATA-1 produced less IFNγ compared with either control- or GATA-3–expressing memory T cells (Figure 5). Given the similar expression levels of GATA-1 and GATA-3 observed in transduced cells (Supplemental Figure 1), these data suggest that the quantitative differences in cytokine production are not due to differential expression of the TFs. Collectively, these findings indicate that, akin to GATA-3, ectopic expression of GATA-1 in human T-cell subsets is sufficient to direct Th2 cell differentiation. However, GATA-1 is more effective in down-regulating IFNγ and slightly less potent than GATA-3 in inducing Th2 cytokines.
Figure 5.
Ectopic GATA-1 expression in human TN,TCM, and TEM cells induces a Th2 cytokine profile. CD45RO–RA+ naive and CD45RO+RA– memory T cells were purified from adult blood PBMCs of healthy donors. Memory T cells were further sorted by FACS for expression of CCR7 into TCM (CCR7+) and TCM (CCR7–) cells. The T-cell populations were TCR activated and transduced with HDV, HDV.GATA-1, or HDV.GATA-3 at the time of activation (see “Materials and methods”). Following 8 to 10 days of expansion, intracellular expression of the cytokines IFNγ, IL-4, IL-5, and IL-13 were determined by flow cytometry. For these experiments transduced cells were left unsorted, but mCD24+ cells were identified and gated on by costaining with an anti-mCD24 antibody. These data are representative of 3 separate experiments using T-cell subsets purified from different adult donors.
It is possible, however, that GATA-1–directed Th2 programming was an indirect effect via the induction of GATA-3 expression. Indeed, a previous report demonstrated that expression of heterologous GATA TFs in murine T cells activates GATA-3 expression, thereby promoting Th2 differentiation.40 However, in our studies we did not detect GATA-3 protein expression in human T cells ectopically expressing GATA-1 (Supplemental Figure 1). Quantitative real-time PCR analyses comparing GATA-3 expression levels in primary human T cells expressing GATA-1 with Th1 and Th2 cells corroborated these results. We found that high-level GATA-3 mRNA expression was highly restricted to Th2 cells, and GATA-1 expression in human T cells did not induce GATA-3 (Supplemental Figure 3). These findings strongly suggest that GATA-1, similar to GATA-3, can directly program Th2 cytokine gene expression in primary human T cells.
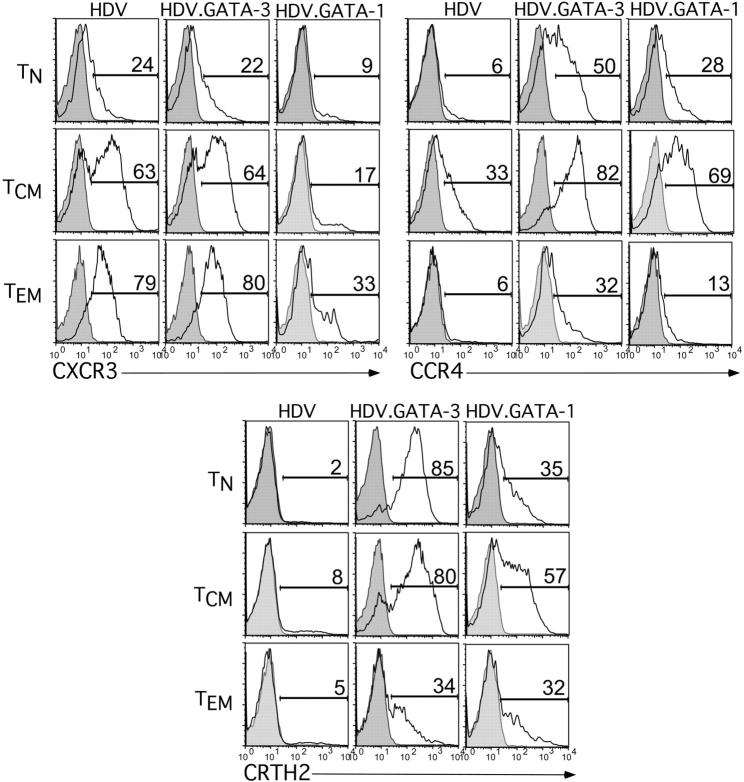
Expression of GATA-1 in primary human naive and memory T cells reprograms chemotactic receptor expression patterns to a Th2 profile
We found that expression of GATA-1 in human T cells could functionally replace GATA-3 to program Th2 cytokine gene expression. However, given the differential regulation of CCR5 by GATA-1 and GATA-3, we next determined whether GATA-1 expression in human T cells also influenced the cell-surface expression of Th1- or Th2-associated chemotactic receptors analogous to GATA-3.31 Similar to the effects of GATA-3 expression in all 3 T-cell subsets, ectopic expression of GATA-1 was associated with induction of the Th2 chemotactic receptors CCR4 and CRTH2 (Figure 6). However, expression of GATA-3 in memory T-cell subsets failed to suppress expression of the Th1-biased chemokine receptor CXCR3, whereas GATA-1 expression greatly reduced CXCR3 cell-surface expression in TCM and TEM cells (Figure 6). These results provided further evidence that GATA-1 and GATA-3 can induce both Th2 effector function and a Th2 pattern of chemotactic receptor expression. However, GATA-1 is significantly more potent at repressing the expression of Th1-associated effector molecules relative to GATA-3. Together, these findings suggest that GATA-1 and GATA-3 share incomplete functional redundancy in the context of T-cell differentiation.
Figure 6.
Expression of GATA-1 in human TN,TCM, and TEM cell subsets induces expression of Th2-associated chemokine receptors. Human TN, TCM, and TEM cells were purified, activated, and transduced with HDV alone, HDV.GATA-1, or HDV.GATA-3 as described in caption for Figure 5. Following expansion, unsorted cells were stained for chemotactic receptors as indicated or an isotype control in conjunction with an anti-mCD24 antibody to gate on transduced populations. The percentage of cells positive for the appropriate chemokine receptor is shown, with the gray, tinted peak representing isotype control staining. Results are representative of 3 experiments using T-cell subsets from distinct adult donors.
Discussion
Despite its critical importance in the pathogenesis of HIV-1, there is a significant knowledge gap in our understanding of the molecular determinants that control CCR5 expression in the physiologic cellular targets of HIV-1. By concurrently comparing the effects of GATA-3 and GATA-1 on CCR5 expression, we demonstrate that these 2 closely related TFs mediate markedly distinct regulatory effects on CCR5 expression, but they also display overlapping functions with respect to their effects on Th1 and Th2 gene expression during T-cell differentiation (summarized in Table 1).
Table 1.
Summary of GATA-1 and GATA-3 effects on gene expression in T cells
|
Induction of Th2 genes
|
Suppression of Th1 genes
|
|||||||
|---|---|---|---|---|---|---|---|---|
| IL-4 | IL-5 | IL-13 | CCR4 | CRTH2 | IFNγ | CXCR3 | CCR5 | |
| GATA-3 | ++++ | ++++ | +++ | +++ | +++++ | + | +/- | +/- |
| GATA-1 | ++ | +++ | ++ | ++ | ++++ | +++ | ++++ | ++++ |
Each + represents a 20% change in T-cell subsets expressing the indicated protein as monitored by flow cytometry compared with HDV-expressing T cells.
Given that GATA-3 is highly expressed on Th2 cells, and that these cells display lower levels of CCR5, we had hypothesized that ectopic expression of GATA-3 would suppress CCR5 expression levels. However, this was not the case, and, by contrast, GATA-1 potently repressed CCR5 expression at both the protein and transcript levels in primary human T-cell subsets and DCs. These effects are, in part, mediated by the inhibition of CCR5 transcription, and they support the notion that GATA TF binding to their cognate cis-elements within the CCR5 promoter is functional13,14 (Figure 4C). Interestingly, GATA-1 was shown to trans-activate CCR5 promoter activity in a transformed T-lymphoid cell line.15 However, our results clearly demonstrate that GATA-1 is a potent repressor of CCR5 expression in primary human T cells and DCs. This discrepancy in CCR5 regulation by GATA-1 in primary versus transformed cell lines may reflect unique combinations or levels of TFs present within constitutively proliferating cells that do not faithfully mimic those in physiologic primary cells. Substantiating this possibility, we recently found that the CCR5 promoter 2 is highly active in primary cells compared with cell lines that serve as surrogates for T cells (eg, Jurkat)39 (S.M., Lisa M. Adams, S.E.V., Adam S. Bellinger, Mrunal Kalkonde, Jose Camargo, Gregory Bonello, Hiromi Tagoh, Seema S. Ahuja, D.U., and S.K.A., manuscript in preparation). Moreover, the differential effects of GATA-1 in T-cell lines and primary T cells may also be attributed to other differences in these cell types, such as cell-type–specific differences in the epigenetic architecture of the CCR5 locus.
The precise mechanism by which GATA-1 represses CCR5 gene transcription is not yet clear, and 2 possible mechanisms can be envisaged. First, GATA-1 might influence CCR5 transcription independently or, as reported in other gene systems, in concert with other TFs that bind to cis-elements within the CCR5 promoter.13,14,41,42 Second, GATA-1 may epigenetically remodel the CCR5 promoter. This possibility is based on the finding that GATA-1–responsive loci are epigenetically modified during hematopoiesis.18 If, in fact, GATA-1 restructures chromatin within the CCR5 locus, this could cause a transcriptionally silent imprint in HSCs during hematopoiesis that may be maintained in developing T cells. Indeed, TN cells that develop in the thymus and are maintained in the periphery do not express CCR5 and require extracellular activation signals to induce its expression.1 Whether GATA-1 can regulate an epigenetically silent imprint at the CCR5 promoter in hematopoietic cell lineages is an area that we are currently investigating. The suppression of CCR5 in human T cells by GATA-1 further reinforces growing evidence that GATA-1 can act as a selective and potent transcriptional repressor, in addition to its more classic role as a transcriptional activator.43,44
It is likely that the functional correlate of reduced CCR5 expression mediated by GATA-1, namely the inhibition of R5-tropic HIV-1 infection and viral replication in T cells, will be evident in other HIV-1 target cells. However, the expression of GATA-1 is highly restricted during hematopoiesis and is not physiologically expressed in peripheral T cells, NKT cells, or DCs (data not shown). However, we hypothesize that GATA-1 expression may be transiently induced in these cell types by specific activation signals, which could then influence the expression of CCR5. In another scenario, GATA-1, which is clearly expressed in HSCs and is silenced on their differentiation to CCR5-expressing DCs (Figure 1), may play a physiologic role in modulating HIV-1 susceptibility of these cell types during hematopoietic development, as well as mast-cell progenitors, which are all potential targets of viral infection in vivo.45-47 A better understanding of the mechanism whereby GATA-1 represses CCR5 expression may help unravel the genetic regulation of CCR5 in HIV-1 target cells and may be useful to devise novel approaches to antagonize CCR5 in individuals with HIV-1 infection.
In addition to the CCR5 inhibitory activity, ectopic expression of GATA-1 was more potent at down-regulating IFNγ and CXCR3 expression compared with GATA-3, even in lineage-committed TEM cells. In a broader context, these findings highlight a remarkable ability to reprogram the expression of Th1 cytokines and chemokine receptors in human TEM cells, which are relevant in the context of T-cell–mediated pathologies such as in autoimmune diseases and allograft rejection.11 Mechanistically, these findings suggest that the divergent N- and C-terminal portions of GATA-1 and GATA-3 have specific effects on the expression of Th1-associated cytokines and chemokine receptors.
We speculate that 3 potential mechanisms may explain the differential activity of GATA-1 and GATA-3. First, the nonconserved trans-activation domains of GATA-1 may serve as additional protein interaction interfaces,17 which could allow it to selectively recruit repressive cofactors to the IFNγ and CXCR3 promoters. Second, the TF called friend of GATA (FOG-1), which can interact with both GATA-1 and GATA-3, might differentially regulate their function. Whereas FOG-1 interaction with GATA-1 is required for its activity during erythropoiesis and megakaryopoiesis,48 it has been shown to inhibit the function of GATA-3 during T-cell differentiation.49 Therefore, the interaction of these GATA TFs with FOG-1 may impart differential regulation of Th1-associated gene expression. Finally, it is also plausible that GATA-1 and GATA-3 may be differentially phosphorylated, acetylated, or methylated within these trans-activation domains, resulting in unique regulation of Th1 gene expression.50-52 Future structure/function studies using chimeric GATA TFs may be useful to uncover specific GATA-regulatory domains and auxiliary transcription factors that determine the expression of Th1 chemokine receptors and cytokines.
Although ectopic expression of GATA-1 is much more potent than GATA-3 in repressing the expression of CCR5 and other Th1 effector molecules, it is highly similar to GATA-3 in inducing Th2 cytokines (IL-4, IL-5, IL-13) and chemotactic receptors (CCR4 and CRTH2) (Table 1). In contrast to a previous study performed in murine T cells,40 we found that the regulation of Th2 cytokines and chemokine receptors was a direct function of GATA-1, occurring without trans-activation of GATA-3 expression. Thus, it seems likely that GATA-3 expression is differentially regulated in mouse and human T cells. Our findings strongly suggest that the conserved DNA binding zinc-finger domain shared between these GATA TFs is largely sufficient to program Th2 effector functions and lymphoid homing propensities.
In summary, we have found that GATA-1 is a potent repressor of CCR5 expression in multiple human cell types that are physiologic targets of HIV-1 in vivo. Decoding the mechanisms underlying the functional similarities and differences between GATA-1 and GATA-3 in programming human Th2 differentiation and repressing CCR5 expression has implications in understanding the molecular regulation of CCR5, as well as other loci that contain GATA-responsive cis-sites. Importantly, these findings raise the unique possibility of harnessing the mechanisms by which GATA-1 mediates the repression of CCR5 as a therapeutic modality to render human T cells refractory to HIV-1 infection.
Supplementary Material
Acknowledgments
We thank Kyra Oswald-Richter for assistance with p24 ELISAs and critical reading. We thank Dr Victor Torres, Dr Eugene Oltz, Dr Wasif Khan, Dr Luc Van Kaer, Dr Vineet KewalRamani, and Dr Chris Arendt for critical reading and Erika Sundrud for help with statistical analyses.
Prepublished online as Blood First Edition Paper, August 9, 2005; DOI 10.1182/blood-2005-03-0857.
Supported by grants from the National Institutes of Health (R01-AI043279) (S.K.A.) and (R01-AI054206-02) (D.U.), and by the Merit Review Entry Program of the Department of Veterans Affairs (S.M.).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Oppermann M. Chemokine receptor CCR5: insights into structure, function, and regulation. Cell Signal. 2004;16: 1201-1210. [DOI] [PubMed] [Google Scholar]
- 2.Moore JP. Coreceptors: implications for HIV pathogenesis and therapy. Science. 1997;276: 51-52. [DOI] [PubMed] [Google Scholar]
- 3.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382: 722-725. [DOI] [PubMed] [Google Scholar]
- 4.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86: 367-377. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273: 1856-1862. [DOI] [PubMed] [Google Scholar]
- 6.Mummidi S, Ahuja SS, Gonzalez E, et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4: 786-793. [DOI] [PubMed] [Google Scholar]
- 7.Wu L, Paxton WA, Kassam N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185: 1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonecchi R, Bianchi G, Bordignon PP, et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187: 129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima C, Mukai T, Yamaguchi N, et al. Induction of the chemokine receptor CXCR3 on TCR-stimulated T cells: dependence on the release from persistent TCR-triggering and requirement for IFN-gamma stimulation. Eur J Immunol. 2002; 32: 1792-1801. [DOI] [PubMed] [Google Scholar]
- 10.Nagata K, Tanaka K, Ogawa K, et al. Selective expression of a novel surface molecule by human Th2 cells in vivo. J Immunol. 1999;162: 1278-1286. [PubMed] [Google Scholar]
- 11.Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu Rev Immunol. 2000;18: 593-620. [DOI] [PubMed] [Google Scholar]
- 12.Clerici M. Cell-mediated immunity in HIV infection. AIDS. 1993;7(suppl 1): S135-S140. [PubMed] [Google Scholar]
- 13.Mummidi S, Ahuja SS, McDaniel BL, Ahuja SK. The human CC chemokine receptor 5 (CCR5) gene: multiple transcripts with 5′-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J Biol Chem. 1997;272: 30662-30671. [DOI] [PubMed] [Google Scholar]
- 14.Moriuchi H, Moriuchi M, Fauci AS. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159: 5441-5449. [PubMed] [Google Scholar]
- 15.Moriuchi M, Moriuchi H, Fauci AS. GATA-1 transcription factor transactivates the promoter for CCR5, a coreceptor for human immunodeficiency virus type 1 entry. Blood. 1999;93: 1433-1435. [PubMed] [Google Scholar]
- 16.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13: 3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21: 3368-3376. [DOI] [PubMed] [Google Scholar]
- 18.Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87: 4025-4039. [PubMed] [Google Scholar]
- 19.Rothenberg EV. T-lineage specification and commitment: a gene regulation perspective. Semin Immunol. 2002;14: 431-440. [DOI] [PubMed] [Google Scholar]
- 20.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2: 933-944. [DOI] [PubMed] [Google Scholar]
- 21.Zhou M, Ouyang W. The function role of GATA-3 in Th1 and Th2 differentiation. Immunol Res. 2003;28: 25-37. [DOI] [PubMed] [Google Scholar]
- 22.Orkin SH, Shivdasani RA, Fujiwara Y, McDevitt MA. Transcription factor GATA-1 in megakaryocyte development. Stem Cells. 1998;16(suppl 2): 79-83. [DOI] [PubMed] [Google Scholar]
- 23.Simon MC. Transcription factor GATA-1 and erythroid development. Proc Soc Exp Biol Med. 1993;202: 115-121. [DOI] [PubMed] [Google Scholar]
- 24.Pevny L, Simon MC, Robertson E, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349: 257-260. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda M, Sakamoto N, Fukumaki Y. Deltathalassemia caused by disruption of the site for an erythroid-specific transcription factor, GATA-1, in the delta-globin gene promoter. Blood. 1992; 80: 1347-1351. [PubMed] [Google Scholar]
- 26.Patmasiriwat P, Fraizer GC, Claxton D, Kantarjian H, Saunders GF. Expression pattern of WT1 and GATA-1 in AML with chromosome 16q22 abnormalities. Leukemia. 1996;10: 1127-1133. [PubMed] [Google Scholar]
- 27.Mehaffey MG, Newton AL, Gandhi MJ, Crossley M, Drachman JG. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood. 2001;98: 2681-2688. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi S, Shimizu R, Suwabe N, et al. GATA factor transgenes under GATA-1 locus control rescue germline GATA-1 mutant deficiencies. Blood. 2000;96: 910-916. [PubMed] [Google Scholar]
- 29.Tsai FY, Browne CP, Orkin SH. Knock-in mutation of transcription factor GATA-3 into the GATA-1 locus: partial rescue of GATA-1 loss of function in erythroid cells. Dev Biol. 1998;196: 218-227. [DOI] [PubMed] [Google Scholar]
- 30.Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J Exp Med. 1999;189: 1735-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sundrud MS, Grill SM, Ni D, et al. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J Immunol. 2003;171: 3542-3549. [DOI] [PubMed] [Google Scholar]
- 32.Motsinger A, Haas DW, Stanic AK, Van Kaer L, Joyce S, Unutmaz D. CD1d-restricted human natural killer T cells are highly susceptible to human immunodeficiency virus 1 infection. J Exp Med. 2002;195: 869-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74: 10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald-Richter K, Grill SM, Shariat N, et al. HIV infection of naturally occurring and genetically reprogrammed human regulatory T-cells. PLoS Biol. 2004;2: E198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caux C, Ait-Yahia S, Chemin K, et al. Dendritic cell biology and regulation of dendritic cell trafficking by chemokines. Springer Semin Immunopathol. 2000;22: 345-369. [DOI] [PubMed] [Google Scholar]
- 36.Pope M. Mucosal dendritic cells and immunodeficiency viruses. J Infect Dis. 1999;179(suppl 3): S427-S430. [DOI] [PubMed] [Google Scholar]
- 37.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401: 708-712. [DOI] [PubMed] [Google Scholar]
- 38.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human V(alpha)24 natural killer T cells. J Exp Med. 2002;195: 637-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mummidi S, Bamshad M, Ahuja SS, et al. Evolution of human and non-human primate CC chemokine receptor 5 gene and mRNA: potential roles for haplotype and mRNA diversity, differential haplotype-specific transcriptional activity, and altered transcription factor binding to polymorphic nucleotides in the pathogenesis of HIV-1 and simian immunodeficiency virus. J Biol Chem. 2000; 275: 18946-18961. [DOI] [PubMed] [Google Scholar]
- 40.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in Th2 development. Mol Cell Biol. 2001;21: 2716-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prieschl EE, Gouilleux-Gruart V, Walker C, Harrer NE, Baumruker T. A nuclear factor of activated T cell-like transcription factor in mast cells is involved in IL-5 gene regulation after IgE plus antigen stimulation. J Immunol. 1995;154: 6112-6119. [PubMed] [Google Scholar]
- 42.Masuda A, Yoshikai Y, Kume H, Matsuguchi T. The interaction between GATA proteins and activator protein-1 promotes the transcription of IL-13 in mast cells. J Immunol. 2004;173: 5564-5573. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez P, Bonte E, Krijgsveld J, et al. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005;24: 2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104: 3136-3147. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz ME, Cicala C, Arthos J, et al. Peripheral blood-derived CD34+ progenitor cells: CXC chemokine receptor 4 and CC chemokine receptor 5 expression and infection by HIV. J Immunol. 1998;161: 4169-4176. [PubMed] [Google Scholar]
- 46.Freedman AR, Gibson FM, Fleming SC, Spry CJ, Griffin GE. Human immunodeficiency virus infection of eosinophils in human bone marrow cultures. J Exp Med. 1991;174: 1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bannert N, Farzan M, Friend DS, et al. Human mast cell progenitors can be infected by macrophagetropic human immunodeficiency virus type 1 and retain virus with maturation in vitro. J Virol. 2001;75: 10808-10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsang AP, Visvader JE, Turner CA, et al. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90: 109-119. [DOI] [PubMed] [Google Scholar]
- 49.Zhou M, Ouyang W, Gong Q, et al. Friend of GATA-1 represses GATA-3-dependent activity in CD4+ T cells. J Exp Med. 2001;194: 1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crossley M, Orkin SH. Phosphorylation of the erythroid transcription factor GATA-1. J Biol Chem. 1994;269: 16589-16596. [PubMed] [Google Scholar]
- 51.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396: 594-598. [DOI] [PubMed] [Google Scholar]
- 52.Chen CH, Zhang DH, LaPorte JM, Ray A. Cyclic AMP activates p38 mitogen-activated protein kinase in Th2 cells: phosphorylation of GATA-3 and stimulation of Th2 cytokine gene expression. J Immunol. 2000;165: 5597-5605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.