Abstract
The clinical efficacy of evaluating genetic anomalies in metaphase cells versus interphase nuclei for multiple myeloma (MM) is poorly understood. Therefore, survival for 154 patients with newly diagnosed untreated MM was compared with results from analysis of metaphase and interphase cells. Metaphases were studied by conventional cytogenetics and fluorescent-labeled DNA probes (fluorescence in situ hybridization [FISH]), whereas inter-phase nuclei were evaluated only by FISH. All FISH studies were done using DNA probes to detect t(4;14)(p16;q32), t(11;14)(q13;q32), t(14;16)(q32;q23), del(17) (p13.1), and chromosome 13 anomalies. Metaphases were abnormal by cytogenetics and/or metaphase FISH in 61 (40%) patients. Abnormal interphase nuclei were observed in 133 (86%) patients, including each patient with abnormal metaphases. FISH was a necessary adjunct to cytogenetics to detect t(4;14) and t(14;16) in metaphase cells. Patient survival was especially poor for patients with greater than 50% abnormal interphase nuclei, although this result was more likely due to level of plasma cells than specific chromosome anomalies. For metaphase data, patients with t(4;14), t(14;16), del(17) (p13.1), and/or chromosome 13 anomalies (primarily monosomy 13) had poor survival. A different outcome was observed for interphase data as patients with t(4;14) or t(14;16) had poor survival, whereas patients with chromosome 13 anomalies had intermediate survival: interphase FISH did not substitute for metaphase analysis.
Introduction
Metaphase cells can be examined by conventional cytogenetic methods to detect a wide variety of chromosome anomalies in proliferating cells from patients with multiple myeloma (MM).1,2 Metaphase cells can also be evaluated by fluorescence in situ hybridization using DNA probes (metaphase FISH) to detect chromosome anomalies associated with MM such as t(11;14)(q13; q32), t(4;14)(p16;q32), t(14;16)(q32;q23), del(17)(p13.1), and chromosome 13 anomalies. In addition, FISH can be used to study interphase nuclei (interphase FISH) to detect these same anomalies in unselected nonproliferating nuclei to establish the overall proportion of normal and abnormal nuclei.3-5 Alternatively, FISH can be used with immunophenotyping techniques to detect chromosome anomalies in plasma cells (plasma cell FISH).6
The clinical efficacy of studying metaphase cells versus interphase nuclei in patients with MM is not known. Therefore, we studied 154 patients with newly diagnosed untreated MM and compared survival with results from conventional cytogenetics, metaphase FISH, and interphase FISH. We attempted to answer 5 questions concerning genetic testing for MM. Is metaphase FISH a useful adjunct to conventional cytogenetics? Does examination of metaphase cells provide prognostic information that is not apparent from analysis of interphase nuclei? Do chromosome 13 anomalies have a different significance when observed in metaphase cells than in interphase nuclei? Is interphase FISH an inexpensive substitute for conventional cytogenetics and/or plasma cell FISH? Does a hierarchy of chromosome anomalies detected in metaphase and/or interphase cells provide useful prognostic information?
Patients, materials, and methods
Patients
This study was performed with the approval of the Mayo Foundation Institutional Review Board and informed consent was provided according to the Declaration of Helsinki. We studied 154 patients with newly diagnosed untreated MM, including 90 men and 64 women; median age was 63 years (range, 33-92 years). Overall, 101 patients have died and 53 are alive: median follow-up was 26.2 months (range, 2 days to 107.6 months). Each patient was referred for conventional cytogenetic studies at the Mayo Clinic Rochester at diagnosis of MM between March 1989 and October 2002. Bone marrow specimens from each patient were collected within 30 days of diagnosis.
Conventional cytogenetic studies
For each patient, bone marrow was processed by standard cytogenetic methods using short-term and long-term cultures; up to 20 Giemsa trypsin Leishman (GTI)– and/or quinacrine fluorescence (QFQ)–banded metaphases were analyzed.7
FISH studies
Left-over bone marrow from conventional cytogenetic studies was stored in 3:1 methanol:glacial acetic acid fixative at -70°C until FISH studies were performed, all in the first 6 months of 2004. Interphase and metaphase FISH were done using probes for immunoglobin heavy chain (IGH) (14q32), fibroblast growth factor receptor-3 (FGFR3) (4p16), cyclin D1 (CCND1) (11q13), macrophage activating factor (MAF) (16q23), p53 (17p13.1), D17Z1 (17cen), D13S319 (13q14), and lysosome-associated membrane protein-1 (LAMP1) (13q34) to detect t(4;14)(p16;q32), t(11;14)(q13;q32), t(14;16)(q32;q23), del(17)(p13.1), and chromosome 13 anomalies, respectively. Normal cutoff values for each probe set were determined for 200 unselected bone marrow interphase nuclei from a series of healthy individuals in a separate study (S.F., S.S., and S.P., unpublished results, June 2003). The normal cutoff was defined as the upper boundary of a one-sided 95% confidence interval to detect the maximum number of false-positive cells in healthy individuals.8 The normal cutoff for translocations was 1.5%, and ranged from 1.5% to 12% for detection of numeric anomalies. For each patient, up to 10 metaphases were analyzed for each probe set. A metaphase FISH study was successful if at least 9 normal metaphases or an abnormal clone was detected. Results were abnormal when at least 2 metaphases had an abnormal signal pattern.
Probes for FGFR3 and MAF were made and validated at Mayo Clinic. Remaining probes were purchased from Vysis, Downers Grove, IL, and validated at Mayo Clinic. Validation was performed using standard procedures for FISH.9 Specimens were analyzed randomly in a blinded fashion by 4 of the authors (Y.K.L., S.F., S.S., and S.P.). For each patient, interphase FISH was done on 200 consecutive nuclei for each probe set: results were abnormal when the percentage of cells with any given chromosome anomaly exceeded the normal cutoff.
Statistical studies
Survival for patient groupings based on metaphase and interphase results was compared by using the Kaplan-Meier method, and differences in survival curves were compared by using log-rank tests. To estimate relative risk of death as a function of a continuous variable (percentage of abnormal nuclei), smoothing splines were used within a Cox model.10 Metaphase and interphase analyses along with other potential risk factors were evaluated by multivariable analysis using Cox proportional hazards models. Variables included in the final model were identified by using stepwise selection and backward elimination. Recursive partitioning was also used to identify prominent predictors of survival. All variable selection approaches yielded similar results.
Results
Among the 154 patients studied, 133 (86%) had an abnormal clone detected in metaphase and/or interphase cells (Table 1).
Table 1.
Results for metaphase and interphase cells in 154 patients at diagnosis of myeloma
|
Metaphase
|
||
|---|---|---|
| Interphase | Normal, n (%) | Abnormal, n (%) |
| Normal | 21 (14) | 0 (0) |
| Abnormal | 72 (47) | 61 (40) |
Metaphase results are based on the combined outcome of conventional cytogenetic and metaphase FISH studies. Interphase results are based on FISH studies only.
Analysis of metaphase cells
Results of cytogenetics and/or metaphase FISH were abnormal for 61 (40%) patients and normal for 93 (60%) (Table 2). Cytogenetics was abnormal for 43 (28%) patients, although loss of Y in 6 patients and constitutional der(13;14)(q10;q10) in 1 patient were not considered clonal abnormalities. Results of metaphase FISH were abnormal for 46 (30%) patients, including 18 (12%) that were normal by cytogenetics. Metaphase FISH was normal for 45 patients, normal but incomplete (<9 normal metaphases for ≥ 1 FISH probes) for 44 and unsuccessful for 19.
Table 2.
Results for metaphases analyzed by conventional cytogenetics and by metaphase FISH for 154 patients
| Parameter | Patients, n (%) |
|---|---|
| Overall Results | |
| Abnormal cytogenetics and/or metaphase FISH | 61 (40) |
| Abnormal cytogenetics | 43 (28) |
| Abnormal metaphase FISH | 46 (30) |
| Cytogenetics versus metaphase FISH | |
| Normal cytogenetics and normal metaphase FISH | 41 (27) |
| Abnormal cytogenetics and normal metaphase FISH | 4 (3) |
| Normal cytogenetics and abnormal metaphase FISH | 18 (12) |
| Abnormal cytogenetics and abnormal metaphase FISH | 28 (18) |
| Normal cytogenetics and incomplete metaphase FISH* | 52 (34) |
| Abnormal cytogenetics and incomplete metaphase FISH* | 11 (7) |
| Detection of common chromosome anomalies in myeloma | |
| t(11;14) detected by cytogenetics and/or metaphase FISH for IGH/CCND1 | 12 (7.8) |
| t(4;14) detected by cytogenetics and/or metaphase FISH for IGH/FGFR3 | 10 (6.5) |
| t(14;16) detected by cytogenetics and/or metaphase FISH for IGH/MAF | 3 (1.9) |
| Metaphase FISH detected loss of D13S319 and LAMP1, or loss of D13S319 | 23 (14.9) |
| Metaphase FISH detected fewer than CCND1 signals | 20 (13.0) |
| Metaphase FISH detect absence of p53 | 9 (5.8) |
Incomplete indicates fewer than 9 metaphases were found to analyze with 1 or more FISH probes
A t(11;14)(q13;q32) and/or fusion of IGH and CCND1 FISH signals was detected by cytogenetics and/or metaphase FISH in 12 (7.8%) patients. Cytogenetics detected t(11;14) in 3 patients; metaphase FISH detected fusion of IGH and CCND1 signals in 2 of these patients, whereas no metaphases were found for analysis with this FISH probe set in the other patient. Metaphase FISH for fusion of IGH and CCND1 signals was abnormal for another 9 patients; 6 of these patients were normal by cytogenetics and 3 had complex karyotypes without apparent t(11;14).
A t(4;14)(p16;q32) is characterized by fusion of IGH and FGFR3 FISH signals. Metaphase FISH detected fusion of IGH and FGFR3 signals in 10 (6.5%) patients: by cytogenetics, 2 of these patients were normal and 8 had complex karyotypes without apparent t(4;14).
A t(14;16)(q32;q23) is characterized by fusion of IGH and MAF FISH signals. Three (1.9%) patients had fusion of IGH and MAF signals by metaphase FISH: each of these patients was abnormal by cytogenetics, but t(14;16) was not apparent.
Metaphase FISH detected loss of D13S319 and/or LAMP1 from chromosome 13 in 23 (14.9%) patients: by cytogenetics, 5 were normal and 18 had complex karyotypes. For 22 of these patients, D13S319 and LAMP1 were both absent, suggesting monosomy 13: each of these specimens lacked a chromosome 13 by cytogenetics. The remaining patient had a single metaphase by cytogenetics with a deletion in chromosome 13 q-arm; this corresponded with loss of D13S319 but not LAMP1 by metaphase FISH. Seven patients were abnormal for chromosome 13 by cytogenetics that were normal by metaphase FISH in 5 and incomplete in 2.
Multiple CCND1 signals were observed by metaphase FISH in 20 (13.0%) patients. By cytogenetics, 8 of these patients were normal and 12 were abnormal, including 6 with trisomy 11, 2 with tetrasomy 11, 2 with unbalanced chromosome 11 translocations, and 2 without apparent chromosome 11 anomalies.
Absence of p53 at 17p13.1 was detected by metaphase FISH in 9 (5.8%) patients. Eight patients lacked p53 but retained the chromosome 17 centromere; this observation is consistent with 17p deletions. In the remaining patient, FISH showed loss of p53 and chromosome 17 centromere; this observation is consistent with monosomy 17. Cytogenetics showed a complex karyotype in each of these 9 patients, but an abnormal chromosome 17 was detected in only 5 patients.
Metaphase FISH detected abnormal metaphases that were not observed by cytogenetics in 18 patients. Cytogenetics detected abnormal metaphases in 15 patients that were normal or incomplete by metaphase FISH. Therefore, in the remainder of this article we combined results of cytogenetics and metaphase FISH. Thus, a patient that was abnormal by either cytogenetics or metaphase FISH is regarded as a patient with abnormal metaphases.
Analysis of interphase cells
Interphase FISH results were abnormal for 133 (86%) of the 154 patients. The median percentage of abnormal nuclei was 17.5% (mean and standard deviation was 26.9% ± 21.6%; range, 2.5%-89.0%). Each of the 61 patients with abnormal metaphases was also abnormal by interphase FISH. Thus, 72 (54%) patients had abnormal interphase FISH but normal metaphases. Patients with various chromosome anomalies are summarized in Table 3.
Table 3.
Summary of interphase FISH results from 154 patients
| Parameter | Patients, n (%) |
|---|---|
| FISH anomalies, no. | |
| 1 | 55 (41) |
| 2 | 46 (35) |
| 3 | 25 (19) |
| 4 | 7 (5) |
| IGH/CCND1 fusion | 26 (20) |
| IGH/FGFR3 fusion | 14 (11) |
| IGH/MAF fusion | 6 (5) |
| Loss of D13S319 with or without loss of LAMP1 | 72 (54) |
| Loss of p53 with or without loss of CEN17 | 19 (14) |
| More than 2 CCND1 | 61 (46) |
The results are from using 5 sets of FISH probes.
Percentage of plasma cells was defined as the higher percentage of plasma cells in a bone marrow aspirate, a bone marrow biopsy, or bone marrow differential. Median percentage of bone marrow plasma cells was 57.0% (mean and standard deviation was 54.7% ± 24.6%; range, 3.0%-99.0%). The percentage of plasma cells was greater than the percentage of abnormal nuclei by interphase FISH for all but 3 patients but was distributed evenly between percentage of abnormal nuclei and 100%. Interphase FISH results were normal for 21 patients: 4 had less than 7.5% plasma cells and 17 had 8% to 80% plasma cells. Interphase FISH results were abnormal for 132 (89%) of 148 patients with at least 10% plasma cells, and 127 (91%) of patients with at least 20% plasma cells.
Chromosome anomalies and survival
Survival results were analyzed 4 ways: (1) interphase FISH, (2) metaphase analysis, (3) metaphase versus interphase emphasizing chromosome 13 anomalies, and (4) hierarchies of chromosome anomalies for metaphase or interphase results (Table 4; Figure 1).
Table 4.
Statistical results for metaphase and interphase analyses
| Study (figure no.), cytogenic result | Cell stage | Patients, n | Deaths, n | Median survival, mo | 95% CI, mo |
|---|---|---|---|---|---|
| Percentage of abnormal nuclei versus survival (1B) | |||||
| Greater than 50% abnormal nuclei | Interphase | 20 | 18 | 6.7 | 2.4-18.9 |
| Less than 50% abnormal nuclei | Interphase | 134 | 83 | 45.0 | 36.7-53.1 |
| Normal metaphases versus abnormal metaphases (1C) | |||||
| Abnormal | Metaphase | 61 | 48 | 19.6 | 14.2-37.8 |
| Normal | Metaphase | 93 | 50 | 46.8 | 42.1-64.2 |
| 13q anomalies in interphase or metaphase (1D) | |||||
| 13q- | Metaphase | 30 | 29 | 12.7 | 5.9-23.9 |
| All tests normal | Interphase and metaphase | 21 | 10 | 45.0 | 37.6-NA |
| 13q- | Interphase | 38 | 26 | 46.8 | 24.1-64.2 |
| Abnormal without 13q- | Metaphase | 31 | 20 | 53.1 | 17.2-NA |
| Normal metaphase and abnormal interphase no 13q- | Interphase and metaphase | 34 | 16 | 55.3 | 36.7-NA |
| Hierarchy of chromosome anomalies and survival*(1E) | |||||
| t(4;14), t(14;16), 17p-, or 13q- | Metaphase | 33 | 30 | 13.9 | 5.9-23.9 |
| t(11;14) without t(4;14), t(14;16), 17p-, or 13q- | Metaphase | 6 | 5 | 31.0 | 17.0-NA |
| Normal | Metaphase | 93 | 52 | 46.7 | 42.1-64.2 |
| Remaining patients | Metaphase | 22 | 14 | 41.6 | 17.2-NA |
| Hierarchy of chromosome anomalies and survival*(1F) | |||||
| t(4;14), t(14;16) | Interphase | 20 | 18 | 13.3 | 6.3-37.8 |
| 13q- or 17p- without t(4;14), t(14;16) | Interphase | 59 | 44 | 33.9 | 19.3-55.5 |
| Normal | Interphase | 21 | 10 | 45.0 | 37.6-NA |
| More than 2 CCND1 and other | Interphase | 39 | 21 | 53.1 | 30.1-NA |
| t(11;14) without t(4;14), t(14;16), 17p-, or 13q- | Interphase | 15 | 8 | 55.3 | 31.0-NA |
CI indicates confidence interval; NA, not achieved.
Hierarchic grouping of patients by their chromosomal anomalies was based on hazard ratios from univariable and multivariable survival models
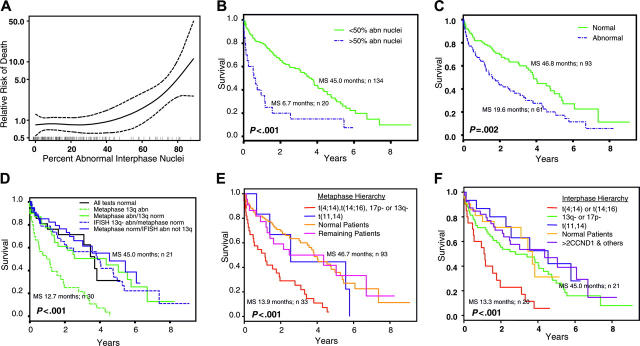
Figure 1.
Prognostic significance for interphase and metaphase results. (A) Estimated relative risk of death (—), along with 95% confidence intervals (-/-/-), for patients with various percentages of abnormal interphase nuclei. Estimated risk is relative to the total set of patients in the study; hence the average risk is 1. Vertical marks on the x-axis correspond to observed data. (B) Patients with greater than 50% abnormal interphase nuclei have poor survival. (C) Abnormal metaphase cells are associated with poor survival. (D) Anomalies of chromosome 13q in metaphase cells is a poor prognosis, but not when seen in interphase cells (IFISH). (E) A hierarchy of chromosome anomalies in metaphase cells is useful to stratify patients into prognostic groups. (F) A hierarchy of chromosome anomalies in interphase cells is useful to stratify patients into prognostic groups. Abn indicates abnormal; norm, normal; MS, median survival.
Relative risk of patient death is strongly correlated with the percentage of abnormal interphase nuclei regardless of FISH anomaly (Figure 1A). The point on the curve that exceeded a hazard ratio of 1.0 (average risk for these patients as a whole) corresponded to approximately 50% abnormal interphase nuclei. Median survival for 20 patients with greater than 50% abnormal interphase nuclei was 6.7 months compared with 45 months for 134 patients with less than 50% abnormal interphase nuclei (P < .001) (Figure 1B). No specific chromosome anomalies were associated with these patients: 18 had abnormal metaphases, whereas 2 had normal metaphases; 12 had t(4;14), t(14;16), or 17p deletions, whereas 4 had t(11;14), and 15 had chromosome 13 anomalies.
Figure 1C shows patient survival based on the presence of abnormal metaphases regardless of anomaly observed. Median survival for 61 patients with abnormal metaphases was 19.6 months compared with 46.8 months for 93 patients with normal metaphases (P = .002).
Patients were classified into 5 groups according to their metaphase and interphase results to study survival associated with chromosome 13 anomalies (Figure 1D). Thirty patients who had metaphases with chromosome 13 anomalies had the poorest survival (median, 12.7 months) (P < .001). In contrast, 38 patients with normal metaphases, but who had interphase nuclei with chromosome 13 anomalies, fared better (median survival, 46.8 months). This latter result was similar to patients with normal metaphases and normal interphase nuclei, for patients who had abnormal metaphases without chromosome 13 anomalies, and for patients with normal metaphases and abnormal interphase nuclei without chromosome 13 anomalies (Table 4).
The best hierarchic grouping of patients by their chromosomal anomalies was determined from hazard ratios for univariable and multivariable survival models. For interphase data, both modeling methods suggest that (1) patients with t(4;14) or t(14;16) had poor survival (hazard ratio, 2.2-2.8), (2) patients with del(17)(p13.1) and/or chromosome 13 anomalies had intermediate survival (hazard ratio, 1.6-1.9), and (3) patients with t(11;14) had favorable survival (hazard ratio, 0.8-1.0) (Table 4). For metaphase data, a slightly different hazard ratio pattern was observed because patients with t(4;14), t(14;16), del(17)(p13.1), and/or chromosome 13 anomalies had poor survival, whereas patients with t(11;14) had intermediate survival. Multivariable results for metaphase suggest that patients with chromosome 13 anomalies (hazard ratio, 2.8) had the strongest association with worsening survival, whereas all other anomalies (hazard ratio, 1.1-1.7) had intermediate survival. A strong association between chromosome 13 anomalies and t(4;14), t(14;16), and/or del(17)(p13.1) was observed in both metaphase and interphase data. Of all patients with chromosome 13 anomalies, 57% by metaphase and 39% by interphase had t(4;14), t(14;16), and/or del(17)(p13.1). From this information, one hierarchy of chromosome anomalies was used to study survival for metaphase data (Figure 1E; Table 4) and another hierarchy was used for interphase data (Figure 1F; Table 4).
Thirty-five of 39 patients who had interphase nuclei with anomalies other than t(4;14), t(14;16), or del(17)(p13.1), and/or chromosome 13 anomalies had more than 2 CCND1 signals (Table 3). Thus, patients in Figure 1F in the more than 2 CCND1 and other category are primarily patients with multiple CCND1 signals.
Metaphase and interphase results along with other potential risk factors (hemoglobin level < 100 g/L [< 10 g/dL], serum calcium ≥ 2.5 mM [11 mg/dL], serum creatinine ≥ 176.8 μM [2 mg/dL], plasma cell labeling index, and percentage of bone marrow plasma cells) were evaluated by multivariable analysis using a Cox proportional hazards model. Only observation of chromosome 13 anomalies in metaphases (P < .001) or greater than 50% abnormal interphase nuclei (P = .01) were significant independent predictors of patient survival.
Discussion
Usefulness of conventional cytogenetic studies
In 1985 it was reported that patients with newly diagnosed MM and abnormal metaphases by conventional cytogenetics had active disease and reduced survival compared with patients who had only normal metaphases.1 In 1995, patients with MM who had metaphase cells with monosomy 13 and/or structural anomalies of chromosome 13 that were detected by conventional cytogenetics were reported to have even greater diminished survival.11 Results of the current investigation and others12,13 verify the conclusions of those studies and demonstrate that detection of abnormal metaphases is associated with significant shortening of survival in MM. In other investigations, chromosome studies have also been useful to identify therapy-related leukemia associated with treatment of MM.1,14
Usefulness of metaphase FISH
This investigation demonstrates the value of metaphase FISH as an adjunct to conventional cytogenetics. In our study, metaphase FISH was abnormal in 18 patients who were normal by conventional cytogenetics. Metaphase FISH was useful to examine cells with complex karyotypes, study metaphases with poor chromosome morphology, and detect cryptic rearrangements. Metaphase FISH detected t(11;14), t(4;14), or t(14;16) in 24 patients; none of these translocations were detected by cytogenetics. Consequently, we believe that t(14;16)(q32;q23) or t(4;14)(p16;q32) is best detected by fusion of IGH and FGFR3 or fusion of IGH and MAF using FISH.15,16
Chromosome 13 controversy
Several studies have shown that patients with chromosome 13 anomalies in their metaphase cells have poor prognosis.11,12 Reduced patient survival has also been associated with observation of chromosome 13 anomalies (monosomy/deletions) by plasma cell FISH.17-19 More recent reports suggest that chromosome 13 anomalies may not be as important to estimate survival as the observation of t(4;14), t(14;16), or del(17)(p13.1).13,15 Because other reports indicate a strong association between chromosome 13 anomalies and t(4;14), t(14;16), or del(17)(p13.1),13,20-22 the specific relationship between chromosome 13 anomalies and prognosis is unclear. Results of the present investigation help to resolve these issues.
In this investigation, the detection of chromosome 13 anomalies in metaphase cells was indeed associated with poor prognosis (P < .001) (Figure 1D-E). In contrast, patients with chromosome 13 anomalies in their interphase nuclei had intermediate survival. Therefore, detection of chromosome 13 anomalies in interphase is not as strong an indicator of survival as observation of chromosome 13 anomalies in metaphase cells. Because t(4;14) is cryptic and t(14;16) is subtle by conventional chromosome studies, their correlation with chromosome 13 anomalies in metaphases was previously not possible. This investigation used FISH to accurately study the correlation of these chromosome anomalies in metaphase cells. In our study, of all patients with chromosome 13 anomalies, 57% by metaphase and 39% by interphase had t(4;14), t(14;16), and/or del(17)(p13.1).
Efficacy of hierarchic chromosome anomalies to estimate prognosis
We previously established a hierarchy of chromosome anomalies in plasma cells that correlated with patient prognosis. Patients with t(4;14), t(14;16), and/or del(17)(p13.1) had a poor prognosis.15 Patients with chromosome 13 anomalies without t(4;14), t(14;16), and/or del(17)(p13.1) had an intermediate prognosis. Patients with other chromosome anomalies, including most patients with t(11;14), had a favorable prognosis. From hazard ratios for univariable and multivariable survival analyses, the data of the present study indicate a somewhat different hierarchy for metaphase (Figure 1E) and interphase data (Figure 1F). For metaphase, patients with t(4;14), t(14;16), del(17)(p13.1), and/or chromosome 13 anomalies had poorer survival than patients with t(11;14) and other anomalies. For interphase data, patients with t(4;14) or t(14;16) had poorer survival than patients with chromosome 13 anomalies, del(17)(p13.1), t(11;14), more than 2 CCND1, and other anomalies.
The prognostic significance of more than 2 CCND1 FISH signals in interphase nuclei is uncertain.12,21 In our hierarchy model for interphase, 35 patients had more than 2 CCND1 FISH signals without t(4;14), t(14;16), del(17)(p13.1), or chromosome 13 anomalies, and they had favorable survival (Figure 1F). This outcome is similar to patients with t(11;14) or normal results. Reportedly, patients with MM who have a clone with a hyperdiploid karyotype without translocations involving IgH have a favorable prognosis.13,23 Thus, we considered the possibility that observation of more than 2 CCND1 signals might be a surrogate marker for hyperdiploidy. Among our 61 patients with more than 2 CCND1 in interphase nuclei, 26 had translocations involving IgH and/or chromosome 13 anomalies. Thus, the observation of multiple CCND1 signals is not a surrogate observation for the kind of hyperdiploid patients with MM who have favorable prognosis hyperdiploidy.
Interphase FISH is a good adjunct to analysis of metaphases
In our investigation, 86% of patients had an abnormal clone that was detectable by interphase FISH. All patients who had abnormal metaphases were also abnormal by interphase FISH. Yet survivals differed among patients depending on whether certain chromosome anomalies were detected in both metaphase and interphase cells, or only interphase nuclei (Figure 1D). This outcome is likely due to active disease, which is associated with proliferation of abnormal metaphases.1 Thus, interphase FISH is not a substitute for metaphase analysis.
Nevertheless, interphase FISH can add to the workup of patients with MM and can be readily performed on the same specimens used for metaphase analyses. Results of interphase FISH studies establish the percentage of abnormal nuclei (tumor burden) and the presence of clinically significant chromosome anomalies.3-5 In addition, interphase FISH defines a subset of patients with greater than 50% abnormal nuclei who have poor prognosis, most likely because of level of plasma cells. However, in our experience interphase FISH is not as likely to detect chromosome anomalies in patients with low levels of plasma cells as plasma cell FISH.13
Survival
At the time of diagnosis for MM, this study shows that patients with chromosome 13 anomalies, t(4;14), t(14;16), and/or del(17)(p13.1) in metaphase cells have a median survival of 13.9 months, whereas survival of patients with other anomalies or normal metaphases ranged from 31.0 to 46.8 months (Table 4). For interphase cells, median survival for patients with t(4;14) or t(14;16) was 13.3 months, whereas survival of patients with other anomalies or normal results ranged from 33.9 to 55.3 months. For comparison, Fonseca et al17 reported a median survival based on plasma cell FISH of 24.7 months for t(4;14), t(14;16), del(17)(p13.1); 42.3 months for chromosome 13 anomalies; and 50.5 months for other anomalies. Interphase FISH assesses both tumor burden and prognostic chromosome anomalies, whereas plasma cell FISH only evaluates prognostic chromosome anomalies. This is the most likely explanation for the apparent different survivals associated with the same prognostic chromosome anomalies detected by interphase FISH and plasma cell FISH.
Algorithm for genetic testing at diagnosis
Our results suggest at diagnosis of MM that it is useful to first perform conventional cytogenetic studies. The results help to detect active proliferating disease and metaphases with poor prognostic anomalies such as chromosome 13 anomalies. Analysis of metaphases can be significantly enhanced by performing metaphase FISH to detect cryptic t(4;14) and t(14;16) and accurately identify chromosome 13 anomalies; this methodology adds significant prognostic information for patients with MM. If results of metaphase analyses are normal, then it is useful to perform interphase FISH studies using probes to detect t(11;14), t(4;14), t(14;16), del(17)(p13.1), or chromosome 13 anomalies. These results provide important prognostic information and establish the percentage of neoplastic cells to help assess tumor burden. This assay can be performed on leftover cells from metaphase studies. Our results show that observation of normal metaphases and normal interphase nuclei is a favorable prognosis, but it does not rule out MM. If results for metaphase cells and interphase nuclei are normal, then it may be useful to do plasma cell FISH to help diagnose MM and to establish the presence of prognostic chromosome anomalies.
Prepublished online as Blood First Edition Paper, July 19, 2005; DOI 10.1182/blood-2005-05-1981.
Supported in part by a grant from Vysis, Downers Grove, IL (G.D.); the International Waldenström Macroglobulinemia Foundation; the National Cancer Institute (grants R01 CA83724-01, Specialized Program of Research Excellence [SPORE] P50 CA100707-01, and P01 CA62242); and the Fund to Cure Myeloma.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66: 380-390. [PubMed] [Google Scholar]
- 2.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic findings in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82: 41-49. [DOI] [PubMed] [Google Scholar]
- 3.Wiktor A, Van Dyke DL. Combined cytogenetic testing and fluorescence in situ hybridization analysis in the study of chronic lymphocytic leukemia and multiple myeloma. Cancer Genet Cytogenet. 2004;153: 73-76. [DOI] [PubMed] [Google Scholar]
- 4.Drach J, Schuster J, Nowotny H, et al. Multiple myeloma: high incidence of chromosomal aneu-ploidy as detected by interphase fluorescence in situ hybridization. Cancer Res. 1995;55: 3854-3859. [PubMed] [Google Scholar]
- 5.Zandecki M, Lai JL, Facon T. Multiple myeloma: almost all patients are cytogenetically abnormal. Br J Haematol. 1996;94: 217-227. [DOI] [PubMed] [Google Scholar]
- 6.Ahmann GJ, Jalal SM, Juneau AL, et al. A novel three-color, clone-specific fluorescence in situ hybridization procedure for monoclonal gammopathies. Cancer Genet Cytogenet. 1998;101: 7-11. [DOI] [PubMed] [Google Scholar]
- 7.Dewald GW, Broderick DJ, Tom WW, Hagstrom JE, Pierre RV. The efficacy of direct, 24-hour culture, and mitotic synchronization methods for cytogenetic analysis of bone marrow in neoplastic hematologic disorders. Cancer Genet Cytogenet. 1985;18: 1-10. [DOI] [PubMed] [Google Scholar]
- 8.Dewald GW, Wyatt WA, Juneau AL, et al. Highly sensitive fluorescence in situ hybridization method to detect double BCR/ABL fusion and monitor response to therapy in chronic myeloid leukemia. Blood. 1998;91: 3357-3365. [PubMed] [Google Scholar]
- 9.Dewald G, Brockman SR, Paternoster SF. Molecular cytogenetic studies in hematological malignancies. In: Finn W, Peterson L, eds. Hemato-pathology in Oncology. Boston, MA: Kluwer Academic Publications; 2004: 69-112. [DOI] [PubMed]
- 10.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, NY: Springer-Verlag; 2000.
- 11.Tricot G, Barlogie B, Jagannath S, et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood. 1995;86: 4250-4256. [PubMed] [Google Scholar]
- 12.Gutierrez NC, Hernandez JM, Garcia JL, et al. Correlation between cytogenetic abnormalities and disease characteristics in multiple myeloma: monosomy of chromosome 13 and structural abnormalities of 11q are associated with a high percentage of S-phase plasma cells. Haematologica. 2000;85: 1146-1152. [PubMed] [Google Scholar]
- 13.Fonseca R, Barlogie B, Bataille R, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64: 1546-1558. [DOI] [PubMed] [Google Scholar]
- 14.Morrison-DeLap SJ, Kuffel DG, Dewald GW, Letendre L. Unbalanced 1;7 translocation and therapy-induced hematologic disorders: a possible relationship. Am J Hematol. 1986;21: 39-47. [DOI] [PubMed] [Google Scholar]
- 15.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101: 4569-4575. [DOI] [PubMed] [Google Scholar]
- 16.Debes-Marun CS, Dewald GW, Bryant S, et al. Chromosome abnormalities clustering and its implications for pathogenesis and prognosis in myeloma. Leukemia. 2003;17: 427-436. [DOI] [PubMed] [Google Scholar]
- 17.Fonseca R, Harrington D, Oken MM, et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: an Eastern Cooperative Oncology Group study. Cancer Res. 2002;62: 715-720. [PubMed] [Google Scholar]
- 18.Kaufmann H, Kromer E, Nosslinger T, et al. Both chromosome 13 abnormalities by metaphase cytogenetics and deletion of 13q by interphase FISH only are prognostically relevant in multiple myeloma. Eur J Haematol. 2003;71: 179-183. [DOI] [PubMed] [Google Scholar]
- 19.Avet-Louseau H, Daviet A, Sauner S, Bataille R. Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br J Haematol. 2000;111: 1116-1117. [DOI] [PubMed] [Google Scholar]
- 20.Fonseca R, Oken MM, Greipp PR. The t(4; 14)(p16.3;q32) is strongly associated with chromosome 13 abnormalities in both multiple myeloma and monoclonal gammopathy of undetermined significance [letter]. Blood. 2001;98: 1271. [DOI] [PubMed] [Google Scholar]
- 21.Magrangeas F, Lode L, Wuilleme S, Minvielle S, Avet-Loiseau H. Genetic heterogeneity in multiple myeloma. Leukemia. 2005;19: 191-194. [DOI] [PubMed] [Google Scholar]
- 22.Avet-Loiseau H, Facon T, Grosbois B, et al. Oncogenesis of multiple myeloma: 14q32 and 13q chromosomal abnormalities are not randomly distributed, but correlate with natural history, immunological features, and clinical presentation. Blood. 2002;99: 2185-2191. [DOI] [PubMed] [Google Scholar]
- 23.Wuilleme S, Robillard N, Lode L, et al. Ploidy, as detected by fluorescence in situ hybridization, defines different subgroups in multiple myeloma. Leukemia. 2005;19: 275-278. [DOI] [PubMed] [Google Scholar]