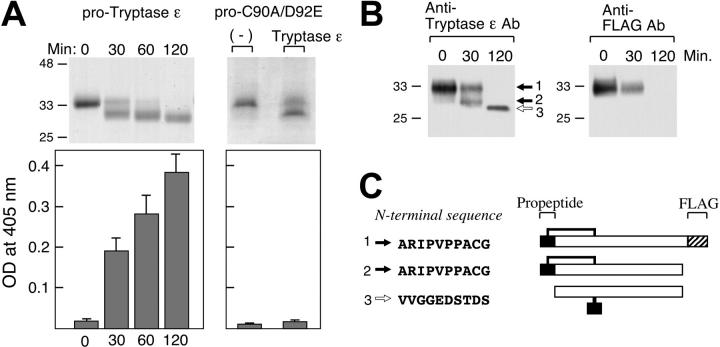
Figure 2.
Activation of purified pro–tryptase ε with active tryptase ε. (A) Insect cell–expressed pro–tryptase ε (left column) or its C90A/D92E mutant (right column) were incubated with spontaneously activated tryptase ε in an approximately 20:1 ratio at 37°C for up to 120 minutes. Aliquots of the resulting digests were subjected to SDS-PAGE under reducing conditions. Shown are the Coomassie blue–stained gels (top row). Other aliquots were evaluated for their enzymatic activities using H-D-Leu-Thr-Arg-pNA (bottom row). In a control experiment for the left-column experiments, pro–tryptase ε was not activated if incubated with the C90A/D92E mutant that lacks its propeptide (data not shown). (B) An excess of pro–tryptase ε was incubated with active tryptase ε for 0, 30, or 120 minutes. SDS-PAGE/immunoblots were probed with anti–tryptase ε antibody (left panel) or anti-FLAG antibody (right panel). (C) The N-terminal amino acid sequences of protein bands 1, 2, and 3 in panel B are shown, as well as a schematic model of the 3 tryptase ε products identified in this study. The black (▪) domain in each right panel is the protease's propeptide, which is covalently linked to the catalytic domain presumably via the Cys-9-Cys112 disulfide bond. The hatched ( ) domain at the protein's C terminus corresponds to the FLAG peptide that is found only in the unprocessed tryptase ε zymogen.
) domain at the protein's C terminus corresponds to the FLAG peptide that is found only in the unprocessed tryptase ε zymogen.