Abstract
Translocations of the anaplastic lymphoma kinase (ALK) gene have been described in anaplastic large-cell lymphomas (ALCLs) and in stromal tumors. The most frequent translocation, t(2;5), generates the fusion protein nucleophosmin (NPM)–ALK with intrinsic tyrosine kinase activity. Along with transformation, NPM-ALK induces morphologic changes in fibroblasts and lymphoid cells, suggesting a direct role of ALK in cell shaping. In this study, we used a mass-spectrometry–based proteomic approach to search for proteins involved in cytoskeleton remodeling and identified p130Cas (p130 Crk-associated substrate) as a novel interactor of NPM-ALK. In 293 cells and in fibroblasts as well as in human ALK-positive lymphoma cell lines, NPM-ALK was able to bind p130Cas and to induce its phosphorylation. Both of the effects were dependent on ALK kinase activity and on the adaptor protein growth factor receptor–bound protein 2 (Grb2), since no binding or phosphorylation was found with the kinase-dead mutant NPM-ALKK210R or in the presence of a Grb2 dominant-negative protein. Phosphorylation of p130Cas by NPM-ALK was partially independent from Src (tyrosine kinase pp60c-src) kinase activity, as it was still detectable in Syf-/- cells. Finally, p130Cas-/- (also known as Bcar1-/-) fibroblasts expressing NPM-ALK showed impaired actin filament depolymerization and were no longer transformed compared with wild-type cells, indicating an essential role of p130Cas activation in ALK-mediated transformation.
Introduction
Nucleophosmin–anaplastic lymphoma kinase (NPM-ALK) is an oncogenic protein generated by the t(2;5)(p23;q35) translocation1 that characterizes most anaplastic large-cell lymphomas (ALCLs). Each of the 2 fusion-derived fractions that form NPM-ALK plays a role in the constitutive tyrosine kinase activity of the chimeric protein. NPM is a nucleolar phosphoprotein involved in the ribosome genesis,2 in the centrosome duplication,3 and in the regulation of the basal level of p53.4 NPM promoter drives the constitutive expression of NPM-ALK in lymphoid cells; the oligomerization domain of NPM allows the dimerization5 and hence the autophosphorylation of NPM-ALK. ALK is a 210-kDa tyrosine kinase (TK) receptor belonging to the insulin growth factor receptor superfamily, which is widely expressed in the nervous system during embryogenesis but only focally in the adult brain.1,6,7 Although the physiologic role of ALK receptor is still unknown, it seems to be involved in neuronal cell differentiation as suggested by the ability to induce neurite outgrowth in vitro8 and by its fundamental role in synapse formation shown in Caenorhabditis elegans.9
The constitutive NPM-ALK activity transforms rat-1 fibroblast10 and is responsible for the malignant transformation of lymphoid cells.11 Furthermore, NPM-ALK has the ability to induce B- and T-cell lymphomas in vivo.12,13 The signaling pathways downstream from NPM-ALK follow a transduction pattern shared with most of deregulated tyrosine kinases, leading to increased proliferation, resistance to apoptosis, and morphologic changes. The enhancement of cell growth in NPM-ALK–positive cells results from the activation of the rat sarcoma viral oncogene homolog (Ras)/extracellular signal-related kinase (ERK) pathway through the recruitment of the adaptors SHC (Src homology 2 domain containing protein) and insulin receptor substrate 1 (IRS-1).14 In addition, NPM-ALK controls cellular mitogenity by binding to phospholipase C γ (PLC-γ).15
In parallel, the inhibition of apoptosis in the NPM-ALK signal transduction derives from the activation of signal transducer and activator of transcription 3 (Stat3),16 leading to an increased expression of the antiapoptotic factor Bcl-x(L) and from the stimulation of the phosphatidylinositol 3–kinase (PI3K)/Akt pathway,17 which confers resistance to apoptosis combined with a deregulated proliferation rate due to the transcription factor forkhead box O3a (FOXO3a).18
In contrast to these fairly well-characterized pathways, minimal data are available regarding the effects of ALK tyrosine kinase activity on cell shape. Upon transduction in Hodgkin lymphoma cell lines, NPM-ALK induces in these cells morphologic changes that make them similar to ALCL cells.19 Moreover, in PC12 cells, ectopic expression of ALK induces a neurite outgrowth that has been shown to depend on the mitogen-activated protein (MAP) kinase pathway.8 In C elegans, the degradation of ALK is required for synapse stabilization,9 whereas in Drosophila, ALK is activated by its ligand Jelly Belly to specify visceral mesoderm migration and differentiation into muscle cells.20,21 Moreover, ALK ectopic expression in Drosophila hemocytes promotes the formation of lamellocytes, specialized cells that participate in the encapsulation and killing of parasites, a phenotype that is coupled with the activation of both the Janus kinase (JAK)/STAT and the Jun kinase pathways through Rho guanosine triphosphatase (GTPase) Rac1.22
In this paper we investigated molecules involved in cell morphology that could also be relevant for the NPM-ALK–mediated transformation process. We started from a mass spectrometry–based screening of proteins interacting with NPM-ALK and involved in the cytoskeleton morphology. Through this analysis we identified p130Cas (p130 Crk-associated substrate) and then demonstrated that NPM-ALK is able to bind and phosphorylate p130Cas. The p130Cas activation is mediated by the adaptor protein growth factor receptor–bound protein 2 (Grb2) and is partially independent from Src. Importantly, we show that p130Cas activation is required for NPM-ALK–mediated cell shape modifications and actin filament depolymerization as well as for cell transformation, but not for cell migration, induced by NPM-ALK.
Taken together, these results suggest not only that p130Cas acts as one of the effectors of the cytoskeleton modifications induced by NPM-ALK but also that morphologic and growth cues are deeply embedded in NPM-ALK–mediated oncogenic processes.
Materials and methods
Cell lines and culture
Human lymphoid cells TS, DHL, and Karpas (NPM-ALK–positive) and CEM, K562, and Namalwa (NPM-ALK–negative) were obtained from New York University and maintained in RPMI 1640 (BioWhittaker, Verviers, Belgium) containing 10% fetal calf serum, 2 mM glutamine (Eurobio Biotechnology, Les Ulis, France), 100 U/mL penicillin, and 100 μg/mL streptomycin (Eurobio Biotechnology).
Human embryonal kidney cells 293T, 293GP, and 293 T-Rex Tet-On (Invitrogen, Carlsbad, CA) and murine fibroblasts MEF, NIH3T3, Syf -/-, Cas-/- (also known as Bcar1-/-), and MEF Tet-Off (Clontech, Palo Alto, CA) were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum, 2 mM glutamine (Eurobio Biotechnology), 100 U/mL penicillin, and 100 μg/mL streptomycin (Eurobio Biotechnology). In MEF and 293 Tet systems, the working concentration of tetracycline or doxycycline in the medium was 1 μg/mL.
Inducible ALK-shRNA interference TS cells were obtained by cotransduction with pLVTH vector containing the H1 promoter ALK-shRNA cassette and pLV-tTRKRAB vector,23 as described by Piva et al.53 These cells undergo NPM-ALK silencing when 1 μg/mL of doxycycline is added to the medium for at least 72 hours.
Cell lysis, immunoprecipitation, and immunoblotting
Total cellular proteins were extracted with 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA (ethylenediaminetetraacetic acid), 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM NaF, 1 mM Na3VO4, and protease inhibitors (Roche, Mannheim, Germany). Cell lysates were centrifuged at 10 000g (13 000 rpm) for 15 minutes and the supernatants were collected and assayed for protein concentration using the Bio-Rad protein assay method. Thirty micrograms of proteins was run on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions.
For immunoprecipitation experiments, 300 μg to 800 μg protein was immunoprecipitated with the appropriate antibody for 1 hour at 4°C followed by 30 μL protein A–Sepharose beads (Amersham, Freiburg, Germany). After SDS-PAGE, proteins were transferred to nitrocellulose, incubated with the specific antibody, and then detected with peroxidase-conjugated secondary antibodies and enhanced chemiluminescent reagent (Amersham).
Reagents and antibodies
PP2 Src-family kinase inhibitor was purchased from Calbiochem (Darmstadt, Germany). The following antibodies were used: polyclonal anti-ALK (1:2000; Zymed, Basel, Switzerland), monoclonal anti-ALK (1:4000; Zymed), monoclonal anti-p130Cas (1:1000; Becton Dickinson, Mountain View, CA), monoclonal anti–phospho-Tyr (PY20; 1:1000; Transduction Laboratories, Lexington, KY), polyclonal anti-Grb2 (1:2000; Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti-Crk (1:1000; Transduction Laboratories), polyclonal anti-Src (1:1000; Santa Cruz Biotechnology), monoclonal antipaxillin (1:1000; Transduction Laboratories), monoclonal antiactin (1:1000; Santa Cruz Biotechnology), and monoclonal anti–focal adhesion kinase (anti-FAK; 1:1000; hybridome immunoglobulin G1 [IgG1] clone 4-4A, kindly gifted by Prof Silengo, University of Turin, Italy). Secondary antimouse or antirabbit peroxidase-conjugated antibodies were purchased from Amersham.
Immunoprecipitation and mass spectrometry analysis
For anti-ALK immunoprecipitation, 293 T-Rex Tet-on cells (Invitrogen) transfected with a wild-type (wt) NPM-ALK and a kinase-dead mutant control NPM-ALKK210R were grown to confluence and induced with 1 μg/mL of tetracycline for 24 hours. Cells were lysed in 20 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 0.1% Triton X-100 in the presence of protease and phosphatase inhibitors. Ten milligrams of cleared cell lysate was mixed with 100 μg monoclonal anti-ALK coupled to agarose beads (Amersham) and rotated at 4°C for 2 hours. Precipitated immunocomplexes were washed 3 times with lysis buffer, eluted, and boiled in Laemli buffer for 5 minutes. Samples were resolved on SDS-PAGE and silver stained. Bands of interest were excised from the gel and subjected to in-gel reduction, alkylation, and digestion with trypsin (Promega, Madison, WI) and analyzed by matrix-assisted laser desorption ionization/time of flight (MALDI-TOF) mass spectrometry. Spectra were obtained using either a Reflex III MALDI-TOF mass spectrometer or an Ultraflex TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany), operating in the positive ion delayed extraction reflector mode. Ions were generated by irradiation of analyte/matrix deposits by a nitrogen laser at 337 nm and analyzed with an accelerating voltage of 20 kV. Each MALDI-TOF spectrum was generated by accumulating data corresponding to more than 400 laser shots. Mass calibration in the range mass over electronic charge (m/z) 800 to 4000 was performed by using a bovine beta-lactoglobulin tryptic peptide mixture. Postacquisition internal calibration was applied by using theoretic masses of trypsin autodigestion peptides. Peptide mass spectra were analyzed using Mascot Peptide Mass Fingerprinting software.24
Some samples were analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) using a nanoflow–high-performance liquid chromatography (HPLC) system (Ultimate; LC Packings, Amsterdam, The Netherlands) interfaced to electrospray quadrupole time-of-flight (Q-TOF) tandem mass spectrometers (QTOF Ultima or QTOF Micro; Waters/Micromass, Manchester, United Kingdom). Protein identification via peptide MS/MS spectra was achieved by using Mascot software24 for searching the National Center for Biotechnology Information (NCBI) nonredundant protein database.25
For antiphosphotyrosine immunoprecipitation, 293 T-Rex Tet-On cells were grown in nonadherent conditions on Poly(2-hydroxyethylmethacrylate) (Poly-HEME; Sigma, St Louis, MO) coated plates, starved for 12 hours, and induced with 1 μg/mL of tetracycline for 24 hours, pelleted, and stored at -80°C. Cells were lysed in 20 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, and 1% Nonidet P-40 in the presence of protease and phosphatase inhibitors. Ten milligrams cleared cell lysate was mixed with 50 μg agarose-conjugated monoclonal 4G10 (Upstate Biotechnology, Lake Placid, NY) and 10 μg monoclonal PY20 (Transduction Laboratories) and incubated rotating at 4°C overnight. Samples were processed as described in “Cell lysis, immunoprecipitation, and immunoblotting.”
Immunofluorescence staining
Cells were grown for 12 hours on glass coverslips pretreated with fibronectin (10 μg/mL phosphate-buffered saline [PBS]) at 37°C for 1 hour to facilitate cell adhesion. Samples were fixed in PBS containing 4% paraformaldehyde at room temperature for 10 minutes and permeabilized with PBS containing 0.3% Triton X-100 for 5 minutes. Coverslips were incubated with PBS containing 3% bovine serum albumin (BSA) for 1 hour at room temperature and then stained with primary antibody for 1 hour followed by fluorescein isothiocyanate (FITC)– or tetramethyl rhodamine isothiocyanate (TRITC)–conjugated secondary antibody (1:200; Sigma) for 1 hour at room temperature.
Primary polyclonal anti-ALK (Zymed) and monoclonal antipaxillin (Transduction Laboratories) antibodies were diluted 1:400 before use. Phycoerythrin (PE)–conjugated phalloidin (1:200 PBS; Sigma) was used to stain actin filaments. Nuclei were stained 10 minutes at room temperature with HOECHST (300 ng/mL; Sigma). Coverslips were mounted in antifading solution and viewed using a Leica TCS SP2 laser-scanning confocal microscope driven by Leica Confocal Software; the images were acquired at room temperature by means of a 63×/1.32 PL APO objective (Leica, Heidelberg, Germany). Brightfield images were acquired on a Leica DM IRE2 microscope using a DC300F camera and were analyzed with IM 50 software.
DNA constructs
Wild-type NPM-ALK or the kinase-dead mutant NPM-ALKK210R16 was cloned in the plasmid vector pcDNA5TO (Invitrogen) at HindIII/XhoI sites and stably transfected into 293 T-Rex Tet-On cells using Effectene reagents as described by manufacturer (Qiagen, Valencia, CA). Single cell–derived clones were selected for NPM-ALK or NPM-ALKK210R expression levels. Wild-type NPM-ALK, digested to obtain a HindIII/XhoI-blunted fragment, was cloned in the plasmid vector pBIEGFP (Clontech) at MluI-blunted sites and then stably transfected into MEF Tet-Off. Murine p130Cas was cloned in the plasmid vector pEGFP-C1 (Becton Dickinson) at EcoRI/BamHI sites or digested to obtain an AseI/BamHI-blunted fragment and cloned in the plasmid vector Pallino26 at EcoRI/XhoI-blunted ends. Wild-type Grb2 or the dominant-negatives Grb2-P49L (mutated in the N-terminal SH3 domain) and Grb2-R86K (mutated in the SH2 domain) were cloned in the plasmid vector pRK5 (a kind gift from Dr A. Pellicer, New York University). Pallino vectors containing NPM-ALK or NPM-ALKK210R or ATIC-ALK (aminoimidazole-4-carboxamide ribonucleotide formyltransferase/inosine 5′-monophosphate [IMP] cyclohydrolase–ALK) were previously described.16 Full-length human ALK receptor was purchased from ATCC (Manassas, VA).
Retrovirus production and cell infection
NPM-ALK and p130Cas retroviruses were obtained by cotransfection of Pallino expression vector containing wild-type NPM-ALK or p130Cas with pMD2VSV-G plasmid into the 293GP packaging cell line (Invitrogen). Retroviruses released in culture medium were collected 24 hours after transfection. Five hundred microliters filtered (pore size 0.45 mm) supernatants from GP cells was supplemented with 8 μg/mL polybrene (Sigma) and added to 5 × 104 to 10 × 104 target cells. After 12 hours of incubation, 1 mL complete medium was added and the cells were cultured for an additional 2 days. The percentages of transduced cells were analyzed for GFP expression by fluorescence-activated cell sorter (FACS). Cells were analyzed for GFP content on a FACSCalibur flow cytometer (Becton Dickinson). Calibration of the instrument was performed using Calibration beads (CALIBrite; Becton Dickinson). The CELLQuest software (Becton Dickinson) was used for the data acquisition and analysis. For cell sorting, cells were suspended at the concentration of 10 × 106/mL in basic sorting buffer (5 mM EDTA, 25 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 7.0], 1% heat-inactivated fetal bovine serum [FBS]) and then sorted for GFP expression on a MoFlo High-Performance cell sorter (DAKO Cytomation, Glostrup, Denmark).
Soft-agar assay
For clonogenic assay, 5 × 105 cells/well were seeded in 0.35% agar on the top of a base layer containing 0.7% agar into 6-well plates. After 3 to 5 weeks of cultivation, colonies were counted under a phase-contrast microscope. All of the experiments were done using triplicate plates for experimental point.
Cell-cycle analysis
For DNA content determination, cells were fixed for 1 hour in 70% ethanol at 4°C. After washing, cells were treated with RNase (0.25 mg/mL) and stained with propidium iodide (50 μg/mL). The S-phase fraction was calculated using the Modfit program from Becton Dickinson.
Chemotaxis assay
Cell migration was evaluated using 24-well, 5-μm pore size Transwell system plates (Costar, Cambridge, MA). Purified cells were washed once in RPMI 1640 (BioWhittaker) containing 0.1% fetal calf serum, 2 mM glutamine (Eurobio Biotechnology), 100 U/mL penicillin, and 100 μg/mL streptomycin (Eurobio Biotechnology) and then adjusted to 1 × 106 cells/mL in the same medium. Cells (1 × 105 in 100 μL) were placed on the top of the Transwell. Stromal-derived factor-1 α (SDF-1α; R&D Systems, Abingdon, United Kingdom), prepared at the indicated concentrations in the same RPMI medium (600 μL total volume), was added to the bottom of the Transwell system. After 2 hours incubation at 37°C in a 5% CO2 atmosphere, the inserts of the transwell chambers were removed and the number of cells that had migrated into the lower well was counted by staining with a DIF-Quick Kit (IMEB, San Marcos, CA).
Matrigel migration assay
Cells were seeded at a density of 1 × 105 cells onto Matrigel (Becton Dickinson)–coated inserts (100 μg/insert) in 24-well, 8-μm pore size Transwell system plates (Costar). After a 48-hour incubation, the inserts were removed, the cells on the upper side of the inserts were removed with a cotton swab, and the number of cells that migrated to the lower side of the filters was counted under a phase-contrast microscope by staining with DIF-Quick Kit (IMEB).
Results
NPM-ALK expression modifies the cell cytoskeleton and the migration pattern
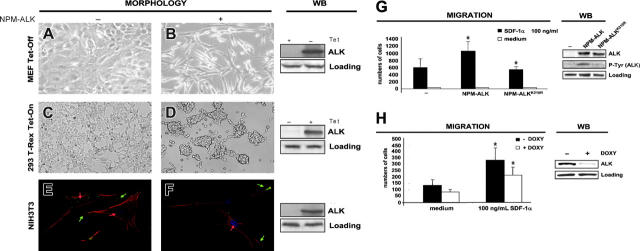
We selected different cell lines to test the effects of NPM-ALK on cell morphology, adhesion, and migration. We used 2 tetracycline-inducible cell lines: MEF Tet-Off fibroblasts were grown in DMEM medium containing tetracycline (1 μg/mL) and were induced to express NPM-ALK whenever tetracycline was depleted from the medium. The 293 T-Rex cells were induced to express NPM-ALK when 1 μg/mL of tetracycline was added to the medium; NPM-ALK was detectable by Western blot (WB) as early as 6 hours after tetracycline induction. NIH3T3 fibroblasts were retrovirally infected to stably express NPM-ALK. Upon NPM-ALK expression, MEF Tet-Off and NIH3T3 fibroblasts assumed the typical transformed phenotype with spindle-shaped cell morphology (Figure 1A-B), whereas 293 T-Rex cells detached from the culture plate (Figure 1C-D).
Figure 1.
Morphologic changes induced by NPM-ALK. (A-B) MEFs Tet-Off show spindle-transformed cell shape when NPM-ALK is expressed. MEFs Tet-Off were stably transfected with the inducible vector NPM-ALK pBIEGFP and forced to express NPM-ALK when grown in medium deprived of tetracycline for 48 hours. (C-D) The 293 T-Rex Tet-On cells undergo changes in adherence when NPM-ALK is induced by adding tetracycline to the culture medium for 24 hours. Contrast-phase images; 40×/0.55 objective lens. (E-F) NIH3T3 fibroblasts were infected with Pallino NPM-ALK retrovirus and then incubated with monoclonal antipaxillin primary antibody followed by FITC-conjugated secondary antibody (green) and with PE-conjugated phalloidin to stain the actin filaments (red). Noninfected NIH3T3 cells (E) show a spread morphology, with clearly detectable actin filaments ending in the focal contacts (red arrows) and with organized paxillin clusters (green arrows), whereas NIH3T3 NPM-ALK cells (F) show only few actin filaments (red arrow) and rare paxillin clusters (green arrows). Images were taken with the Leica confocal microscope using a 63×/1.32 objective lens. Right panels show NPM-ALK expression by WB in the corresponding cells. Samples were blotted with anti-ALK monoclonal antibody. (G) CEM cells were infected with retroviruses expressing NPM-ALK or the kinase-dead mutant NPM-ALKK210R as control and then sorted for GFP expression. The histograms represent the numbers of cells migrated in response to SDF-1α in a transwell assay. The histograms summarize the results of 3 independent experiments using triplicate wells for experimental point. The right panels show NPM-ALK expression and phosphorylation by WB in the corresponding cells. Samples were blotted with anti-ALK and anti-PY20 monoclonal antibodies. (H) NPM-ALK silencing by ALK-shRNA decreases the migration rate of TS cells. TS cells were cotransduced with pLVTH ALK-shRNA and pLV-tTRKRAB vectors to obtain a doxycycline-dependent inducible NPM-ALK silencing. Cells were grown in the presence of doxycycline (1 μg/mL) for 72 hours. The histograms represent the numbers of cells migrated in response to SDF-1α in a transwell assay. The histograms are from 3 independent experiments using triplicate wells for experimental point. The right panels show NPM-ALK expression by WB in the corresponding cells. Samples were blotted with anti-ALK monoclonal antibody. *Statistically significant as analyzed by Student t test.
In fibroblasts, the transformed morphology corresponded to a disruption of the actin cytoskeleton in immunofluorescence (Figure 1E-F). Together with actin, we stained for the scaffolding protein paxillin as a marker of the cellular structural organization because it is found at the interface between the plasma membrane and the actin cytoskeleton.27,28 Noninfected NIH3T3 cells were characterized by a strong paxillin signal at the focal adhesions, whereas NIH3T3 NPM-ALK cells showed few residual paxillin clusters at the edge of the elongated cellular processes (Figure 1E-F green arrows). Moreover, the actin cytoskeleton turned from a polarized organization to a scattered pattern. The actin filaments were clearly visible in noninfected NIH3T3 cells, whereas only a few were detectable in NIH3T3 NPM-ALK (Figure 1E-F red arrows).
For cellular migration we used T-lymphoblastoid CD4+ CEM cells, which are described to migrate in response to SDF-1α, the ligand of the chemokine receptor CXCR4, predominantly expressed on naive CD4+ and CD8+ T cells.29 CEM cells were infected with Pallino retroviruses containing NPM-ALK, or the kinase-dead mutant (NPM-ALKK210R) as control, and then sorted for the GFP expression. In a chemotaxis assay, upon stimulation with the chemokine SDF-1α, the migration rate of CEM NPM-ALK was significantly higher than CEM wt or CEM NPM-ALKK210R (Figure 1G). Similarly, in inducible ALK-shRNA interference TS cells both the spontaneous and the SDF-1α–induced migration rates were significantly reduced when NPM-ALK expression was ablated (Figure 1H). Taken together, these data indicate a role of NPM-ALK in the migration process of transformed lymphoid cells.
Identification of NPM-ALK–interacting partners by mass spectrometry
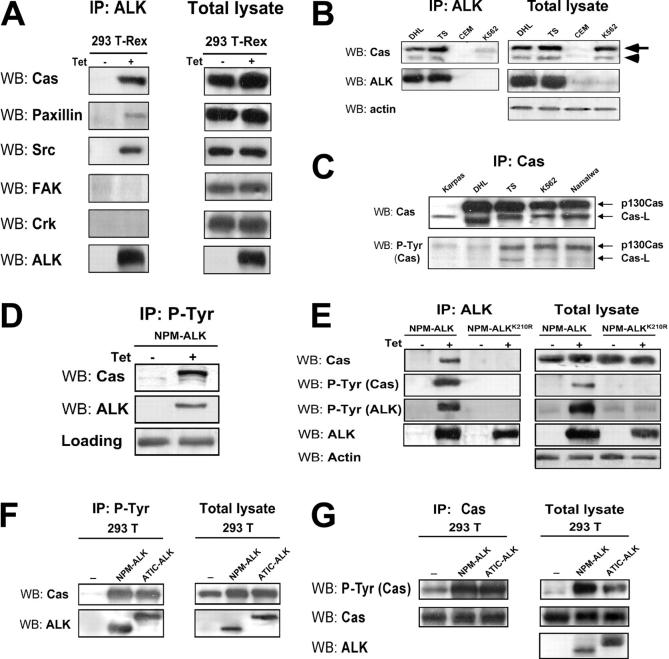
Since few data are available on the signaling pathways that NPM-ALK activates to influence cytoskeleton organization and cell migration, we decided to apply mass spectrometry to identify new interactors or substrates that are tyrosine phosphorylated after NPM-ALK expression. To this end, we performed immunoprecipitations with a monoclonal anti-ALK antibody or with a mixture of antiphosphotyrosine antibodies, respectively. The 293T-Rex cells were induced with tetracycline for 24 hours to express NPM-ALK or NPM-ALKK210R as control. For antiphosphotyrosine immunoprecipitations, cells were grown on Poly-HEME in order to reduce the basal phosphorylation due to adhesion-mediated signaling. After comparing the patterns of NPM-ALK and NPM-ALKK210R samples, the bands present only in the NPM-ALK sample were cut from the gel (Figure 2). Proteins were identified by peptide mass mapping by MALDI tandem mass spectrometry and by peptide sequencing by LC-MS/MS. From the list of the identified proteins we focused on proteins known to play a role in cytoskeleton reorganization or cell migration. Table 1 lists the identified proteins related to the cytoskeleton. Most of the proteins found were structural proteins, in keeping with recently published data.30 One of these proteins was the p130Cas that is an adaptor protein involved in many cellular processes related to the cytoskeleton organization, migration, and transformation.31
Figure 2.
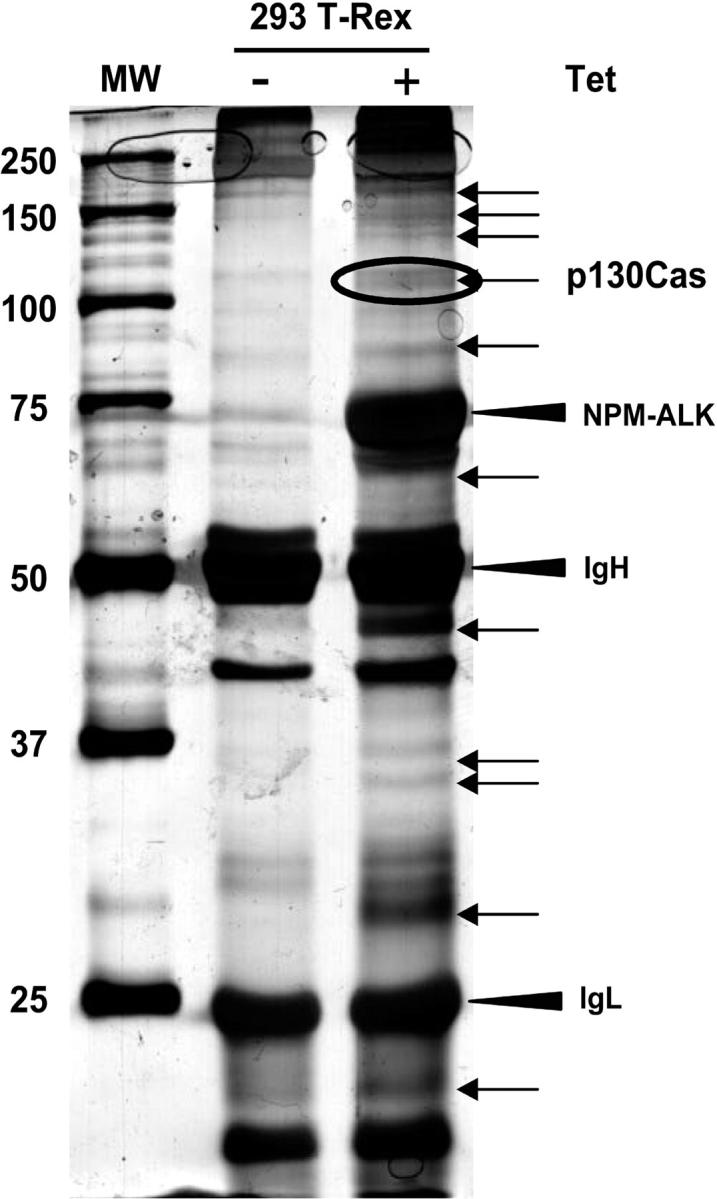
Identification of p130Cas from silver-stained SDS-PAGE. The 293 T-Rex Tet-On cells were transfected with NPM-ALK and cultured for 24 hours in presence or absence of tetracycline. Thirty milligrams of total cell lysate was immunoprecipitated with a mixture of antiphosphotyrosine antibodies (4G10; Upstate Biotechnology; and PY20; Transduction Laboratories) and resolved on 8% SDS-PAGE. The gel was silver stained and the bands present only in the induced sample (+), as indicated by the arrows, were analyzed by mass spectrometry. Molecular marker is shown on the left. The p130Cas was identified from the circled band. See “Materials and methods” for details.
Table 1.
Identification by mass spectrometry of adaptors and structural proteins involved in cytoskeleton organization in NPM-ALK–positive cells
| NCBI accession no. | Identified protein | No. of peptide matches | Sequence coverage, % | IP | MW, kDa | MALDI identification | MS/MS identification |
|---|---|---|---|---|---|---|---|
| NP_0055382 | p130 Crk-associated substrate (p130Cas) | 3 | 6 | P-Tyr | 93 | – | + |
| AAH08633 | Actin, beta | 9 | 29 | P-Tyr | 41 | + | + |
| AAC16672 | Alpha-actinin-2-associated LIM protein | 6 | 21 | P-Tyr | 34 | + | – |
| AAA61279 | Vimentin | 15 | 32 | ALK | 53 | + | + |
| P05217 | Tubulin, beta | 9 | 28 | ALK | 48 | + | + |
| AAC51654 | Myosin VI | 8 | 8 | P-Tyr | 147 | + | + |
| NP_005955 | Myosin heavy chain, nonmuscle type B | 7 | 5 | ALK | 227 | + | + |
| NP_002464 | Myosin, heavy polypeptide 9, nonmuscle | 22 | 16 | ALK | 229 | + | – |
| NP_006650 | Tubulin, gamma complex-associated protein 2 | 2 | 2 | ALK | 103 | – | + |
| NP_004915 | Actinin alpha 4 | 2 | 7 | ALK | 104 | – | + |
| NP_055140 | Coronin 1C | 2 | 4 | ALK | 53 | – | + |
| NP_002009 | Flightless 1 homolog | 3 | 2 | ALK | 144 | – | + |
| NP_056065 | Coiled-coil protein BICD2 | 7 | 10 | P-Tyr | 93 | – | + |
| NP_006089 | RACK1 | 3 | 9 | P-Tyr | 35 | – | + |
IP indicates immunoprecipitation; MW, molecular weight.
NPM-ALK binds and phosphorylates p130Cas
We confirmed that NPM-ALK was able to associate with p130Cas in 293 T-Rex cells by immunoprecipitation (Figure 3A). Next, we extended our analysis to other molecules known to be often in complex with p130Cas: paxillin and Src were shown to coprecipitate with NPM-ALK, whereas FAK and Crk were almost undetectable (Figure 3A). Importantly, the binding between NPM-ALK and p130Cas was also demonstrated in NPM-ALK–positive cells derived from human lymphomas (Figure 3B). In lymphoid cell lines, NPM-ALK associates not only with p130Cas (Figure 3B arrow) but also with Cas lymphocyte-type (Cas-L), a p105 kDa member of the Cas family exclusively expressed in lymphocytes32,33 (Figure 3B arrowhead). In ALK-positive lymphoid cells, both p130Cas and Cas-L are present in a phosphorylated form (Figure 3C).
Figure 3.
NPM-ALK binds and phosphorylates p130Cas. (A) The 293 T-Rex Tet-On cells were cultured with tetracycline for 24 hours; total cell lysate was immunoprecipitated (IP) with anti-ALK monoclonal antibody and blotted with the indicated antibodies by WB. NPM-ALK coprecipitates with p130Cas, paxillin, and Src. (B) NPM-ALK coprecipitates both with p130Cas (arrow) and with Cas-L (arrowhead) in lymphoid cell lines. Total lysates from ALK-positive (DHL, TS) and ALK-negative (CEM, K562) cell lines were immunoprecipitated with anti-ALK monoclonal antibody and blotted with the indicated antibodies. (C) In ALK-positive lymphoid cells, both p130Cas and Cas-L are phosphorylated. Total lysates were immunoprecipitated with anti-p130Cas monoclonal antibody and blotted with the anti-PY20 monoclonal antibody. (D) The adaptor p130Cas is phosphorylated in the presence of NPM-ALK. The 293 T-Rex Tet-On cells expressing NPM-ALK were cultured with tetracycline for 24 hours; cell lysate was immunoprecipitated with anti-PY20 monoclonal antibody and blotted with the indicated antibodies. (E) The binding and the phosphorylation of p130Cas depends on NPM-ALK kinase activity. The 293 T-Rex Tet-On cells expressing the active form of NPM-ALK or the kinase-dead control (NPM-ALKK210R) were cultured with tetracycline for 24 hours; cell lysates were immunoprecipitated with anti-ALK monoclonal antibody and blotted with the indicated antibodies. (F-G) The phosphorylation of p130Cas does not depend on the binding with the NPM portion of NPM-ALK. The 293 T cells were transfected with Pallino p130Cas alone or in combination with Pallino NPM-ALK or Pallino ATIC-ALK. Samples were collected 48 hours after transfection and total lysates were immunoprecipitated and blotted with the indicated antibodies.
Moreover, in the presence of NPM-ALK, p130Cas was phosphorylated (Figure 3D). This effect was specifically due to the tyrosine kinase activity of ALK because the mutated kinase-dead NPM-ALKK210R failed to bind and to phosphorylate p130Cas (Figure 3E). The phosphorylation of p130Cas was detected also expressing the ATIC-ALK fusion protein (Figure 3F-G) as well as the full-length ALK receptor (data not shown), thus suggesting a general mechanism of the ALK kinase domain.
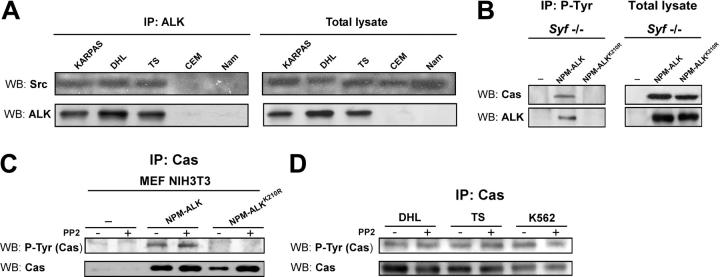
NPM-ALK has recently been reported to associate with Src in Karpas and DHL lymphoid cells.34 Considering that p130Cas is a major substrate of Src,35 we checked the potential role of Src in NPM-ALK–mediated phosphorylation of p130Cas. We confirmed the binding between NPM-ALK and Src both in 293 T-Rex cells (Figure 3A) and in lymphoid cell lines (Figure 4A). We took advantage of Src, Yes1, and Fyn triple knock-out fibroblasts to elucidate a potential role of Src kinases in NPM-ALK–mediated p130Cas phosphorylation. NPM-ALK could induce p130Cas phosphorylation even in the absence of Src, thus demonstrating the presence of a Src-independent pathway (Figure 4B). In addition, no differences in NPM-ALK–dependent p130Cas phosphorylation were found following treatment with the Src family inhibitor PP2 both in MEF NIH3T3 cells and in lymphoid cell lines (Figures 4C-D), thus confirming the results obtained in Src/Yes1/Fyn triple knock-out fibroblasts.
Figure 4.
The phosphorylation of p130Cas by NPM-ALK is independent from Src tyrosine kinase activity. (A) NPM-ALK coprecipitates with Src in lymphoid cells. Total lysates from ALK-positive (Karpas, DHL, TS) and ALK-negative (CEM, Namalwa) cell lines were immunoprecipitated with anti-ALK monoclonal antibody and blotted with anti-Src polyclonal antibody. (B) The NPM-ALK–mediated phosphorylation of p130Cas is Src independent. Syf triple knock-out fibroblasts were cotransfected with Pallino p130Cas and Pallino NPM-ALK or Pallino NPM-ALKK210R as a control. Samples were collected 48 hours after transfection and total lysates were immunoprecipitated with anti-PY20 monoclonal antibody and blotted with the indicated antibodies. (C) Src inhibition does not limit p130Cas phosphorylation. MEF NIH3T3 cells were cotransfected with Pallino p130Cas and Pallino NPM-ALK or Pallino NPM-ALKK210R as a control. Forty-eight hours after transfection, samples were cultivated in the presence of 30 μM PP2 for 1 hour and then collected. The adaptor p130Cas phosphorylation levels were detected by immunoprecipitation with anti-p130Cas monoclonal antibody followed by blotting with anti-PY20 monoclonal antibody. (D) ALK-positive (DHL and TS) and ALK-negative (K562) cells were cultivated in the presence of 30 μM PP2 for 1 hour and then collected. The adaptor p130Cas phosphorylation levels were detected by immunoprecipitation with anti-p130Cas monoclonal antibody followed by blotting with anti-PY20 monoclonal antibody.
NPM-ALK needs the adaptor Grb2 to bind p130Cas
Since p130Cas has no SH2 domains for a direct binding with phosphoproteins,31 we looked for an adaptor protein that could mediate the association between NPM-ALK and p130Cas. Grb2 has previously been shown to bind both NPM-ALK36 and p130Cas37 and we confirmed the association between NPM-ALK and Grb2 in ALK-positive lymphoid cell lines (Figure 5A). Moreover, the R86K-mutated Grb2 has been reported to disrupt the association between p130Cas and Grb2.37 By using 2 dominant-negative forms of Grb2, R86K and P49L, respectively mutated in the SH2 and in the N-terminus SH3 domain, we were able to show that Grb2 was involved in the binding of NPM-ALK to p130Cas. In fact the R86K Grb2 construct was able to disrupt the interaction between NPM-ALK and p130Cas, whereas the P49L mutation had no effect, indicating that the SH2 domain of Grb2 was fundamental for the association between NPM-ALK and p130Cas (Figure 5B). Moreover, as a result of the releasing of NPM-ALK from the complex, p130Cas was no longer phosphorylated (Figure 5C).
Figure 5.
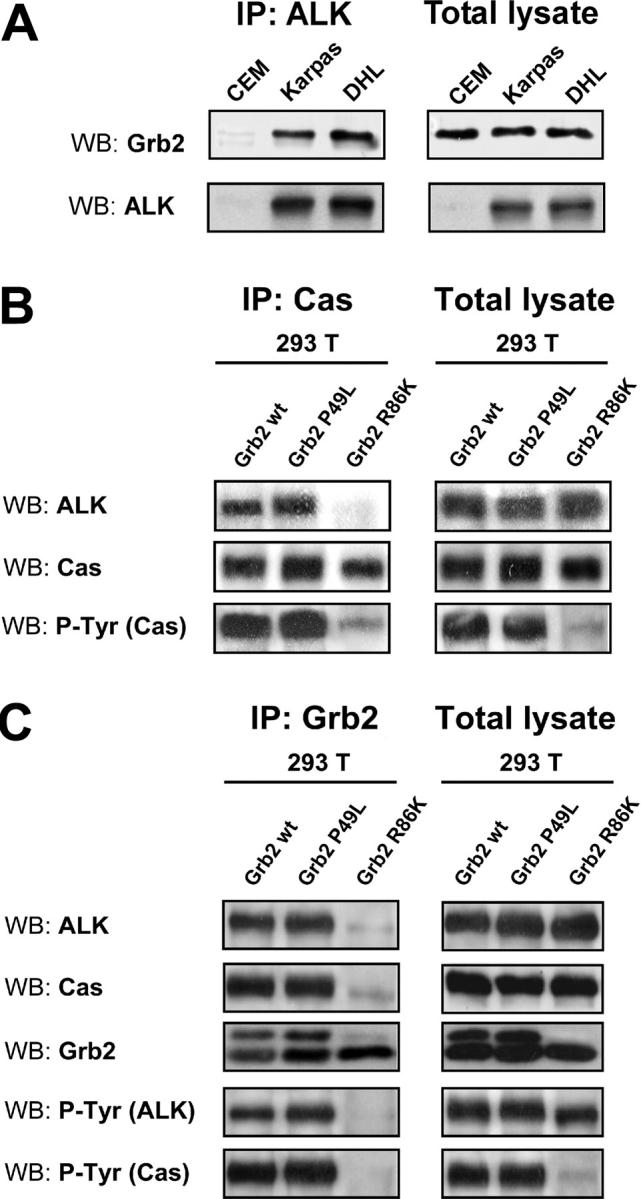
NPM-ALK binds p130Cas through the SH2 domain of Grb2. (A) NPM-ALK binds Grb2 in lymphoid cell lines. Total lysates were immunoprecipitated with anti-ALK monoclonal antibody and blotted with the indicated antibodies. (B-C) The 293 T cells were transfected with Pallino NPM-ALK, Pallino p130Cas, pRK5 Grb2, and the dominant-negative Grb2 constructs pRK5 P49L (mutated in the SH3 domain) or pRK5 R86K (mutated in the SH2 domain) as indicated. Samples were collected 48 hours after transfection and total lysates were immunoprecipitated with anti-Grb2 polyclonal antibody and blotted with the indicated antibodies. Grb2 R86K was able to disrupt the binding of p130Cas to NPM-ALK (B) as well as p130Cas phosphorylation (C).
p130Cas is required for NPM-ALK–mediated actin remodeling and cell transformation
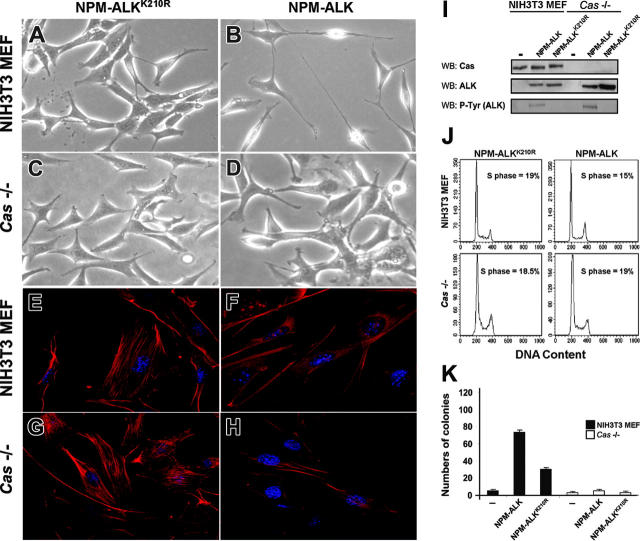
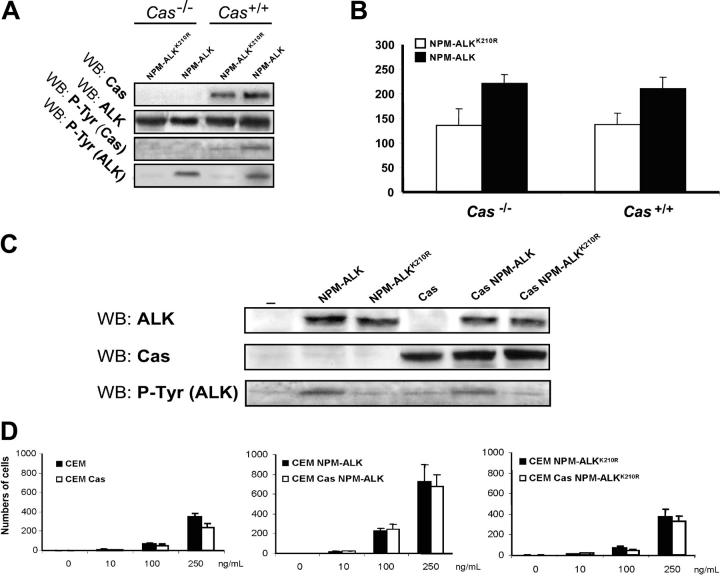
Regarding the morphologic features, we asked if p130Cas could influence NPM-ALK–induced cytoskeleton modifications. We showed that the expression of NPM-ALK in NIH3T3 MEF cells led to spindle-shaped morphology and the outgrowth of elongated processes (Figure 6A-B), but both of the effects were not evident in Cas-/- MEFs (Figure 6C-D). In immunofluorescence, NIH3T3 MEFs expressing NPM-ALK displayed a clear loss of the actin structure (Figure 6F) in contrast to Cas-/- fibroblasts in which the actin filaments were still detectable (Figure 6H), thus suggesting an important role of p130Cas in NPM-ALK–mediated actin filament remodeling.
Figure 6.
The adaptor p130Cas is required for NPM-ALK–mediated transformation and actin filaments organization. NIH3T3 MEFs and Cas-/- MEFs were infected with Pallino NPM-ALK or Pallino NPM-ALKK210R retroviruses and sorted to obtain greater than 95% GFP-positive cells. (A-B) The expression of NPM-ALK in NIH3T3 MEFs led to spindle-shape morphology and to the outgrowth of cellular processes. NPM-ALKK210R was used as a control. (C-D) Cas-/- cells infected with Pallino NPM-ALK did not show evident morphologic differences in comparison with the control NPM-ALKK210R. Phase-contrast images acquired with a 40×/0.55 objective lens. (E-H) Cas-/- fibroblasts expressing NPM-ALK retain the organization of the actin cytoskeleton. Compared with NPM-ALK NIH3T3 MEFs that undergo an evident loss of the actin structure (F), NPM-ALK Cas-/- fibroblasts show a phenotype resembling Cas-/- infected with the control NPM-ALKK210R (G-H). Cells were incubated with PE-conjugated phalloidin to stain the actin filaments (red). Images were taken with the Leica confocal microscope equipped with a 63×/1.32 oil immersion objective lens. (I) Protein expression levels were analyzes by WB as indicated. (J) Cell-cycle analysis was performed by DNA content evaluation on cells in logarithmic growth phase. (K) NIH3T3 and Cas-/- MEFs, infected as indicated, were plated in soft agar and cultured for 3 weeks. The histograms represent the average numbers of colonies from the indicated cells and constructs. Data are from 1 of 3 independent experiments, each including triplicates for experimental point.
Next, we investigated the functional role of p130Cas in some NPM-ALK–mediated biologic effects, such as cell transformation and migration. First, we tested the colony-forming ability of NPM-ALK–expressing cells by soft-agar assay. We infected NIH3T3 MEFs and Cas-/- MEFs with retroviruses expressing NPM-ALK and NPM-ALKK210R as control (Figure 6I). NPM-ALK was efficient in transforming NIH3T3 MEFs as expected but failed to transform Cas-/- fibroblasts, thus underlining the importance of p130Cas in mediating the transforming properties of NPM-ALK (Figure 6K).
To exclude the possibility of an intrinsic proliferative defect of Cas-/- fibroblasts, we performed a cell-cycle analysis on nonconfluent MEFs in logarithmic growth phase and found no proliferative differences between Cas-/- cells and NIH3T3 MEFs (Figure 6J). In addition, we rescued Cas-/- fibroblasts by retroviral infection with Pallino p130Cas. In soft-agar assay the ectopic hyperexpression of p130Cas was sufficient to transform Cas-/- fibroblasts (data not shown) in accordance to data previously described.38
Finally, we asked whether the overexpression of p130Cas could increase the cellular migration rate. For this purpose, we infected both Cas-/- and Cas+/+ fibroblasts with Pallino NPM-ALK retrovirus (Figure 7A), sorted them for green fluorescent protein (GFP) content, and then performed a Matrigel migration assay. The expression of NPM-ALK was sufficient to increase cell migration as expected, although p130Cas expression did not provide an additional effect on cell migration (Figure 7B). Nevertheless, we also investigated the role of p130Cas in lymphoid cells migration; we overexpressed p130Cas together with either NPM-ALK or NPM-ALKK210R in CEM cells via retroviral infection, and GFP-positive cells were enriched by cell sorting. Despite the strong expression levels of p130Cas and NPM-ALK obtained in CEM cells (Figure 7C) in a chemotaxis assay with SDF-1α, the synergistic effect of SDF-1α and NPM-ALK on cell migration was not further increased by the overexpression of p130Cas, thus suggesting that p130Cas could not be a fundamental player in this process (Figure 7D).
Figure 7.
The p130Cas is not required for NPM-ALK–mediated migration. (A) Cas-/- and Cas+/+ rescued fibroblasts were infected with Pallino NPM-ALK retrovirus and then sorted for GFP content. Protein expression levels were verified by WB as indicated. (B) Cells were plated on a Matrigel-coated insert and the number of migrated cells was evaluated after 48 hours. The histograms represent the average number of migrated cells from 3 independent experiments using triplicate wells for experimental point. (C) CEM lymphoblastoid cells were infected with Pallino retroviruses containing NPM-ALK or NPM-ALKK210R together with retrovirus for p130Cas. Protein expression levels were verified by WB as indicated. (D) The histograms represent the average numbers of migrated cells in response to increasing concentrations of SDF-1α as indicated. Data are from 1 of 2 independent experiments. Error bars indicate standard deviation. *Statistically significant analysis as measured by the Student t test.
Discussion
Many canonic steps of malignant transformation in which the fusion protein NPM-ALK is directly involved have been fairly well described and include increased mitogenity, protection from apoptosis, and growth in the absence of adhesion or growth factors.10,39,40 The molecular mechanisms underlying these processes are similar to those employed by other tyrosine kinases and rely on the activation of signaling molecules such as PI3K, PLC-γ, Stat3, and MAPK.15-17,39,41
In line with these oncogenic properties, NPM-ALK is also capable of changing cell morphology in a similar fashion to other well-known oncogenes. In fibroblasts, NPM-ALK induces an elongated and birifrangent morphology and in lymphoid cells is able to increase the size of the cells as well as to drive anaplastic features.19
In the present study we started from a proteomic approach to identify molecules involved in these effects induced by NPM-ALK. By mass spectrometry on 293 T-Rex cells expressing a tetracycline-inducible form of NPM-ALK, we identified some cytoskeleton-associated proteins that precipitate or are phosphorylated as an effect of the tyrosine kinase activity of NPM-ALK. Interestingly, similar proteins, such as beta actin and tubulin, have also been found to be phosphorylated in anaplastic lymphoma cell lines (Karpas and DHL) with a recently described method of immunoaffinity profiling of tyrosine phosphorylation,30 thus rendering our approach based on inducible 293 T-Rex cells a reliable method. Together with structural proteins, in the MALDI analysis of precipitated bands we found p130Cas, a 130-kilodalton (kDa) phosphotyrosine (pTyr)–scaffolding protein that normally associates with 2 oncoproteins, pp60v-src (v-Src) and p47gag-crk (v-Crk).35,42
The fundamental role of p130Cas in actin cytoskeleton remodeling and cell migration has been suggested by several lines of evidence including its phosphorylation following integrin engagement and its presence in the focal adhesions that form the molecular bridges between the extracellular matrix and the actin cytoskeleton.43 In agreement with these functions, Cas-/- fibroblasts show normal focal adhesion formation but have defects in cell migration.38,44 In these events, the phosphorylation of p130Cas is mediated by FAK, Src, and protein tyrosine kinase 2β (Pyk2) in an integrin-dependent fashion.45,46
Here we show that p130Cas precipitates with the oncogenic tyrosine kinase NPM-ALK both in 293 T-Rex cells and in human-derived anaplastic lymphoma cells Karpas, DHL, and TS. In addition, p130Cas is phosphorylated when NPM-ALK is expressed and this phosphorylation is strictly dependent on the intrinsic tyrosine kinase activity of ALK since a kinase-dead mutant is no longer able to bind and phosphorylate p130Cas. Similarly to NPM-ALK, the ATIC-ALK fusion protein and the full-length ALK receptor phosphorylate p130Cas, thus excluding an essential role of the NPM portion of the molecule and indicating a general mechanism most likely shared by all ALK fusion proteins. Therefore, these findings add ALK to other tyrosine kinase receptors known to phosphorylate p130Cas such as EGF, fibroblast growth factor (FGF), insulin-like growth factor 1 (IGF-1), nerve growth factor (NGF), and platelet-derived growth factor (PDGF)31 and, more interestingly, are comparable to another fusion protein involved in hematopoietic malignancies, break point cluster region–Abelson (Bcr-Abl), which was demonstrated to bind and phosphorylate p130Cas in Bcr-Abl–positive cell lines and in samples obtained from chronic myeloid leukemia (CML) and acute lymphoblastic leukemia (ALL) patients.47,48
The phosphorylation of p130Cas at a molecular level has been shown in vitro to be mediated by 4 different kinases (ie, Src, Fak, Pyk2, and Abl). Since it has recently been described that Src kinase can bind to and be activated by NPM-ALK,34 we considered the possibility that p130Cas phosphorylation by NPM-ALK required Src as intermediate kinase. However, in triple knock-out Src, Yes1, and Fyn cells NPM-ALK was still able to phosphorylate p130Cas at least to some extent and p130Cas phosphorylation was still detectable both in MEF NIH3T3 fibroblasts and in lymphoid cell lines even after the treatment with the Src-family inhibitor PP2, thus indicating the existence of a Src-independent pathway in p130Cas activation. This pathway could pass through the adaptor protein Grb2, a molecule previously known to bind both NPM-ALK36 and p130Cas.37 Indeed, we show in this paper that in 293 T-Rex cells, an SH2 Grb2 mutant, acting as a dominant-negative, impaired both the NPM-ALK binding to and the phosphorylation of p130Cas, thus revealing Grb2 as a key regulator of this interaction.
The role of p130Cas in ALK-mediated cytoskeleton organization was evident by studying the polymerization of the actin filaments. Deregulated ALK kinase activity forces cells to acquire a transformed phenotype with reduced adherence to the plate and a spindle, more birifrangent shape. In addition, ALK-transformed MEFs show long processes, in a way similar to the described neurite outgrowth induced by ALK in PC12 cells,8 indicative of a direct role of ALK in cytoskeleton organization. Indeed, ALK-transformed MEFs showed a prominent actin depolymerization similar to that induced by other oncogenic fusion proteins such as Bcr-Abl.49 Interestingly, these effects were clearly impaired in Cas-/- MEFs, indicating an essential role of p130Cas in ALK-mediated actin depolymerization. It still remains to be clarified if p130Cas could be involved also in the cellular shaping of NPM-ALK–expressing lymphoid cells.
Besides the functions on organizing the cell cytoskeleton, p130Cas is involved in broader oncogenic effects that include cell proliferation and apoptosis, indicating p130Cas as a major player in the complex network that is connecting cell adhesion and migration to cell growth and survival.31 Indeed, p130Cas overexpression is sufficient to induce an anchorage-independent growth in cells,38,50 possibly through the activation of the c-Jun N-terminal kinase (JNK)/c-jun pathway,51 as well as protection from apoptosis.31 In this study we tested the role of p130Cas in NPM-ALK–mediated transformation, showing that Cas-/- fibroblasts can no longer be transformed by NPM-ALK. These effects on anchorage-independent growth could derive from a combination of cytoskeleton modifications and proliferative signals both provided by NPM-ALK in wt MEFs but impaired in the absence of p130Cas. Overall the data are suggesting that NPM-ALK could control a Src-dependent and a Src-independent transformation pathway that both use p130Cas as pivotal adaptor. The Src-independent pathway may require Grb2 as key adaptor molecule, as previously discussed. In this scenario, it is possible to further speculate that NPM-ALK activates both Src and p130Cas, where phosphorylated p130Cas may further enhance Src activity in a sort of activating loop similar to what has already been described in other cancers.52
In this paper, we also describe a novel function of NPM-ALK (ie, its capability of increasing the migration rate of T lymphocytes). CEM lymphoblastoid cells have previously been proven to migrate in a gradient of SDF-1α, which is the ligand for CXCR4, a chemokine receptor expressed in T cells.29 Upon retroviral expression of NPM-ALK, CEM cells almost doubled their migration rate, indicating that NPM-ALK tyrosine kinase activity influences the activation state of molecules involved in this process. However, in contrast to transformation, we could not find a clear effect of p130Cas overexpression in NPM-ALK–induced migration. Regarding lymphoid cells, this could be explained by considering that in T lymphocytes a major role in beta1 integrin– and T-cell receptor (TCR)–mediated migration has been shown for Cas-L, a p105-kDa protein that is a member of the Cas family selectively expressed in lymphocytes.32,33 Indeed, we showed that NPM-ALK is capable of phosphorylating Cas-L in lymphocytes. It is therefore possible that NPM-ALK in lymphocytes may promote migration through the activation of different molecules than p130Cas. Further studies in lymphocytes are needed to solve this issue.
In conclusion, in this study we show p130Cas as a novel downstream interactor of NPM-ALK capable of modulating its transforming properties. This role could involve not only the p130Cas effects on cytoskeleton remodeling but also its involvement in important proliferative and survival pathways. Further studies are required to unravel this differential requirement of ALK for p130Cas or its family members in different cell subtypes.
Prepublished online as Blood First Edition Paper, August 16, 2005; DOI 10.1182/blood-2005-03-1204.
Supported by National Institutes of Health (NIH) R01-CA64033, Ministero dell'Università e Ricerca Scientifica (MIUR), Compagnia di San Paolo, Torino (Progetto Oncologia), Regione Piemonte (Ricerca Sanitaria Finalizzata and Ricerca Scientifica), and Associazione Italiana per la Ricerca sul Cancro (AIRC).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Morris SW, Kirstein MN, Valentine MB, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263: 1281-1284. [DOI] [PubMed] [Google Scholar]
- 2.Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56: 379-390. [DOI] [PubMed] [Google Scholar]
- 3.Okuda M. The role of nucleophosmin in centrosome duplication. Oncogene. 2002;21: 6170-6174. [DOI] [PubMed] [Google Scholar]
- 4.Kurki S, Peltonen K, Latonen L, et al. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5: 465-475. [DOI] [PubMed] [Google Scholar]
- 5.Chan PK, Chan FY. Nucleophosmin/B23 (NPM) oligomer is a major and stable entity in HeLa cells. Biochim Biophys Acta. 1995;1262: 37-42. [DOI] [PubMed] [Google Scholar]
- 6.Morris SW, Naeve C, Mathew P, et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin's lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene. 1997;14: 2175-2188. [DOI] [PubMed] [Google Scholar]
- 7.Iwahara T, Fujimoto J, Wen D, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14: 439-449. [DOI] [PubMed] [Google Scholar]
- 8.Souttou B, Carvalho NB, Raulais D, Vigny M. Activation of anaplastic lymphoma kinase receptor tyrosine kinase induces neuronal differentiation through the mitogen-activated protein kinase pathway. J Biol Chem. 2001;276: 9526-9531. [DOI] [PubMed] [Google Scholar]
- 9.Liao EH, Hung W, Abrams B, Zhen M. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 2004;430: 345-350. [DOI] [PubMed] [Google Scholar]
- 10.Wellmann A, Doseeva V, Butscher W, et al. The activated anaplastic lymphoma kinase increases cellular proliferation and oncogene up-regulation in rat 1a fibroblasts. FASEB J. 1997;11: 965-972. [DOI] [PubMed] [Google Scholar]
- 11.Ma Z, Cools J, Marynen P, et al. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95: 2144-2149. [PubMed] [Google Scholar]
- 12.Kuefer MU, Look AT, Pulford K, et al. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90: 2901-2910. [PubMed] [Google Scholar]
- 13.Chiarle R, Gong JZ, Guasparri I, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101: 1919-1927. [DOI] [PubMed] [Google Scholar]
- 14.Fujimoto J, Shiota M, Iwahara T, et al. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5). Proc Natl Acad Sci U S A. 1996;93: 4181-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai RY, Dieter P, Peschel C, Morris SW, Duyster J. Nucleophosmin-anaplastic lymphoma kinase of large-cell anaplastic lymphoma is a constitutively active tyrosine kinase that utilizes phospholipase C-gamma to mediate its mitogenicity. Mol Cell Biol. 1998;18: 6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zamo A, Chiarle R, Piva R, et al. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects haematopoietic cells from cell death. Oncogene. 2002;21: 1038-1047. [DOI] [PubMed] [Google Scholar]
- 17.Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signalling pathway. Blood. 2000;96: 4319-4327. [PubMed] [Google Scholar]
- 18.Gu TL, Tothova Z, Scheijen B, Griffin JD, Gilliland DG, Sternberg DW. NPM-ALK fusion kinase of anaplastic large-cell lymphoma regulates survival and proliferative signalling through modulation of FOXO3a. Blood. 2004;103: 4622-4629. [DOI] [PubMed] [Google Scholar]
- 19.Horie R, Watanabe M, Ishida T, et al. The NPM-ALK oncoprotein abrogates CD30 signalling and constitutive NF-kappaB activation in anaplastic large cell lymphoma. Cancer Cell. 2004;5: 353-364. [DOI] [PubMed] [Google Scholar]
- 20.Lee HH, Norris A, Weiss JB, Frasch M. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature. 2003;425: 507-512. [DOI] [PubMed] [Google Scholar]
- 21.Englund C, Loren CE, Grabbe C, et al. Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature. 2003;425: 512-516. [DOI] [PubMed] [Google Scholar]
- 22.Zettervall CJ, Anderl I, Williams MJ, et al. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci U S A. 2004;101: 14192-14197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77: 8957-8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matrix Science. Mascot: Peptide Mass Fingerprint. http://www.matrixscience.com/search_form_select.html. Accessed June 5, 2004.
- 25.National Center for Biotechnology Information. Nonredundant protein database. Available at http://www.ncbi.nlm.nih.gov/BLAST. Accessed June 10, 2004.
- 26.Grignani F, Kinsella T, Mencarelli A, et al. High-efficiency gene transfer and selection of human haematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 1998;58: 14-19. [PubMed] [Google Scholar]
- 27.Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol. 2000;2: E231-E236. [DOI] [PubMed] [Google Scholar]
- 28.Turner CE. Paxillin interactions. J Cell Sci. 2000;113(Pt 23): 4139-4140. [DOI] [PubMed] [Google Scholar]
- 29.Rabin RL, Park MK, Liao F, Swofford R, Stephany D, Farber JM. Chemokine receptor responses on T cells are achieved through regulation of both receptor expression and signalling. J Immunol. 1999;162: 3840-3850. [PubMed] [Google Scholar]
- 30.Rush J, Moritz A, Lee KA, et al. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat Biotechnol. 2005;23: 94-101. [DOI] [PubMed] [Google Scholar]
- 31.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene. 2001;20: 6448-6458. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi Y, Iwata S, Kamiguchi K, Morimoto C. Tyrosine phosphorylation of Crk-associated substrate lymphocyte-type is a critical element in TCR- and beta 1 integrin-induced T lymphocyte migration. J Immunol. 1999;163: 3727-3734. [PubMed] [Google Scholar]
- 33.van Seventer GA, Salmen HJ, Law SF, et al. Focal adhesion kinase regulates beta1 integrin-dependent T cell migration through an HEF1 effector pathway. Eur J Immunol. 2001;31: 1417-1427. [DOI] [PubMed] [Google Scholar]
- 34.Cussac D, Greenland C, Roche S, et al. Nucleophosmin-anaplastic lymphoma kinase of anaplastic large-cell lymphoma recruits, activates, and uses pp60c-src to mediate its mitogenicity. Blood. 2004;103: 1464-1471. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda M, Mayer BJ, Fukui Y, Hanafusa H. Binding of transforming protein, P47gag-crk, to a broad range of phosphotyrosine-containing proteins. Science. 1990;248: 1537-1539. [DOI] [PubMed] [Google Scholar]
- 36.Crockett DK, Lin Z, Elenitoba-Johnson KS, Lim MS. Identification of NPM-ALK interacting proteins by tandem mass spectrometry. Oncogene. 2004;23: 2617-2629. [DOI] [PubMed] [Google Scholar]
- 37.Hakak Y, Martin GS. Cas mediates transcriptional activation of the serum response element by Src. Mol Cell Biol. 1999;19: 6953-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Honda H, Nakamoto T, Sakai R, Hirai H. p130(Cas), an assembling molecule of actin filaments, promotes cell movement, cell migration, and cell spreading in fibroblasts. Biochem Biophys Res Commun. 1999;262: 25-30. [DOI] [PubMed] [Google Scholar]
- 39.Slupianek A, Nieborowska-Skorska M, Hoser G, et al. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 2001;61: 2194-2199. [PubMed] [Google Scholar]
- 40.Armstrong F, Duplantier MM, Trempat P, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene. 2004;23: 6071-6082. [DOI] [PubMed] [Google Scholar]
- 41.Turner SD, Tooze R, Maclennan K, Alexander DR. Vav-promoter regulated oncogenic fusion protein NPM-ALK in transgenic mice causes B-cell lymphomas with hyperactive Jun kinase. Oncogene. 2003;22: 7750-7761. [DOI] [PubMed] [Google Scholar]
- 42.Reynolds AB, Roesel DJ, Kanner SB, Parsons JT. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol. 1989;9: 629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harte MT, Hildebrand JD, Burnham MR, Bouton AH, Parsons JT. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J Biol Chem. 1996;271: 13649-13655. [DOI] [PubMed] [Google Scholar]
- 44.Honda H, Oda H, Nakamoto T, et al. Cardiovascular anomaly, impaired actin bundling and resistance to Src-induced transformation in mice lacking p130Cas. Nat Genet. 1998;19: 361-365. [DOI] [PubMed] [Google Scholar]
- 45.Schlaepfer DD, Hunter T. Focal adhesion kinase overexpression enhances ras-dependent integrin signalling to ERK2/mitogen-activated protein kinase through interactions with and activation of c-Src. J Biol Chem. 1997;272: 13189-13195. [DOI] [PubMed] [Google Scholar]
- 46.Schlaepfer DD, Broome MA, Hunter T. Fibronectin-stimulated signalling from a focal adhesion kinase-c-Src complex: involvement of the Grb2, p130cas, and Nck adaptor proteins. Mol Cell Biol. 1997;17: 1702-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Jong R, van Wijk A, Haataja L, Heisterkamp N, Groffen J. BCR/ABL-induced leukemogenesis causes phosphorylation of Hef1 and its association with Crkl. J Biol Chem. 1997;272: 32649-32655. [DOI] [PubMed] [Google Scholar]
- 48.Salgia R, Pisick E, Sattler M, et al. p130CAS forms a signalling complex with the adapter protein CRKL in haematopoietic cells transformed by the BCR/ABL oncogene. J Biol Chem. 1996;271: 25198-25203. [DOI] [PubMed] [Google Scholar]
- 49.Cheng K, Kurzrock R, Qiu X, et al. Reduced focal adhesion kinase and paxillin phosphorylation in BCR-ABL-transfected cells. Cancer. 2002;95: 440-450. [DOI] [PubMed] [Google Scholar]
- 50.Burnham MR, Bruce-Staskal PJ, Harte MT, et al. Regulation of c-SRC activity and function by the adapter protein CAS. Mol Cell Biol. 2000;20: 5865-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oktay M, Wary KK, Dans M, Birge RB, Giancotti FG. Integrin-mediated activation of focal adhesion kinase is required for signalling to Jun NH2-terminal kinase and progression through the G1 phase of the cell cycle. J Cell Biol. 1999;145: 1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burnham MR, Harte MT, Richardson A, Parsons JT, Bouton AH. The identification of p130cas-binding proteins and their role in cellular transformation. Oncogene. 1996;12: 2467-2472. [PubMed] [Google Scholar]
- 53.Piva R, Chiarle R, Manzanna A, et al. Ablation of oncogenic ALK is a viable therapeutic approach for anaplastic large-cell lymphomas. Blood. 2005; in press. [DOI] [PMC free article] [PubMed]