Abstract
We have previously shown that the interleukin 10 (IL-10)/-592*A allele of the recipient is associated with less severe acute graft-versus-host disease (GVHD) and a lower risk of nonrelapse mortality after hematopoietic cell transplantation (HCT) from an HLA-identical sibling. In the present study, we examined variation in the IL-10 receptor β gene as a further test of the hypothesis that the IL-10 pathway regulates the risk of acute GVHD. A single nucleotide polymorphism (A/G) at cDNA position 238 of the IL-10 receptor β gene (IL10RB/c238) was genotyped in 953 HC transplant recipients and their HLA-identical sibling donors. IL-10/-592 and IL10RB/c238 genotypes were tested for association with GVHD by multivariable analysis. The IL-10/-592*A allele of the recipient and IL10RB/c238*G allele of the donor were significantly associated with a lower risk of grades III-IV acute GVHD (trend P < .001 and P = .02, respectively). The donor IL10RB/c238*G allele provided protection among patients with the IL-10/-592 A/C or A/A genotypes but not among patients with the high-risk IL-10/-592 C/C genotype. These data suggest an interaction of the patient IL-10/-592 and donor IL10RB/c238 genotypes on risk of GVHD, further supporting the hypothesis that the IL-10 pathway plays an important role in controlling the severity of acute GVHD.
Introduction
Outcome of allogeneic hematopoietic cell transplantation (HCT) is affected by several variables including disease and disease status, patient and donor age, and patient and donor HLA matching.1-3 In addition to HLA matching, genetic diversity among patients and donors may contribute to differences in individual responses to tissue injury, inflammation, and severity of acute graft-versus-host disease (GVHD).4,5 Previous studies have implicated polymorphisms in cytokine genes as factors affecting GVHD and survival.6-12 Variation in the promoter region of the interleukin 10 (IL-10) gene has been associated with risk of GVHD6,9,11 and risk of transplant-related mortality.11,12 IL-10 is a potent suppressor of tumor necrosis factor α (TNFα), IL-1α, IL-1β, IL-6, IL-12, and interferon γ (IFNγ) production.13 IL-10 downregulates the expression of major histocompatibility complex (MHC) and costimulatory molecules and by these mechanisms may attenuate alloreactive T-cell responses.14,15 IL-10 is also an important effector molecule for CD4+CD25+ T-regulator cells and has been shown to enhance the generation of type I regulatory T cells that mediate anergy and facilitate tolerance.16-19
We have previously shown that a single-nucleotide polymorphism (SNP) at position -592 in the promoter region of the IL-10 gene was predictive of outcome following transplantation of hematopoietic cells from an HLA-identical sibling.11 The IL-10/-592*A allele of the recipient was associated with less severe GHVD and improved survival. We have extended our study of the IL-10 pathway to an analysis of a polymorphism in the gene encoding the IL-10 receptor β chain (IL10RB). The IL-10RB SNP maps to position 238 in the IL10RB cDNA (IL10RB/c238) and results in a change from nucleotide A to G at codon 47 (cDNA sequence first described by Lutfalla et al52), corresponding to an amino acid change from Lys to Glu.20 In the current study, a synergistic effect was observed between the IL-10/-592 genotype of the patient and the IL10RB/c238 genotype of the donor. The presence of the IL-10/-592*A allele in the patient and the IL10RB/238*G allele in the donor was associated with a significantly reduced risk of severe acute GVHD.
Patients, materials, and methods
Study populations
The study population has been previously genotyped for SNPs in the IL-10 gene.11 Briefly, DNA samples were available for 953 patients who received T-cell–replete grafts from an HLA-identical sibling. Inclusion criteria were the availability of pretransplantation blood samples, the use of methotrexate and cyclosporine for prophylaxis against GVHD, and the availability of acute GVHD grading scores before this study began. The ethnicity of the patients and their sibling donors was self-described as 814 white, 24 Asian, 17 African American, 46 Hispanic, 19 Native American, and 33 of other or unknown origin (Table 1). In this cohort, the incidence of severe grades III-IV GVHD was lower among Asian (8%) and Hispanic (11%) patients compared with white patients (19%). All patients and donors gave written informed consent according to protocols approved by the Fred Hutchinson Cancer Research Center Institutional Review Board. Healthy controls included 71 volunteer Taiwanese donors who gave consent according to a protocol approved by the National Taiwan University Hospital Institutional Review Board.
Table 1.
Characteristics of transplant recipients and donors, n = 953
| African American | Native American | Asian | White | Hispanic | Other/unknown | Combined | |
|---|---|---|---|---|---|---|---|
| No. | 17 | 19 | 24 | 814 | 46 | 33 | 953 |
| Age, y, median (range) | 36 (4-58) | 38 (1-68) | 42 (8-60) | 39 (16-60) | 37 (9-57) | 36 (3-65) | 38 (1-68) |
| Transplantation year, no. (%) | |||||||
| 1981-1991 | 6 (35) | 5 (28) | 6 (25) | 338 (42) | 9 (20) | 10 (30) | 374 (39) |
| 1992-2000 | 11 (65) | 14 (74) | 18 (75) | 476 (58) | 37 (80) | 23 (70) | 579 (61) |
| Sex, patient/donor, no. (%) | |||||||
| Male/male | 4 (24) | 11 (58) | 7 (29) | 268 (33) | 19 (41) | 13 (29) | 322 (34) |
| Male/female | 6 (35) | 4 (21) | 7 (29) | 199 (24) | 9 (20) | 10 (30) | 235 (25) |
| Female/male | 5 (29) | 1 (5) | 5 (21) | 160 (20) | 9 (20) | 2 (6) | 182 (19) |
| Female/female | 2 (12) | 3 (16) | 5 (21) | 187 (23) | 9 (20) | 8 (24) | 214 (22) |
| Total body irradiation, no. (%) | |||||||
| Yes | 9 (53) | 10 (53) | 16 (67) | 361 (44) | 25 (54) | 16 (48) | 438 (46) |
| No | 8 (47) | 9 (47) | 8 (33) | 453 (56) | 21 (46) | 17 (52) | 515 (54) |
| Disease group, no. (%) | |||||||
| Nonmalignant disease* | 2 (12) | 4 (21) | 5 (21) | 137 (17) | 12 (26) | 7 (21) | 167 (18) |
| Low-risk malignance† | 8 (47) | 8 (42) | 14 (58) | 407 (50) | 24 (52) | 13 (39) | 474 (50) |
| High-risk malignance‡ | 7 (41) | 7 (37) | 5 (21) | 270 (33) | 10 (22) | 13 (39) | 312 (33) |
| Cumulative incidence, % | |||||||
| Acute GVHD§ | 18 | 26 | 8 | 19 | 11 | 15 | 19 |
| Chronic GVHD∥ | 41 | 53 | 42 | 41 | 35 | 27 | 40 |
| Nonrelapse mortality∥ | 24 | 37 | 21 | 25 | 28 | 21 | 25 |
| Overall survival∥ | 41 | 58 | 71 | 57 | 56 | 61 | 57 |
Patients received a myeloablative hematopoietic cell transplant from an HLA genetically identical sibling donor. This population included 33 patients of other or unknown origin.
Nonmalignant disease includes aplastic anemia, myelodysplastic syndrome, and paroxysmal nocturnal hematuria
Low-risk malignancy includes acute lymphoblastic leukemia (ALL), acute myelogenous leukemia (AML), or non-Hodgkin lymphoma (NHL) in remission and chronic myelogenous leukemia (CML) in chronic phase
High-risk malignancy includes ALL, AML, chronic lymphocytic leukemia, or NHL in relapse, CML in other than chronic phase, multiple myeloma, and Hodgkin disease
Cumulative incidence rates of grades III-IV acute GVHD at day 100
Cumulative incidence rates of chronic extensive GVHD, nonrelapse mortality, and survival at 3 years
Genotyping
The strategy and design of mismatched polymerase chain reaction and restriction fragment-length polymorphism (PCR/RFLP) has been described previously.21,22 The IL10RB/c238 genotype was determined by an XcmI-based mismatched PCR/RFLP assay using the forward primer IL10RB2fXcm (5′-ttg tct taC CAt cat agc att gg-3′) and the reverse primer IL10RB2rXcmI (5′-tga gct gtg aaa gCc agg ttc cct-3′) for amplification. The “CCA” nucleotides in the forward primer incorporate a common XcmI recognition motif in the amplicon amplified from either the c238 A allele or G allele. This provides a convenient internal control for endonuclease restriction reactions. The “C” nucleotide in the reverse primer incorporates an XcmI recognition motif in the presence of c238 A allele.
PCR reactions were performed in a 10 μL total volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.2 mM deoxyribonucleotides, 4 pmol of each primer, 2.0 mM MgCl2, and 0.25 units AmpliTaq Gold DNA polymerase (Applied Biosystem, Foster City, CA). Samples containing 1 to 5 ng DNA were subjected to 40 cycles of denaturation (94°C, 30 s), annealing (54°C, 30 s), and elongation (72°C, 60 s). In a total volume of 9 μL, 2 μL of the IL10RB amplicon was digested with 3 units of restriction endonuclease XcmI (New England Biolabs, Beverly, MA) in buffer 2 at 37°C for overnight reaction and then analyzed by electrophoresis in 3% agarose gel. The presence of a 133-bp fragment indicates incomplete digestion. The restricted pattern was 118-bp and 15-bp fragments for the IL10RB/C238 G/G genotype; 118-bp, 98-bp, 20-bp, and 15-bp fragments for the A/G genotype; and 98-bp, 20-bp, and 15-bp fragments for the A/A genotype.
To control for reagent specificity of mismatched PCR/RFLP assay, 24 DNA samples from the International Histocompatibility Working Group (IHWG) cytokine gene polymorphism reference panel23 were tested by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry (Sequenom, San Diego, CA).24,25 These 24 DNA samples were unrelated parents from Centre d'Étude du Polymorphisme Humain (CEPH) families. Briefly, uniplex PCR was carried out by the following PCR primers: 5′-acg ttg gat gca ttc tac agt ggg agt cac-3′ and 5′-acg ttg gat gac ctt agg tac tga gct gtg-3′. After primer extension, the purified DNA fragments were spotted onto a 384-element silicon chip and analyzed in the Bruker Biflex III MALDI-TOF SpectroREADER mass spectrometer (Sequenom), and the resulting spectra were processed with SpectroTYPER (Sequenom). Genotyping for IL10RB/c238 was also determined among 71 unrelated Taiwanese volunteers by using MALDI-TOF mass spectrometry.
Analysis of clinical outcomes
Acute and chronic GHVD were diagnosed and graded according to standard criteria.26,27 Nonrelapse mortality was defined as death before recurrent malignancy. Cumulative incidence of acute GVHD, chronic GVHD, and nonrelapse mortality were estimated according to the methods of Andersen et al.28 Death was considered to be a competing risk for analysis of acute and chronic GVHD, and recurrent malignancy was considered to be a competing risk for analysis of chronic GVHD and nonrelapse mortality.
Statistical analysis
The associations of SNP-defined IL-10 and IL10RB genotypes with clinical outcome were evaluated with proportional hazards regression models, adjusting for known risk factors. Outcomes were censored at the time of a competing event. Analysis of acute GHVD was adjusted for patient age at transplantation (continuous), donor-recipient sex mismatch, use of total body irradiation in the conditioning regimen, and disease risk group. Transplantation year (1981-1991 vs 1992-2000) was included in the multivariable analysis of acute GVHD because the sensitivity for diagnosing gut GVHD increased after 1991. Patients received either marrow (n = 817) or growth factor–mobilized blood cells (n = 136). This variable did not affect risk of grades III-IV acute GVHD and therefore was not included in the multivariable analysis. Analysis of extensive chronic GVHD was adjusted for the same risk factors, except for transplantation year. Analysis of nonrelapse mortality was adjusted for patient age at transplantation, time from diagnosis to transplantation (continuous), and disease risk group.
All P values are 2-sided and derived from likelihood ratio statistics from the proportional hazards regression models. Hazard ratios for the M/m and m/m genotypes were compared with the M/M genotype, the designated reference group, where “M” represents the more frequent (major) allele and “m” represents the less frequent (minor) allele in the studied population. Tests for trend were carried out by assigning the ordinal values 1, 2, and 3 to the genotypes M/M, M/m, and m/m, respectively, and testing the association of the resulting variable with outcome.
Results
Association of IL-10 and IL10RB genetic variation with clinical outcome
As previously reported, the IL-10/-592*A allele of the patient was significantly associated with a lower incidence of severe acute GVHD (trend P < .001; Table 2).11 The incidence rates of grades III-IV GVHD for the C/C, A/C, and A/A genotypes were 22%, 16%, and 10%, respectively. The donor IL-10/-592*A allele was also significantly associated with a lower incidence of severe acute GVHD (trend P = .03). After adjustment for the patient IL-10/-592 genotype, however, the donor genotype association with severe acute GVHD was no longer significant (trend P = .71).
Table 2.
Association of IL-10/-592 and IL10RB/c238 genotypes with grades III-IV acute GVHD
| Genotype (number) | Incidence of GVHD, % | Hazard ratio | Trend P* |
|---|---|---|---|
| Patient genotype | |||
| IL-10/-592 | <.001 | ||
| C/C (504) | 22 | Reference | |
| A/C (368) | 16 | 0.7 (0.5-0.9) | |
| A/A (81) | 10 | 0.4 (0.2-0.8) | |
| IL10RB/c238 | .31 | ||
| A/A (494) | 21 | Reference | |
| A/G (385) | 16 | 0.8 (0.6-1.0) | |
| G/G (74) | 20 | 1.0 (0.6-1.7) | |
| Donor genotype | |||
| IL-10/-592 | .03 | ||
| C/C (492) | 21 | Reference | |
| A/C (371) | 17 | 0.8 (0.6-1.1) | |
| A/A (90) | 13 | 0.6 (0.3-1.0) | |
| IL10RB/c238 | .02 | ||
| A/A (515) | 21 | Reference | |
| A/G (356) | 17 | 0.8 (0.5-1.0) | |
| G/G (82) | 12 | 0.6 (0.3-1.1) |
Multivariable proportional hazard regression analysis adjusted for age at transplantation, transplantation year, sex mismatch, use of total body irradiation in the conditioning regimen, and disease risk group
The donor IL10RB/c238 genotype was also significantly associated with risk of severe acute GVHD (trend P = .02; Table 2). The incidence rates of grades III-IV GVHD for the A/A, A/G, and G/G genotypes were 21%, 17%, and 12%, respectively. There was no significant association between acute GVHD and IL10RB genotype of the patient, and no significant association between donor IL10RB genotype and risk of extensive chronic GVHD or nonrelapse mortality. Cumulative incidence rates of extensive chronic GVHD at 3 years for recipients with the donor IL10RB A/A, A/G, and G/G genotypes were 41%, 38%, and 44%, respectively, and the cumulative incidence rates of nonrelapse mortality at 3 years for recipients with the donor IL10RB A/A, A/G, and G/G genotypes were 27%, 22%, and 22%, respectively.
Variable IL-10 and IL10RB gene frequencies in different ethnic groups
IL10RB/c238 genotypes were determined among 24 unrelated white patients by MALDI-TOF mass spectrometry and mismatched PCR/RFLP. The results of IL10RB/c238 genotyping by these 2 assays were identical. The frequency of the G allele among these 24 white patients was 37.5% and the frequency of A allele was 62.5%. In contrast, the frequency of the G allele among the 71 Taiwanese volunteers was 64.1% and the frequency of the A allele was 35.9%. There were also significant differences in IL-10/-592 and IL10RB/c238 SNP-defined allele frequencies among the transplantation patient population according to ethnicity (Table 3). The IL-10/-592*A allele frequency ranged from 24.7% among white patients to 68.8% among Asian patients. The IL10RB/c238*G allele frequency ranged from 25.6% among white patients to 47.9% among Asian patients.
Table 3.
IL-10/-592 and IL10RB/c238 allele and genotype frequencies according to ethnic group
| Allele and genotype frequencies | African American | Native American | Asian | White | Hispanic | Combined population | P* |
|---|---|---|---|---|---|---|---|
| Patients | |||||||
| No. | 17 | 19 | 24 | 814 | 46 | 953 | |
| IL-10/-592 allele, % | < .001 | ||||||
| C | 52.9 | 60.5 | 31.3 | 75.3 | 67.4 | 72.2 | |
| A | 47.1 | 39.5 | 68.8 | 24.7 | 32.6 | 27.8 | |
| IL-10/–592 genotype, no. (%) | < .001 | ||||||
| C/C | 5 (29) | 8 (42) | 2 (8) | 460 (57) | 21 (46) | 504 (53) | |
| A/C | 8 (47) | 7 (37) | 11 (46) | 306 (38) | 20 (40) | 368 (39) | |
| A/A | 4 (24) | 4 (21) | 11 (46) | 48 (6) | 5 (11) | 81 (8) | |
| IL10RB/c238 allele, % | .001 | ||||||
| A | 76.5 | 44.7 | 52.1 | 74.4 | 67.4 | 72.0 | |
| G | 23.5 | 55.3 | 47.9 | 25.6 | 32.6 | 38.0 | |
| IL10RB/c238 genotype, no. (%) | < .001 | ||||||
| A/A | 10 (59) | 4 (21) | 8 (33) | 447 (55) | 18 (39) | 494 (52) | |
| A/G | 6 (35) | 9 (47) | 9 (38) | 318 (39) | 26 (57) | 385 (40) | |
| G/G | 1 (6) | 6 (32) | 7 (29) | 49 (6) | 2 (4) | 74 (8) | |
| Donors | |||||||
| IL-10/-592 allele, % | < .001 | ||||||
| C | 38.2 | 63.2 | 37.5 | 74.3 | 64.1 | 71.1 | |
| A | 61.8 | 36.8 | 62.5 | 25.7 | 35.9 | 28.9 | |
| IL-10/-592 genotype, no. (%) | < .001 | ||||||
| C/C | 2 (12) | 8 (42) | 1 (4) | 454 (56) | 19 (41) | 492 (52) | |
| A/C | 9 (53) | 8 (42) | 16 (67) | 302 (37) | 21 (46) | 371 (39) | |
| A/A | 6 (35) | 3 (16) | 7 (29) | 58 (7) | 6 (13) | 90 (9) | |
| IL10RB/c238 allele, % | < .001 | ||||||
| A | 82.4 | 55.3 | 45.8 | 74.8 | 67.4 | 72.7 | |
| G | 17.6 | 44.7 | 54.2 | 25.2 | 32.6 | 27.3 | |
| IL10RB/c238 genotype, no. (%) | < .001 | ||||||
| A/A | 11 (65) | 6 (32) | 7 (29) | 459 (56) | 20 (43) | 515 (54) | |
| A/G | 6 (35) | 9 (47) | 8 (33) | 300 (37) | 22 (48) | 356 (37) | |
| G/G | 0 (0) | 4 (21) | 9 (38) | 55 (7) | 4 (8) | 82 (9) |
Comparison between white patients and Asian patients by chi-square test or Fisher exact test
Association of IL-10 and IL10RB polymorphisms with grades III-IV GVHD among white patients
The incidence of grades III-IV GVHD among white cases with patient IL-10/-592 C/C (n = 460), A/C (n = 306), and A/A (n = 48) genotypes were 22%, 16%, and 10%, respectively (trend P = .001; hazard ratio for the A/C and A/A genotypes, 0.6 and 0.4, respectively; data not shown). The incidence rates of grades III-IV GVHD among white cases with donor IL10RB/c238 A/A (n = 459), A/G (n = 300), and G/G (n = 55) genotypes were 21%, 18%, and 15%, respectively (trend P = .14; hazard ratio for the A/G and G/G genotypes, 0.8 and 0.7, respectively; data not shown). Although these results for the IL10RB/c238 G/G genotype did not reach statistical significance in the white patient–only subset, the trend is nevertheless similar to those of the combined population.
Interaction between the IL-10 and IL10RB polymorphisms and risk of severe acute GVHD
The potential interaction between the IL-10 and IL10RB SNPs and their association with acute GVHD was examined by stratifying the combined cohort of 953 cases into 9 groups according to patient IL-10/-592 and donor IL10RB/c238 genotypes. No grades III-IV severe acute GVHD occurred among the 16 pairs with a patient IL-10 A/A genotype and a donor IL10RB G/G genotype compared with a 22% incidence among pairs with a patient IL-10 C/C genotype and a donor IL10RB A/A genotype (P = .007; Table 4). The hazard ratios were intermediate and ranged from 0.4 to 0.6 for 141 pairs with a patient IL-10 A/C genotype and a donor IL10RB A/G genotype, 37 pairs with a patient IL-10 A/C genotype and a donor IL10RB G/G genotype, 33 pairs with a patient IL-10 A/A genotype and a donor IL10RB A/A genotype, and 32 pairs with a patient IL-10 A/A genotype and a donor IL10RB A/G genotype (Table 4). Similar results were observed when the analysis was restricted to white patients (Table 4).
Table 4.
Association of patient IL-10/-592 and donor IL10RB/c238 genotypes with risk of grades III-IV acute GVHD
|
Genotype
|
White patients
|
Combined patient population
|
|||||
|---|---|---|---|---|---|---|---|
| Patient IL-10/-592 | Donor IL10RB | Incidence of GVHD, % (no. of cases) | Hazard ratio*(95% CI) | P† | Incidence of GVHD, % (no. of cases) | Hazard ratio (95% CI)* | P† |
| C/C | A/A | 22 (269) | Reference | 22 (292) | Reference | ||
| C/C | A/G | 25 (164) | 1.1 (0.8-1.7) | .53 | 22 (183) | 1.0 (0.7-1.5) | .96 |
| C/C | G/G | 26 (27) | 1.1 (0.5-2.5) | .75 | 24 (29) | 1.1 (0.5-2.2) | .96 |
| A/C | A/A | 22 (165) | 0.9 (0.6-1.4) | .78 | 22 (190) | 0.9 (0.6-1.4) | .77 |
| A/C | A/G | 9 (118) | 0.4 (0.2-0.7) | .001 | 11 (141) | 0.4 (0.2-0.7) | .001 |
| A/C | G/G | 4 (23) | 0.2 (0-1.4) | .03 | 8 (37) | 0.4 (0.1-1.2) | .05 |
| A/A | A/A | 12 (25) | 0.5 (0.2-1.5) | .17 | 15 (33) | 0.6 (0.2-1.5) | .25 |
| A/A | A/G | 11 (18) | 0.5 (0.1-2.2) | .33 | 9 (32) | 0.4 (0.1-1.3) | .09 |
| A/A | G/G | 0 (5) | 0 (0) | .12 | 0 (16) | 0 (0) | .006 |
CI indicates confidence interval. For white patients, n = 814; for the combined patient population, n = 953.
Compared with cases with a patient IL-10/–592 C/C genotype and a donor IL10RB/c238 A/A genotype
Multivariable proportional hazard regression analysis adjusted for age at transplantation, transplantation year, sex mismatch, use of total body irradiation in the conditioning regimen, and disease risk group
Effects of patient IL-10/-592 genotype on the association of donor IL10RB genotype with GVHD
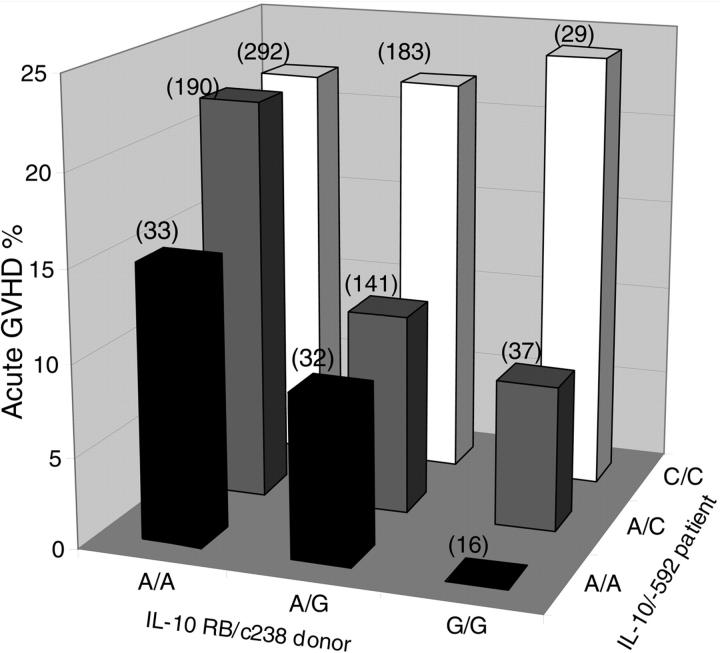
When the analysis was limited to pairs where the patient had the IL-10 C/C genotype, there was no evidence for a protective effect of the IL10RB/c238*G allele. In this situation, the incidence rates of severe acute GVHD were similar for pairs when the donor had IL10RB A/A, A/G, and G/G genotypes (range, 22%-24%; trend P = .82; Table 4; Figure 1). When the analysis was limited to pairs where the patient had the IL-10 A/C genotype, the incidence rates of severe acute GVHD were 22%, 11%, and 8% for pairs where the donor had the IL10RB A/A, A/G, and G/G genotypes, respectively (trend P = .005). When the analysis was limited to pairs where the patient had the IL-10 A/A genotype, the incidence rates of severe acute GVHD were 15%, 9%, and 0% for pairs where the donor had the IL10RB A/A, A/G, and G/G genotypes, respectively (trend P = .06). Results were similar when the analysis was restricted to white patients (Table 4), but the smaller number of white cases with the IL-10/-592 A/A genotype precluded a significant trend analysis when comparing the 3 different donor IL10RB genotypes (A/A, A/G, and G/G). These results suggest that the protective effect of the donor IL10RB/c238*G allele on GVHD is operative only among recipients with the IL-10/-592*A allele and that there is no beneficial effect of the donor IL10RB*G allele in cases where the recipient is homozygous for the IL-10/-592*C allele.
Figure 1.
Cumulative incidence of grades III-IV acute GVHD at day 100 following transplantation according to the IL-10/-592 genotype of the recipient and IL10RB/c238 genotype of the donor (n = 953). The number of patients in each group is indicated in parentheses.
Discussion
We and others have previously shown that genetic polymorphism in the IL-10 promoter region of the recipient has a significant impact on the outcome of HCT from an HLA-identical sibling donor.6-9,11 Keen et al12 have reported that the presence of donor R2-G-C-C haplotype (defined by a microsatellite polymorphism and 3 SNPs at positions -1082, -819, and -592 in the IL-10 promoter region) was associated with increased risk of transplant-related mortality following unrelated donor transplantation, whereas the presence of donor R3-G-C-C haplotype was associated with reduced risk of transplant-related mortality. In our previous study,11 we genotyped SNPs in the 5′ flanking promoter regions of the IL-10 gene at positions -3575, -2763, -1082, and -592 and identified the -592*A allele as a specific marker for the T-C-A-T-A haplotype. This previous study has also shown that the -592*A allele is associated with a favorable outcome after HCT. When SNPs at positions -1082, -2763, or -3575 were analyzed in a multivariable model, it appeared that they did not independently affect the risk of severe acute GVHD. A trend for an association of severe GVHD with the IL-10/-592 genotype of donor was observed in the current study but this was not significant when adjusted for the IL-10/-592 genotype of the patient. It is unclear in this study of sibling donor transplantations if the trend for the donor IL-10 genotype with GVHD is real or simply due to correlated genotypes between patient and sibling donor pairs who share the same parents. Definitive examination of a possible association between the donor IL-10 genotype and acute GVHD will require studies of patients with unrelated donors. In the current study, we report for the first time that a nonsynonymous polymorphism in exon 2 of the donor IL10RB gene also affects the incidence of severe acute GVHD. The finding that IL-10 and IL-10RB genetic polymorphisms are both associated with GVHD further strengthens previous guidance suggesting that IL-10 plays an important role in the development of acute GVHD. Based on the results of published in vivo and in vitro studies and the clinical correlations reported here, we hypothesize that the IL-10 pathway is regulated by functional polymorphisms that result in the variable production of IL-10 by recipient and possibly donor cells and variable ligand binding or signal transduction through the IL-10 receptor on donor cells.
IL-10 downregulates immune responses, induces anergy, and facilitates the induction of tolerance.13-19 IL-10 maintains CD4+CD25+ regulatory T cells, induces type 1 regulatory T cells, and appears to be an important effector molecule for these 2 types of regulatory T cells.16 Both CD4+CD25+ and type 1 regulator T cells can suppress GVHD.16-19 Higher IL-10 production by ex vivo–stimulated recipient cells before allogeneic HCT and increased IL-10 gene expression in recipient-derived monocytes is associated with a reduced incidence of GVHD and reduced risk of early death.29,30 On the other hand, administration of exogenous IL-10 in murine models has variable effects on GVHD,31-33 and increased IL-10 serum levels after transplantation have been associated with an increased risk of GVHD and fatal outcome.34,35 It is not known, however, whether blood levels reflect the biologic activity of IL-10 or the critical interactions between regulatory and effector T cells in peripheral lymphoid tissues, skin, and gut, where the outcome of the allograft reaction is determined.
Conflicting results have been reported regarding the genetic control of IL-10 production.36-39 Previous studies have reported increased IL-10 production in vitro associated with presence of the -1082*G allele or G-C-C haplotype (defined by 3 SNPs at positions -1082, -819, and -592 of the IL-10 gene).36,37 The association of the IL-10/-592*C allele and -1082*G allele, or G-C-C haplotype, in patients with GVHD or autologous GVHD40 and the assumption that the IL-10 G-C-C haplotype is associated with higher production of IL-10 has lead to the conclusion that IL-10 may promote the development of alloimmunity. Keijsers et al38 and Gibson et al,39 however, reported that the -1082*G allele and A-A-G-C-C haplotype (defined by 5 SNPs at positions -3575, -2763, -1082, -819, and -592) are associated with a lower IL-10 production. Our previous haplotype analysis showed that -592*A was a specific marker for a T-C-A-T-A haplotype (-3575/-2763/-1082/-819/-592) and also a marker for a low risk of GVHD, suggesting that the IL-10/-592*C allele or G-C-C haplotype is associated with lower IL-10 production.
Inconsistent results have also been reported regarding the association of IL-10 production and IL-10 blood levels with transplantation outcome.41,42 Higher IL-10 production by ex vivo–stimulated recipient's cells before allogeneic HCT has been associated with reduced risks of acute GVHD and nonrelapse mortality.30,43 On the other hand, increased IL-10 serum levels after transplantation has been associated with an increased risk of acute GVHD and nonrelapse mortality.34,35 Miura et al40 reported that the IL-10/-1082 G/G (or -592 C/C) genotype was associated with both an increased risk of the autologous GVHD and higher IL-10 production following autologous transplantations. Higher IL-10 blood levels however could also be the consequence of intense T-cell activation and inflammation, which potentially may override genetic control. It is also unknown if cytokine blood levels reflect the bioactivity of cytokines in the microenvironment where critical cell-to-cell interactions occur. Further studies will be needed to clarify the genetic control of IL-10 production and the mechanism by which IL-10 regulates the in vivo alloimmune response.
The IL-10 receptor is composed of 2 subunits, IL-10Rα and IL10RB.44 IL10RB contributes little to IL-10 binding affinity but acts as an accessory subunit for signaling when IL-10 binds to IL-10Rα.44-47 Its principle function appears to be recruitment of a Jak kinase (Tyk2) into the signaling pathway. IL10RB is also a subunit of IL-22, IL-26, IL-28A, IL-28B, and IL-29.47 The substitution of nucleotide A by nucleotide G at position cDNA 238 results in an amino acid substitution at codon 47 of Lys by Glu. The functional consequence of this substitution in IL10RB is currently unknown.
Ethnicity in this analysis was found to contribute to risk of GVHD as previously reported by others.48-50 The incidence of grades III-IV acute GVHD ranged from 8% in patients of Asian origin to 19% in white patients. This trend in our Asian patients is consistent with the lower incidence of acute GVHD reported for Japanese patients.48-50 The IL-10/-592*A allele is found more frequently in Japanese (67%)51 than white patients (25%), suggesting that the higher frequency of -592*A in Japanese patients may explain the lower incidence of GVHD observed in Japanese patients. We also show in this study that the frequency of the IL10RB/238 allele is different in white patients and Taiwanese patients. The IL10RB/c238*G allele was associated with a lower incidence of severe GVHD and was found more frequently in the Asian population.
In summary, the results of this study indicate that the IL-10/-592 genotype of the patient and IL10RB/c238 genotype of the donor synergistically affect the incidence of severe grades III-IV acute GVHD. These data are consistent with the hypothesis that genetic regulation of IL-10 during the early posttransplantation period affects the intensity of the alloimmune response. Determination of functional genotypes in the IL-10 pathway may have clinical utility for risk assessment, counseling, and treatment planning before transplantation. Genotypes associated with higher risk might serve as an indication for modification of dose-intensive conditioning regimens or more intensive immune suppression therapy. Further insight into the mechanism underlying the regulation and function of IL-10 might also suggest new strategies for preventing GVHD and facilitating induction of tolerance.
Acknowledgments
The authors thank Craig Nisperos for technical assistance and Susan Greenman for manuscript preparation.
Prepublished online as Blood First Edition Paper, August 18, 2005; DOI 10.1182/blood-2004-11-4338.
Supported by grants AI33484, AI49213, CA18029, CA15704, and CA18221 from the National Institutes of Health; and grants NSC93-3112-B-002-004 and IBMS-CRC92-T04, Taiwan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Ferrara JLM, Deeg HJ. Graft-versus-host disease. N Engl J Med. 1991;324: 667-674. [DOI] [PubMed] [Google Scholar]
- 2.Beatty PG, Clift RA, Mickelson EM, et al. Marrow transplantation from related donors other than HLA-identical siblings. N Engl J Med. 1985;313: 765-771. [DOI] [PubMed] [Google Scholar]
- 3.Perreault C, Decary F, Brochu S, Gyger M, Belanger R, Roy D. Minor histocompatibility antigens. Blood. 1990;76: 1269-1280. [PubMed] [Google Scholar]
- 4.Krenger W, Hill GR, Ferrara JLM. Cytokine cascades in acute graft-versus-host disease. Transplantation. 1997;64: 553-558. [DOI] [PubMed] [Google Scholar]
- 5.Bidwell, Keen L, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1: 3-19. [DOI] [PubMed] [Google Scholar]
- 6.Cavet J, Middleton PG, Segall M, Noreen H, Davies SM, Dickinson AM. Recipient tumor necrosis factor-alpha and interleukin-10 gene polymorphisms associate with early mortality and acute graft-versus-host disease severity in HLA-matched sibling bone marrow transplants. Blood. 1999;94: 3941-3946. [PubMed] [Google Scholar]
- 7.Cullup H, Dickinson AM, Jackson GH, Taylor PR, Cavet J, Middleton PG. Donor interleukin 1 receptor antagonist genotype associated with acute graft-versus-host disease in human leucocyte antigen-matched sibling allogeneic transplants. Br J Haematol. 2001;113: 807-813. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi H, Furukawa T, Hashimoto S, et al. Contribution of TNF-alpha and IL-10 gene polymorphisms to graft-versus-host disease following allo-hematopoietic stem cell transplantation. Bone Marrow Transplant. 2000;26: 1317-1323. [DOI] [PubMed] [Google Scholar]
- 9.Socie G, Loiseau P, Tamouza R, et al. Both genetic and clinical factors predict the development of graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Transplantation. 2001;72: 699-706. [DOI] [PubMed] [Google Scholar]
- 10.Cavet J, Dickinson AM, Norden J, Taylor PR, Jackson GH, Middleton PG. Interferon-gamma and interleukin-6 gene polymorphisms associate with graft-versus-host disease in HLA-matched sibling bone marrow transplantation. Blood. 2001;98: 1594-1600. [DOI] [PubMed] [Google Scholar]
- 11.Lin MT, Storer B, Martin PJ, et al. Relation of an interleukin-10 promoter polymorphism to graft-versus-host disease and survival after hematopoietic-cell transplantation. N Engl J Med. 2003;349: 2201-2210. [DOI] [PubMed] [Google Scholar]
- 12.Keen LJ, Defor TE, Bidwell JL, Davies SM, Bradley GA, Hows JM. Interleukin-10 and tumor necrosis factor alpha region haplotypes predict transplant-related mortality after unrelated donor stem cell transplantation. Blood. 2004;103: 3599-3602. [DOI] [PubMed] [Google Scholar]
- 13.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147: 3815-3822. [PubMed] [Google Scholar]
- 14.Ding L, Linsley PS, Huang LY, Germain RN, Shevach EM. IL-10 inhibits macrophage costimulatory activity by selectively inhibiting the up-regulation of B7 expression. J Immunol. 1993;151: 1224-1234. [PubMed] [Google Scholar]
- 15.Groux H, Bigler M, de Vries JE, Roncarolo MG. Interleukin-10 induces a long-term antigen-specific anergic state in human CD4+ T cells. J Exp Med. 1996;184: 19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal Malefyt R, Roncarolo MG. IFN-á and IL-10 induce the differentiation of human type 1 regulatory cells. J Immunol. 2001;166: 5530-5539. [DOI] [PubMed] [Google Scholar]
- 17.Zeller JC, Panoskaltsis-Mortari A, Murphy WJ, et al. Induction of CD4+ T cell alloantigen-specific hyporesponsiveness by IL-10 and TGF-beta. J Immunol. 1999;163: 3684-3691. [PubMed] [Google Scholar]
- 18.Taylor PA, Lees CJ, Blazar B. The infusion of ex vivo activated and expanded CD4+CD25+ immune regulatory cells inhibits grafted-versus-host disease lethality. Blood. 2002;99: 3493-3499. [DOI] [PubMed] [Google Scholar]
- 19.Hoffmann P, Ermann J, Edinger M, Fathman CG, Strober S. Donor type CD4+CD25+ regulatory T cells suppress lethal acute graft-versus-host disease after allogeneic bone marrow transplantation. J Exp Med. 2002;196: 389-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UW-FHCRC. SeattleSNPs: NHLBI Program for Genomic Application. http://pga.mbt.washington.edu. Accessed December 2002.
- 21.Tseng L-H, Chen P-J, Lin M-T, et al. Simultaneous genotyping of single nucleotide polymorphisms in the IL-6, TNFα and TNFβ genes. Tissue Antigens. 2002;59: 280-286. [DOI] [PubMed] [Google Scholar]
- 22.Tseng L-H, Chen P-J, Lin M-T, et al. Simultaneous genotyping of single nucleotide polymorphisms in the IL-1 gene complex by multiplex-polymerase chain reaction-restriction fragment length polymorphism. J Immunol Methods. 2002;267: 151-156. [DOI] [PubMed] [Google Scholar]
- 23.Mytilineos J. Cytokine polymorphisms. www.ihwg.org/components/cytokine/cytover.htm. Accessed September 2, 2002.
- 24.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296: 2225-2229. [DOI] [PubMed] [Google Scholar]
- 25.Tang K, Fu DJ, Julien D, Braun A, Cantor CR, Koster H. Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci U S A. 1999;6: 10016-10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Przepiorka D, Weisdorf D, Martin P, et al. Consensus conference on acute GVHD grading. Bone Marrow Transplant. 1995;15: 825-828. [PubMed] [Google Scholar]
- 27.Sullivan KM, Agura E, Anasetti C, et al. Chronic graft-versus-host disease and other late complication of bone marrow transplantation. Semin Hematol. 1991;28: 250-259. [PubMed] [Google Scholar]
- 28.Andersen PK, Borgan O, Gill RD, Keiding N. Statistical models based on counting processes. New York, NY: Springer-Verlag; 1993.
- 29.Bacchetta R, Bigler M, Touraine JL, et al. High levels of interleukin 10 production in vivo are associated with tolerance in SCID patients transplanted with HLA mismatched hematopoietic stem cells. J Exp Med. 1994;179: 493-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holler E, Roncarolo MG, Hintermeier-Knabe R, et al. Prognostic significance of increased IL-10 production in patients prior to allogeneic bone marrow transplantation. Bone Marrow Transplant. 2000;25: 237-241. [DOI] [PubMed] [Google Scholar]
- 31.Smith SR, Terminelli C, Pennline KJ, Kenworthy-Bott L, Donkin J, Calzetta A. Inhibitory effects of recombinant human interleukin 10 on disease manifestation in a P?F1 model of acute graft versus host disease. Transplantation. 1995;59: 890-896. [PubMed] [Google Scholar]
- 32.Blazar BR, Taylor PA, Smith S, Vallera DA. Interleukin-10 administration decreases survival in murine recipients of major histocompatibility complex disparate donor bone marrow grafts. Blood. 1995;85: 842-851. [PubMed] [Google Scholar]
- 33.Blazar BR, Taylor PA, Panoskaltsis-Mortari A, et al. Interleukin-10 dose-dependent regulation of CD4+ and CD8+ T cell-mediated graft-versus-host disease. Transplantation. 1998;66: 1220-1229. [DOI] [PubMed] [Google Scholar]
- 34.Hempel L, Korholz D, Nussbaum P, Bonig H, Burdach S, Zintl F. High interleukin-10 serum levels are associated with fatal outcome in patients after bone marrow transplantation. Bone Marrow Transplant. 1997;20: 365-368. [DOI] [PubMed] [Google Scholar]
- 35.Takatsuka H, Takemoto Y, Okamoto T, et al. Predicting the severity of graft-versus-host disease from interleukin-10 levels after bone marrow transplantation. Bone Marrow Transplant. 1999;24: 1005-1007. [DOI] [PubMed] [Google Scholar]
- 36.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24: 1-8. [DOI] [PubMed] [Google Scholar]
- 37.Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P. Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42: 1101-1108. [DOI] [PubMed] [Google Scholar]
- 38.Keijsers V, Verweij CL, Westendorp RGJ, Breedveld FC, Huizinga TWJ. IL-10 polymorphisms in relation to production and rheumatoid arthritis. Arthritis Rheum. 1997;40(suppl 9): S179. [Google Scholar]
- 39.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166: 3915-3922. [DOI] [PubMed] [Google Scholar]
- 40.Miura Y, Thoburn CJ, Bright EC, Chen W, Nakao S, Hess AD. Cytokine and chemokine profiles in autologous graft-versus-host disease (GVHD): interleukin 10 and interferon gamma may be critical mediators for the development of autologous GVHD. Blood. 2002;100: 2650-2658. [DOI] [PubMed] [Google Scholar]
- 41.Kida Y. Interleukin-10 and graft-versus-host disease [comment]. New Engl J Med. 2004,350: 1361-1362. [DOI] [PubMed] [Google Scholar]
- 42.Lin M-T, Martin P, Hansen J. Interleukin-10 and graft-versus-host disease [author reply]. New Engl J Med. 2004,350: 1361-1362. [DOI] [PubMed] [Google Scholar]
- 43.Baker KS, Roncarolo MG, Peters C, Bigler M, DeFor T, Blazar BR. High spontaneous IL-10 production in unrelated bone marrow transplant recipients is associated with fewer transplant-related complications and early deaths. Bone Marrow Transplant. 1999;23: 1123-1129. [DOI] [PubMed] [Google Scholar]
- 44.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleumin-10 receptor. Annu Rev Immunol. 2001;19: 683-765. [DOI] [PubMed] [Google Scholar]
- 45.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16: 5894-5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donnelly RP, Sheikh F, Kotenko SV, Dickensheets H. The expanded family of class II cytokines that share the IL-10 receptor-2 (IL-10R2) chain. J Leuk Biol. 2004;76: 314-321. [DOI] [PubMed] [Google Scholar]
- 47.Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22: 929-979. [DOI] [PubMed] [Google Scholar]
- 48.Morishima Y, Morishita Y, Tanimoto M, et al. Low incidence of acute graft-versus-host disease by the administration of methotrexate and cyclosporine in Japanese leukemia patients after bone marrow transplantation from human leukocyte antigen compatible siblings; possible role of genetic homogeneity: The Nagoya Bone Marrow Transplantation Group. Blood. 1989;74: 2252-2256. [PubMed] [Google Scholar]
- 49.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor: Japan Marrow Donor Program. N Engl J Med. 1998;339: 1177-1185. [DOI] [PubMed] [Google Scholar]
- 50.Oh H, Loberiza FR Jr, Zhang MJ, et al. Comparison of graft-versus-host disease and survival after HLA-identical sibling bone marrow transplantation in ethnic population. Blood. 2005;105: 1408-1416. [DOI] [PubMed] [Google Scholar]
- 51.Tegoshi H, Hasegawa G, Obayashi H, et al. Polymorphisms of interferon-gamma gene CA-repeat and interleukin-10 promoter region (-592A/C) in Japanese type I diabetes. Hum Immunol. 2002;63: 121-128. [DOI] [PubMed] [Google Scholar]
- 52.Lutfalla G, Gardiner K, Uze G. A new member of the cytokine receptor gene family maps on chromosome 21 at less than 35 kb from IFNAR. Genomics. 1993;16: 366-373. [DOI] [PubMed] [Google Scholar]