Abstract
Germ line PTPN11 mutations cause 50% of cases of Noonan syndrome (NS). Somatic mutations in PTPN11 occur in 35% of patients with de novo, nonsyndromic juvenile myelomonocytic leukemia (JMML). Myeloproliferative disorders (MPDs), either transient or more fulminant forms, can also occur in infants with NS (NS/MPD). We identified PTPN11 mutations in blood or bone marrow specimens from 77 newly reported patients with JMML (n = 69) or NS/MPD (n = 8). Together with previous reports, we compared the spectrum of PTPN11 mutations in 3 groups: (1) patients with JMML (n = 107); (2) patients with NS/MPD (n = 19); and (3) patients with NS (n = 243). Glu76 was the most commonly affected residue in JMML (n = 45), with the Glu76Lys alteration (n = 29) being most frequent. Eight of 19 patients with NS/MPD carried the Thr73Ile substitution. These data suggest that there is a genotype/phenotype correlation in the spectrum of PTPN11 mutations found in patients with JMML, NS/MPD, and NS. This supports the need to characterize the spectrum of hematologic abnormalities in individuals with NS and to better define the impact of the PTPN11 lesion on the disease course in patients with NS/MPD and JMML. (Blood. 2005;106:2183-2185)
Introduction
The PTPN11 proto-oncogene encodes Src-homology tyrosine phosphatase 2 (SHP-2), a protein tyrosine phosphatase with a role in signal transduction and hematopoiesis.1,2 Somatic PTPN11 mutations exist in 35% of juvenile myelomonocytic leukemia (JMML) specimens and are less frequent in other leukemias.3-6 SHP-2 relays signals from activated growth factor receptors to Ras. PTPN11, KRAS2, NRAS, and NF1 mutations are found in mutually exclusive subsets of patients with JMML.3,4 These data support the hypothesis that hyperactive Ras signaling plays a central role in JMML.
Germ-line PTPN11 mutations cause approximately 50% of cases of Noonan syndrome (NS),7,8 a congenital disorder characterized by facial anomalies, short stature, and heart defects.9 Whereas NS is frequently inherited as an autosomal dominant condition, almost half of the constitutional PTPN11 mutations found in NS arise sporadically. Germ-line PTPN11 mutations are also found in patients with multiple lentigene syndrome (LS), a rare developmental disorder clinically related to NS.9 Infants with NS are predisposed to developing a myeloproliferative disorder (NS/MPD), which may regress without treatment or follow an aggressive clinical course similar to JMML.10-14 By contrast, cases of JMML that arise in patients without NS have a poor prognosis without hematopoietic stem cell transplantation.15-18 Recent studies show that children with JMML have improved outcomes when they are treated aggressively early in the course of disease.18 Therefore, differentiating JMML from NS/MPD and identifying patients with NS/MPD who will require aggressive treatment are important clinical questions. We identify PTPN11 mutations in 77 newly reported patients with JMML and NS/MPD, and compare the mutational spectrum in JMML, NS/MPD, and NS/LS to determine if genotype-phenotype correlations exist that may help guide diagnosis and clinical management.
Study design
Tissue samples (bone marrow, peripheral blood, and, rarely, buccal swab and skin fibroblasts) from patients with JMML and NS/MPD were collected under Institutional Review Board-approved protocols at each institution and with informed consent. DNA was extracted and analyzed for mutations in Freiburg, Germany; New York, New York; Rome, Italy; Nagoya, Japan; and San Francisco, CA. Since the data accumulated thus far have demonstrated that PTPN11 mutations in myeloproliferative disorders occur in exons 3 (∼90%) or 13 (∼10%),3,4 these were the only exons screened in many of the European and American cases as previously described.3,4,8 Mutations in the JMML cases from Nagoya were detected by analyzing exons 3, 8, and 13 employing standard cloning and sequencing techniques.
Results and discussion
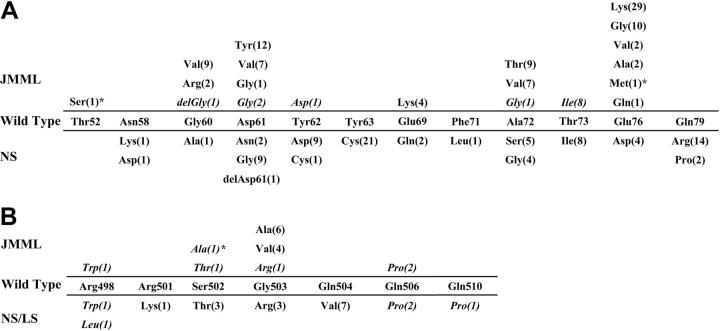
Results of the PTPN11 mutational screening performed on the 77 newly reported patients (JMML = 69, NS/MPD = 8) are listed in Table 1. Two mutations, 155C > T (Thr52Ser) and 226-227GA > AT (Glu76Met), had not been previously documented in JMML or other malignancies.3-6,19,20 Figure 1 shows an updated compendium of PTPN11 mutations previously documented in JMML or NS/MPD combined with the current cohort of 77 newly reported patients. The series includes 107 JMML cases, 19 patients with NS/MPD, and 243 patients with NS or LS. While other exons are commonly mutated in the germ line of patients with NS and LS, only cases with exon 3 and 13 mutations are listed in Figure 1.
Table 1.
PTPN11 mutations in 77 newly reported children with JMML or NS/MPD
| Cohort, no. cases | Nucleotide substitution | Amino acid substitution |
|---|---|---|
| JMML, N = 69 | ||
| 1 | 155C > G* | Thr52Ser |
| 1 | 178G > C | Gly60Arg |
| 9 | 179G > T | Gly60Val |
| 6 | 181G > T | Asp61Tyr |
| 6 | 182A > T | Asp61Val |
| 1 | 182A > G | Asp61Gly |
| 5 | 214G > A | Ala72Thr |
| 4 | 215C > T | Ala72Val |
| 20 | 226G > A | Glu76Lys |
| 6 | 227A > G | Glu76Gly |
| 1 | 226-227GA > AT* | Glu76Met |
| 5 | 1508G > C | Gly503Ala |
| 4 | 1508G > T | Gly503Val |
| NS/MPD, N = 8 | ||
| 2 | 182A > G | Asp61Gly |
| 1 | 215C > G | Ala72Gly |
| 2 | 218C > T | Thr73lle |
| 1 | 1492C > T | Arg498Trp |
| 1 | 1504T > G* | Ser502Ala |
| 1 | 1517A > C | Gly506Pro |
Novel mutation.
Figure 1.
PTPN11 mutations in JMML, NS/MPD, and NS/LS. The middle sections of both panels show wild-type SHP2 amino acid residue at each position. (A) Residues located within the N-SH2 domain encoded by exon 3. (B) Residues located within the portion of the catalytic domain encoded by exon 13. Amino acid substitutions documented in JMML and NS/MPD (italics), and in NS and LS (italics) are shown above and below the wild-type SHP2 sequence, respectively. Del indicates a deletion of this amino acid. Digits in parentheses indicate the numbers of individuals with JMML, NS/MPD, or NS carrying a specific mutation. Novel mutations are identified by asterisks. Whereas virtually all mutations in JMMLand NS/MPD are located within these confined regions, mutations associated with NS alone alter other residues of SHP2 in approximately 50% of the cases.9 Our data, updated to January 2005, includes 107 cases with JMML, 19 with NS/MPD, 181 with NS, and 42 with LS.
All defects with the exception of 2 result in an amino acid substitution. While there is overlap with respect to the substitutions seen in patients with NS/MPD and NS (Figure 1; Asp61Gly, Tyr62Asp, Ala72Gly, Thr73Ile, Ser502Thr, and Gly503Arg) or LS (Arg498Trp, Gln506Pro), only the Asp61Gly mutation is shared among patients with JMML, NS/MPD, and NS. Remarkably, 45 (42%) of the mutations found in JMML cases alter codon 76. Mutations affecting Glu76 are rare in individuals with NS (< 3% of cases), but are conservative (Glu76Asp) when they occur.
We identified a 214G > A (Ala72Thr) mutation in the bone marrow specimen of a 5-month-old girl with JMML for whom umbilical cord was collected at birth. Remarkably, the cord blood contained the same PTPN11 mutation, which was absent in the child's buccal cells and parental DNA. Despite the assumption that JMML arises in utero, this is, to our knowledge, one of the first demonstrations that a somatically acquired JMML-associated PTPN11 mutation occurred before birth.
In this newly reported cohort of 77 patients (Table 1), 6 missense changes, including 1 novel mutation affecting exon 13, 1504T > G (Ser502Ala), were identified in 8 NS/MPD specimens. We had buccal swab DNA (n = 2) and skin fibroblast DNA (n = 1) available for analysis from 3 patients and confirmed the presence of the PTPN11 mutation in each case, supporting the germ-line origin of the lesion. Together with previous data, the Thr73Ile substitution was identified in 8 out of 19 NS/MPD cases (Figure 1), representing the most common mutation in patients with NS/MPD. This mutation is uncommon in NS patients without MPD, and has not been observed among patients with JMML.
Analysis of DNA from the unaffected parents of 3 children with NS/MPD with different PTPN11 mutations (Asp61Gly, Ser502Ala, and Gly506Pro) demonstrated the sporadic origin of each lesion. Consistent with these results, all previous children with NS/MPD carrying a mutated PTPN11 gene have been documented to be sporadic cases.3,14 These findings suggest that NS/MPD is associated with mutations that might be associated with decreased fertility or have more severe consequences on fetal survival. This is consistent with the observation that mutations associated with NS/MPD have only rarely been identified in patients with familial NS cases, and that mutants detected in families transmitting NS are unlikely to be associated with NS/MPD.7,21-25
We identified 1 patient with JMML carrying a somatic 182A > G (Asp61Gly) mutation previously associated with NS.7 The mutation was found in hematopoietic cells but was absent in skin fibroblasts. The same mutation was found in 2 patients with NS/MPD. In one of the latter patients the germ-line nature of this lesion was confirmed by analyzing buccal cells. To our knowledge, the 182A > G (Asp61Gly) mutation is the only PTPN11 mutation associated with JMML, NS/MPD, and NS. The finding of Asp61Gly in JMML and NS/MPD is intriguing as this substitution was used to generate a mouse knock-in model of NS.26 When homozygous, the Asp61Gly mutant is embryonic lethal, whereas heterozygotes have decreased viability. Of note, while the JMML-like picture in infants with NS usually develops at or shortly after birth, Ptpn11Asp61 Gly/+ mice show signs of a mild myeloproliferative disorder and splenomegaly by 5 months of age.
Similar to what is observed in NS, the vast majority of mutations identified in JMML and NS/MPD alter residues located at the interface between the N-terminal Src homology 2 (N-SH2) and catalytic (protein tyrosine phosphatase [PTP]) domains.27 They are predicted to promote SHP-2 gain of function by impairing the switch between the active and inactive conformation of the protein, favoring a shift in the equilibrium toward the former. One exception is the Thr52Ser substitution, which affects the N-SH2 phosphotyrosyl-binding site.27 It has been hypothesized that the genotype-phenotype relationships observed in patients with somatic and germ line mutations of PTPN11 are due to distinct gain-of-function effects on the protein product SHP-2.3 Consistent with this view, somatically acquired JMML-associated PTPN11 mutations are predicted to have a strong gain-of-function effect that might otherwise affect embryonic/fetal development if transmitted in the germ line, explaining why these mutations are not seen in NS. In contrast, germ-line mutations identified in NS are predicted to have weaker hematologic effects. It is also possible that the rare germ-line PTPN11 lesions observed in NS/MPD can exhibit intermediate effects. Indeed, in vitro and in vivo experiments on primary hematopoietic cells and cell lines show that somatic mutants confer more pronounced effects on cell growth than common mutants only found in NS,28-30 and exhibit an increased phosphatase activity basally.3
Our data raise a number of new questions. First, are somatic PTPN11 mutations sufficient to initiate MPD and, if not, what are the cooperating molecular lesions? Second, do specific PTPN11 mutations have different consequences depending on whether they involve the entire hematopoietic compartment or arise as clonal events? Third, do some patients with NS and PTPN11 mutations develop transient myeloproliferation, which is unrecognized? Finally, does the nature of a PTPN11 mutation (germ-line versus somatic) and the specific amino acid substitution detected provide information that can be used to guide decisions regarding the need to initiate aggressive treatment in infants presenting with JMML? These questions will be answered through ongoing collaborations between laboratory researchers and clinical investigators who are evaluating and treating children with these disorders.
Acknowledgments
The authors wish to thank all of the patients, families, and referring physicians who contributed samples to the investigators. We wish to acknowledge the National Children's Cancer Foundation on behalf of the samples collected in the United States.
Prepublished online as Blood First Edition Paper, May 31, 2005; DOI 10.1182/blood-2005-02-0531.
Supported by the Deutsche José Carreras Leukämie Stiftung e.V. (C.P.K., C.K.), and by a Translational Research Award (6021-99) from the Leukemia and Lymphoma Society (R.P.C.), Telethon-Italy grant GGP04172 (M.T.), by the Cancer Research Coordinating Committee, the Frank A. Campini Foundation and the Hope Street Kids Foundation (M.L.L.), and by US Public Health Service grants R01 CA95621 (P.D.E., R.P.C., M.L.L.), K24 CA80916 (P.D.E.), HL71207, HD01294, HL074728 (B.D.G.), P30 CA82103, and R01 CA104282 (M.L.L.) from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Neel BG, Gu H, Pao L. The `Shp'ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28: 284-293. [DOI] [PubMed] [Google Scholar]
- 2.Tartaglia M, Niemeyer CM, Shannon KM, Loh ML. SHP-2 and myeloid malignancies. Curr Opin Hematol. 2004;11: 44-50. [DOI] [PubMed] [Google Scholar]
- 3.Tartaglia M, Niemeyer CM, Fragale A, et al. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet. 2003;34: 148-150. [DOI] [PubMed] [Google Scholar]
- 4.Loh ML, Vattikuti S, Schubbert S, et al. Mutations in PTPN11 implicate the SHP-2 phosphatase in leukemogenesis. Blood. 2004;103: 2325-2331. [DOI] [PubMed] [Google Scholar]
- 5.Loh ML, Reynolds MG, Vattikuti S, et al. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children's Cancer Group. Leukemia. 2004;18: 1831-1834. [DOI] [PubMed] [Google Scholar]
- 6.Tartaglia M, Martinelli S, Cazzaniga G, et al. Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood. 2004;104: 307-313. [DOI] [PubMed] [Google Scholar]
- 7.Tartaglia M, Mehler EL, Goldberg R, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29: 465-468. [DOI] [PubMed] [Google Scholar]
- 8.Tartaglia M, Kalidas K, Shaw A, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet. 2002;70: 1555-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. In press. 2005. [DOI] [PubMed]
- 10.Bader-Meunier B, Tchernia G, Mielot F, et al. Occurrence of myeloproliferative disorder in patients with Noonan syndrome. J Pediatr. 1997;130: 885-889. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M, Horibe K, Miyajima Y, Matsumoto K, Nagashima M. Spontanous remission of juvenile chronic myelomonocytic leukemia in an infant with Noonan syndrome. J Pediatr Hematol Oncol. 1997;19: 177-179. [DOI] [PubMed] [Google Scholar]
- 12.Choong K, Freedmann MH, Chitayat D, Kelly EN, Taylor G, Zipusky A. Juvenile myelomonocytic leukemia and Noonan syndrome. J Pediatr Hematol Oncol. 1999;21: 523-537. [PubMed] [Google Scholar]
- 13.Silvio F, Carlo L, Elena B, Nicoletta B, Daniela F, Roberto M. Transient abnormal myelopoiesis in Noonan syndrome. J Pediatr Hematol Oncol. 2002;24: 763-764. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida R, Miyata M, Nagai T, Yamazaki T, Ogata T. A 3-bp deletion mutation of PTPN11 in an infant with severe Noonan syndrome including hydrops fetalis and juvenile myelomonocytic leukemia. Am J Med Genet. 2004;128A: 63-66. [DOI] [PubMed] [Google Scholar]
- 15.Niemeyer CM, Arico M, Basso G, et al. Chronic myelomonocytic leukemia in childhood: a retrospective analysis of 110 cases. European Working Group on Myelodysplastic Syndromes in Childhood (EWOG-MDS). Blood. 1997;89: 3534-3543. [PubMed] [Google Scholar]
- 16.Niemeyer CM, Kratz C. Juvenile myelomonocytic leukemia. Curr Oncol Rep. 2003;5: 510-515. [DOI] [PubMed] [Google Scholar]
- 17.Emanuel PD. Juvenile myelomonocytic leukemia. Curr Hematol Rep. 2004;3: 203-209. [PubMed] [Google Scholar]
- 18.Locatelli F, Nollke P, Zecca M, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood. 2005;105: 410-419. [DOI] [PubMed] [Google Scholar]
- 19.Loh ML, Martinelli S, Cordeddu V, et al. Acquired PTPN11 mutations occur rarely in adult patients with myelodysplastic syndromes and chronic myelomonocytic leukemia. Leuk Res. 2005;29: 459-462. [DOI] [PubMed] [Google Scholar]
- 20.Bentires-Alj M, Paez JG, David FS, et al. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 2004;64: 8816-8820. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida R, Hasegawa T, Hasegawa Y, et al. Protein-tyrosine phosphatase, nonreceptor type 11 mutation analysis and clinical assessment in 45 patients with Noonan syndrome. J Clin Endocrinol Metab. 2004;89: 3359-3364. [DOI] [PubMed] [Google Scholar]
- 22.Musante L, Kehl HG, Majewski F, et al. Spectrum of mutations in PTPN11 and genotype-phenotype correlation in 96 patients with Noonan syndrome and five patients with cardio-facio-cutaneous syndrome. Eur J Hum Genet. 2003;11: 201-206. [DOI] [PubMed] [Google Scholar]
- 23.Kosaki K, Suzuki T, Muroya K, et al. PTPN11 (protein-tyrosine phosphatase, nonreceptor-type 11) mutations in seven Japanese patients with Noonan syndrome. J Clin Endocrinol Metab. 2002;87: 3529-3533. [DOI] [PubMed] [Google Scholar]
- 24.Zenker M, Buheitel G, Rauch R, et al. Genotypephenotype correlations in Noonan syndrome. J Pediatr. 2004;144: 368-374. [DOI] [PubMed] [Google Scholar]
- 25.Maheshwari M, Belmont J, Fernbach S, et al. PTPN11 mutations in Noonan syndrome type I: detection of recurrent mutations in exons 3 and 13. Hum Mutat. 2002;20: 298-304. [DOI] [PubMed] [Google Scholar]
- 26.Araki T, Mohi MG, Ismat FA, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10: 849-857. [DOI] [PubMed] [Google Scholar]
- 27.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92: 441-450. [DOI] [PubMed] [Google Scholar]
- 28.Mohi MG, Williams IR, Dearolf CR, et al. Prognostic, therapeutic, and mechanistic implications of a mouse model of leukemia evoked by Shp2 (PTPN11) mutations. Cancer Cell. 2005;7: 179-191. [DOI] [PubMed] [Google Scholar]
- 29.Chan RJ, Leedy MB, Munugalavadla V, et al. Human somatic PTPN11 mutations induce hematopoietic cell hypersensitivity to granulocyte-macrophage colony stimulating factor. Blood. 2005;105: 3737-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubbert S, Lieuw K, Rowe SL, et al. Functional analysis of leukemia-associated PTPN11 mutations in primary hematopoietic cells. Blood. Prepublished on March 10, 2005, as DOI 10.1182/blood-2004-11-4207. ( [DOI] [PMC free article] [PubMed]; Now available as Blood. 2005;106: 311-317).15761018 [Google Scholar]