Abstract
Epstein-Barr virus (EBV) is associated with B-cell lymphomas such as Hodgkin lymphoma, Burkitt lymphoma, and post-transplantation lymphoma, which originate from clonal germinal center (GC) B cells. During the process of somatic hypermutation, GC B cells can acquire deleterious or nonsense mutations in the heavy and light immunoglobulin genes. Such mutations abrogate the cell surface expression of the B-cell receptor (BCR), which results in the elimination of these nonfunctional B cells by immediate apoptosis. EBV encodes several latent genes, among them latent membrane protein 1 (LMP1) and LMP2A, which are regularly expressed in EBV-positive Hodgkin lymphoma and posttransplantation lymphomas. Since LMP1 and LMP2A mimic the function of 2 key receptors on B cells, CD40 and BCR, respectively, we wanted to learn whether EBV infection can rescue proapoptotic GC B cells with crippling mutations in the heavy chain immunoglobulin locus from apoptosis. We show here that BCR-negative GC B cells readily enter the cell cycle upon infection with EBV in vitro and yield clonal lymphoblastoid cell lines that are incapable of expressing a functional BCR because the rearranged and formerly functional heavy chain immunoglobulin alleles carry deleterious mutations. Our findings imply an important role for EBV in the process of lymphomagenesis in certain cases of Hodgkin lymphoma and posttransplantation lymphomas.
Introduction
The hallmark of the germinal center (GC) reaction is the process of somatic hypermutation (SH), which leads to B cells with a highaffinity B-cell receptor (BCR) and the abrogation of the default apoptosis pathway of GC B cells.1 Since SH in the V genes of the rearranged immunoglobulin heavy (HC) and light chain (LC) alleles is largely random, most mutations will have a negative impact on the BCR to bind its cognate antigen. Some mutations, such as generation of a stop codon or a frame shift mutation caused by insertions or deletions, will abrogate proper translation of the immunoglobulin genes, which make up the BCR. Such mutations are lethal for the cells that harbor them: these cells are eliminated by apoptosis either because they cannot compete with cells that bind antigen more strongly or because they no longer express a BCR on their surface and therefore fail to signal.2
It is therefore surprising that about 25% of Hodgkin/Reed-Sternberg (HRS) cells, which are the characteristic cells of classic Hodgkin lymphoma, carry crippling mutations in the rearranged HC or LC alleles, which preclude the expression of a functional BCR.3 The Hodgkin tumors are clonal and derived from GC B cells because they carry originally functional light and heavy chain immunoglobulin alleles with typical modifications introduced by SH.4,5 It is therefore likely that HRS cells with crippling mutations in the immunoglobulin genes have acquired additional transforming event(s), which enabled the cells to escape the negative selection in the GC reaction.
One of the most prominent candidates in supporting this transforming event is Epstein-Barr virus (EBV), which is categorized as a class 1 carcinogen by the World Health Organization and has been found associated with B-cell lymphomas such as Hodgkin lymphoma, Burkitt lymphoma, posttransplantation lymphomas, and certain carcinomas, among others.6 In all EBV-infected tumor cells the virus adopts a latent state, which is accompanied by the expression of a restricted number of latent viral genes. In vitro, EBV infects resting human B lymphocytes and transforms them into lymphoblastoid cell lines (LCLs); a process that is termed growth transformation and a hallmark of this virus. EBV encodes several proteins, which could play a crucial role in rescuing GC B cells from apoptosis. These B cells are inherently prone to die, and to survive must receive specific survival signals such as activation of the B cell and CD40 receptors by contact with antigen and T helper cells, respectively. These signals could be replaced by the viral proteins LMP2A and LMP1, both of which are expressed in EBV-infected HRS B cells and posttransplantation lymphomas.7-9 Whereas LMP1 in many aspects resembles a constitutively active CD40 receptor,10-12 the cytoplasmic tail of LMP2A harbors an immunoreceptor tyrosine-based activation motif (ITAM).13 This motif has originally been identified in the coreceptors CD79A and CD79B, which make up the BCR, together with the assembled heavy and light chains.14,15 LMP2A expression itself stimulates ITAM-specific tyrosine kinases and mimics the presence of a BCR in transgenic mice.16 It is therefore possible that LMP2A, when expressed in GC B cells, may simulate an activation signal by using receptor-proximal signaling components similar to the BCR. This molecular mimicry might constitute an important survival signal for GC B cells, since LMP2A expression could rescue proapoptotic B cells with a nonfunctional or absent BCR from the default apoptosis pathway.
We wanted to test the hypothesis, whether EBV can rescue GC B cells with a crippled BCR, in in vitro experiments. To this end, we isolated BCR-negative (BCR-) GC cells and infected them with a green fluorescent protein (GFP)–encoding wild-type EBV. We found that EBV's capacity to transform primary human B lymphocytes includes GC B cells that are incapable of expressing a functional immunoglobulin heavy chain due to stop codons, frame shift mutations, or devastating deletions introduced during somatic hypermutation. Thus, our experiments mimic what might be one step in the pathogenesis of Hodgkin lymphoma and posttransplantation lymphomas. Our results also suggest that an EBV infection can rescue BCR- GC B cells from apoptosis in an in vivo situation, indicating that EBV can be directly involved in the initial steps of lymphomagenesis of GC-derived B-cell lymphomas.
Materials and methods
Isolation, separation, and infection of primary B lymphocytes
Human primary B cells were isolated from adenoids, depleted from T cells, purified by Ficoll density centrifugation, and characterized by fluorescence-activated cell-sorter scanner (FACS) analysis with α–λ–LC–phycoerythrin (PE) and α–κ-LC–fluorescein isothiocyanate (FITC) (Dako Cytomation, Carpinteria, CA), α–CD3–allophycocyanin (APC) (BD Biosciences, San Jose, CA), α–CD77-FITC (BD Biosciences), and α–CD21-FITC (Chemicon, Temecula, CA) antibodies. For separation into a BCR+ and a BCR- fraction, the cells were stained with α–λ and κ-LC antibodies (BD Biosciences). B-cell preparations were depleted using magnetic cell sorting (MACS) columns and α–mouse immunoglobulin G1 (IgG1) MicroBeads (Miltenyi Biotec, Auburn, CA) to obtain BCR- cells. As an alternative, BCR- cells were depleted for CD3 cells (α-CD3; Miltenyi Biotec) and enriched for CD77 surface expressing B cells (α-FITC; Miltenyi Biotec) by magnetic sorting in certain experiments. A variant of the maxi-EBV p2089 was prepared from a stable cell line as described:17,18 1 × 105 B cells were infected and initially plated on irradiated CD40 ligand feeder layer19 in the presence of 2 ng/mL interleukin-4 (IL-4) (PAN Biotech, Karlsruhe, Germany) and 1 μg/mL cyclosporine A (Novartis, Basel, Switzerland). Four days after infection the cell proliferation assay was performed using the BrdU Flow Kit (BD Biosciences). Limiting dilution assays were performed with BCR- cells to obtain single-cell clones.
Screening for BCR- single-cell clones
Single-cell clones were analyzed by FACS for LC surface expression with α–λ-LC–PE (BD Biosciences) and α–κ-LC–APC antibodies (Caltag Laboratories, Burlingame, CA). The LC negative cells were further analyzed for HC expression by Western blot using an α–human IgA + IgG + IgM (H+L) antibody directly coupled with peroxidase (Dianova, Hamburg, Germany). Purified immunoglobulin (Beriglobin, Aventis-Behring, Marburg, Germany) was used in Western blots as a positive control. For detection of EBNA2, a combination of α-EBNA2a and α-EBNA2b antibodies was used with an α–rat peroxidase–coupled secondary antibody (Dianova).
PCR amplification and analysis of the IgH gene loci
Genomic DNA was isolated and IgVH gene rearrangements were amplified with the second round VH polymerase chain reaction (PCR) primer sets20 and sequenced on both strands. Analysis was performed with the V BASE database (www.dnaplot.de) and blast searches against the human germ line sequence of the complete HC gene locus (PubMed accession ng_001019). In order to discriminate between a completely (VDJH) versus an incompletely (DJH) rearranged HC locus versus a possible HC germ line situation, we used 2 sets of PCR primers: D7-2721 combined with the JH1.4.5 primers,20 with which the intergenic sequence between the last DH and the first JH segment can be amplified. To discriminate between a completely (VDJH) versus an incompletely (DJH) rearranged HC locus, we used a primer pair that amplifies part of the intergenic region in between the most 3′ VH gene and the first DH segment (GLfront: 5′-AGAGCAGACTCCAGGGACG-3′ with GLback: 5′-CCACACCAAGGTCATCATTGTAG-3′).
Results
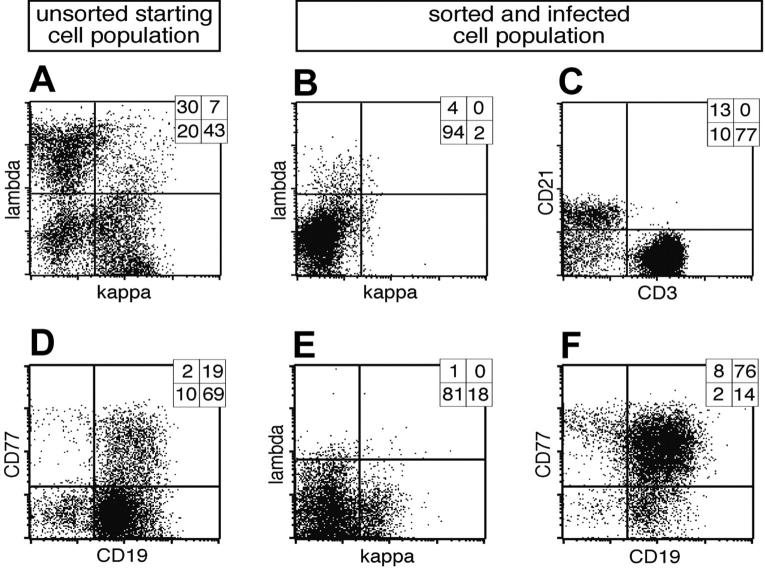
B cells prepared from adenoids revealed 3 discernable subpopulations: light chain (LC) lambda positive (λ+), kappa positive (κ+), and LC-negative cells, which are BCR- (Figure 1A). In order to isolate BCR- cells, λ+/κ+ cells were depleted with LC-specific antibodies coupled indirectly to magnetic beads (Figure 1B). Although the resulting BCR- fraction mostly consists of remnant CD3+ T cells, about 15% of all BCR- cells are CD21+ B cells (Figure 1C). The BCR- and control BCR+ cell populations were infected with EBV in the presence of cyclosporine A to abrogate an EBV-specific T-cell response. The EBV strain used in this study is based on a modified maxi-EBV genome, which encodes the GFP gene cloned onto the B95.8 prototype EBV genome.17 Four days after infection GFP-positive (GFP+) cells could be detected among BCR- cells as well as BCR+ cells (Figure 2A, middle and bottom panels), whereas in the uninfected population, GFP was detectable only at background levels (Figure 2A, upper panel).
Figure 1.
Cell surface expression of λ and κ light chains of primary lymphocytes and preparation of BCR- B cells. (A) Primary lymphocytes were isolated from nasal adenoids and stained for cell surface expression of the immunoglobulin light chains λ and κ. Most cells were either single λ- or κ-positive BCR+ B cells, whereas a smaller fraction was double negative for both immunoglobulin light chains. (B) The lymphocyte preparation was depleted by magnetic sorting with λ- and κ-specific antibodies indirectly coupled to magnetic beads, and the BCR- fraction was again analyzed for cell surface expression of λ and κ light chains. (C) The BCR- population consisted of a smaller CD21+ B-lymphocyte fraction, whereas CD3+ T lymphocytes dominated. CD3-/CD21- cells are probably monocytes, which were not further analyzed. (D) In a second approach primary lymphocytes from adenoids were analyzed for surface expression of CD19 and CD77, revealing the initial fraction of GC B cells. (E) The lymphocyte preparation was depleted with CD3, λ- and κ-specific antibodies, enriched for CD77 surface-positive GC B cells, and again analyzed for light chain cell surface expression. (F) Prior to EBV infection, the cells were characterized for their expression of CD77 and CD19, indicating the germinal center origin of the surface BCR- B-cell preparation. Quadrant statistics based on intact cells in the lymphocyte gate by forward and sideward scatter criteria are depicted in the top right-hand corner of each FACS plot.
Figure 2.
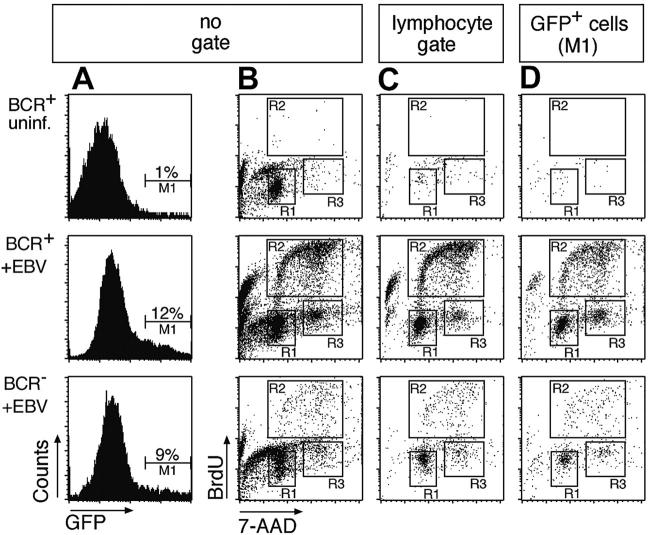
BCR+ as well as BCR- B cells infected with EBV enter the S phase of the cell cycle and proliferate in vitro. (A) An EBV genome, which encodes GFP,17 was used to infect the BCR- and BCR+ cell populations (middle and bottom panels) or BCR+ cells were left uninfected (top row panels). GFP+ cells were found in BCR+ and BCR- cells, indicating that EBV can infect cells in both populations. (B) To analyze the cell cycle distribution of the uninfected and infected BCR- and BCR+ bulk populations, the cells were labeled with the nucleotide analog BrdU 4 days after infection for 2 hours. Dual parameter flow cytometry of the cells with an APC-conjugated α-BrdU antibody and with 7-AAD to reveal the cellular DNA content indicated that the EBV-infected B-cell pool entered S phase and proliferated irrespective of their BCR surface status (middle and bottom panels, gates R2 and R3). Cells in the uninfected population enter neither S phase (top panel, region R2) nor M phase (region R3), as indicated by the lack of BrdU incorporation and predominance of G1/G0 phase (gate R1). (C) Dual parameter flow cytometry of EBV-infected B cells gated for typical forward/sideward scatter lymphocyte morphology. (D) Triple parameter flow cytometry of GFP+/EBV-infected B cells (gate M1 in panel A). BrdU incorporation and 7-AAD staining showed a similar cell cycle distribution in panels B, C, and D. Since only a fraction of EBV-infected primary B cells express GFP at a detectable level, the number of GFP-positive cells in the BCR+ and BCR- populations is markedly lower than in panel C. Also in contrast to panel B, most GFP+ cells in D are not apoptotic, as indicated by a minority of cells with sub-G1 DNA content.
To analyze the cell cycle status of infected and uninfected cells, BrdU incorporation assays were performed. Two-parameter FACS analysis revealed cycling cells in the EBV-infected BCR- and BCR+ cell populations but not in uninfected cells (Figure 2B). Cycling cells are typical lymphocytes by forward and sideward scatter criteria (Figure 2C and data not shown), whereas a large fraction of the entire cell population is apoptotic or has disintegrated 4 days after preparation (Figure 2B). Since EBV infects B cells exclusively, GFP+ cells are indicative of selectively infected B cells (Figure 2A), which enter the cell cycle irrespective of their surface BCR status (Figure 2D). The lower absolute number of proliferating cells in the BCR- cell population compared to BCR+ cells merely reflects the different absolute numbers of B cells in the 2 cell populations. Whereas more than 90% of BCR+ cells are also CD21+/CD19+ (data not shown), only about 15% of the cells in the BCR- preparation (Figure 1C) are B cells, which EBV infects. Taking the GFP+ cells into account (Figure 2D), BCR- cells were only slightly if at all compromised in entering S phase in comparison to BCR+/GFP+ cells as indicated by their relative distributions in the different phases of the cell cycle (Table 1). Our data indicate that EBV infection can rescue BCR- B cells from early apoptosis and induce their initial proliferation in vitro.
Table 1.
Relative cell cycle distribution
|
No gate
|
Lymphocyte gate
|
GFP+ cells (M1)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cells | G0/G1 | S | G2/M | G0/G1 | S | G2/M | G0/G1 | S | G2/M |
| BCR+ uninfected | 93 | 0 | 7 | 53 | 5 | 42 | 72 | 0 | 28 |
| BCR+ + EBV | 48 | 39 | 13 | 42 | 49 | 9 | 40 | 43 | 17 |
| BCR- + EBV | 68 | 16 | 16 | 50 | 30 | 20 | 45 | 32 | 23 |
Values are percentages of total cells.
Since BCR- B cells readily entered the cell cycle upon infection with EBV, we wished to establish BCR- single-cell lymphoblastoid cell clones in order to analyze their immunoglobulin genes at the sequence level, eventually. In a pilot experiment, we isolated BCR- cells from patient A. From 3 × 108 cells in the first preparation step, about 2 × 106 BCR- cells were obtained after depletion, and 1 × 105 were infected with EBV. In the presence of the calcineurin inhibitor cyclosporine A, the EBV-infected cells were initially plated in a 6-well cluster plate on irradiated CD40L feeder L-cell layers in the presence of IL-4 to support the survival of B cells.1,19,22 After several weeks, proliferating cells were again enriched for BCR- cells by depletion and plated in 96-well cluster plates in limiting dilutions to obtain clonal LCLs. Cells in 136 wells, which fulfilled the statistical criteria of single-cell clones, were further expanded and analyzed by FACS for surface expression of the BCR again with LC-specific antibodies.
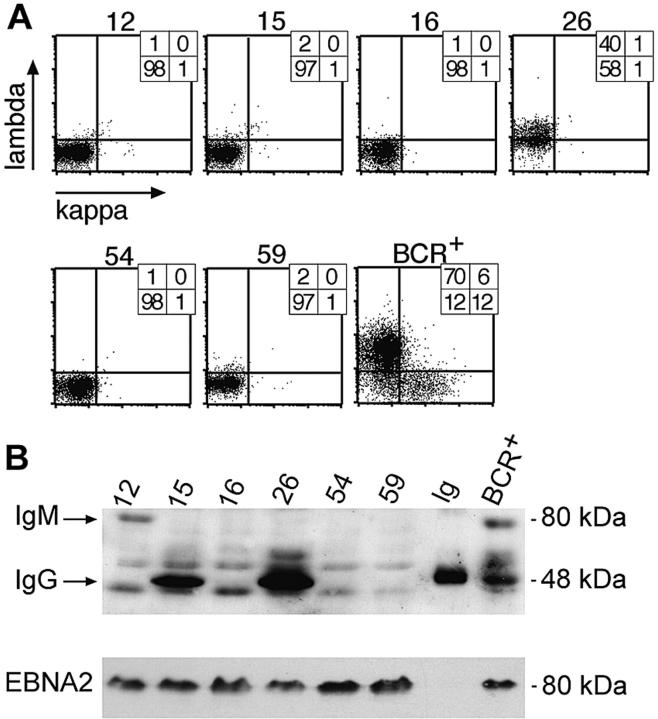
One hundred seven cell clones were found to be BCR- (Figure 3A), and 33 were further analyzed for HC expression by Western blotting (Figure 3B). Of the 33 cell clones, 22 cell clones did not express the immunoglobulin HC detectably in Western blot analysis, and 18 clones were further subjected to PCR amplification and DNA sequencing of the rearranged VH gene with VH family-specific forward and a set of JH-specific backward primers as described.20 All 18 sequenced VH loci showed multiple deviations from the germ line VH gene sequences, as expected due to somatic hypermutation, but the VH sequences were derived from only 3 individual B-cell clones. The VH sequence of clone A54 shown in Figure 4A was independently identified in 11 cell clones, whereas the VH sequence from clone A16 in Figure 4B was identified twice. Five identical VH sequences did not show any obvious nonfunctional mutational defects (data not shown). We interpret this finding to indicate that the bulk culture was made up of descendants of a very limited number of individual B cells at the time of plating at limiting dilution.
Figure 3.
Analysis of clonal LCLs for expression of immunoglobulin chains by FACS and Western blot immunodetection. (A) Single cell clones were obtained from limiting dilution assays and tested for surface BCR expression with light chain–specific antibodies by FACS. Six LCL clones are shown as representative examples, together with a control bulk population obtained from EBV-infected BCR+ B cells (BCR+). The numbers indicate the percentile of cells in the respective quadrants. (B) Single-cell clones no. 16, 54, and 59, which all fail to express λ or κ light chains at their cell surfaces, as shown in panel A, also are negative with regard to intracellular expression of the IgG-HC (50 kDa) as revealed by Western blotting. Clones no. 15 and 26 were immunoglobulin HC positive in Western blotting, but only no. 26 expressed a surface BCR, as shown in panel A. Clone no. 12, which does not express a surface BCR, is shown to be IgG-HC negative but IgM-HC (75 kDa) positive in Western blotting. The lane labeled “Ig” is a positive control with a purified commercial human IgG preparation. Another positive control is a pool of BCR+/EBV-infected LCLs. All LCLs are EBV infected, since they express the latent viral gene product EBNA2, which also served as a loading control.
Figure 4.
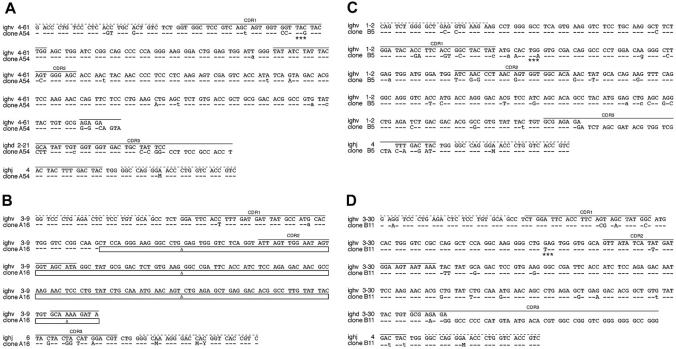
Sequence analysis of clonal LCLs derived from EBV-infected BCR- B cells. The sequences of 4 single-cell LCL clones are shown, which carry a crippled heavy chain allele and fail to express a functional BCR. A detailed comparison of VDJH gene regions of the 4 sequenced clones (bottom lines) to the most homologous germ line genes (top lines) is shown. Sequence identity is indicated by dashes, and nucleotide exchanges introduced by somatic hypermutation are indicated. Uppercase characters denote replacement mutations, and lowercase characters describe silent mutations. (A) In clone A54 an in-frame stop codon (marked with asterisks) in CDR1 (solid lines) abrogates HC expression. (B) In clone A16 only the variable and the joining segment could be determined, since this rearranged heavy chain allele suffers from a massive internal deletion, which is indicated by boxes. The deletion also causes a frame shift mutation such that the C segments are out of phase. The clones B5 (C) and B11 (D) are crippled due to in-frame stop codons (asterisks). The forward and backward primer binding sites (dotted lines) contain wobbled bases: M (adenosine or a cytidine), and Y (cytidine or a thymidine).
We therefore changed our strategy to avert the clonal dominance seen in the first experiment. From 2 patients, CD77+ GC B cells were prepared, λ+/κ+ cells were depleted along with remaining T cells, and about 105 BCR- CD77+ cells were infected with EBV (Figure 1D-F). The infected cells were cultivated on plastic in the absence of CD40L/IL-4 stimulation in bulk cultures for 3 weeks. Surviving cells were enriched for BCR- cells and plated in limiting dilution experiments. A total number of 185 proliferating B-cell clones were analyzed by FACS for LC surface expression, of which 104 clones were found to be BCR-. Of 67 clones analyzed by Western blotting, 14 did not express the HC detectably. The VH sequence of the PCR products of these B-cell clones showed either a stop codon (patient B, clone 5 and clone 11; Figure 4C-D) or a frame shift mutation (patient C; data not shown).
All crippled B-cell clones were analyzed for the status of the second HC allele. Using 2 primer pairs, which either detect the germ line composition or discriminate between DJ or VDJ rearrangements in the HC locus, we found PCR products characteristic of DHJH rearrangements in all 5 cases, but no germ line situation or additional VHDHJH rearrangements, indicating that the second HC alleles are nonfunctional (data not shown).
In all experiments we found 5 proliferating B-cell clones, which were genotypically BCR-. Their VH sequences display deviations from the germ line V gene database entries (Figure 4); a finding that is a hallmark of somatic hypermutation and underlines the GC origin or passage of the isolated B cells. The clone A54 shows a C to G transversion in CDR1, which leads to an in-frame translational stop codon (Figure 4A). The VH gene of the LCL A16 is characterized by a drastic deletion of all of CDR2 and parts of CDR3, which also causes a frame shift mutation. The 2 clones from patient B are sterile for BCR expression due to in-frame stop codons (Figure 4C-D). One clone derived from patient C suffers from a frame shift mutation presumably in the region of the D segment, such that the C segments of the immunoglobulin HC gene are out of phase (data not shown). Taken together, the sequence analysis of 5 immunoglobulin HC genes indicates that none of the isolated clonal LCLs can express a functional BCR. Since the second alleles of the 5 B-cell clones showed only a DHJH rearrangement, we conclude that the completely rearranged and sequenced immunoglobulin genes had been functional in the naive B-cell status before the cells underwent somatic hypermutation in a GC reaction where they all experienced loss of function mutations.
Discussion
The rescue of proapoptotic germinal center B cells by EBV could be the crucial initial step in the evolution of B cells to Hodgkin Reed-Sternberg cells and similarly involved in lymphomagenesis of posttransplantation lymphomas. A certain fraction of these malignancies are characterized by tumor B cells, which fail to express a surface BCR because the immunoglobulin genes are either transcriptionally silent or inactivated by somatic mutations.23,24 Although numerous reports describe the establishment of LCLs from human pre- and pro-B cells in vitro,25-27 and EBV equally infects and growth transforms naive and memory B cells,28 the survival of B cells depends on a functional BCR.2 Therefore, it was unclear whether EBV can infect proapoptotic germinal center B cells ex vivo and rescue those that are destined to undergo programmed cell death because they are BCR-. This is a relevant issue since germinal center B cells have a very limited life span ex vivo, whereas the expression of counteracting viral genes occurs relatively late after B-cell infection. The relative survival of germinal center B cells in cell culture was determined to be between 10% and 25% of the initially seeded cells 16 to 24 hours after isolation,1,29,30 and the ex vivo lifespan of BCR- cells is likely to be shorter than the BCR+ subpopulation. This likely assumption is critical since the 2 viral genes LMP1 and LMP2A are expressed later during EBV infection.
For several reasons, we expect LMP1 and LMP2A to be crucially involved in rescuing proapoptotic GC B cells. LMP1 expression induces a number of antiapoptotic cellular genes and leads to NF-κB activation18 similar to a ligand-activated CD40 receptor.10,31 LMP2A acts as a constitutive activated receptor and has been recognized to mimic physiologic functions of a “trickling” B-cell receptor32 in vitro and in a transgenic mouse model.33 It is therefore obvious to postulate that LMP2A expression in GC B cells might rescue proapoptotic B cells since LMP2A constitutes a fully functional BCR analog in vivo.34 Several publications suggest a correlation between crippling mutations and LMP2A-expressing HRS cells because most cases of Hodgkin lymphoma with crippled BCR are EBV positive.3,5,35,36 In fact, preliminary experiments indicate that LMP2A is essential to growth transform primary B cells, which are phenotypically BCR-. To our knowledge, the expression kinetic of LMP2A has not been investigated, but LMP1 is known to become expressed near the levels ordinarily found in LCLs 2 to 3 days after infection.37-39 This is because LMP1 as well as LMP2A are indirectly but concomitantly up-regulated by EBNA2, whose expression precedes that of LMP1.6 In contrast, 2 additional viral genes, which are known or suspected to have antiapoptotic functions, are maximally expressed within the first 24 hours of B-cell infection such that the late onset of LMP1 and LMP2A expression might be compensated by very early viral functions.40
Our findings are consistent with the hypothesis that EBV infection can rescue proapoptotic GC B cells in vivo and are in accordance with 2 similar reports.41,42 As a consequence, the resultant pool of EBV-positive rescued cells is that from which EBV-positive Hodgkin lymphoma and posttransplantation lymphoma subsequently arise.24 However, the in vivo phenotype of EBV-associated malignancies such as Hodgkin tumors differs from the cell lines established here, which are classic lymphoblastoid cell lines (Figure 3 and data not shown). Whereas EBV adopts a transcription program in LCLs, which is termed latency III and includes the expression of 11 EBV genes, HRS cells are characterized by a restricted set of expressed EBV latent genes, termed latency II.24 This is characterized by expression of EBNA1 mRNA and the LMP1, LMP2A, and LMP2B transcripts, among others.43 At the single-cell level, monoclonal antibody staining has confirmed the expression of EBNA144 and high levels of both LMP1 and LMP2A7,8,45 in the HRS cells. These findings suggest that proapoptotic GC cells could be rescued through the expression of LMP1 and/or LMP2A. Indeed, it has been shown that germinal center cells in healthy carriers of the virus already express typical latency II genes, including LMP2A.46 Since LMP2A is already known to substitute for the BCR in transgenic animals,16 it is a likely candidate that might play a critical role in rescue of BCR-defective GC B cells in vivo and in vitro.
Taken together, our data formally demonstrate that EBV actively subverts the physiologic, rapid death of proapoptotic GC B cells even among those that have lost the capacity to encode a functional BCR and are therefore prone to die. It is the focus of ongoing work to examine the role of LMP2A and other EBV latent genes, which are expressed during latency II, and determine whether they are essential for growth transformation and proliferation of BCR-defective GC B cells.
Acknowledgments
We are grateful to R. Küppers, U. Strobl, and B. Sugden for suggestions and comments.
Prepublished online as Blood First Edition Paper, August 2, 2005; DOI 10.1182/blood-2005-06-2341.
Supported by grant SFB455 of the Deutsche Forschungsgemeinschaft and by Public Health Service Grant CA70723.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Liu YJ, Joshua DE, Williams GT, Smith CA, Gordon J, MacLennan IC. Mechanism of antigen-driven selection in germinal centres. Nature. 1989;342: 929-931. [DOI] [PubMed] [Google Scholar]
- 2.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117: 787-800. [DOI] [PubMed] [Google Scholar]
- 3.Marafioti T, Hummel M, Foss HD, et al. Hodgkin and Reed-Sternberg cells represent an expansion of a single clone originating from a germinal center B-cell with functional immunoglobulin gene rearrangements but defective immunoglobulin transcription. Blood. 2000;95: 1443-1450. [PubMed] [Google Scholar]
- 4.Bräuninger A, Küppers R, Strickler JG, Wacker HH, Rajewsky K, Hansmann ML. Hodgkin and Reed-Sternberg cells in lymphocyte predominant Hodgkin disease represent clonal populations of germinal center-derived tumor B cells. Proc Natl Acad Sci U S A. 1997;94: 9337-9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanzler H, Küppers R, Hansmann ML, Rajewsky K. Hodgkin and Reed-Sternberg cells in Hodgkin's disease represent the outgrowth of a dominant tumor clone derived from (crippled) germinal center B cells. J Exp Med. 1996;184: 1495-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieff E, Rickinson AB. Epstein-Barr virus and its replication. In: Knipe DM, Howley PM, Griffin DE, Straus SE, eds. Fields' Virology. Vol 2. 4th ed. Philadelphia, PA: Lippincott-Williams & Wilkins; 2001: 2511-2573. [Google Scholar]
- 7.Herbst H, Dallenbach F, Hummel M, et al. Epstein-Barr virus latent membrane protein expression in Hodgkin and Reed-Sternberg cells. Proc Natl Acad Sci U S A. 1991;88: 4766-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niedobitek G, Kremmer E, Herbst H, et al. Immunohistochemical detection of the Epstein-Barr virus–encoded latent membrane protein 2A in Hodgkin's disease and infectious mononucleosis. Blood. 1997;90: 1664-1672. [PubMed] [Google Scholar]
- 9.Brink AA, Dukers DF, van den Brule AJ, et al. Presence of Epstein-Barr virus latency type III at the single cell level in post-transplantation lymphoproliferative disorders and AIDS related lymphomas. J Clin Pathol. 1997;50: 911-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kilger E, Kieser A, Baumann M, Hammerschmidt W. Epstein-Barr virus–mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998;17: 1700-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammerschmidt W, Sugden B. Epstein-Barr virus sustains Burkitt's lymphomas and Hodgkin's disease. Trends Mol Med. 2004;10: 331-336. [DOI] [PubMed] [Google Scholar]
- 12.Gires O, Zimber-Strobl U, Gonnella R, et al. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 1997;16: 6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235: 241-251. [DOI] [PubMed] [Google Scholar]
- 14.Reth M. Antigen receptor tail clue. Nature. 1989;338: 383-384. [PubMed] [Google Scholar]
- 15.Cambier JC. Antigen and Fc receptor signaling: the awesome power of the immunoreceptor tyrosine-based activation motif (ITAM). J Immunol. 1995;155: 3281-3285. [PubMed] [Google Scholar]
- 16.Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9: 405-411. [DOI] [PubMed] [Google Scholar]
- 17.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci U S A. 1998;95: 8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dirmeier U, Hoffmann R, Kilger E, et al. Latent membrane protein 1 of Epstein-Barr virus coordinately regulates proliferation with control of apoptosis. Oncogene. 2005;24: 1711-1717. [DOI] [PubMed] [Google Scholar]
- 19.Banchereau J, de Paoli P, Valle A, Garcia E, Rousset F. Long-term human B cell lines dependent on interleukin-4 and antibody to CD40. Science. 1991;251: 70-72. [DOI] [PubMed] [Google Scholar]
- 20.Küppers R. Molecular single-cell PCR analysis of rearranged immunoglobulin genes as a tool to determine the clonal composition of normal and malignant human B cells. In: Gu H, Rajewsky K, eds. Methods Molecular Biology. Vol 271. Totowa, NJ: Humana Press; 2004: 225-238. [DOI] [PubMed] [Google Scholar]
- 21.Müschen M, Rajewsky K, Bräuninger A, et al. Rare occurrence of classical Hodgkin's disease as a T cell lymphoma. J Exp Med. 2000;191: 387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimber-Strobl U, Kempkes B, Marschall G, et al. Epstein-Barr virus latent membrane protein (LMP1) is not sufficient to maintain proliferation of B cells but both it and activated CD40 can prolong their survival. EMBO J. 1996;15: 7070-7078. [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas RK, Re D, Wolf J, Diehl V. Part I: Hodgkin's lymphoma–molecular biology of Hodgkin and Reed-Sternberg cells. Lancet Oncol. 2004;5: 11-18. [DOI] [PubMed] [Google Scholar]
- 24.Küppers R. B cells under influence: transformation of B cells by Epstein-Barr virus. Nat Rev Immunol. 2003;3: 801-812. [DOI] [PubMed] [Google Scholar]
- 25.Katamine S, Otsu M, Tada K, et al. Epstein-Barr virus transforms precursor B cells even before immunoglobulin gene rearrangements. Nature. 1984;309: 369-372. [DOI] [PubMed] [Google Scholar]
- 26.Hansson M, Falk K, Ernberg I. Epstein-Barr virus transformation of human pre-B cells. J Exp Med. 1983;158: 616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory CD, Kirchgens C, Edwards CF, et al. Epstein-Barr virus–transformed human precursor B cell lines: altered growth phenotype of lines with germ-line or rearranged but nonexpressed heavy chain genes. Eur J Immunol. 1987;17: 1199-1207. [DOI] [PubMed] [Google Scholar]
- 28.Ehlin-Henriksson B, Gordon J, Klein G. B-lymphocyte subpopulations are equally susceptible to Epstein-Barr virus infection, irrespective of immunoglobulin isotype expression. Immunology. 2003;108: 427-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangeney M, Richard Y, Coulaud D, Tursz T, Wiels J. CD77: an antigen of germinal center B cells entering apoptosis. Eur J Immunol. 1991;21: 1131-1140. [DOI] [PubMed] [Google Scholar]
- 30.Lebecque S, de Bouteiller O, Arpin C, Banchereau J, Liu YJ. Germinal center founder cells display propensity for apoptosis before onset of somatic mutation. J Exp Med. 1997;185: 563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zarnegar B, He JQ, Oganesyan G, Hoffmann A, Baltimore D, Cheng G. Unique CD40-mediated biological program in B cell activation requires both type 1 and type 2 NF-kappaB activation pathways. Proc Natl Acad Sci U S A. 2004;101: 8108-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381: 751-758. [DOI] [PubMed] [Google Scholar]
- 33.Merchant M, Swart R, Katzman RB, et al. The effects of the Epstein-Barr virus latent membrane protein 2A on B cell function. Int Rev Immunol. 2001;20: 805-835. [DOI] [PubMed] [Google Scholar]
- 34.Casola S, Otipoby KL, Alimzhanov M, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5: 317-327. [DOI] [PubMed] [Google Scholar]
- 35.Muschen M, Rajewsky K, Brauninger A, et al. Rare occurrence of classical Hodgkin's disease as a T cell lymphoma. J Exp Med. 2000;191: 387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuppers R. Molecular biology of Hodgkin's lymphoma. Adv Cancer Res. 2002;84: 277-312. [DOI] [PubMed] [Google Scholar]
- 37.Martin JM, Veis D, Korsmeyer SJ, Sugden B. Latent membrane protein of Epstein-Barr virus induces cellular phenotypes independently of expression of Bcl-2. J Virol. 1993;67: 5269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allday MJ, Crawford DH, Griffin BE. Epstein-Barr virus latent gene expression during the initiation of B cell immortalization. J Gen Virol. 1989;70: 1755-1764. [DOI] [PubMed] [Google Scholar]
- 39.Alfieri C, Birkenbach M, Kieff E. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology. 1991;181: 595-608. [DOI] [PubMed] [Google Scholar]
- 40.Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. In press. [DOI] [PMC free article] [PubMed]
- 41.Bechtel D, Kurth J, Unkel C, Kuppers R. Transformation of BCR-deficient germinal center B cells by EBV supports a major role of the virus in the pathogenesis of Hodgkin and post transplant lymphoma. Blood. Prepublished on August 30, 2005, as DOI 10.1182/blood-2005-06-2342. [DOI] [PubMed]
- 42.Chaganti S, Bell AI, Begue-Pastor N, et al. Epstein-Barr virus infection in vitro can rescue germinal centre B cells with inactivated immunoglobulin genes. Blood. Prepublished on August 25, 2005, as DOI 10.1182/blood-2005-06-2327. [DOI] [PubMed]
- 43.Deacon EM, Pallesen G, Niedobitek G, et al. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177: 339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grasser FA, Murray PG, Kremmer E, et al. Monoclonal antibodies directed against the Epstein-Barr virus–encoded nuclear antigen 1 (EBNA1): immunohistologic detection of EBNA1 in the malignant cells of Hodgkin's disease. Blood. 1994;84: 3792-3798. [PubMed] [Google Scholar]
- 45.Pallesen G, Hamilton-Dutoit SJ, Rowe M, Young LS. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet. 1991;337: 320-322. [DOI] [PubMed] [Google Scholar]
- 46.Babcock GJ, Hochberg D, Thorley-Lawson AD. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000;13: 497-506. [DOI] [PubMed] [Google Scholar]