Abstract
Acute myeloid leukemia cells have constitutive activation of phosphatidylinositol 3(PI3) kinase and require PI3 kinase activation for survival; however, the function of the PI3 kinase pathway in the survival of leukemic cells is poorly defined. We have studied the role of one PI3 kinase substrate, mTOR (mammalian target of rapamycin), in primary leukemic cells. In initial experiments, we have defined a novel growth medium that improves survival of acute myeloid leukemia (AML) blasts in long-term suspension culture and the survival of leukemic stem cells in short-term cultures. Inhibition of mTOR using rapamycin leads to a modest decrease in cell survival after 2 days of incubation with more significant decrease in survival after 7 days of culture. However, when rapamycin is added to etoposide in 2-day cultures, there is a dramatic increase in the cytotoxicity of etoposide against AML blasts. Furthermore, etoposide consistently decreased the engraftment of AML cells in nonobese diabetic/severe combined immunodeficient (NOD/SCID) animals, and this effect was enhanced by coincubation with rapamycin, demonstrating that mTOR regulates survival of AML stem cells after etoposide treatment. These results suggest that rapamycin in combination with etoposide-based chemotherapy may be efficacious in the treatment of AML.
Introduction
Acute myeloid leukemia is a malignant hematopoietic disease characterized by the accumulation of immature myeloblasts which are blocked in cellular differentiation and resistant to cell death.1 Work from several laboratories has demonstrated that acute myeloid leukemia (AML) arises from leukemic stem cells which have the phenotype lineage negative (lin-), CD34+, CD38-.2,3 Injection of these cells into immunocompromised nonobese diabetic/severe combined immunodeficient (NOD/SCID) animals leads to a transplantable disease with phenotypic and molecular characteristics of the human disease. Despite extensive study, the molecular pathogenesis of AML is still only partially understood. A recent model suggests that development of AML requires multiple genetic changes which alter different cellular programs.4 Many transcription factor fusion proteins such as AML1/ETO (acute myeloid leukemia 1/eight twenty one) or MLL/AF9 (mixed lineage leukemia/ALL1 fused gene from chromosome 9) block myeloid differentiation, providing one necessary event for leukemogenesis.5,6 Disordered cell growth, such as that generated by the BCR/ABL (breakpoint cluster region/abelson murine leukemia) oncogene in chronic myeloid leukemia (CML) is a proposed necessary second event. However, the mechanism of altered cell growth in AML is poorly understood. Several genetic mutations in growth regulatory genes, including Flt3, Ras, and c-kit, have been described.7-10 In some patient samples inhibition of Flt3 causes decreased survival of blasts; however, Flt3 inhibitors as single agents have had only modest efficacy in phase 1 clinical trials.11-13 Similarly, the exact role of Ras and c-kit mutations in maintaining leukemic cell survival is only partially defined. To address this issue, we and others have recently studied the phosphotidylinositide-3-kinase (PI3K) pathway in AML. We have demonstrated that PI3 kinase is constitutively activated in the majority of AML patient samples and that activation of PI3 kinase is necessary for the survival of AML blasts.14-18 However, the downstream effectors of PI3 kinase activity in leukemic cells are unknown.
PI3K activates multiple downstream effector pathways.19 Most of the described actions of PI3K require activation of Akt (protein kinase B [PKB], subsequently referred to as Akt).20,21 Activated Akt may itself lead to activation of NFκB (nuclear factor-κB), bclxl (B-cell leukemia XL), mTOR (mammalian target of rapamycin), and inactivation of GSK-3β (glycogen synthase kinase-3β), p53, and FOXO (Forkhead box class O) family proteins.20 The effects of these pathways are varied in different cell types; however, multiple studies demonstrate that the PI3K/Akt pathway regulates cell survival, proliferation, growth, and other effects, although the exact effects are cell type specific. Of these effectors, NFκB is known to be constitutively activated in AML blasts and required for leukemic cells to survive genotoxic stress.22 A recent report also demonstrated constitutive phosphorylation of FOXO1 and GSK in AML blasts.16 Although not demonstrated, it is likely that activation of these mediators is dependent on PI3 kinase signaling. Thus, several lines of evidence both confirm constitutive activation of PI3 kinase in AML and suggest that it plays a critical role in the survival of AML blasts. However, the role of the different downstream effector pathways in AML biology and whether that information can be targeted clinically remains poorly defined.
To address these issues, we have chosen to study 1 PI3 kinase effector, the mammalian target of rapamycin or mTOR protein. TOR was discovered in yeast as the target of the macrolide antifungal, rapamycin.23 Rapamycin was initially described as a product of a soil bacterium and found to have both antiproliferative and immunosuppressive activities in humans.24,25 These biologic activities are mediated by binding of rapamycin to the carrier protein, FKBP (FK506-binding protein).26 The rapamycin/FKBP complex binds to and inhibits the activity of mTOR.27 mTOR is a large serine threonine kinase with homology to ATM (ataxia telangiectasia mutated) and ATR (telangiectasia and Rad3-related protein) as well as other members of the PIKK (PI3 kinase-related kinase) family of kinases.28 The protein appears to function as a coordinating sensor of nutrient status by responding to both amino acid and glucose deprivation and regulating glucose and amino acid uptake by cells.29 It also modulates the activity of the p70S6 kinase and 4EBP-1 (4E-binding protein-1) and 4EBP-2 proteins essential for the initiation of protein translation.30 Thus, mTOR may act as a checkpoint sensor indicating to cells that there are sufficient nutrients available to proceed through the cell cycle. Importantly, rapamycin is a clinically tested drug that is approved for use as an immunosuppressant and, in appropriate doses, is well tolerated.31 Given the biologic and potential clinical relevance of this pathway, we undertook to study the role of mTOR in the survival of AML blasts.
We have previously demonstrated that p70S6 kinase and 4EBP-1 are constitutively phosphorylated in AML blasts after thawing.14 Furthermore, we observed that in an AML cell line, mTOR inhibition enhanced the toxicity of cytosine arabinoside (Ara-C), a nucleoside analog that inhibits cells by blocking DNA synthesis. Initial attempts to extend these observations to primary cells were difficult because of poor survival of AML blasts in culture after thawing and because of a lack of efficacy of cytosine arabinoside on AML blasts in culture where the cells are not actively cycling. We therefore undertook to develop new culture conditions for AML blasts and to test the response to other chemotherapeutic agents in culture. Here, we demonstrate improved survival of AML blasts in a novel medium containing low serum concentrations and combinations of endothelial cell cytokines. We have confirmed activation of the mTOR pathway in AML blasts in these culture conditions. Treatment of cells with etoposide, a topoisomerase inhibitor that induces double-strand DNA breaks in noncycling cells, showed that the drug induced apoptosis. This effect is enhanced by addition of rapamycin which inhibits the mTOR pathway. This inhibition of survival extends to the stem cell component of AML cells because incubation of AML blasts in combinations of etoposide and rapamycin inhibits the ability of the cells to engraft in NOD/SCID animals. These results demonstrate that mTOR is activated in AML blasts and that inhibition of mTOR alters the apoptotic threshold of the cells in the presence of etoposide.
Materials and methods
Cells
Leukemia samples were obtained from the Stem Cell and Leukemia Core Facility at the University of Pennsylvania Cancer Center. Samples were obtained from patients presenting with acute leukemia seen at the Hospital of the University of Pennsylvania after informed consent in accordance with institutional guidelines. Bone marrow or peripheral blood samples were collected, and samples were prepared by Ficoll gradient centrifugation. Mononuclear cells were frozen as viable cells in fetal calf serum and 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen. Percentage of blasts in the unmanipulated samples as determined morphologically and by flow cytometry was recorded. Samples were chosen with greater than 70% blasts.
CD34 cells were obtained from the National Heart, Lung, and Blood Institute (NHLBI) Programs of Excellence in Gene Therapy Cell Processing Core (http://www.med.cornell.edu/pegt/pegtNSC_HSCPC.html). Purity testing demonstrated the samples to be consistently greater than 80% CD34+. All cells were frozen in 10% DMSO and stored in liquid nitrogen.
Tissue culture
Patient samples were rapidly thawed at 37°C and diluted with Dulbecco modified Eagle medium (DMEM). Cells were then respun and plated at 1 × 106 cells/mL. For growth curves, small aliquots were removed at indicated intervals, and viable cell number was demonstrated by trypan blue staining. For toxicity experiments, relative cell survival was compared using an XTT assay, a colorimetric assay for viable cell number, according to the manufacturer's recommendations (Molecular Probes, Eugene, OR). Results were measured in triplicate using absorption at 515 nm. For characterization of culture conditions for AML cells, cells were incubated either in a serum-free medium with hematopoietic cytokines (BIT 9500; Stem Cell Technologies, Vancouver, Canada) with interleukin-3 (IL3; 5 ng/mL), IL6 (10 ng/mL), and stem cell factor (SCF; 50 ng/mL) (all from R&D Systems, Minneapolis, MN), endothelial growth media (EGM) or EGM2 (Cambrex BioScience, Walkersville, MD). EGM contains bovine brain extract (BBE), heparin, and epidermal growth factor (EGF). EGM2 contains 2% fetal calf serum, hydrocortisone, heparin, antibiotics, EGF, human fibroblast growth factor (hFGF-B), insulin-like growth factor (R3-IGF-1), vascular endothelial growth factor (VEGF), and ascorbic acid. Cells were plated at 1 × 106 to 2 × 106 cells/mL for described experiments and incubated in 5% CO2. Rapamycin was purchased from Sigma-Aldrich (St Louis, MO) and diluted in DMSO.
Human umbilical vein endothelial cells (HUVECs) were purchased from Cambrex and thawed and cultured according to their recommendations. Cells were grown either in EGM or EGM2. HUVEC-conditioned medium was generated by growing HUVECs to confluence. Medium was then removed and replaced with additional medium. Cells were grown for an additional 2 days and medium was harvested. Medium was filtered through a 0.45-μM filter to exclude passage of cells. Medium was divided into aliquots and frozen prior to use.
Western blotting
Cells were pelleted by centrifugation, washed once in cold phosphate-buffered saline (PBS) and repelleted. Cells were lysed in lysis buffer containing 1% Triton-X 100, 150 mM NaCl, 50 mM Tris (tris(hydroxymethyl)aminomethane)-HCl with protease and phosphatase inhibitors (10 μg/mL aprotinin, 10 μg/mL leupeptin, 1 μg/mL pepstatin, 5 mM EDTA [ethylenediaminetetraacetic acid], 5 mM sodium orthovanadate, 50 mM NaF, 50 mM Na-pyrophosphate, 150 μM PMSF [phenylmethylsulfonyl fluoride]) for 5 minutes on ice. Total cell lysate (100 μg) per sample was loaded on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and separated by electrophoresis. Proteins were blotted onto nitrocellulose using a semidry transfer apparatus (Bio-Rad, Mississauga, ON, Canada). Blots were blocked in 5% dried milk solution diluted in TBST (Tris-buffered saline/0.05% Tween 20), incubated with indicated antibody, and signal detected with an horseradish peroxidase (HRP)-conjugated secondary antibody and enhanced chemiluminescence (ECL) detection (Amersham Biosciences, Uppsala, Sweden). Blots were stripped with 2% SDS at 50°C for 30 minutes and reprobed. All antibodies were from Cell Signaling (Beverly, MA).
Annexin V assay
Cells were incubated in DMSO alone, etoposide, rapamycin, or a combination of etoposide and rapamycin as indicated for 24 to 48 hours. Cells were pelleted by centrifugation and incubated with anti-annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI). Single-cell suspensions were analyzed by fluorescence-activated cell scanner (FACScan; Becton Dickinson, San Jose, CA). Early apoptotic cells were scored as annexin V positive, PI negative to exclude necrotic cells.
NOD/SCID engraftment studies
Six- to 8-week-old animals were sublethally irradiated 1 day prior to injection with 300 cGy. For each animal, 5 to 10 × 106 total mononuclear cells from leukemic samples were allowed to recover in medium for 2 hours and then either immediately injected or incubated overnight in appropriate conditions prior to injection. Animals were monitored for signs of illness and humanely killed 7 weeks after injection. Both femurs were flushed with normal saline to collect bone marrow cells. Bone marrow red blood cells were removed using hypotonic buffer, and remaining cells were stained with directly conjugated human antigen-specific antibodies (CD45, CD33, CD19; all from Pharmingen, San Diego, CA). Cells were analyzed on a FACS Calibur (Becton Dickinson, San Jose, CA) using Cellquest or FlowJo software. (FlowJo, Ashland, OR) Studies were approved by the University of Pennsylvania animal care committee.
Results
Patient sample characteristics
Samples were collected from all patients seen at the University of Pennsylvania Hospital for a diagnosis of acute myelogenous leukemia who consented to have blood or bone marrow taken for research purposes. Most samples were obtained at the time of diagnosis and prior to initiation of therapy. The classification system for AML has been recently changed, but these samples were classified according to the French-American-British (FAB) system. Samples represented the following subtypes of AML: M1, M2, M4, and M5, although the majority of samples were M1 or M4 subtype (Table 1). M3 samples were excluded, and no M6 or M7 samples were tested. Samples were selected from the tissue bank if they contained an adequate number of cells to complete multiple studies and had at least 70% blasts in the initial isolate prior to manipulation.
Table 1.
Characteristics of patient samples
| Row no. | Sample no. | FAB subtype | Blasts, % | Sample type |
|---|---|---|---|---|
| 1 | 134 | NOS | 99 | Pheresis |
| 2 | 162 | M4 | 99 | Pheresis |
| 3 | 228 | M4 | 96 | PB |
| 4 | 258 | M4 | 99 | Pheresis |
| 5 | 321 | M0 | 92 | PB |
| 6 | 325 | NOS | 99 | Pheresis |
| 7 | 329 | M4/5 | 76 | PB |
| 8 | 330 | M4 | 99 | Pheresis |
| 9 | 346 | M1 | 94 | PB |
| 10 | 350 | M2 | 99 | PB |
| 11 | 353 | M1 | 79 | PB |
| 12 | 364 | M1 | 99 | Pheresis |
| 13 | 374 | M4 | 95 | PB |
| 14 | 401 | M1 | 91 | PB |
| 15 | 416 | M1 | 91 | PB |
| 16 | 434 | M1 | 70 | BM |
PB indicates peripheral blood; BM, bone marrow.
Characterization of novel growth conditions for primary AML blasts
Maintenance and growth of primary AML cells in culture has been problematic; however, a recent report suggested that cocultivation of AML cells with human umbilical vein endothelial cells (HUVECs) would improve survival.32 HUVECs have been used as stromal cells in several cell systems and are known to produce multiple cytokines.33 We studied the effect of cocultivation with HUVECs on the survival of primary AML cells. Growth of a typical sample in serum-free medium with interleukin 3 (IL3), interleukin 6 (IL6), and stem cell factor (SCF) maintained viable cells for 7 to 10 days on average (Figure 1A, □) but there was no substantial growth of cells in these conditions. Cocultivation of AML cells with HUVECs provided a modest increase in cell growth in several samples (Figure 1A, ⋄; data not shown). To determine whether HUVECs were providing a secretable factor or providing an appropriate surface for growth, we also compared the growth of AML cells in HUVEC-conditioned medium. This medium was harvested from confluent HUVECs as described under “Materials and methods.” These conditions consistently generated increased cell numbers in 10-day cultures, and we elected to study these conditions further. We further tested HUVEC-conditioned medium in longer-term cultures (Figure 1; data not shown). As shown in Figure 1B (○) HUVEC-conditioned medium was able to sustain viable AML cells for approximately 2 weeks. For these experiments we used a commercially developed medium for maintenance of HUVECs. This medium, designated EGM for endothelial growth medium can be purchased as an undefined medium containing bovine brain extract or a defined medium (EGM2) containing several growth factors as shown under “Materials and methods.” As controls, we studied the growth of AML cells in EGM and EGM2. Unexpectedly, we found that both EGM and EGM2 maintained AML cells better than HUVEC-conditioned medium (Figure 1B-C). With EGM and EGM2 we consistently saw improved survival compared with serum-free medium with hematopoietic cytokines or HUVEC-conditioned medium (CM; n = 5). In some samples, survival was sustained only a few additional days (data not shown), although in 2 of the 5 samples tested, cells were able to survive for 4 weeks as shown (Figure 1B-C). Because EGM2 is the more defined reagent, further characterization was performed using EGM2 medium alone. To determine whether growth in EGM2 medium affected differentiation, we examined cells by flow cytometry and by morphology. Morphologically, cells continued to appear blastlike with large cells and a large nuclear to cytoplasmic ratio (data not shown). By flow cytometry, most samples showed a decrease in the percentage of cells expressing the immature hematopoietic marker, CD34, and an increase in expression of the myeloid differentiation marker, CD13 (data not shown). These studies suggest that AML blasts in EGM2 medium may have some slight evidence of maturation but largely remain as undifferentiated cells. These studies demonstrate that a medium containing only endothelial cell growth factors, not hematopoietic cytokines, is capable of sustaining primary AML cells in culture.
Figure 1.
EGM and EGM2 media support the survival of leukemic stem cells. (A-C) Primary AML cells were thawed and plated at 1 × 106 cells/mL in indicated medium. Viable cell number was counted by trypan blue exclusion. (D) Primary AML cells were thawed and allowed to recover for 2 hours. Viable cells were counted, and 5 million cells per animal were injected immediately (day 0) or after 1 to 2 days of incubation in EGM2 medium. No adjustments to cell numbers were made after incubation. Cells were injected into sublethally irradiated NOD/SCID mice, and animals were monitored for 7 weeks. Animals were humanely killed and bilateral femurs were harvested. Percentage shown is average of human CD45+/CD33+ cells in the bone marrow. Each dot indicates 1 animal, and the central bar is the mean of the average engraftment.
EGM2 medium maintains SCID leukemia-initiating cells in culture
Previous studies have shown that AML exists as a hierarchy of cells arising from leukemic stem cells that can be assayed by xenotransplantation into immunocompromised NOD/SCID mice.2 Such stem cells are designated SCID leukemia-initiating cells or SL-ICs. We and others have previously shown that SL-IC numbers decrease after even short-term culture in serum-free medium with hematopoietic cytokines.34 To determine whether EGM2 medium would sustain SL-ICs, we thawed AML cells and injected cells that were freshly thawed or cells cultured in EGM for 1 or 2 days (Figure 1D). Cells were then injected into NOD/SCID animals, and animals were analyzed for engraftment 6 weeks after injection. Five million cells were injected per animal based on initial cell counts with no adjustment after incubation. As can be seen in Figure 1D, no significant change in the levels of engraftment were seen comparing cells injected immediately after thawing with those cultured for 1 to 2 days in EGM2 medium. Similar results were seen with 2 other patient samples (data not shown; cf, Figure 6B, DMSO-treated cells compared with freshly injected cells). These results suggest that EGM2 medium provides improved survival of SL-ICs in culture for up to 2 days. From these results, we have used these culture conditions to study AML survival in the experiments described here.
Figure 6.
AML stem cells require mTOR for survival of genotoxic stress. Cells were thawed, and 5 × 106 cells per animal were injected immediately or incubated overnight in the presence of indicated compound. Etoposide was used at 5 μM and rapamycin at 10 μM. Cells were injected into sublethally irradiated mice and monitored for 7 weeks. Animals were humanely killed, bilateral femurs were harvested and analyzed for the presence of human cells. Percentage engraftment is indicated as percentage of human CD45+/CD33+ cells present. (C) Purified CD34 cells (1 × 106) per animal were incubated overnight in the indicated conditions. Cells were injected in NOD/SCID mice as above and analyzed as above. In this case, analysis confirmed expression of CD45+CD33+ cells and CD45+CD19- cells, indicating myeloid and lymphoid engraftment. For comparison, values shown are the percentage of CD45+CD33+ cells, but similar effects were seen comparing CD19+ cells or total CD45+ populations. Note that because of morbidity, some conditions show results of only 2 animals, and standard deviations in this experiment could not be calculated.
mTOR is constitutively activated in AML blasts
We had previously demonstrated that multiple downstream substrates of PI3 kinase, including the mTOR substrates, p70S6 kinase and 4EBP-1, were activated in AML blasts. However, these experiments were performed in serum-free medium without or with hematopoietic cytokines. Because signaling pathways are responsive to extracellular stimuli, we reexamined expression and phosphorylation of p70S6 kinase and 4EBP-1 in AML cells incubated in EGM2 medium for 16 hours. As shown (Figure 2A), the majority of AML blasts have activation of p70S6 kinase. Overall, 3 of 17 samples analyzed did not show phosphorylation of p70S6 kinase (Figure 2A, samples 325, 401, and 434; data not shown). Curiously, several samples showed detectable phosphorylation, although expression of the protein was minimal (Figure 2A, samples 329, 346l, and 416). In contrast, all samples showed phosphorylation of 4EBP-1, although in 2 of 17 samples this was only weakly detectable (Figure 2A, sample 353; cf, Figure 4C, sample no. 134). There was no correlation between samples that did not show phosphorylation of 4EBP-1 and p70S6 kinase. Therefore, we conclude that there is evidence of activation of the mTOR signaling pathway in 17 of 17 samples analyzed. This activation is apparently independent of culture conditions, although we have not compared the level of activation directly in different medium.
Figure 2.
AML cells require mTOR for long-term survival in vitro. (A) Samples were thawed and immediately lysed for analysis. Samples were lysed and analyzed by SDS-PAGE and Western blotting for the indicated proteins. U937 cells treated with rapamycin (lane 1, +) or not treated were used as control samples. Note that in U937 samples, inhibition of phosphorylation of 4EBP-1 is not complete after treatment with rapamycin. (B) Samples were thawed, allowed to recover for 2 hours, counted, and plated at 2 × 106 cells/mL in 96-well plates. Cells were incubated in EGM2 medium for 48 hours in indicted concentrations of rapamycin, and relative cell survival was measured using the XTT assay. Results were standardized so that untreated samples were assigned a value of 1.0. (C) Cells were processed and cultured as for panel B, but plates were incubated for 7 days before analysis by XTT assay. (D) Normal human CD34 cells were cultured for 2 or 7 days in serum-free medium with hematopoietic cytokines, and relative cell survival was measured using the XTT assay described under “Materials and methods.”
Figure 4.
Rapamycin inhibits mTOR in primary AML samples. (A-B) Patient samples were thawed and incubated for 16 hours in different concentrations of rapamycin as shown. As controls, U937 cells were similarly incubated. After incubation, cells were harvested, lysed, and analyzed for expression and phosphorylation of p70S6 kinase by Western blotting. (C-D) Patient samples were thawed and incubated for 16 hours in nothing (-), 0.1% DMSO (D), 10 μM etoposide (E), 10 μM rapamycin (R), or a combination of etoposide and rapamycin at the same concentrations (E + R). Cells were harvested, lysed, and analyzed by Western blotting.
AML cells do not require mTOR activation for short-term survival in vitro but do require mTOR for long-term survival
To test the role of mTOR in the survival of AML cells, cells were incubated in the mTOR inhibitor rapamycin for 48 hours, and survival was compared with untreated cells (Figure 2B). As can be seen, rapamycin induced only a small decrease in survival in samples tested (Figure 2B). Some samples (eg, no. 330) showed toxicity at 100 μg/mL rapamycin, but the significance of this is unclear. This result is in contrast to recent results by Recher et al35; however, these investigators examined survival in a 14-day leukemia colony-forming assay. We hypothesized that the difference may reflect the duration of exposure to rapamycin and retested the same samples in a 7-day liquid culture assay (Figure 2C). Consistent with the results of Recher et al,35 2 of 4 samples tested were sensitive to rapamycin even at 10 nM drug (Figure 2C, sample 330, filled squares; data not shown). Two other samples tested showed some sensitivity to the drug at low doses, suggesting that over this time frame, mTOR is required for optimum AML survival (Figure 2C, sample 364, filled circles; data not shown). Finally, to determine whether normal hematopoietic stem cells require mTOR for survival, we studied CD34 cells from healthy donors in 2-day and 7-day assays. Normal CD34 cells showed no alteration in survival at either time point (Figure 2D). Taken together, these data suggest that AML cells are heterogeneous in their sensitivity to rapamycin and that this sensitivity requires prolonged exposures. In addition, these data suggest that AML cells are more sensitive to the compound than normal hematopoietic cells, indicating that there should be a therapeutic index for use of the compound in AML.
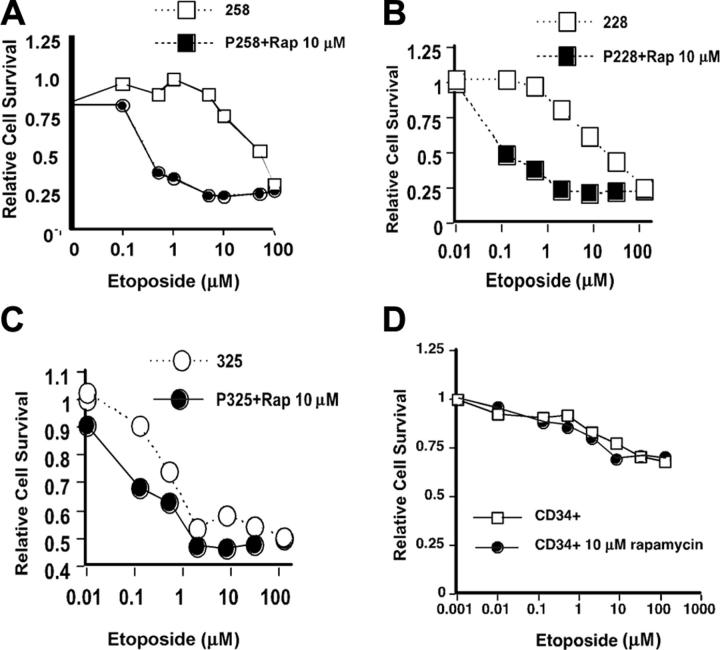
Rapamycin enhances the toxicity of etoposide against AML blasts
We have previously reported that rapamycin enhanced the toxicity of cytosine arabinoside (Ara-C) against the human myeloid leukemia cell line U937. We initially attempted to study this drug combination on primary AML cells. However, although cytosine arabinoside is a potent antileukemic drug in the clinic, we did not find toxicity of Ara-C against AML cells in the 48-hour in vitro assay (data not shown). Likely this is because the cells do not complete passage through the 2 S phases required for the toxicity of the nucleoside analog Ara-C. Therefore, we choose to study the combination of etoposide and rapamycin (Figure 3) in EGM2 medium. Etoposide is a topoisomerase 2 inhibitor which is also active against AML. AML cells showed a typical dose response to etoposide with an ED50 (effective dose 50%) between 5 and 10 μM in this assay (Figure 3A-B) for most samples. One sample tested (no. 325) was sensitive to etoposide at lower concentrations with an ED50 less than 1 μM (Figure 3C). We then tested the effect of adding rapamycin to etoposide on the survival of AML blasts (Figure 3A-C, closed shapes). As shown, rapamycin shifted the dose response to etoposide in almost all samples tested (Figure 3A-B). The only sample in which this effect was not seen was that of patient no. 325 (Figure 3C), which was more sensitive to etoposide than other samples. These results demonstrate that rapamycin can enhance the cytotoxic effect of etoposide even in short-term cultures. Importantly, when CD34 cells from a healthy donor were challenged with etoposide, a modest toxicity was seen at equivalent concentrations. This toxicity was not enhanced by addition of rapamycin (Figure 3D). This suggests that combinations of rapamycin and etoposide would be effective against AML cells without damaging normal hematopoietic stem cells.
Figure 3.
AML cells require mTOR for survival of genotoxic stress. (A-C) Primary patient samples were thawed and plated in 96-well plates at 2 × 106 cells/mL in increasing concentrations of etoposide for 48 hours. Relative cell survival was measured using an XTT assay. All values are expressed as a fraction of the untreated controls. Cells were either incubated in increasing concentrations of etoposide alone (□) or increasing concentrations of etoposide with 10 μM rapamycin present (▪). (D) Normal purified CD34+ hematopoietic stem cells were thawed, plated at 1 × 106 cells/mL in 96-well dishes, and analyzed as described under “Materials and methods.” Cells were incubated in different concentrations of etoposide in the absence (□) or presence (▪) of rapamycin.
Rapamycin inhibits mTOR in primary AML cells
To determine whether the effects described were from inhibition of mTOR, we examined phosphorylation of mTOR substrates in AML cells after a 16-hour incubation in rapamycin. As seen, phosphorylation of p70S6K was consistently blocked by 10 nM rapamycin in U937 cells (Figure 4A, lanes 7-12) confirming activity of the compound. Interestingly, some AML samples showed inhibition of p70S6K at 100 nM rapamycin (sample no. 330, Figure 4B compare lane 3 with lane 4), whereas in one sample p70S6K phosphorylation was only partially inhibited even at 10 μM rapamycin (Sample 162, Figure 4A, lanes 3-6). The majority of samples showed inhibition of p70S6 kinase between 100 nM and 1 μM but not at 10 nM (data not shown). The reason for this is unclear, but it has been reported that rapamycin is exported from cells through ATP (adenosine triphosphate)-binding cassette (ABC) type transporters that mediate multidrug resistance,36 and AML cells are known to express several of these transporters.37 Importantly, because of its unique mechanism of action with binding of drug to the intermediate, FKBP, required for inhibition of mTOR, rapamycin appears to be quite specific for mTOR inhibition. Nonetheless, we cannot exclude the possibility of off-target effects at these concentrations. Taken together, these results show that rapamycin inhibits mTOR in AML blasts.
We next studied the effects of etoposide and combinations of etoposide and rapamycin on the activation of mTOR substrates (Figure 4B). In the majority of samples (3 of 4 studied), etoposide had no effect on p70S6K phosphorylation or 4EBP-1 phosphorylation (Figure 4C-D, samples 330, 134, and 364). In one sample, etoposide appears to stimulate p70S6K phosphorylation (Figure 4D, lane 2 versus 3), although it did not effect 4EBP-1 phosphorylation substantially. This is one of the few samples that did not show spontaneous activation of p70S6 kinase, and the significance of this is unknown. In all samples tested, p70S6K and 4EBP-1 phosphorylation were suppressed by rapamycin whether etoposide was present or not. These data suggest that it is the inhibition of mTOR by rapamycin that leads to the combined effect of the drug combination seen in Figure 3.
Rapamycin enhances etoposide-induced apoptosis in primary AML cells
It has recently been reported that constitutive activation of Akt generates chemotherapy resistance in an animal model of lymphoma which can be altered by treatment of cells with rapamycin.38 Because we have previously demonstrated activation of Akt in AML cells14 and because rapamycin enhances chemosensitivity in these cells, we examined the induction of apoptosis in these cells to see whether rapamycin is altering the apoptotic threshold after etoposide treatment in AML cells. To perform this analysis, we examined cells treated with etoposide and rapamycin for expression of the apoptotic marker, annexin V (Figure 5). Cells were again treated with etoposide alone or etoposide with rapamycin for 24 hours. As shown (Figure 5A), early apoptotic cells are annexin V positive and PI negative (boxed region). In this experiment, addition of rapamycin to etoposide increased the percentage of early apoptotic cells (Figure 5B) in 2 of 3 samples, demonstrating that rapamycin alters the apoptotic threshold in these cells after challenge with etoposide. In the third sample (black bars in Figure 5), substantial apoptosis was induced by rapamycin and an additional effect was not seen in this sample.
Figure 5.
Rapamycin enhances apoptosis in primary AML cells. Primary patient samples were thawed and incubated as for Figure 3. Cells were harvested, pelleted, and incubated with anti-annexin V-FITC and PI. Single-cell suspensions were analyzed by FACScan (Becton Dickinson). Early apoptotic cells were scored as annexin V positive, PI negative to exclude necrotic cells. (A) FACS data from a representative sample. (B) Graph of percentage early apoptotic cells for 3 different patient samples indicated by white, hatched, or black bars.
Rapamycin enhances the cytotoxicity of etoposide on AML stem cells
The previous experiments were all done on total blast populations; however, it is critical in AML therapy to target the leukemic stem cell which gives rise to the disease. To confirm that the effect seen above is true in AML stem cells, we cultured AML cells overnight in the solvent DMSO, etoposide alone, or in etoposide with rapamycin and assayed the survival of SL-ICs by injection into NOD/SCID animals (Figure 6). Animals were injected with an initial cell dose of 5 × 106 cells per animal; no adjustments were made for possible changes in viable cell number in overnight incubations. Animals were injected, observed for 7 weeks, and then humanely killed. Bone marrow was harvested from both femurs, and the percentage of the murine bone marrow repopulated with human leukemic cells was assayed by analyzing the percentage of cells expressing human CD45 and human CD33 (a marker for myeloid lineage). Consistent with the results above, overnight incubation of cells in EGM medium with DMSO added did not affect engraftment (Figure 6B, compare fresh cells with DMSO). When cells were incubated with 5 μM etoposide for 16 hours, engraftment was decreased, confirming the toxicity of etoposide against SL-ICs (note results are shown in a log scale). When 10 μM rapamycin was added to etoposide, engraftment was further decreased as shown. This experiment confirms that rapamycin enhances the toxicity of etoposide on AML stem cells in 2 separate patient samples analyzed (Figure 6A-B). For comparison, we compared treatment of one patient sample (no. 364, Figure 6A) with etoposide and rapamycin to treatment with the PI3 kinase inhibitor, LY294002. In this sample, both LY294002 and the combination of etoposide and rapamycin decreased engraftment more than 20-fold. To determine whether the effects seen are specific to leukemic stem cells or also apply to normal hematopoietic stem cells, we studied normal CD34+ hematopoietic cells, representing a partially purified hematopoietic stem cell compartment. CD34+ cells were thawed and incubated in equivalent concentrations of etoposide, rapamycin, or the combination. Under these conditions, with 1 × 106 cells initially plated, the percentage of engraftment was approximately 10% (Figure 6C). This was decreased modestly by etoposide alone and rapamycin alone. In this case, the combination decreased the average engraftment to 3.87% (3-fold lower than the untreated cells). Thus, the toxicity of etoposide and rapamycin against normal hematopoietic stem cells is less than that against leukemic stem cells, again, suggesting that the effects described are specific to leukemic stem cells.
Discussion
We have studied survival of AML cells in culture. As shown, we have found that AML cells, particularly AML stem cells, survive better in a culture medium with 2% fetal calf serum and a combination of cytokines used to maintain endothelial cells than in serum-free medium with hematopoietic cytokines added. We have used these medium conditions to study the role of mTOR in the survival of AML blasts. Consistent with our previous results, we have demonstrated that AML blasts do not require mTOR for survival in a 48-hour in vitro assay, but they do require mTOR for survival for 7 days in culture. In addition, combinations of etoposide and rapamycin show enhanced toxicity against AML blasts and decrease the ability of AML cells to engraft in NOD/SCID animals. These results demonstrate an important role for the mTOR signaling pathway in regulating survival in AML cells.
Culture of primary AML cells has been problematic for many years. We serendipitously observed that AML cells survive well in a medium containing endothelial cell cytokines. There are other reports that endothelial cell cytokines can support the growth or survival of normal hematopoietic stem cells and speculation that the kdr (kinase insert domain-containing receptor) receptor expressed on AML cells may have a role in mediating survival of leukemic blasts.39,40 To our knowledge, however, this is the first detailed report of culture of AML cells in a medium without hematopoietic cytokines. Further characterization of this result is clearly necessary. We have not determined which of the several elements of EGM2 is critical for survival of leukemic blasts. In addition, we do not have quantitative analysis of leukemic stem cell survival in these conditions, and we have not tested the effect of combinations of endothelial cell and hematopoietic cytokines. These remain areas for further investigation. Nonetheless, the medium is readily available and has provided an improved system for analysis of functional effects of rapamycin and chemotherapy on SL-ICs.
We have further demonstrated discrete effects of rapamycin on survival of AML cells. Inhibition of mTOR with rapamycin leads to modest and variable decrease in survival in 2-day cultures. However, with longer-term culture, rapamycin is toxic to primary AML cells. The molecular mechanism of this effect is unclear at this point. As recently reviewed, mTOR regulates multiple functions in yeast and mammalian cells.41,42 These functions include cell-cycle progression, protein translation, and nutrient uptake. It is possible that rapamycin is leading to down-regulation of nutrient uptake or protein biosynthesis that effectively starves the cell of necessary metabolites after prolonged exposure. Furthermore, it is important to note that recent data support the existence of functions of mTOR that are not inhibited by rapamycin.43 It is unclear at this point whether such signaling pathways occur in AML cells; however, it is possible that the effects described are applicable only to the rapamycin-sensitive functions of mTOR. Perhaps normal hematopoietic stem cells are better able to tolerate this effect than leukemic cells. In addition, we have documented that in 2-day cultures, rapamycin decreases the survival of AML cells treated with etoposide. The temporal kinetics of this response suggests the possibility that this may occur through a discrete mechanism of action. A recent report44 demonstrates that rapamycin sensitizes lung cancer cells to cisplatin through a TP53 and p21Waf1-dependent mechanism. We have not studied this mechanism in AML cells, but these are important areas for future studies.
Our results and those of Recher et al35 both suggest that rapamycin treatment may be efficacious in the therapy of AML. Recher et al35 report responses in 2 patients treated with rapamycin alone. Our results in long-term culture experiments support the biologic basis for this study design. However, from the documented interaction with etoposide shown here, we have recently initiated a phase 1 trial to test the safety of rapamycin with the etoposide-containing chemotherapy regimen, MEC (mitoxantrone, etoposide, and cytarabine) in patients with relapsed AML. These important clinical studies should help define the best approach to using rapamycin in the therapy of AML, either as a single agent, perhaps best administered for a period of weeks to months, or for a short period as a chemosensitizer in combination with chemotherapy.
Acknowledgments
We thank the patients and staff of the Hematologic Malignancies unit at the Hospital of the University of Pennsylvania who helped make these studies possible.
Prepublished online as Blood First Edition Paper, September 8, 2005; DOI 10.1182/blood-2004-11-4468.
Supported by the National Heart, Lung, and Blood Institute (grant K08-HL73977) (J.T.), by a Leukemia and Lymphoma Society of America grant. M.C. is a recipient of the Leukemia and Lymphoma Society of America Clinical Scholar Award.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Smith M, Barnett M, Bassan R, Gatta G, Tondini C, Kern W. Adult acute myeloid leukaemia. Crit Rev Oncol Hematol. 2004;50: 197-222. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3: 730-737. [DOI] [PubMed] [Google Scholar]
- 3.Ailles LE, Gerhard B, Hogge DE. Detection and characterization of primitive malignant and normal progenitors in patients with acute myelogenous leukemia using long-term coculture with supportive feeder layers and cytokines. Blood. 1997;90: 2555-2564. [PubMed] [Google Scholar]
- 4.Kelly LM, Gilliland DG. Genetics of myeloid leukemias. Annu Rev Genomics Hum Genet. 2002;3: 179-198. [DOI] [PubMed] [Google Scholar]
- 5.Yuan Y, Zhou L, Miyamoto T, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci U S A. 2001;98: 10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20: 5695-5707. [DOI] [PubMed] [Google Scholar]
- 7.Furitsu T, Tsujimura T, Tono T, et al. Identification of mutations in the coding sequence of the protooncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest. 1993;92: 1736-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beghini A, Peterlongo P, Ripamonti CB, et al. C-kit mutations in core binding factor leukemias. Blood. 2000;95: 726-728. [PubMed] [Google Scholar]
- 9.Nakao M, Yokota S, Iwai T, et al. Internal tandem duplication of the flt3 gene found in acute myeloid leukemia. Leukemia. 1996;10: 1911-1918. [PubMed] [Google Scholar]
- 10.Shen WP, Aldrich TH, Venta-Perez G, Franza BR Jr, Furth ME. Expression of normal and mutant ras proteins in human acute leukemia. Oncogene. 1987;1: 157-165. [PubMed] [Google Scholar]
- 11.Levis M, Tse KF, Smith BD, Garrett E, Small D. A FLT3 tyrosine kinase inhibitor is selectively cytotoxic to acute myeloid leukemia blasts harboring FLT3 internal tandem duplication mutations. Blood. 2001;98: 885-887. [DOI] [PubMed] [Google Scholar]
- 12.Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105: 22-30. [DOI] [PubMed] [Google Scholar]
- 13.Smith BD, Levis M, Beran M, et al. Single-agent CEP-701, a novel FLT3 inhibitor, shows biologic and clinical activity in patients with relapsed or refractory acute myeloid leukemia. Blood. 2004;103: 3669-3676. [DOI] [PubMed] [Google Scholar]
- 14.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003;102: 972-980. [DOI] [PubMed] [Google Scholar]
- 15.Min YH, Eom JI, Cheong JW, et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia. 2003;17: 995-997. [DOI] [PubMed] [Google Scholar]
- 16.Cheong JW, Eom JI, Maeng HY, et al. Phosphatase and tensin homologue phosphorylation in the C-terminal regulatory domain is frequently observed in acute myeloid leukaemia and associated with poor clinical outcome. Br J Haematol. 2003;122: 454-456. [DOI] [PubMed] [Google Scholar]
- 17.Kubota Y, Ohnishi H, Kitanaka A, Ishida T, Tanaka T. Constitutive activation of PI3K is involved in the spontaneous proliferation of primary acute myeloid leukemia cells: direct evidence of PI3K activation. Leukemia. 2004;18: 1438-1440. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Konopleva M, Cabreira-Hansen M, et al. Inhibition of phosphatidylinositol 3-kinase dephosphorylates BAD and promotes apoptosis in myeloid leukemias. Leukemia. 2004;18: 267-275. [DOI] [PubMed] [Google Scholar]
- 19.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296: 1655-1657. [DOI] [PubMed] [Google Scholar]
- 20.Thompson JE, Thompson CB. Putting the rap on Akt. J Clin Oncol. 2004;22: 4217-4226. [DOI] [PubMed] [Google Scholar]
- 21.Coffer PJ, Woodgett JR. Molecular cloning and characterisation of a novel putative protein: serine kinase related to the cAMP-dependent and protein kinase C families. Eur J Biochem. 1991;201: 475-481. [DOI] [PubMed] [Google Scholar]
- 22.Guzman ML, Swiderski CF, Howard DS, et al. Preferential induction of apoptosis for primary human leukemic stem cells. Proc Natl Acad Sci U S A. 2002;99: 16220-16225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic, I: taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo). 1975;28: 721-726. [DOI] [PubMed] [Google Scholar]
- 24.Martel RR, Klicius J, Galet S. Inhibition of the immune response by rapamycin, a new antifungal antibiotic. Can J Physiol Pharmacol. 1977;55: 48-51. [DOI] [PubMed] [Google Scholar]
- 25.Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo). 1984;37: 1231-1237. [DOI] [PubMed] [Google Scholar]
- 26.Bierer BE, Mattila PS, Standaert RF, et al. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc Natl Acad Sci U S A. 1990;87: 9231-9235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabers CJ, Martin MM, Brunn GJ, et al. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270: 815-822. [DOI] [PubMed] [Google Scholar]
- 28.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18: 1926-1945. [DOI] [PubMed] [Google Scholar]
- 29.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem. 1995;270: 2320-2326. [DOI] [PubMed] [Google Scholar]
- 30.Aoki M, Blazek E, Vogt PK. A role of the kinase mTOR in cellular transformation induced by the oncoproteins P3k and Akt. Proc Natl Acad Sci U S A. 2001;98: 136-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahan BD, Julian BA, Pescovitz MD, Vanrenterghem Y, Neylan J. Sirolimus reduces the incidence of acute rejection episodes despite lower cyclosporine doses in caucasian recipients of mismatched primary renal allografts: a phase II trial. Rapamune Study Group. Transplantation. 1999;68: 1526-1532. [DOI] [PubMed] [Google Scholar]
- 32.Liesveld JL, Rosell KE, II CM, Lancet JE, Abboud CN. Endothelial coculture consistently increases acute myelogenous leukemia blast survival in vitro [abstract]. Blood. 2002; 100. Abstract no. 2175.
- 33.Yamaguchi H, Ishii E, Tashiro K, Miyazaki S. Role of umbilical vein endothelial cells in hematopoiesis. Leuk Lymphoma. 1998;31: 61-69. [DOI] [PubMed] [Google Scholar]
- 34.Ailles LE, Gerhard B, Kawagoe H, Hogge DE. Growth characteristics of acute myelogenous leukemia progenitors that initiate malignant hematopoiesis in nonobese diabetic/severe combined immunodeficient mice. Blood. 1999;94: 1761-1772. [PubMed] [Google Scholar]
- 35.Recher C, Beyne-Rauzy O, Demur C, et al. Antileukemic activity of rapamycin in acute myeloid leukemia. Blood. 2005;105: 2527-2534. [DOI] [PubMed] [Google Scholar]
- 36.Miller DS, Fricker G, Drewe J. p-Glycoprotein-mediated transport of a fluorescent rapamycin derivative in renal proximal tubule. J Pharmacol Exp Ther. 1997;282: 440-444. [PubMed] [Google Scholar]
- 37.van der Kolk DM, de Vries EG, Muller M, Vellenga E. The role of drug efflux pumps in acute myeloid leukemia. Leuk Lymphoma. 2002;43: 685-701. [DOI] [PubMed] [Google Scholar]
- 38.Wendel HG, De Stanchina E, Fridman JS, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428: 332-337. [DOI] [PubMed] [Google Scholar]
- 39.List AF, Glinsmann-Gibson B, Stadheim C, Meuillet EJ, Bellamy W, Powis G. Vascular endothelial growth factor receptor-1 and receptor-2 initiate a phosphatidylinositide 3-kinase-dependent clonogenic response in acute myeloid leukemia cells. Exp Hematol. 2004;32: 526-535. [DOI] [PubMed] [Google Scholar]
- 40.Santos SC, Dias S. Internal and external autocrine VEGF/KDR loops regulate survival of subsets of acute leukemia through distinct signaling pathways. Blood. 2004;103: 3883-3889. [DOI] [PubMed] [Google Scholar]
- 41.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23: 3151-3171. [DOI] [PubMed] [Google Scholar]
- 42.Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16: 29-37. [DOI] [PubMed] [Google Scholar]
- 43.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14: 1296-1302. [DOI] [PubMed] [Google Scholar]
- 44.Beuvink I, Boulay A, Fumagalli S, et al. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120: 747-759. [DOI] [PubMed] [Google Scholar]