Abstract
Reactive oxygen species (ROS) are toxic compounds produced by normal metabolic processes. Their reactivity with cellular components is a major stress for aerobic cells that results in lipid, protein, and DNA damage. ROS-mediated DNA damage contributes to spontaneous mutagenesis, and cells deficient in repair and protective mechanisms have elevated levels of spontaneous mutations. In Escherichia coli a large number of genes are involved in the repair of oxidative DNA damage and its prevention by detoxification of ROS. In humans, the genes required for these processes are not well defined. In this report we describe the human OXR1 (oxidation resistance) gene discovered in a search for human genes that function in protection against oxidative damage. OXR1 is a member of a conserved family of genes found in eukaryotes but not in prokaryotes. We also outline the procedures developed to identify human genes involved in the prevention and repair of oxidative damage that were used to identify the human OXR1 gene. This procedure makes use of the spontaneous mutator phenotype of E. coli oxidative repair-deficient mutants and identifies genes of interest by screening for antimutator activity resulting from cDNA expression.
Reactive oxygen species (ROS) are formed as by-products of normal metabolism of aerobic organisms and react with DNA to produce damage (1). Cells protect themselves from ROS by detoxification mechanisms and by mechanisms that repair the damage ROS produce (2–6). In humans oxidative damage results in mutagenesis, triggers apoptosis, and has been implicated as a contributing cause of a number of human diseases, including cancer and neurodegenerative diseases. Oxidative damage has also been implicated as a contributing factor to the aging process (3, 7, 8). For example, mutations in genes affecting the cell's ability to repair oxidative damage, such as BRCA1 and ATM, have been shown to predispose patients to cancer (9) and mutations in the superoxide dismutase gene, which affects the cell's ability to detoxify reactive oxygen species, predisposes patients to amyotrophic lateral sclerosis (10). Protective mechanisms also interfere with cancer therapies, preventing or repairing oxidative DNA damage produced by radiation treatments and other therapies (11). Our understanding of ROS in human cells is limited, and the biological consequences of oxidative damage are complex. The mechanisms that provide protection from ROS are more clearly understood in Escherichia coli.
In wild-type prokaryotic and eukaryotic cells spontaneous mutagenesis by reactive oxygen species is held in check by enzymes that detoxify ROS and by enzymes that repair ROS damage to DNA. Imbalances in these processes can increase the spontaneous levels of mutation and increase sensitivity to exogenous sources of ROS (2–6). We have constructed a series of mutant strains of E. coli defective in repair pathways acting on oxidative DNA damage for use in searches for human oxidation protection genes. These E. coli mutants carry various combinations of mutations in fpg, mutY, nth, nei, and mutH. All of these mutations confer sensitivity to exogenous peroxide treatments or oxidative mutagenesis, except nei, which increases the peroxide sensitivity of nth, fpg, and mutH strains but has no detectable effect in an otherwise wild-type cell (refs. 12–15 and M.R.V., J. Wyrzykowski, and L. Fan, unpublished data). Most of these mutant strains also exhibit a spontaneous mutator phenotype that results largely, or exclusively, from their inability to repair spontaneous oxidative damage (refs. 12–14 and M.R.V., J. Wyrzykowski, and L. Fan, unpublished data). Thus, by screening cDNA libraries for genes that counteract the spontaneous oxidation-dependent mutator phenotype of the above E. coli mutants, it is possible to identify genes that either prevent or repair oxidative DNA damage.
Materials and Methods
Bacterial Strains.
MV3884 is a mutH472∷Tn10 nth-1∷kan,ble derivative of MV1161 (16). It was constructed by sequential introduction of the mutH allele from strain CGSC7254 (Coli Genetic Stock Center, Yale University) and the nth allele from strain BW372 (17) by P1 transduction selecting for the appropriate drug resistance, and testing for the mutator phenotype and peroxide sensitivity resulting from each mutation. The cDNA library used to transform the test strain was a gift from E. Perkins and M. Resnick (National Institute of Environmental Sciences, Research Triangle Park, NC; ref. 18).
Culture Media.
LB ampicillin plates were standard LB medium (16) containing 100 μg/ml ampicillin. Semi-enriched medium containing E salts (ref. 19; ESEM) plates are standard SEM plates in which the salts solution has been replaced by E salts (16, 19). The low level of arginine supplied is sufficient to allow a background growth of arginine-requiring cells to reach a growth ceiling of approximately 5 × 109 cells per plate. Once the arginine is exhausted, only Arg+ revertants will continue to grow to form colonies (20). Standard yeast extract/peptone/dextrose (YEPD) plates and broth were used for routine growth of yeast. Minimal drop-out medium lacking uracil was used to select for Ura3+ recombinants (21). All yeast incubations were performed at 30°C; all bacterial incubations were performed at 37°C.
Screening for Human Antimutator Genes.
Competent MV3884 cells were transformed with 600 ng of cDNA present in the pSE380 vector, which contains an isopropyl-β-d-thiogalactoside (IPTG)-inducible synthetic promoter that functions in E. coli (18). Transformants were selected on LB ampicillin plates. Transformants were then picked, inoculated into 96-well microtiter trays containing 250 μl LB ampicillin, and grown overnight for subsequent testing. Trays were spotted onto two ESEM-ampicillin plates with a multiprong device; one plate contained IPTG (2 mM) for induction of cDNA transcription. Spontaneous mutation frequencies were estimated, and spots showing an IPTG-inducible decrease in spontaneous mutagenesis were identified, purified, and tested further. Quantitative levels of Arg− mutations in the presence or absence of IPTG induction were determined (Fig. 1), and clones showing a clear decrease in mutagenesis were selected, and their plasmids were purified and retransformed into a fresh isolate of the MV3884 mutant strain to confirm that the antimutator phenotype was due to the presence of the cDNA. To eliminate clones that either interfered with the Arg reversion assay or had nonspecific effects on mutagenesis, cDNA clones were transformed into either an ung or a dnaQ mutant strain, two mutator strains that also have an increased spontaneous mutation frequency similar to that of MV3884, but for reasons other than oxidative repair deficiencies. Clones showing antimutator activities in the ung or dnaQ mutants similar to those seen in MV3884 were presumed to affect steps in the Arg+ mutagenesis process subsequent to the production and processing of the initial DNA damage and were eliminated from the screen.
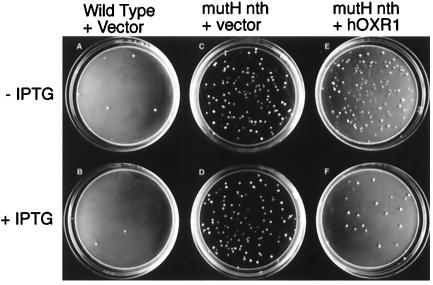
Figure 1.
Effect of hOXR1 expression on Arg+ mutagenesis in E. coli. (A and B) MV3626 (wild type/pTrc99A; Amersham Pharmacia). (C and D) MV4174 (mutH nth/pTrc99A). (E and F) MV4300 [mutH nth/pMV420 (hOXR1)]. (A, C, and E) No IPTG induction. (B, D, and F) IPTG (1 mM) induction.
Cloning of Saccharomyces cerevisiae OXR1.
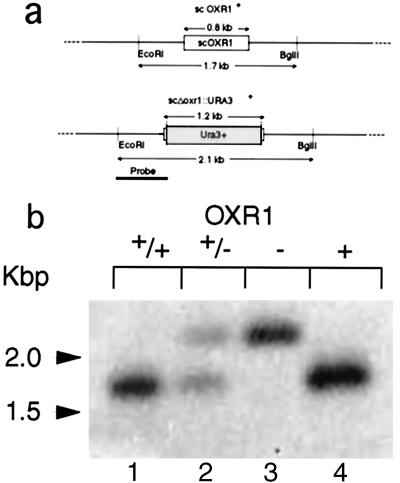
The S. cerevisiae OXR1 gene (scOXR1) was cloned via a PCR approach. Primers 1 and 2 (see below) were used to amplify the scOXR1 coding sequence and 300 bp of flanking DNA on each side of the wild-type yeast R117 (22). These primers also included new restriction sites that allowed cloning of the amplified sequence into the EcoRI and BamHI sites of the vector pTrc99A (Amersham Pharmacia) to produce plasmid pMV600. A second PCR reaction was performed using the pMV600 plasmid as a template to clone the two flanking regions in separate reactions, using primers 1 and 4 to clone the upstream flanking region and primers 5 and 6 to clone the downstream flanking region. Each flanking region contained either the first or last three codons of scOXR1 and introduced restriction sites compatible with the SacI XbaI sites needed to insert the Ura3+ cassette between the two flanking DNA regions. The three fragments were assembled to produce the plasmid pMV605, in which the Ura3+ DNA sequences replaced all but the first three and last three codons of scOXR1 and are flanked on each side by 300 bp of scOXR1 flanking DNA. This fragment (shown in Results, Fig. 4) was then purified as a single PvuII fragment. Competent ura3− mutant yeast cells (strain R117) were then transformed by Li acetate transformation with the URA3+-carrying PvuII fragment purified from the pMV605 plasmid, and URA+ colonies were selected by growth in the absence of uracil. Two URA+ transformants were purified and designated strains N1 and N2. N1 and N2 were sporulated in acetate medium, and tetrads were dissected. Two haploid strains N1–9 (URA3+) and N1–4 (ura3−) were selected for further study. Genetic structures of the mutants were confirmed by Southern blotting. The primers used were: primer 1, ATCATCGAATTCATATGACCGGACTCGTAAT; primer 2, ATCATCGGATCCTTTTTTTTCACATTGGGAG-3′; primer 4, ATCATCGAGCTCTCCAAACATTGTCGCTCC; primer 5, ATCATCCCCGGGGTAGGATAGTGTCATCTA, and primer 6, ATCATCCTGCAGTTTTTTTTCACATTGGGAG.
Figure 4.
Analysis of a yeast scΔoxr1∷URA3 mutant. (a) Sizes of the wild-type OXR1- and scΔoxr1∷URA3+-containing DNA restriction fragments. Also shown is the probe used in b. (b) Hybridization analysis of scΔoxr1∷URA3+ and OXR1 wild-type strains. Lane 1, Wild-type diploid parent strain, R117; lane 2, OXR1/Δoxr1∷URA3+ diploid strain, N1; lane 3, Δscoxr1∷URA3+ haploid segregant strain, N1–9; lane 4, OXR1+ ura3− haploid segregant N1–4.
Southern Hybridizations.
Yeast genomic DNA was prepared as described by Adam et al. (21). Standard hybridization methods were used to measure the size of the OXR1 or URA3 replacement allele carrying DNA fragments (23). The 300-bp EcoRI–SacI fragment from pMV603, corresponding to the upstream OXR1 flanking DNA (shown in Results, Fig. 4), was used as a probe.
Yeast Strains.
Yeast strains used in this study are derivatives of R117 (22), a strain congenic to 381G (24). Strains were grown in standard YEPD medium at 30°C. Additional strains constructed in this study were derivatives of R117 and carry the following additional genetic markers: N1, MATa/MATα Δoxr1∷URA3/OXR1; N1–9, MATa Δoxr1∷URA3; and N1–4, MATa OXR1.
Peroxide Sensitivity Testing.
Peroxide sensitivity tests were performed as described by Ramotar et al. (1). Briefly, overnight cultures of yeast strains were diluted to an OD600 of approximately 0.3 in standard YEPD medium and grown with aeration to an OD600 of approximately 0.8–1. Cells were harvested, washed once in sterile water, and resuspended in PBS (pH 7.4). Samples were treated with H2O2 at the indicated concentrations for 1 h at 30°C with aeration. After treatment, cells were diluted in PBS and titered on YEPD plates. Experiments were repeated at least three times; representative data are shown.
DNA Sequencing.
DNA sequencing was performed by the MIT Center for Cancer Research, the University of Massachusetts Medical Center, or the Iowa State University DNA sequencing facilities. DNA and predicted protein sequences were analyzed using Blast sequence searches (25).
Results
Screening for Oxidation Protection Genes.
To identify DNA oxidation protection genes, we transformed E. coli oxidation repair-defective spontaneous mutator strains with a human cDNA library and screened transformants for a reduction in mutator activity. Genes exhibiting antimutator activity were then subjected to a variety of tests to confirm that the reduction in mutagenesis was a consequence of reduced oxidative mutagenesis, rather than nonspecific effects on the mutagenesis assay system (see Materials and Methods). Of the approximately 10,000 cDNAs tested in the initial screen, several reduced spontaneous mutagenesis in the oxidation-specific mutator strains. Of particular interest was the oxidation resistance gene, which we named OXR1. This gene was initially identified using the E. coli mutH nth double mutant strain as the mutagenesis indicator strain. This strain is highly sensitive to peroxide treatments, and both mutations contribute to this phenotype (refs. 12 and 15, and M.R.V., J. Wyrzykowski, and L. Fan, unpublished data). Fig. 1 compares spontaneous Arg+ mutagenesis in wild-type and mutH nth double-mutant strains and shows that IPTG induction of the vector has no effect. However, when the human OXR1 (hOXR1) gene is induced by IPTG it causes about a 5-fold reduction in spontaneous Arg+ mutagenesis in the mutH nth strain (Fig. 1F) without a detectable effect on growth (data not shown). Thus, hOXR1 functions as an antimutator in this E. coli genetic background.
Analysis of the hOXR1 DNA Sequence.
DNA sequence analysis shows that the hOXR1-expressing plasmid pMV520 carries a cDNA insert of approximately 1.7 kbp encoding a previously unidentified human gene that matches several human and mouse expressed sequence tag database sequences and shares homology with genes found in S. cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, and Drosophila. Homologues are not found in E. coli or other bacterial species. Thus hOXR1 appears to be a member of a conserved family of genes present in a wide variety of eukaryotic species. Fig. 2 compares the predicted hOXR1 protein sequence with the corresponding regions of its homologues and shows a consensus sequence for the OXR1 family of genes. The highest degree of homology is in the carboxyl-terminal half of the protein, and several consensus motifs are identifiable in this region (Fig. 2). Two additional regions conserved primarily among the higher eukaryotes are found in the region corresponding to hOXR1 amino acid residues 100–200 (Fig. 2). The conserved motifs of OXR1 do not correspond to motifs of known function; thus their functions cannot currently be predicted. The hOXR1-related genes are the S. cerevisiae OXR1 homologue (scOXR1), which is 27% identical and 43% similar to the hOXR1 gene. It is known only as the ORF YPL196w, and no phenotype has been ascribed to this gene. Similarly, the S. pombe and C. elegans OXR1 homologues have been identified by genomic sequencing efforts and are known only as open reading frames. Only the Drosophila gene has been characterized to date (see Drosophila below).
Figure 2.
Alignment of OXR1 homologues. Numbering corresponds to the human OXR1 protein sequence. It is compared with its homologues from C. elegans (AAD31551), beginning with residue 425; Drosophila melanogaster L82C (AAD28510), beginning with residue 421; and the entire sequences of S. cerevisiae YPL196w (CAA97909) and S. pombe (CAB16289) alleles. Highly conserved, identical residues are indicated in bold type with a dark background; conserved residues and their conservative substitutions are in plain type with a light gray background. The consensus sequence is indicated on the bottom line; regions containing conservative substitutions are indicated by dots.
Genomic hOXR1 Structure and Locus.
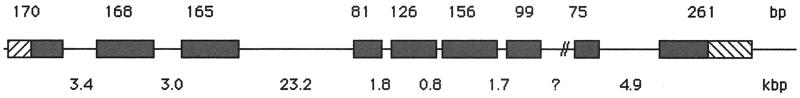
The current release of the human genome sequence indicates that the hOXR1 gene is located on chromosome 8 (q23). Its genomic structure is shown in Fig. 3. It comprises nine exons. The first exon includes 74 bp of upstream untranslated sequence present in the cDNA, and the last exon includes 156 bp of downstream untranslated DNA sequence. The full length of the genomic hOXR1 DNA cannot be predicted from the existing data because of a sequencing gap of unknown length between exons 7 and 8. A second homologous sequence is present on chromosome 15 (q21). This region of chromosome 15 corresponds to the region of hOXR1 shown in Fig. 2 beginning with amino acid 204. It is likely to be a pseudogene, based on its lack of introns and the presence of a frameshift mutation early in the OXR1 coding sequence that disrupts the ORF, leaving only a small portion of the OXR1 gene as a potential ORF.
Figure 3.
Genomic structure of OXR1. The OXR1-containing region of chromosome 8q23 is shown (not to scale). The sizes (in bp) of exons (black boxes) are shown above the line; the sizes of introns (in kbp) are shown below the line. The region between exons 7 and 8, indicated by the question mark, contains a sequencing gap of unknown size. Untranslated regions that are also present in the cDNA clone are shown as striped boxes.
Yeast OXR1 Mutants Are Sensitive to Hydrogen Peroxide Treatments.
To determine whether the OXR1 gene affects oxidative damage resistance in a eukaryotic organism, we constructed a S. cerevisiae strain deleted for the scOXR1 (YPL196w) ORF. scOXR1 was cloned along with approximately 300 bp of both upstream and downstream flanking sequences, and deletion was constructed by the use of PCR methods to replace all but six codons of the OXR1 coding sequence with a cassette that expresses the URA3+ gene. The yeast strain R117 (22) was transformed with the linear DNA fragment carrying the URA3+ gene surrounded by the scOXR1 sequences, and URA3+ recombinants were selected. URA3+ diploid recombinants were then sporulated and tetrads were analyzed. URA3+ haploid segregants were viable, indicating that scOXR1 is not an essential gene (data not shown). Fig. 4A shows the expected scOXR1+ and scΔoxr1∷URA3+ gene structures, and Fig. 4B shows that scOXR1 restriction fragments of the appropriate sizes are present in the OXR1+ ura3− strain and the scΔoxr1∷URA3+ mutant haploid strains.
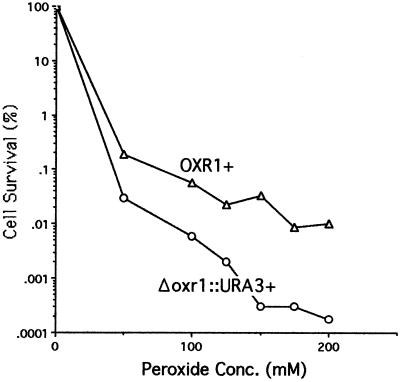
To determine whether mutation of oxr1 adversely affects oxidation sensitivity in yeast, cells were grown to mid-log phase and treated with up to 200 mM H2O2, then plated on YEPD agar to determine viable cell numbers. Fig. 5 compares the hydrogen peroxide sensitivity of wild-type and oxr1 mutant haploid yeast strains and shows that mutation of oxr1 results in increased sensitivity to hydrogen peroxide, thus demonstrating that the wild-type scOXR1 provides protection against the deleterious effects of oxidation. Similar results were obtained with an independent Δoxr1∷URA3 isolate, and introduction of the wild-type scOXR1 gene cloned, along with its upstream DNA sequence, onto the yeast vector pRS315 (26) restores wild-type resistance to the oxr1 deletion mutant, confirming that the sensitivity is due to the loss of OXR1 function (data not shown). The level of peroxide sensitivity resulting from the oxr1 mutation is greater than that resulting from mutations inactivating yeast oxidative repair genes such as ogg1, ntg1, ntg2, apn1, rad1, rev3, or rad52 (27, 28). Individually, these mutations have no adverse effect on peroxide resistance, and even the combination of ntg1 ntg2 apn1 and rev3 mutations has no detectable effect on peroxide sensitivity (28). A level of peroxide sensitization similar to that conferred by the scΔoxr1∷URA3+ mutation requires either the combination of ntg1 ntg2 apn1 with rad52 or rad1, or the combination of rev3 with rad52. These observations indicate that the OXR1 gene plays an important role in oxidative protection in yeast and, presumably, other eukaryotes that have OXR1 homologues.
Figure 5.
Hydrogen peroxide resistance of wild-type (○) and scΔoxr1∷URA3 (▵) haploid strains of yeast. The titers of surviving cells were determined by plating on YEPD agar plates that were incubated for 2–3 days at 30°C. Representative data from at least four experiments are shown, all showing similar levels of difference between the wild type and mutant strains. Similar results were also seen with an independent oxr1 deletion mutant strain.
Discussion
Interspecies Complementation of Antimutator Activity.
The use of spontaneous oxidation as the DNA damaging treatment provides a high degree of sensitivity compared with methods used by others (18, 29), because mutagenic oxidative damage is constantly occurring, thereby allowing mutations to accumulate in these sensitive strains of E. coli. Expression of cDNAs that result in either a small reduction in the production of DNA damage or a small increase in DNA repair activity reduces the number of spontaneous mutations. Genes counteracting the low constant rate of oxidative damage are likely to be important for protection against the low, spontaneous level of oxidative damage normally produced within cells. The use of this approach resulted in the discovery of the human OXR1 gene as an antimutator mutation that reduced oxidative mutagenesis in an E. coli mutator strain.
OXR1 Homologues.
OXR1 is conserved among eukaryotes, and homologues have been identified in a number of different species. Comparison of the various known forms of OXR1 indicates that the most highly conserved region of the gene is its the carboxyl-terminal half. However, because the gene has been identified primarily in sequencing projects rather than genetic studies of DNA repair, little is known about its physiological function.
The S. cerevisiae OXR1 gene is composed primarily of the highly conserved carboxyl-terminal domain of the human OXR1 gene (Fig. 2). The presence of OXR1 in S. cerevisiae allowed the use of yeast genetics to construct an OXR1 deletion mutant and to analyze its function in oxidative protection. This mutant was found to be sensitized to treatments with exogenous hydrogen peroxide, thus demonstrating that OXR1 is required for normal levels of resistance to oxidative damage and that this function is contained within the most conserved region of the OXR1 family.
Genetic Analysis of the Drosophila OXR1 Homologue.
Genetic studies of the Drosophila OXR1 homologue (30) implicate it in other cellular processes in addition to oxidation protection. The Drosophila homologue is encoded by the L82 gene, which produces seven different known isoforms, L82A through L82G (30). The hOXR1 homology region of L82 is contained within its carboxyl-terminal exon, and all known isoforms contain this exon. The largest isoform of L82 encodes a protein of 1270 amino acids, whereas the smallest encodes only the 192-amino acid protein that makes up just the OXR1 homology region. Mutants of L82 are defective in eclosion and, therefore, fail to release adults from pupae (30). This developmental defect can be complemented by expression of the largest isoform, L82A (30). Because other isoforms have not been tested, it is uncertain whether the developmental deficiency of the L82 mutant strain results from loss of the OXR1 region alone, or if the other upstream domains of L82A are important for this process. This stage of Drosophila development is associated with dramatic increases in catalase and superoxide dismutase expression (31, 32), suggesting that oxidative stress may increase at this stage of development and that Drosophila induces protective mechanisms to counteract this stress. These observations raise the possibility that one function of L82 gene expression during eclosion may be to contribute to a general increase in protection against oxidative damage to DNA.
The identification of the OXR1 family is a step toward identifying all of the genes that contribute to protection against ROS in humans. The functional genomic approach we have taken has the potential to support the identification of genes that complement OXR1 family members, interact with them, or provide alternative pathways for response to ROS. The elucidation of these pathways should be of importance in understanding human disease processes.
Acknowledgments
We thank Richard Baker for advice and assistance with yeast genetics experiments; Ed Perkins and Michael Resnick for the cDNA library; Mary Berlyn (Coli Genetic Stock Center, Yale University) for bacterial strains; and Michael Resnick, Graham Walker, Tony Poteete, Martin Marinus, and Ben Van Houten for critical reading of the manuscript. M.R.V. thanks the members of the Housman laboratory for advice and assistance in the early stages of this project conducted in the laboratory during a sabbatical leave. This work was funded by grants to M.R.V. from the American Cancer Society, Massachusetts Division, Inc., the Howard Hughes Medical Research Foundation, and the Worcester Foundation for Biomedical Research, and by grants from the National Institutes of Health to D.H.
Abbreviations
- ROS
reactive oxygen species
- YEPD
yeast extract/peptone/dextrose
- IPTG
isopropyl-β-d-thiogalactoside
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF309387).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260495897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260495897
References
- 1.Ramotar D, Popoff S C, Gralla E B, Demple B. Mol Cell Biol. 1991;11:4537–4544. doi: 10.1128/mcb.11.9.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohr V A, Dianov G L. Biochimie. 1999;81:155–160. doi: 10.1016/s0300-9084(99)80048-0. [DOI] [PubMed] [Google Scholar]
- 3.Croteau D L, Bohr V A. J Biol Chem. 1997;272:25409–25412. doi: 10.1074/jbc.272.41.25409. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham R P. Mutat Res. 1997;383:189–196. doi: 10.1016/s0921-8777(97)00008-6. [DOI] [PubMed] [Google Scholar]
- 5.Demple B, Harrison L. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- 6.Henle E S, Linn S. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 7.Loft S, Poulsen H E. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 8.Marnett L J. Carcinogenesis. 2000;21:361–370. doi: 10.1093/carcin/21.3.361. [DOI] [PubMed] [Google Scholar]
- 9.Gowen L C, Avrutskaya A V, Latour A M, Koller B H, Leadon S A. Science. 1998;281:1009–1012. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 10.Orrell R W. Neuromusc Disord. 2000;10:63–68. doi: 10.1016/s0960-8966(99)00071-1. [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky M F, Engelbergs J, Thomale J, Schweer T. Recent Res Cancer Res. 1998;154:127–146. doi: 10.1007/978-3-642-46870-4_7. [DOI] [PubMed] [Google Scholar]
- 12.Jian D, Hatahet Z, Blaisdell J O, Melamede R J, Wallace S S. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saito Y, Uraki F, Hakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. J Bacteriol. 1997;179:3782–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaisdell J O, Hatahet Z, Wallace S W. J Bacteriol. 1999;181:6396–6402. doi: 10.1128/jb.181.20.6396-6402.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Winkler M E. J Bacteriol. 2000;182:5025–5028. doi: 10.1128/jb.182.17.5025-5028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volkert M R, Nguyen D C. Proc Natl Acad Sci USA. 1984;81:4110–4114. doi: 10.1073/pnas.81.13.4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham R P, Weiss B. Proc Natl Acad Sci USA. 1985;82:474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkins E L, Sterling J F, Hashem V I, Resnick M A. Proc Natl Acad Sci USA. 1999;96:2204–2209. doi: 10.1073/pnas.96.5.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel H J, Bonner D M. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 20.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 22.Baker R E, Harris K, Zhang K. Genetics. 1998;149:73–85. doi: 10.1093/genetics/149.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Frisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 24.Hartwell L H. J Cell Biol. 1980;121:503–512. [Google Scholar]
- 25.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 26.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas D, Scot A D, Barbey R, Padula M, Boiteux S. Mol Gen Genet. 1997;254:171–178. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- 28.Swanson R L, Morey N J, Doetsch P W, Jinks-Robertson S. Mol Cell Biol. 1999;19:2929–2935. doi: 10.1128/mcb.19.4.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Memisoglu A, Samson L. Crit Rev Biochem Mol Biol. 1996;31:405–447. doi: 10.3109/10409239609108724. [DOI] [PubMed] [Google Scholar]
- 30.Stowers R S, Russell S, Garza D. Dev Biol. 1999;213:116–130. doi: 10.1006/dbio.1999.9358. [DOI] [PubMed] [Google Scholar]
- 31.Massie H R, Aiello V R, Williams T R. Mech Ageing Dev. 1980;12:279–286. doi: 10.1016/0047-6374(80)90051-2. [DOI] [PubMed] [Google Scholar]
- 32.Orr W C, Orr E C, Legan S K, Sohal R S. Arch Biochem Biophys. 1996;15:251–258. doi: 10.1006/abbi.1996.0250. [DOI] [PubMed] [Google Scholar]