Abstract
Transcription factors are critical for instructing the development of B lymphocytes from multipotential progenitor cells in the bone marrow (BM). Here, we show that the absence of STAT3 impaired B-cell development. Mice selectively lacking STAT3 in BM progenitor cells displayed reduced numbers of mature B cells, both in the BM and in the periphery. The reduction in the B-cell compartment included reduced percentages and numbers of pro-B, pre-B, and immature B cells in the absence of STAT3, whereas the number of pre–pro-B cells was increased. We found that pro-B and pre-B–cell populations lacking STAT3 were hyporesponsive to IL-7 because of a decreased number of IL-7–responsive cells rather than decreased expression or signaling of IL-7Rα. Moreover, STAT3-deficient mice displayed enhanced apoptosis in the pro-B population when deprived of survival factors, suggesting that at least 2 mechanisms (impaired differentiation and enhanced apoptosis) are involved in the mutant phenotype. Last, BM transplantation confirmed that impaired B lymphopoiesis in the absence of STAT3 was caused by a cell autonomous defect. In sum, these studies defined a specific role for STAT3 in early B-cell development, probably acting at the pre–pro-B transition by contributing to the survival of IL-7–responsive progenitors.
Introduction
B lymphopoiesis involves the expression of multiple factors, including cytokine receptors and transcription factors.1-3 Among the cytokine receptors, Flt3 and IL-7R are known to be critical for early B-cell development. Targeted deletion of Flt3 or Flt3 ligand impairs the development of pro-B and pre-B cells.4-6 Similarly, genetic ablation of the IL-7 or the IL-7Rα chain results in a developmental block in CLP and pro-B stages.7-9 Mice lacking both Flt3L and IL-7Rα completely lose B lymphocytes in the BM, suggesting that these 2 molecules do not have completely overlapping functions in B lymphopoiesis.10 Several transcription factors are also indispensable for B-cell development and function in a sequential and an interdependent manner to instruct the generation of B-cell precursors from hematopoietic progenitors.11 For instance, PU.1 was first proposed to initiate B-cell differentiation by promoting the expression of IL-7R and EBF, another key transcription factor.11,12 However, more thorough studies indicated that PU.1 is not strictly required for B lymphopoiesis because mice lacking PU.1 in the B-cell lineage do not display abnormalities in early B-cell development. Nonetheless, abnormalities occur at more mature stages, including a shift of conventional B2 cells to B1 cells in the spleens of the mutant mice.13-15 The upregulation of EBF in conjunction with E2A further facilitates the formation of pro-B cells, where B-cell–specific genes such as Pax5, Igα, Igβ, λ5, VpreB, RAG1, and RAG2 are subsequently induced. Although the specification of B cells requires EBF and E2A, commitment and maintenance require Pax5.2
Although IL-7R signaling activates STAT1, STAT3, and STAT5, it is generally believed that STAT5 plays the major role in IL-7–mediated B lymphopoiesis.16-20 The expansion of pro-B cells in transgenic mice expressing constitutively active STAT5 correlated with an increase in cyclin D2, pim-1, and bcl-xL expression, suggesting that STAT5 directly affects the proliferation and survival of pro-B cells.19 Overexpression of this form of STAT5 also restored B-cell differentiation in the IL-7R–deficient mice, including VH immunoglobulin rearrangement and appearance of immature and mature B cells.19 In addition, a severe deficiency in B lymphopoiesis was observed in the absence of STAT5a/b, resulting in combined immunodeficiency similar to that caused by lack of IL-7 signaling.17 STAT5 also regulates germline transcription, histone acetylation, and DNA recombination of distal VH gene segments in an IL-7–dependent manner. In the absence of STAT5a/b, these events are greatly impaired.18 Although STAT1 affects the survival and proliferation of T lymphocytes, contributes to the development of adaptive immune responses, and has a differential effect on the expression of MHC class I in T and B cells, it has not been shown to play a direct role in lymphopoiesis.21,22
The functions of STAT3 in vivo have been extensively investigated using conditional gene targeting in mice. These studies showed that STAT3 has multiple functions in different tissues, contributing to survival, apoptosis, migration, and gene expression.23 In addition, STAT3 has been implicated in oncogenesis. Activated STAT3 is observed in many malignancies in humans, and overexpression of a constitutively active form of STAT3 results in the transformation of fibroblasts.24 It is also required for the development of B-cell lymphoma in transgenic mice and for the growth and survival of human and mouse NPM-ALK–transformed B and T cells.25 We have previously shown that STAT3 can be a negative regulator of granulocyte–colony-stimulating factor (G-CSF)–mediated granulopoiesis. Enhanced proliferation and increased granulocyte development were observed in the STAT3-deficient BM.26 However, the role of STAT3 in normal B lymphopoiesis remains to be elucidated.
In this study, we have demonstrated that STAT3 is required for efficient B lymphopoiesis. Mice lacking STAT3 in the BM displayed impaired B-cell development and reduced B-cell lineages, including pro-B, pre-B, immature B, and mature B cells. IL-7–mediated proliferation of BM cells was also significantly reduced, which correlated with a decreased number of IL-7–responsive precursors in the BM. These results indicate that STAT3 is critical for normal early B-cell development and that it acts in conjunction with IL-7 and STAT5 to facilitate the transition from pre–pro-B cells.
Materials and methods
Mice
Generation of mice with a conditional STAT3 allele has been described previously.27 These mice were mated with a transgenic line bearing Cre recombinase driven by the IFN-inducible Mx promoter.28 Littermate mice were generated to be homozygous for the conditional STAT3 alleles (STAT3f/f) with or without the MxCre transgene. Induction of Cre and subsequent deletion of STAT3f/f was accomplished by 2 successive intraperitoneal injections separated by 4 days with 150 μg poly (I:C) per mouse. For the purpose of simplicity, we have indicated STAT3f/f mice that do not express the Cre transgene as control mice and MxCre-STAT3f/f mice as STAT3KO mutants. Controls and mutants received injections of poly (I:C). Although the STAT3 deletion efficiency varied in different organs, STAT3 was almost completely deleted in the BM and liver, judging from polymerase chain reaction (PCR) and protein blots. Neomycin (2 mg/mL) was added to drinking water to control possible bacterial infections.
Genotyping for targeted alleles
Genomic DNA was prepared by boiling tail samples in 300 μL NaOH 50 mM for 2 hours at 95°C followed by neutralization with Tris-Cl, pH 7.5, to final 100 mM. Two microliters of DNA solution was subjected to PCR with the use of 2 forward primers and 1 reverse primer to distinguish wild-type, floxed, and deleted alleles. Primers used were as follows: forward, 5′-GAA GGC AGG TCT CTC TGG TGC TTC-3′; forward, 5′-CAG AAC CAG GCG GCT CGT GTG CG-3′; reverse, 5′-GCT GCC AAC AGC CAC TGC CCC AG-3′.
Flow cytometric analysis
Single-cell suspensions were prepared from bone marrow, spleen, and peripheral blood after erythrocyte depletion. Cells were stained with antibodies, as indicated: anti–B220-PE (RA3-6B2), anti–IL-7Rα-PE (A7R34), anti–CD24-PE-Cy5 (M1/69), anti–BrdU-FITC and streptavidin—APC-Cy7 (eBioscience, San Diego, CA), anti–IgM-FITC (1B4B1), anti–IgD-PE (123), anti–BP-1-FITC (6C3), and streptavidin-APC (Bio-Legend, San Diego, CA), and anti–CD43-biotin (S7) (BD PharMingen, San Diego, CA). Data were collected on FACSCanto or FACSCan and analyzed by FACSDiva or CellQuest software, respectively (BD Biosciences, San Jose, CA).
Statistical analysis
All statistical comparisons were validated using the Student t test.
Colony-formation assay
Colony-formation assay was performed as described.26 Briefly, 2 × 105 BM cells were seeded in complete medium with methylcellulose (MethoCult M3630; StemCell Technologies, Vancouver, BC, Canada) with or without exogenously added rmIL-7 (PeproTech, Rocky Hill, NJ) at 10 and 100 ng/mL for 7 days, followed by enumerating pre-B CFUs by light microscopy.
In vitro proliferation assay
Proliferation assay was performed with either [3H]-thymidine incorporation or BrdU incorporation assay. For [3H]-thymidine incorporation, BM cells were seeded at 2 × 105 cells per well in 96-well plates and were incubated in the absence or presence of rmIL-7 at 5 or 50 ng/mL or in the absence or presence of 1 ng/mL rmIL-3 (all from PeproTech) for 48 hours. Then 1 μCi (0.037 MBq) [3H]-thymidine was added during the final 16 hours before harvest. For BrdU incorporation assays, 2.5 × 106 BM cells without or with rmIL-3 (5 ng/mL) or IL-7 (10 or 50 ng/mL) for 38 hours were incubated with 10 μM BrdU (Sigma) for the final 3 hours. Labeled cells were stained with anti-B220, anti-CD43, and anti-BrdU, followed by FACS analysis. Stimulation index was calculated as percent cytokine stimulated ÷ percent medium alone.
In vitro apoptotic assay
Single-cell suspensions were prepared from BM after erythrocyte depletion and were incubated in vitro in complete medium without cytokines for 0 hours and 3 hours. BM cells were then stained with anti-B220, anti-CD43, and annexin V, and then FACS analysis was performed.
BM transplantation
Five million BM cells from control or STAT3KO mice were transplanted intravenously into RAG1KO mice that were previously irradiated with 950 rads (9.5 Gy). Two months later, cells from BM or spleens of chimeric mice were stained with anti-IgM, anti-B220, and anti-CD43 (for BM cells) or anti-IgM and anti-IgD (for splenocytes), and then FACS analysis was performed.
Intracellular staining of STAT3
BM cells were surface stained with antibodies to B220, CD43, and IgM; this was followed by fixation and permeabilization (Fix&Perm, Caltag, Burlingame, CA) and staining with rabbit anti-STAT3 or phospho-STAT3 antibodies (Cell Signaling, Beverly, MA). Washed cells were incubated with FITC-conjugated secondary antibody to rabbit antibody and then underwent FACS analysis.
Results
Impaired B-cell development in STAT3KO mice
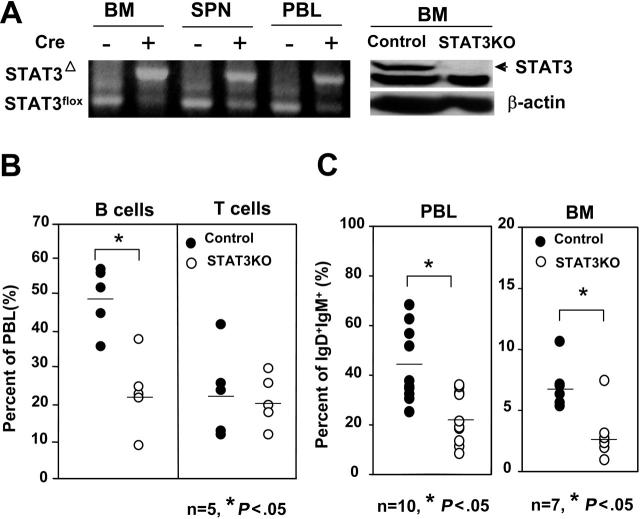
We previously analyzed the role of STAT3 in BM using conditional knockout mice and documented a requirement for STAT3 as a negative regulator for G-CSF–mediated granulopoiesis.26 In the same experiments, we noted reduced production of lymphocytes in the periphery, prompting us to investigate which population of lymphocytes was affected. First, we determined the degree of STAT3 deletion in the BM and periphery. Genomic DNA from BM, spleen, or peripheral blood leukocytes (PBLs) was subjected to PCR amplification for the targeted allele. As shown in Figure 1A (left panel), STAT3 was efficiently deleted in the BM following poly (I:C) injection. However, deletion was less efficient in the spleen and PBL, judging by the residual STAT3flox allele. Deletion of STAT3 in BM was further confirmed through Western blotting, as shown in Figure 1A (right panel). We next analyzed which population of lymphocytes was affected in the STAT3KO mice. PBLs of control or STAT3KO mice were subjected to FACS analysis with the use of anti-CD3 and anti-CD19 antibodies. As shown in Figure 1B, reduced numbers of B cells were observed in the PBLs of STAT3KO mice, whereas numbers of T cells remained normal. Reduced B-cell numbers were confirmed by staining PBL with anti-IgD and anti-IgM antibodies (Figure 1C, left panels). Interestingly, a significantly reduced percentage of mature B cells was observed in the BM (Figure 1C, right panels), suggesting that the defect in the periphery originated from reduced BM progenitors. The phenotype observed in the periphery may be an underestimate of the true requirement for STAT3, given the residual numbers of long-lived wild-type cells resistant to Cre-mediated deletion.
Figure 1.
Decreased numbers of B, but not T, cells in the BM and periphery of STAT3KO mice. (A) STAT3 deletion was complete in BM but was partial in the spleens and PBLs. Genomic DNA prepared from BM, spleen (SPN), and peripheral blood (PBL) of control (cre–) or STAT3KO (cre+) mice was subjected to PCR using primers that differentiated deleted allele (STAT3Δ) and flox allele (STAT3flox). Total cell extracts prepared from the BM of control or STATKO mice were subjected to immunoblot using antibodies to STAT3 or β-actin (right panels). (B) Decreased numbers of B cells in the periphery. Peripheral blood cells of control (•) or STAT3KO (○) mice were stained with anti-CD3 and anti-CD19, and then FACS analysis for B cells (CD19+) or T cells (CD3+) was performed. (C) Decreased numbers of mature B cells in the BM and the periphery. BM or PBL of control (•) or STAT3KO (○) mice were stained with anti-IgD and anti-IgM, then FACS analysis was performed. *P < .05; Student t test. Horizontal bars indicate mean values for each group.
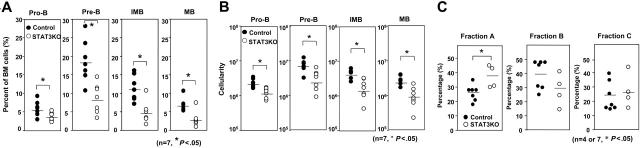
Peripheral B cells are derived from BM progenitors that undergo a developmental series of pro-B, pre-B, immature B, and mature B stages. We asked whether the absence of STAT3 influenced the development of distinct stages of B-cell development. BM from control or STAT3KO mice was stained with antibodies to IgM, B220, and CD43 to enumerate pro-B (IgM–B220+CD43+), pre-B (IgM–B220+CD43–), and immature B (IgM+B220+CD43–) cells or with antibodies to IgM and IgD for mature B (IgM+IgD+) cells. As shown in Figure 2A-B, the percentages and numbers of pro-B, pre-B, immature B, and mature B cells were significantly reduced in the absence of STAT3. Because all the B lineages were affected in the BM of STAT3KO mice, we reasoned that partial developmental arrest must have occurred earlier than at the pro-B stage. We further defined early pro-B stages by staining BM with antibodies to B220, CD43, CD24, and BP-1.29 As shown in Figure 2C, although the percentage of the fraction B or the early pro-B (B220+CD43+BP-1–CD24+) population was moderately reduced, the percentage of the fraction A or the pre–pro-B (B220+CD43+BP-1–CD24–) population was increased in STAT3KO BM. This result suggested that STAT3 is required for efficient transition from the pre–pro-B to the early pro-B stage.
Figure 2.
Impaired development of different B lineages in the BM of STAT3KO mice. BM cells of control (•) or STAT3KO (○) mice were stained with anti-B220, anti-CD43, anti-IgM, and anti-IgD antibodies, then FACS analysis was performed. (A) Reduced percentages of pro-B (B220+CD43+IgM–), pre-B (B220+CD43–IgM–), immature B (IMB; B220+ CD43–IgM+), and mature B (MB; IgD+IgM+) in the BM of STAT3KO mice. (B) Reduced numbers of pro-B, pre-B, IMB, and MB in the BM of STAT3KO mice. Numbers were calculated from the total number of BM multiplied by the percentage of corresponding B lineages. (C) Accumulation of pre–pro-B cells in the BM of STAT3KO mice. BM cells of control (•) and STAT3KO (○) mice were 4-color stained with anti-CD43, anti-B220, anti–BP-1, and anti-CD24, followed by FACS analysis. Individual percentages of fraction A (CD43+B220+BP-1–CD24– or pre-pro B), fraction B (CD43+B220+BP-1–CD24+ or early pro-B), and fraction C (CD43+B220+BP-1+CD24+ or late pro-B) are indicated. *P < .05; Student t test (control mice, n = 7; STAT3KO mice, n = 4). Horizontal bars indicate the mean value for each group.
Reduced response to IL-7 in STAT3KO BM
IL-7R is expressed primarily on common lymphoid progenitors and is important for the development of B and T lymphocytes.30,31 To study whether the reduction in number of B-lineage cells in STAT3KO mice was the result of impaired responses to IL-7, BM from control or STAT3KO mice was treated with IL-7. As shown in Figure 3A, dose-dependent proliferation of control BM cells was observed in response to IL-7, whereas the STAT3KO cells displayed only minimal response. The impaired response was specific for IL-7 because IL-3–mediated proliferation was comparable between control and mutant cells (Figure 3B). To identify the IL-7 responders, BrdU was used to label proliferating cells, which, in conjunction with surface marker staining, could differentiate responses in B or non–B cells. As shown in Figure 3C, though non–B (B220–) cells of both mice incorporated BrdU independently of IL-7 stimulation, B-lineage cells were responsive to IL-7. Interestingly, B cells of STAT3KO mice displayed less BrdU staining than did those of control mice (right panels). Moreover, the main responders to IL-7 were B220+ BM cells, but the main responders to IL-3 were B220– BM cells (Figure 3D-E). B220+ STAT3KO BM displayed reduced proliferation in response to IL-7, whereas the response to IL-3 in B220– BM was almost equivalent to that of control cells, again indicating that impaired response in mutant cells was specific for IL-7.
Figure 3.
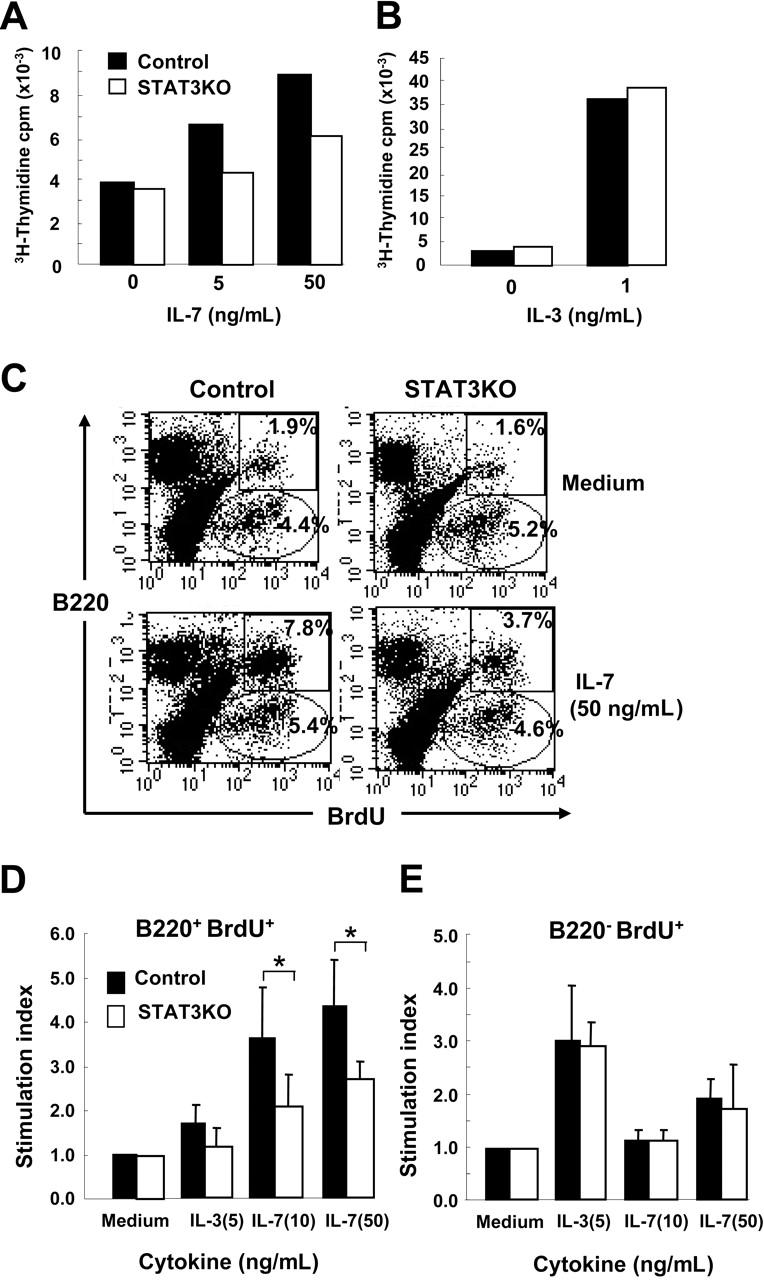
Reduced IL-7–dependent proliferation of BM cells from STAT3KO mice. BM cells of control (▪) or STAT3KO (□) mice were stimulated with or without IL-7 (A) or IL-3 (B) at the indicated doses for 48 hours, and cell proliferation was measured by [3H]-thymidine incorporation. (C) BM cells of control (left panels) or STAT3KO (right panels) mice were stimulated without (top panels) or with (50 ng/mL; bottom panels) IL-7 for 38 hours. Proliferation of different cells was measured by incorporation of BrdU for 3 hours, followed by surface staining of anti-B220 and intracellular staining of anti-BrdU antibodies and FACS analysis. B220+BrdU+ indicated proliferating B cells, and B220–BrdU+ indicated proliferating non-B cells. An identical procedure was performed as described in panel C except that cells were treated without or with IL-3 (5 ng/mL) and IL-7 (10 and 50 ng/mL), respectively. Samples were subjected to FACS analysis, and stimulation indexes for B220+ (D) and B220– (E) were shown. Stimulation index = % cytokine stimulated/% medium alone. *P < .05; Student t test; n = 3. Proliferation results are presented as mean ± SE of replicate samples.
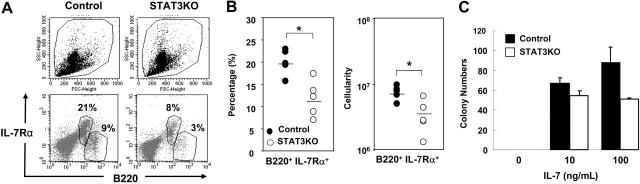
Several mechanisms could account for the reduced response to IL-7. First, because IL-7 activates STAT proteins,16 it is possible that the loss of STAT3 results in impaired IL-7 receptor signaling, preventing STAT phosphorylation. To investigate this possibility, the kinetics of activation of STAT1 and STAT5 in response to IL-7 was measured by immunoblotting. No significant differences in STAT1 or STAT5 protein levels or phosphorylation were observed between control and mutant cells (data not shown), indicating that receptor-proximal signaling was intact. We also asked whether reduced expression of IL-7Rα or reduced numbers of IL-7 responding cells in STAT3KO mice could account for impaired IL-7 responses. BM from control or STAT3KO mice was analyzed by FACS using antibodies to IL-7Rα and B220. As shown in Figure 4A-B, IL-7Rα–expressing cells were mostly B220intermediate, indicative of pro-B and pre-B populations. Interestingly, although IL-7Rα expression (MFI) in STAT3KO BM was comparable to that in control cells, the percentage of IL-7Rα–positive cells was greatly reduced. These results suggested that reduced proliferation of STAT3KO BM cells in response to IL-7 was likely caused by reduced numbers of responders instead of reduced expression or responsiveness of IL-7R.
Figure 4.
Decreased numbers of IL-7 responders in the BM of STAT3KO mice. (A) BM cells of control (left panels) or STAT3KO (right panels) mice were stained with anti-B220 and anti–IL-7Rα followed by FACS analysis. Light scatter patterns are shown in the top panels, and percentages of IL-7Rα+B220intermediate and IL-7Rα–B220high are shown in the bottom panels. (B) Percentage (left panel) and cellularity (right panel) of B220+IL-7Rα+ cells in control (•) and STAT3KO (○) mice were measured. Cellularity was calculated by multiplying the percentage with the total number of BM cells. n = 3; *P < .05; Student t test. (C) Reduced CFU–pre-B progenitor cells in the BM of STAT3KO mice. Colony-formation assay was performed by seeding 2 × 105 BM cells of control (▪) or STAT3KO (□) mice in the complete medium without or with 10 or 100 ng/mL IL-7 for 7 days, followed by counting of the number of the pre-B colonies under a light microscope. Colony-formation assay is presented as mean ± SE of replicate samples.
Because the expression of IL-7Rα marks the development from multiple lineage progenitors to common lymphoid progenitors, it is crucial for determining the fate of B-cell lineages. Therefore, the reduced number of IL-7Rα expressers prompted us to determine whether progenitors can differentiate into pre-B cells in the absence of STAT3. BM cells were seeded in methylcellulose supplemented with different doses of IL-7, and pre-B colonies were scored. As shown in Figure 4C, the number of CFU–pre-B in the control BM cells increased with the enhanced doses of IL-7, whereas reduced numbers of CFU–pre-B were derived from mutant BM. These results indicated that IL-7–responsive progenitors existed in reduced numbers in the absence of STAT3.
Enhanced apoptosis of pro-B cells in the absence of STAT3
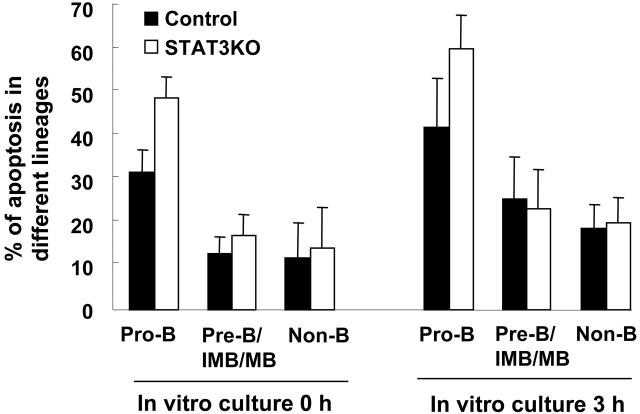
One of the prominent functions of STAT3 is to serve as a survival factor,32 and targeted deletion of STAT3 results in enhanced apoptosis in sensory neurons, motoneurons, and thymic epithelium.33-35 Therefore, we determined whether the reduced B lineages in STAT3KO mice were attributed to enhanced apoptosis. BM cells prepared from control or STAT3KO mice were incubated in vitro for 0 hour or 3 hours in the absence of cytokines, followed by FACS analysis for the apoptotic rate of different B-cell lineages. As shown in Figure 5, though a comparable percentage of annexin V–positive B220– (non-B) or B220+CD43– (pre-B, immature B, and mature B) was found between control and STAT3KO mice, significantly enhanced apoptosis of B220+CD43+ (pro-B) cells at both times was observed in the absence of STAT3, suggesting a critical role of STAT3 for maintaining survival of pro-B cells.
Figure 5.
Enhanced apoptosis of pro-B cells in the BM of STAT3KO mice. BM cells of control (▪) or STAT3KO (□) mice were incubated in the culture medium for 0 hour or 3 hours and were stained with anti-B220, anti-CD43, and annexin V, then FACS analysis was performed. Percentages of apoptotic pro-B (CD43+B220+annexin V+), pre-B/IMB/MB (CD43–B220+ annexin V+), and non-B (B220– annexin V+) were plotted (control mice, n = 8; STAT3KO mice, n = 5). Apoptosis results are presented as mean ± SE of replicate samples.
Cell-autonomous requirement of STAT3 for normal B-cell development
To investigate whether the impaired development of B cells resulted from a cell-autonomous mechanism rather than an indirect effect of STAT3 loss elsewhere in the animal, we performed BM transplantation. Five million BM cells from control or STAT3KO mice were transplanted into lethally irradiated RAG1KO mice. Two months later, repopulated splenocytes or BM cells derived from the RAG1KO mice that had received transplants were subjected to immunoblot analysis for the expression of STAT3 protein and to FACS analysis for different lineages of B cells. As shown (Figure 6A, E), the repopulated BM or splenocytes derived from STAT3KO mice were completely devoid of STAT3. It was also clear that STAT3KO→RAG1KO chimeric mice displayed significantly reduced percentages and cellularities of pro-B, pre-B, immature B, and mature B in the BM as opposed to control→RAG1KO chimeric mice (Figure 6B-C). Interestingly, though an enhanced percentage of T1 transitional B cells (IgDlow-IgMhigh) were observed in the spleens of STAT3KO→RAG1KO chimeric mice, the percentage of T2 transitional B/mature B (IgDhighIgMintermediate-high) was only slightly reduced compared with control→RAG1KO mice (Figure 6D, F). However, the cellularity of T1 and T2/mature B cells was drastically reduced in the STAT3KO→RAG1KO chimeric mice compared with control→RAG1KO mice (Figure 6G-H), which might have reflected a poor reconstitution rate of splenocytes in the absence of STAT3. Taken together, these results further demonstrated that the impaired B lymphopoiesis in BM and spleen was the result of a cell-autonomous requirement of STAT3 in differentiating B-cell progenitors.
Figure 6.
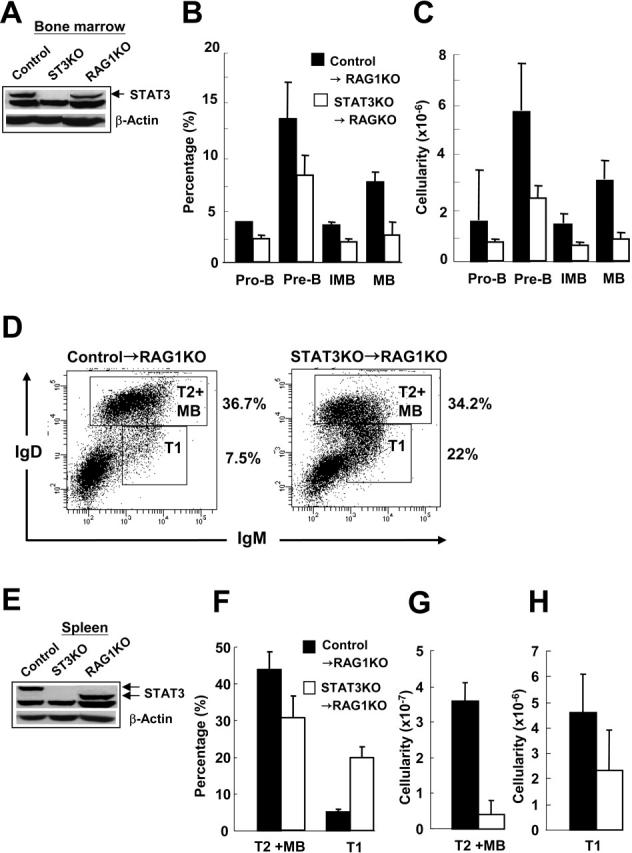
Impairment of B-cell development in the absence of STAT3 is cell autonomous. BM of control (▪) or STAT3KO (□) mice was intravenously transplanted into lethally irradiated RAG1KO mice for 2 months. Total cell extracts prepared from BM (A) or spleen (E) were subjected to immunoblot using antibodies to STAT3 or β-actin. BM cells of chimeric mice were stained with anti-CD43, anti-IgM, and anti-B220 antibodies, followed by FACS analysis. Percentage (B) and cellularity (C) of reconstituted B-cell lineages were determined. (D) Similarly, splenocytes of control→RAG1KO (left panel) or STAT3→RAG1KO (right panel) chimeric mice were isolated after transplantation and were stained with anti-IgM and anti-IgD followed by FACS analysis. Percentages of T1 and T2+MB are shown. Percentage (F) and cellularity (G-H) are shown. IgMhigh IgDlow represent T1 cells. IgMintermediate-high IgDhigh represent T2 or mature B (MB) cells. Results are presented as mean ± SE of replicate samples.
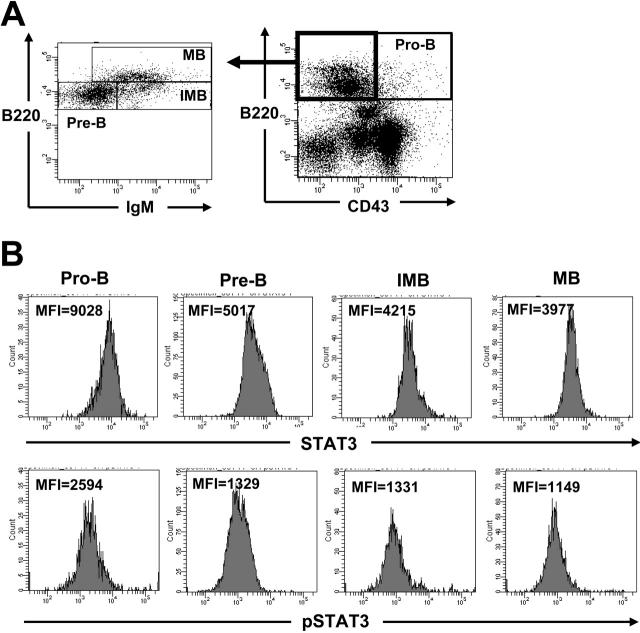
Because the deletion of STAT3 resulted in an increased pre–pro-B population and an impaired development of pro-B, pre-B, immature, and mature B cells, we reasoned that STAT3 might be differentially expressed in different lineages of B cells in the BM. To determine the protein levels of STAT3 in different populations, intracellular staining for STAT3 in conjunction with surface staining for different B lineages of BM cells from control mice was performed. As shown in Figure 7B (upper panels), pro-B cells expressed almost twice as much STAT3 as did pre-B, immature B, or mature B cells. We next determined whether tyrosine phosphorylation of STAT3 was also enhanced in the pro-B population. Indeed, the level of phosphotyrosyl STAT3 was also increased in pro-B cells relative to other lineages of B cells (Figure 7B, lower panels), suggesting that activation of STAT3 is critical for B-cell development at an early stage.
Figure 7.
Enhanced expression and activation of STAT3 in the pro-B lineage of BM cells. Freshly prepared BM cells of control mice were first stained with antibodies to CD43, B220, and IgM, followed by permeabilization and staining with primary antibodies either to STAT3 or to phospho-STAT3 and FITC-conjugated secondary antibody to rabbit immunoglobulin. Stained cells were then analyzed by FACS according to surface markers. Pre-B, IMB, and MB were analyzed by gating on CD43–B220+ cells (A). The intensity (MFI) of STAT3 (B, top panels) or pSTAT3 (bottom panels) gated on pro-B (CD43+B220+), pre-B (CD43–IgM–B220+), immature B (IMB, CD43–IgM+B220+), and mature B (MB, CD43–IgM+B220high) cells is shown.
Discussion
We have previously shown that mice lacking STAT3 in their hematopoietic progenitors developed neutrophilia and that BM cells were hyperresponsive to G-CSF stimulation, suggesting that STAT3 is a negative regulator for G-CSF–mediated granulopoiesis.26 Here, we have demonstrated that STAT3 serves as a positive regulator for B lymphopoiesis. In the absence of STAT3, B but not T cells were significantly reduced in the periphery. The decreased number of peripheral B cells resulted from the impaired development of B-cell lineages in the BM, including reduced numbers of pro-B, pre-B, immature, and mature B cells (Figures 1, 2). Interestingly, an increased percentage of pre–pro-B cells or Hardy fraction A29 was observed in the BM of STAT3KO mice, implying partial developmental arrest at the pre–pro-B stage in the absence of STAT3 (Figure 2C). In addition, it has been shown that STAT3 is constitutively active in B-1, but not B-2, cells.36 We also noticed that the percentage of peritoneal CD5+ B cells was decreased in the STAT3KO mice (data not shown), suggesting that STAT3 might also be required for the development of B-1 cells.
Recently, Fornek et al37 used CD19CreSTAT3f/f, another STAT3 conditional knockout mouse, to demonstrate a role for STAT3 in B lymphocytes. Their STAT3KO mice displayed defects in T-cell–dependent (TD) IgG responses but normal TD IgM, IgE, and IgA responses and T-cell–independent IgM and IgG3 responses. Interestingly, they failed to observe any developmental defects in the B-cell compartment. In agreement with their findings, we also examined CD19CreSTAT3f/f mice and did not detect any defects in B-cell development23 (data not shown). Given that CD19 is not expressed until the pro-B stage,38 after the transition from the pre–pro-B stage where MxCreSTAT3f/f mice started to show defects, it is likely that the critical requirement for STAT3 during B lymphopoiesis commences at this earlier stage. These results imply that loss of STAT3 at the pro-B but not at the pre–pro-B stage can be compensated for by other factors, possibly other STAT proteins. Alternatively, although CD19 starts to express on pro-B cells, the activity of the promoter is not at full strength. This notion is supported by incomplete deletion of floxed alleles in BM pre-B cells.39,40 Therefore, it is likely that the normal B lymphopoiesis in CD19CreSTAT3f/f mice is caused by the effect of residual STAT3 at early stages of B lineages.
One possible candidate for the discrepancy seen in CD19CreSTAT3f/f and MxCreSTAT3f/f mice is Flt3. It is known that Flt3 expression starts in multipotent progenitors and that Flt3–Flt3L interaction is essential for the development of early lineages, such as CLP 5 and ETP.41 Recently, Laouar et al42 showed that STAT3 is required for Flt3-dependent development of dendritic cells. Given the role of Flt3 signaling during B lymphopoiesis, it is likely that impaired B-cell development in MxCreSTAT3f/f mice is affected by diminished response to Flt3L. Alternatively, STAT3 may affect B lymphopoiesis by regulating Notch signals through the interactions with Hes, a downstream Notch mediator. It has been demonstrated that overexpression of a dominant-negative form of STAT3 blocks radial glia and astrocytic differentiation induced by Notch.43 Interestingly, the density of Delta-1, a Notch ligand, can determine the development of hematopoietic stem cells (HSCs) into precursors of T and B cells.44 Delta-1 promotes the generation of Thy1+CD25+ early T precursor at a higher density and enhances the generation of B220+CD43low, a B precursor, in addition to the early T precursors at low density. Therefore, STAT3 may be required to regulate the balance of Notch signaling contributing to B-cell development. In any case, it is notable that the timing of the B-cell defect in the absence of STAT3 corresponds to the stage of highest STAT3 expression and tyrosyl phosphorylation.
By performing BM transplantation, we found that the requirement for STAT3 during B-cell development occurs through a cell-intrinsic mechanism. B cells derived from STAT3-deficient BM cells were indeed devoid of STAT3 at DNA and protein levels (Figure 6; data not shown). In these chimeric mice, we also reproducibly observed reduced percentages and numbers of B cells in the BM and periphery. Surprisingly, we also observed another developmental defect in the spleen, where STAT3KO transitional T1 cells were increased compared with controls. For the moment, it is unclear why T1 cells are arrested at this stage. However, despite the increased percentage of T1 cells in STAT3KO→RAG1KO chimeric mice, the B-cell cellularity was dramatically reduced compared with that of control→RAG1KO chimeric mice. Therefore, STAT3 may influence homeostatic proliferation, in addition to fulfilling its developmental role.
We showed that IL-7–dependent proliferation of BM cells was reduced in the absence of STAT3. This reduction appeared to be caused by a decrease in the number of IL-7–responsive progenitors rather than by a signaling effect in the absence of STAT3 (Figure 4; data not shown). We used intracellular staining but did not observe significantly enhanced tyrosine phosphorylation of STAT3 in the pro-B population in response to IL-7 (data not shown), probably because the main responders were pre-B rather than pro-B cells. Therefore, the cause of hyporesponsiveness without STAT3 is presumably indirect. The other possibility for reduced response to IL-7 was enhanced apoptosis in the absence of STAT3. It is known that the STAT3 downstream molecules Bcl-245 and Mcl-146 are critical for cell survival. Therefore, one possible mechanism for the increased apoptosis is reduced expression of these antiapoptotic proteins in the absence of STAT3. It remains unclear why the pro-B population is particularly susceptible to apoptosis when STAT3 is deleted. STAT5 appears to be absolutely critical for IL-7 signaling in T and B lymphocytes,17 but the loss of STAT3 only impaired B lymphopoiesis. The critical role of STAT3 in early B-cell development without a corresponding requirement in T-cell development remains an enigma.
Acknowledgments
We thank Drs Jeffery J. Y. Yen and Kuo-I. Lin for reading the manuscript, Drs Li-Mei Pai and Ming-Zong Lai for providing reagents, and Wei Zhu for excellent technical assistance.
Prepublished online as Blood First Edition Paper, July 6, 2006; DOI 10.1182/blood-2006-05-024430.
Supported by the National Science Council of Taiwan and National Taiwan University Hospital (C.-K.L.) and by grant R01AI28900 from the National Institutes of Health (D.E.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22: 55-79. [DOI] [PubMed] [Google Scholar]
- 2.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102: 4949-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5: 497-508. [DOI] [PubMed] [Google Scholar]
- 4.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3: 147-161. [DOI] [PubMed] [Google Scholar]
- 5.Sitnicka E, Bryder D, Theilgaard-Monch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17: 463-472. [DOI] [PubMed] [Google Scholar]
- 6.Maraskovsky E, Peschon JJ, McKenna H, Teepe M, Strasser A. Overexpression of Bcl-2 does not rescue impaired B lymphopoiesis in IL-7 receptordeficient mice but can enhance survival of mature B cells. Int Immunol. 1998;10: 1367-1375. [DOI] [PubMed] [Google Scholar]
- 7.Carvalho TL, Mota-Santos T, Cumano A, Demengeot J, Vieira P. Arrested B lymphopoiesis and persistence of activated B cells in adult interleukin 7(–/–) mice. J Exp Med. 2001;194: 1141-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181: 1519-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peschon JJ, Morrissey PJ, Grabstein KH, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180: 1955-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sitnicka E, Brakebusch C, Martensson IL, et al. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198: 1495-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medina KL, Pongubala JM, Reddy KL, et al. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7: 607-617. [DOI] [PubMed] [Google Scholar]
- 12.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16: 297-309. [DOI] [PubMed] [Google Scholar]
- 13.Ye M, Ermakova O, Graf T. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med. 2005;202: 1411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 2005;106: 2083-2090. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki H, Somoza C, Shigematsu H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106: 1590-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin JX, Migone TS, Tsang M, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2: 331-339. [DOI] [PubMed] [Google Scholar]
- 17.Yao Z, Cui Y, Watford WT, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103: 1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertolino E, Reddy K, Medina KL, Parganas E, Ihle J, Singh H. Regulation of interleukin 7-dependent immunoglobulin heavy-chain variable gene rearrangements by transcription factor STAT5. Nat Immunol. 2005;6: 836-843. [DOI] [PubMed] [Google Scholar]
- 19.Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Farrar MA. STAT5 activation underlies IL7 receptor-dependent B cell development. J Immunol. 2004;172: 4770-4778. [DOI] [PubMed] [Google Scholar]
- 20.Sexl V, Piekorz R, Moriggl R, et al. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of stat5. Blood. 2000;96: 2277-2283. [PubMed] [Google Scholar]
- 21.Lee CK, Smith E, Gimeno R, Gertner R, Levy DE. STAT1 affects lymphocyte survival and proliferation partially independent of its role downstream of IFN-gamma. J Immunol. 2000;164: 1286-1292. [DOI] [PubMed] [Google Scholar]
- 22.Lee CK, Gimeno R, Levy DE. Differential regulation of constitutive major histocompatibility complex class I expression in T and B lymphocytes. J Exp Med. 1999;190: 1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109: 1143-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98: 295-303. [DOI] [PubMed] [Google Scholar]
- 25.Chiarle R, Simmons WJ, Cai H, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11: 623-629. [DOI] [PubMed] [Google Scholar]
- 26.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17: 63-72. [DOI] [PubMed] [Google Scholar]
- 27.Raz R, Lee CK, Cannizzaro LA, d'Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96: 2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269: 1427-1429. [DOI] [PubMed] [Google Scholar]
- 29.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173: 1213-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91: 661-672. [DOI] [PubMed] [Google Scholar]
- 31.Sudo T, Nishikawa S, Ohno N, et al. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc Natl Acad Sci U S A. 1993;90: 9125-9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirano T, Ishihara K, Hibi M. Roles of STAT3 in mediating the cell growth, differentiation and survival signals relayed through the IL-6 family of cytokine receptors. Oncogene. 2000;19: 2548-2556. [DOI] [PubMed] [Google Scholar]
- 33.Alonzi T, Middleton G, Wyatt S, et al. Role of STAT3 and PI 3-kinase/Akt in mediating the survival actions of cytokines on sensory neurons. Mol Cell Neurosci. 2001;18: 270-282. [DOI] [PubMed] [Google Scholar]
- 34.Sano S, Takahama Y, Sugawara T, et al. Stat3 in thymic epithelial cells is essential for postnatal maintenance of thymic architecture and thymocyte survival. Immunity. 2001;15: 261-273. [DOI] [PubMed] [Google Scholar]
- 35.Sano S, Itami S, Takeda K, et al. Keratinocytespecific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 1999;18: 4657-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karras JG, Wang Z, Huo L, Howard RG, Frank DA, Rothstein TL. Signal transducer and activator of transcription-3 (STAT3) is constitutively activated in normal, self-renewing B-1 cells but only inducibly expressed in conventional B lymphocytes. J Exp Med. 1997;185: 1035-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 2006;107: 1085-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Nagro CJ, Otero DC, Anzelon AN, Omori SA, Kolla RV, Rickert RC. CD19 function in central and peripheral B-cell development. Immunol Res. 2005;31: 119-131. [DOI] [PubMed] [Google Scholar]
- 39.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25: 1317-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires anti-apoptotic MCL-1. Nature. 2003;426: 671-676. [DOI] [PubMed] [Google Scholar]
- 41.Sambandam A, Maillard I, Zediak VP, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6: 663-670. [DOI] [PubMed] [Google Scholar]
- 42.Laouar Y, Welte T, Fu XY, Flavell RA. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 2003;19: 903-912. [DOI] [PubMed] [Google Scholar]
- 43.Kamakura S, Oishi K, Yoshimatsu T, Nakafuku M, Masuyama N, Gotoh Y. Hes binding to STAT3 mediates crosstalk between Notch and JAK-STAT signalling. Nat Cell Biol. 2004;6: 547-554. [DOI] [PubMed] [Google Scholar]
- 44.Dallas MH, Varnum-Finney B, Delaney C, Kato K, Bernstein ID. Density of the Notch ligand Delta1 determines generation of B and T cell precursors from hematopoietic stem cells. J Exp Med. 2005;201: 1361-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL. Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene. 2002;21: 7611-7618. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Ma Y, Cole SM, et al. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102: 344-352. [DOI] [PubMed] [Google Scholar]