Abstract
New prognostic factors may result in better risk classification and improved treatment of children with acute lymphoblastic leukemia (ALL). Recently, high expression of a gene named OPAL1 (outcome predictor in acute leukemia) was reported to be associated with favorable prognosis in ALL. Therefore, we investigated whether OPAL1 expression was of prognostic importance in 2 independent cohorts of children with ALL treated on Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL)–92/97 (n = 180) and St Jude Total 13 protocols (n = 257). We observed a consistently higher (2.8-fold) expression of OPAL1 in TEL-AML1–positive ALL compared with TEL-AML1–negative ALL in both cohorts, but higher OPAL1 expression was not consistently associated with other favorable prognostic indicators such as age and white blood cell count, or ALL genetic subtype. Lower OPAL1 expression was also not associated with increased in vitro drug resistance. Multivariate analyses including known risk factors showed that OPAL1 expression was not independently related to prognosis in either the COALL or St Jude cohorts. In conclusion, OPAL1 expression may not be an independent prognostic feature in childhood ALL, and its previously reported prognostic impact appears to be treatment dependent.
Introduction
The prognosis of childhood acute lymphoblastic leukemia (ALL) has improved remarkably in the past 4 decades due to the introduction of effective risk-adapted combination chemotherapies. Conventional factors used to stratify patients are clinical and biologic parameters such as age at diagnosis, initial white blood cell (WBC) count, immunophenotype, the presence of specific genetic abnormalities,1 and early response to treatment.2 Newer approaches include in vitro drug resistance profiles3 and measurement of minimal residual disease after induction of initial remission.4,5
The use of DNA microarrays enables investigators to simultaneously assess the expression of thousands of genes. In previous studies of childhood ALL, microarray analysis was successfully applied to identify known genetic and phenotypic subtypes6-8 as well as treatment-specific changes in gene expression9 and genes related to drug resistance.10 Recently, this technology was used to identify 3 novel genes, referred to as G0, G1, and G2, that were highly predictive of outcome in 254 patients with childhood ALL enrolled in Pediatric Oncology Group (POG) treatment protocols.11-13 The top discriminating gene, G0, was fully cloned and named OPAL1 (outcome predictor in acute leukemia 1). The function of OPAL1 is unknown, although the presence of a cytochrome c–like heme-binding site and a transmembrane domain suggested OPAL1 may be involved in the mitochondrial electron transport chain.14 We initially identified this gene as one of the top-ranked class discriminating genes that was overexpressed in ALL cells positive for the TEL-AML1 gene fusion.7,8 In the POG study, OPAL1 was expressed at higher levels in ALL subgroups with a favorable prognosis (ie, ALL with t(12;21)/TEL-AML1, normal and hyperdiploid karyotypes) compared with a subgroup with an unfavorable prognosis (ie, ALL with t(9;22)/BCR-ABL) and another subgroup previously associated with an unfavorable prognosis in (ie, ALL with t(1;19)/E2A-PBX1).12 High OPAL1 expression was shown to be highly predictive of a favorable outcome in the total ALL group, but also in ALL subgroups, such as T-lineage ALL and t(12;21)/TEL-AML1–positive B-lineage ALL. Finally, low OPAL1 was significantly related to induction failures.12
To validate these interesting results independently, we analyzed in depth the expression pattern of OPAL1 in 2 independent cohorts of children with newly diagnosed ALL treated on protocols of the Cooperative Study Group for Childhood Acute Lymphoblastic Leukemia (COALL; n = 180) and the St Jude Children's Research Hospital (St Jude; n = 257). OPAL1 expression was investigated in relation to in vitro resistance to 4 widely used drugs in the treatment of childhood ALL (ie, prednisolone, vincristine, l-asparaginase, and daunorubicin). In addition, OPAL1 expression was tested as a predictor of clinical outcome in childhood ALL, where ALL subtypes as well as other known prognostic factors were included in a multivariate analysis.
Patients, materials, and methods
Leukemia samples
Bone marrow and peripheral blood samples were obtained after informed consent from children with newly diagnosed ALL who were enrolled on either the COALL-92/97 protocol (n = 180)10,15,16 or the St Jude Total Therapy 13 protocol (n = 257).7,8,17,18 These 2 independent trials used similar chemotherapeutic agents. Approval was obtained from the Erasmus MC/Sophia Children's Hospital or St Jude Children's Research Hospital institutional review boards for these studies. If necessary, peripheral blood or diagnostic bone marrow samples were enriched for leukemic blasts to be 90% or more as previously described.10,19
In vitro drug resistance assay
In COALL patients, responsiveness of leukemia cells to prednisolone (PRED; Bufa Pharmaceutical Products, Uitgeest, The Netherlands), vincristine (VCR; TEVA Pharma, Mijdrecht, The Netherlands), l-asparaginase (ASP; Paronal, Christiaens, Breda, The Netherlands), and daunorubicin (DNR; Cerubidine, Rhône-Poulenc Rorer, Amstelveen, The Netherlands) was determined by the 4-day in vitro MTT drug resistance assay.3 The concentration ranges tested for these drugs were: PRED, 0.008 to 250 μg/mL (n = 167); VCR, 0.05 to 50 μg/mL (n = 166); ASP, 0.003 to 10 IU/mL (n = 166); and DNR, 0.002 to 2.0 μg/mL (n = 140). The drug concentration lethal to 50% of the ALL cells (LC50 value) was used as the measure of cellular drug resistance.3,20
Real-time quantitative PCR
The mRNA expression levels of OPAL1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference were determined using quantitative real-time polymerase chain reaction (RTQ-PCR) analysis on the ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA) as previously described.21-24 All PCR reactions were performed with an amplification efficiency of more than 95%. OLIGO 6.22 software (Molecular Biology Insights, Cascade, CO) was used to design primer and probe combinations within OPAL1 (NM_017787) and GAPDH (NM_2046). Primer sequences used were 5′-TCCTTTTGGGTCTTAGACAG-3′ (sense) and 5′-TTGGCAAAAACCTGAAAT-3′ (antisense) for OPAL1 and 5′-GTCGGAGTCAACGGATT-3′ (sense) and 5′-AAGCTTCCCGTTCTCAG-3′ (antisense) for GAPDH. Probe sequences used were 5′-ACAGTCTCAGTGCTGCAACTACTACTATGA-3′ for OPAL1 and 5′-TCAACTACATGGTTTACATGT TCCAA-3′ for GAPDH. For each sample, the comparative cycle time (Ct) value of the OPAL1 PCR was normalized by subtracting the Ct value of the GAPDH PCR (ΔCt).21 From this ΔCt value the relative OPAL1 expression to GAPDH in arbitrary units (AU) was calculated using the following formula: relative mRNA expression = 2–ΔCt × 100%. We observed a significant correlation between OPAL1 mRNA expression assessed by RT-PCR and microarray (rs = 0.35, P = .003, n = 72).
Microarray analysis
Total RNA was hybridized to U133A (COALL) and U95Av2 (St Jude) GeneChip oligonucleotide microarrays, according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). Data analysis was performed as described before and gene expression data of the leukemic samples included in this present study were previously published.8,10 Briefly, gene expression values were scaled to the target intensity of 2500, using Affymetrix Microarray Suite (MAS) 5.0 software, and were log2 transformed. To analyze the expression of OPAL1, we used the U133A probe set 202808_at (COALL), which covers the same DNA sequence (99.8% sequence identity) as the Affymetrix U95Av2 GeneChip probe set 38652_at (St Jude); the latter was also used by Mosquera-Caro et al. and Yeoh et al.8,12 More information on these probe sets and its target sequences is available at Affymetrix NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx). OPAL1 expression determined with both the U133A and U95Av2 arrays was highly correlated and available for a subset of St Jude patients (rs = 0.58, P < .001, n = 92).
Statistics
The duration of disease-free survival (DFS) was defined as the time from diagnosis until the date of leukemia relapse (event), the last follow-up, or secondary events other than relapse (censored). DFS curves were calculated according to the Kaplan-Meier method or a modification thereof in the presence of competing events.25,26 Because no cut-off for OPAL1 expression was provided by Mosquera-Caro et al,12 we performed survival analyses in 2 different ways: OPAL1 expression was treated either as a continuous variable or as a categorical variable (OPAL1 expression was divided into 3 equal-sized groups by the 33rd and 67th percentile of expression; that is, low [bottom third], intermediate [intermediate third], and high [top third]). The predictive value of OPAL1 expression in 3 groups (2 degrees of freedom [df]) and of OPAL1 expression as a continuous variable was analyzed by log-rank test and by a Cox proportional hazard regression model (adjusted for competing events in all analyses of the St Jude cohort).27,28 The association of OPAL1 expression with DFS considering other known prognostic factors was assessed in univariate and in multivariate analyses. The model for multivariate analysis included conventional risk factors (ie, WBC count, age, immunophenotype, and genetic abnormalities). Differences in OPAL1 expression between ALL subgroups were tested using the Mann-Whitney U test. The Spearman correlation test was used to compare the expression of OPAL1 by microarray with the expression data obtained by RTQ-PCR and to relate OPAL1 expression to in vitro drug resistance. All statistical tests were performed at a 2-tailed significance level of .05. When applicable, Bonferroni correction was applied to correct for multiple comparisons.
Results
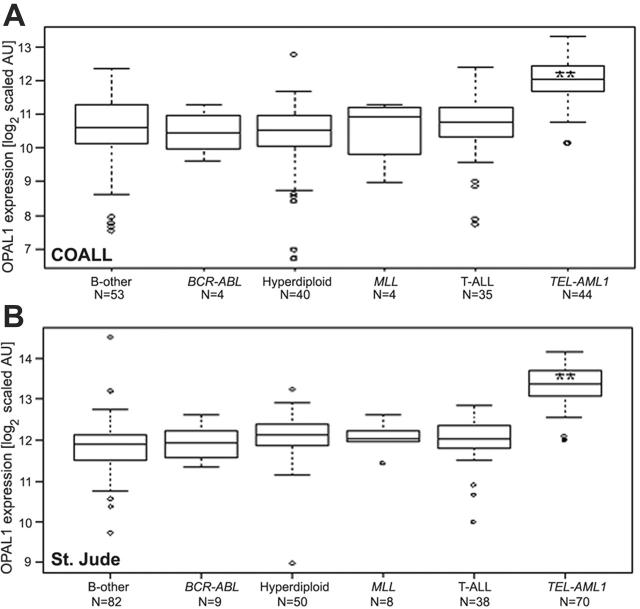
The association of OPAL1 expression with prognostic features in ALL was tested in 2 cohorts of children with ALL. High OPAL1 expression was consistently observed in TEL-AML1–positive ALL in both patient cohorts (Figure 1, Table 1; 2.8-fold, P < .001). There was no further evidence of a significant relation between OPAL1 expression and any other prognostic features in COALL patients, whereas low OPAL1 expression was related to ages older than 10 years, and high OPAL1 expression was related to a hyperdiploid karyotype among St Jude patients (Table 1). The frequency distribution of these risk categories between the 2 cohorts was comparable, except for WBC count, where we observed a trend toward higher counts in the COALL cohort (P = .004; 2-sample Kolmogorov-Smirnov test). The likely reason for these differences between the St Jude and COALL patients is that patients with higher WBC counts were more likely to have enough cells for in vitro drug resistance testing.
Figure 1.
OPAL1 expression in different ALL subtypes. OPAL1 expression was compared in a total of 180 children with ALL treated according to COALL protocols (A), and in a total of 257 children with ALL treated according to St Jude protocols (B). OPAL1 expression is shown in log2-transformed scaled arbitrary units (AU). The medians (horizontal lines), the 25th and 75th percentiles (boxes), the ranges (bars), and the outliers (open circles) are shown. **Indicates P < .001, determined by the Mann-Whitney U test.
Table 1.
OPAL1 expression in prognostic subgroups of ALL at diagnosis
|
COALL
|
St Jude
|
|||||
|---|---|---|---|---|---|---|
| Variable | N | Ratio | P | N | Ratio | P |
| Age | ||||||
| No older than 10 y | 131 | 1.00* | 181 | 1.00* | ||
| Older than 10 y | 49 | 0.86 | .39 | 76 | 0.87 | .001† |
| WBC count | ||||||
| Less than 10 × 109/L | 43 | 1.00* | 90 | 1.00* | ||
| 10 × 109-49 × 109/L | 67 | 0.91 | .97 | 80 | 1.11 | .42 |
| 50 × 109-100 × 109/L | 28 | 1.04 | .93 | 40 | 1.05 | .88 |
| More than 100 × 109/L | 42 | 1.04 | .76 | 47 | 0.85 | .023 |
| Sex | ||||||
| Female | 76 | 1.00* | 95 | 1.00* | ||
| Male | 104 | 1.00 | .74 | 162 | 1.09 | .53 |
| ALL subtype | ||||||
| B-other | 53 | 1.00* | 82 | 1.00* | ||
| BCR-ABL | 4 | 0.89 | .74 | 9 | 1.02 | .63 |
| Hyperdiploid‡ | 40 | 0.94 | .47 | 50 | 1.17 | .002† |
| MLL rearranged | 4 | 1.23 | .79 | 8 | 1.10 | .19 |
| TEL-AML1 | 44 | 2.69 | < .001† | 70 | 2.82 | < .001† |
| T-lineage | 35 | 1.13 | .32 | 38 | 1.10 | .056 |
OPAL1 expression was compared in prognostic ALL subgroups defined by age, WBC count, sex, immunophenotype, and genetic subtype in children treated on COALL and St Jude protocols. Indicated are the number of patients (N) for each group, the ratio of OPAL1 expression in nonreference versus reference groups (fold difference in median scaled OPAL1 expression), and P values comparing the nonreference versus reference group determined by the Mann-Whitney U test.
Reference group.
Significant after Bonferroni correction.
Cytogenetic analysis revealed more than 50 chromosomes.
Because drug resistance is a major cause of treatment failure, we investigated in the COALL cohort whether OPAL1 expression was related to in vitro drug resistance for any of 4 drugs that form an integral component of contemporary chemotherapeutic protocols for children with ALL. No correlation was observed between in vitro drug resistance and OPAL1 expression for PRED (rs < 0.001, P = .99), ASP (rs =–0.02, P = .81), and DNR (rs = 0.05, P = .54). By contrast, VCR resistance showed a significant positive correlation with OPAL1 expression (rs = 0.34, P < .001), which is opposite of what would be expected if high OPAL1 expression is related to a good prognosis, as previously reported.12 Nevertheless, this observation is concordant with a 3.6-fold increased VCR resistance among TEL-AML1–positive ALL compared with non–TEL-AML1 B-lineage ALL (P < .001; median LC50 = 0.697 μg/mL and 0.193 μg/mL, respectively).
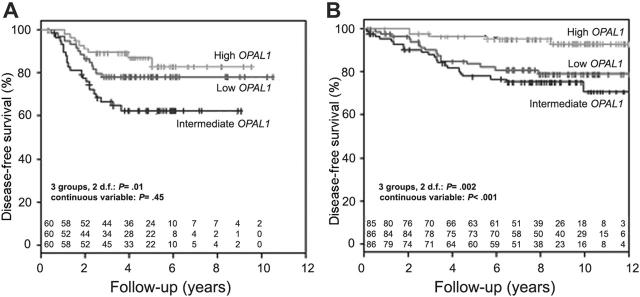
The relation between OPAL1 expression and treatment outcome was subsequently investigated in both cohorts. Of the 180 patients who were part of the COALL cohort, 42 had disease-related events and 6 had competing events (2 secondary malignancies and 4 deaths in remission). Of the 257 patients included in the St Jude study Total Therapy 13, 42 had disease-related events and 20 had competing events (16 secondary malignancies and 4 deaths in remission).18,28 In the COALL cohort, OPAL1 expression was significantly associated with DFS when the patient population was divided into 3 equally sized groups based on the individual rank in OPAL1 expression (2 df, P = .01; Figure 2A) but not when OPAL1 expression was treated as a continuous variable (P = .45), or when the top 33% of patients with high expression were compared with the bottom 33% of patients with low expression (P = .28; Table 2). In contrast, as reported in a preliminary analysis of the St Jude cohort by others,11-13 OPAL1 expression was significantly associated with DFS among the St Jude cohort, whether patients were divided into 3 equally sized groups (2 df, P = .002; Figure 2B), OPAL1 expression was treated as a continuous variable (P < .001), or the top third of patients with high expression were compared with the bottom third with low expression (P = .01; Table 2).
Figure 2.
Disease-free survival according to OPAL1 expression in children with ALL. OPAL1 expression was not associated with disease-free survival among 180 children with newly diagnosed ALL treated on COALL 92/97 (A), but an association was observed among 257 newly diagnosed children with ALL treated on St Jude Total 13 protocols (B).
Table 2.
Univariate analysis of the prognostic value of OPAL1 expression in pediatric ALL
|
COALL
|
St Jude
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Variable | N | 4-y DFS, % ± SE | OPAL1 categorical variable P*(HR) | OPAL1 continuous variable P†(HR) | N | 4-y DFS, % ± SE | OPAL1 categorical variable P*(HR) | OPAL1 continuous variable P†(HR) |
| Total group | ||||||||
| Low‡ | 60 | 78 ± 5 | -(1.00) | 85 | 86 ± 4 | -(1.00) | ||
| Intermediate | 60 | 63 ± 7 | .07 (1.88) | .45 (0.92) | 86 | 82 ± 4 | .33 (1.37) | < .001§ (0.62) |
| High | 60 | 87 ± 5 | .28 (0.61) | 86 | 96 ± 2 | .01§ (0.28) | ||
| B-lineage | ||||||||
| Low‡ | 48 | 79 ± 6 | -(1.00) | 73 | 87 ± 4 | -(1.00) | ||
| Intermediate | 49 | 69 ± 7 | .33 (1.49) | .078 (0.81) | 73 | 89 ± 4 | .68 (1.18) | < .001§ (0.56) |
| High | 48 | 91 ± 5 | .18 (0.48) | 73 | 97 ± 2 | .02 (0.23) | ||
| T-lineage | ||||||||
| Low‡ | 12 | 92 ± 8 | -(1.00) | 12 | 75 ± 13 | -(1.00) | ||
| Intermediate | 11 | 50 ± 16 | .06 (7.90) | .06 (1.9) | 13 | 67 ± 14 | .86 (1.14) | .34 (1.58) |
| High | 12 | 42 ± 14 | .03 (9.80) | 13 | 69 ± 13 | .76 (1.22) | ||
| Non-TEL-AML1 | ||||||||
| Low‡ | 34 | 76 ± 7 | -(1.00) | 49 | 87 ± 5 | -(1.00) | ||
| Intermediate | 33 | 75 ± 8 | .93 (1.04) | .84 (0.97) | 50 | 84 ± 5 | .23 (1.76) | .35 (0.76) |
| High | 34 | 66 ± 9 | .55 (1.33) | 50 | 96 ± 3 | .52 (0.69) | ||
| TEL-AML1 | ||||||||
| Low‡ | 15 | 92 ± 3 | -(1.00) | 23 | 87 ± 7 | -(1.00) | ||
| Intermediate | 14 | 92 ± 7 | .89 (0.82) | .61 (0.70) | 24 | 100 | .17 (0.23) | < .001§ (0.07) |
| High | 15 | 100 | .97 (0.94) | 23 | 100 | ND | ||
Overview of DFS analyses for low, intermediate, and high OPAL1 expression in COALL and St Jude study cohorts in the total group of pediatric ALL patients, within B-lineage and T-lineage ALL patients, and within genetic subgroups of ALL that were associated with OPAL1 expression in COALL patients and St Jude patients. Dashes have been inserted for the P values of the reference groups.
HR indicates hazard ratio; ND, not detected (ie, no events occurred).
Cox univariate analysis adjusted for competing events; OPAL1 expression categorical.
OPAL1 expression continuous variable.
Reference group.
Significant after Bonferroni correction.
The clinical value of OPAL1 expression was further studied within major prognostically important ALL subtypes (Table 2). In both cohorts the opposite (although not statistically significant in St Jude patients) correlation was observed in T-ALL (ie, a low OPAL1 expression correlated with a favorable DFS) (Table 2). This result differs from our results for the total group of ALL patients (Figure 2; Table 2) as well as those obtained for the initial POG cohort,11-13 where high OPAL1 expression was found to be a favorable outcome predictor in patients with T-ALL.
Among patients with B-lineage ALL, higher expression of OPAL1 was only significantly related to a favorable prognosis in the St Jude cohort, both as a continuous variable (P < .001) and when expression was applied as a categorical variable for the top third (P = .02; Table 2).
Among patients with TEL-AML1–positive B-lineage ALL treated on the COALL protocols, OPAL1 expression was not significantly associated with DFS (Table 2; P = .61, P > .89). When all patients with TEL-AML1–positive patients treated on the St Jude protocols (Total Therapy 13A and 13B) were analyzed, the association between high OPAL1 expression and higher DFS was only significant when the expression was analyzed as a continuous variable (P < .001). Interestingly, OPAL1 expression treated as a continuous variable had no prognostic significance among the 36 TEL-AML1–positive patients treated in study 13A (P = .32), but was significant among the 34 TEL-AML1–positive patients treated in study 13B (P < .001). Notably, because of different criteria for assigning patients to the low-risk versus high-risk treatment in protocols Total 13A versus Total 13B, 35 of 36 (97%) patients with TEL-AML1–positive ALL enrolled in the Total 13A, compared with only 10 of 34 (29%) patients in Total 13B, were treated according to the high-risk arm of the respective protocols, suggesting that the prognostic impact of OPAL1 expression may be treatment dependent. When the outcome analyses were limited to the St Jude high-risk protocols, OPAL1 expression was not significantly related to DFS (P > .05). In contrast, among patients treated on the Total 13B low-risk protocol (n = 70), OPAL1 expression was significantly related to DFS in the univariate analysis (P = .021, hazard ratio [HR] = 0.54), and when other prognostic factors were considered, (ie, age, WBC count, hyperdiploid karyotype, and TEL-AML1 gene fusion; P = .017, HR = 0.28). No association of OPAL1 expression and DFS was detected within patients treated according to the COALL high-risk and low-risk protocols. It should be noted that TEL-AML1 status was not a criteria for treatment stratification in COALL protocols.
Most importantly, when known risk factors (ie, age, WBC count, immunophenotype, and genetic subtypes) were included in a multiple regression model, OPAL1 expression was no longer predictive of prognosis in ALL in the COALL or in the St Jude study groups (P > .25, continuous variable; Table 3; P > .10, categorical variable; data not shown).
Table 3.
Multivariate analysis of the prognostic value of OPAL1 expression in pediatric ALL
|
COALL
|
St Jude
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Variable | N | HR | 95% CI | P | N | HR | 95% CI | P |
| Age | ||||||||
| No older than 10 y | 131 | 1.0* | NA | 181 | 1.0* | NA | ||
| Older than 10 y | 49 | 1.24 | 0.64-2.41 | .52 | 76 | 1.37 | 0.67-2.79 | .39 |
| WBC count | ||||||||
| Less than 10 × 109/L | 43 | 1.0* | NA | 90 | 1.0* | NA | ||
| 10 × 109-49 × 109/L | 67 | 0.54 | 0.19-1.49 | .23 | 80 | 0.99 | 0.42-2.34 | .99 |
| 50 × 109-100 × 109/L | 28 | 1.03 | 0.35-3.02 | .95 | 40 | 0.78 | 0.24-2.46 | .68 |
| More than 100 × 109/L | 42 | 1.59 | 0.58-4.32 | .36 | 47 | 1.32 | 0.52-3.34 | .56 |
| ALL subtype | ||||||||
| B-other | 53 | 1.0* | NA | 82 | 1.0* | NA | ||
| BCR-ABL | 4 | 2.32 | 0.51-10.6 | .28 | 9 | 9.09 | 2.48-33.3 | < .001 |
| Hyperdiploid† | 40 | 0.15 | 0.03-0.65 | .01 | 50 | 0.84 | 0.28-2.45 | .74 |
| MLL rearranged | 4 | 9.51 | 2.47-36.5 | .001 | 8 | 1.97 | 0.37-10.3 | .42 |
| TEL-AML1 | 44 | 0.18 | 0.05-0.68 | .01 | 70 | 0.93 | 0.18-4.73 | .93 |
| T-lineage | 35 | 0.79 | 0.33-1.93 | .6 | 38 | 2.73 | 1.11-6.71 | .029 |
| OPAL1 expression | 180 | 1.03 | 0.77-1.34 | .9 | 257 | 0.71 | 0.4-1.28 | .25 |
Cox multivariate proportional hazards analysis computed with known prognostic factors (ie, age, WBC count, immunophenotype, and genetic subtype). OPAL1 expression was treated as a continuous variable.
95% CI indicates 95% confidence interval; NA, not applicable.
Reference group.
Cytogenetic analysis revealed more than 50 chromosomes.
Discussion
Recently, we identified expression signatures associated with cellular drug resistance and outcome in ALL.10 In our prior study, OPAL1 was not among the top 124 most discriminating genes for cellular drug resistance that were also associated with treatment outcome. This, per se, does not exclude a predictive role for OPAL1 in childhood ALL, as this gene may be significant at a lower level than the cut-off P values used for the construction of our resistance signature models, or may be directly related to treatment outcome, as the selection of genes in our earlier study was focused on in vitro drug resistance profiles and not directly on outcome. Therefore, we analyzed the expression patterns of OPAL1 in leukemic cells at initial diagnosis of ALL of 2 independent groups of 180 COALL and 257 St Jude patients, and assessed the relation between the expression of this gene to age, WBC count, sex, immunophenotype, genetic subtype, in vitro drug resistance, and clinical outcome. The COALL and St Jude protocols represent 2 independent protocols that use similar chemotherapeutic agents.
We observed a 2.8-fold higher OPAL1 expression in children with TEL-AML1–positive B-lineage ALL in both patient cohorts. This is consistent with the higher OPAL1 expression levels observed in TEL-AML1–positive cases as initially reported by our group8 and by Mosquera-Caro et al.12 OPAL1 expression was elevated in hyperdiploid B-lineage ALL samples in the St Jude, but not the COALL cohort. This is in disagreement with the previous POG report, which describes a consistently higher expression of OPAL1 in hyperdiploid ALL.12 In the St Jude patient group, higher levels of OPAL1 expression were found in hyperdiploid ALL, and lower levels of OPAL1 expression were found in patients older than 10 years. With the exception of TEL-AML1–positive ALL, OPAL1 expression was not associated with any other prognostic factor in COALL patients. However, interpretation of associations in some subgroups may be difficult due to limited sample size. Taken together, with the exception of TEL-AML1–positive ALL these data suggest that, in contrast to the observation made by Mosquera-Caro et al,12 high OPAL1 expression was not consistently related to ALL subgroups with a favorable prognosis in these 2 cohorts.
In vitro sensitivity to several drugs is related to favorable outcome.3,20,29,30 Based on the previously observed relation between high OPAL1 expression and favorable prognosis,12,13 we tested the relation between high OPAL1 expression and in vitro drug sensitivity. However, in the present study we observed no relation between a high OPAL1 expression and sensitivity to PRED, ASP, and DNR, and only a weak positive correlation with VCR resistance, which is in the opposite direction of what would be expected if high OPAL1 expression is related to a good prognosis. This indicates that OPAL1 may not be a major determinant of cellular drug sensitivity.
We found no evidence of an association between low OPAL1 expression and worse outcome in T-lineage and TEL-AML1–negative B-lineage ALL patients treated according COALL and St Jude protocols. In fact, in our univariate analysis of patients with T-lineage ALL, a worse outcome was consistently associated with high OPAL1 expression. This contradicts the results reported by Mosquera-Caro et al.12 The significant association between high OPAL1 expression and favorable DFS in TEL-AML1–positive ALL patients treated at St Jude may depend on the risk-group stratification applied to these patients. OPAL1 expression was only significant within TEL-AML1–positive ALL patients treated on the Total 13B protocol, where most patients (24 of 34) were stratified in the low-risk treatment arm. In contrast, OPAL1 expression had no predictive value in Total 13A–treated patients, where most (35 of 36) TEL-AML1–positive patients were treated according to a high-risk protocol.
These data indicate that OPAL1 expression may be prognostic in patients with TEL-AML1–positive ALL treated with reduced-intensity chemotherapy. The same remission induction and reinduction treatment was used in all St Jude protocols, but the main difference was the reduced number of antileukemic agents (4 versus 7 [for protocol 13B; 8 for protocol 13A]) used in the continuation phase of the low-risk protocol (ie, mercaptopurine, methotrexate, PRED, VCR) compared with the high-risk protocol (ie, mercaptopurine, methotrexate, dexamethasone [or PRED], VCR, etoposide, cyclophosphamide, cytarabine [and for protocol 13A, ASP]).
No association of DFS and OPAL1 expression in patients treated on either low- or high-risk protocols was found for the COALL group. Overall, both COALL protocols use the same medications for the treatment of low- and high-risk patients, but treatment for low-risk patients was reduced by 1 or 2 doses per drug. Furthermore, in comparison with the 13B low-risk regimen, all COALL patients had an additional intensification phase (6 to 7 antileukemic agents for 4 to 8 weeks). This again points to a relation of OPAL1 expression and DFS only if patients are treated with lower-intensity chemotherapy, (ie, fewer antileukemic agents), such as the low-risk arm of St Jude protocol 13B. Importantly, in both COALL and St Jude cohorts, the relationship of OPAL1 expression with DFS was not independent of known risk factors (ie, age, WBC count, and ALL subtype) in a multivariate analysis. In addition, increased OPAL1 expression was not independently associated with in vitro drug sensitivity of COALL-treated children with ALL (MTT data were not available for St Jude patients).
In conclusion, previously reported prognostic properties of OPAL1 expression in childhood ALL may be related to the relatively modest intensity of treatment given to the patients in whom this observation was previously made and may not be significant when other known risk factors are included or when more intensive chemotherapy is given. The present data thus indicate that OPAL1 expression is not universally predictive of treatment outcome in childhood ALL, particularly in the context of contemporary treatment protocols.
Acknowledgments
We thank Drs Cheng Cheng, Wenjian Yang, and Renée X. de Menezes for helpful discussion, and Susan C. J. M. Peters for her technical assistance. We are indebted to all clinical staff at St Jude and the COALL centers of all who cared for these patients and to the patients and parents for their participation in these studies.
Appendix
Members of the COALL Study Group: Prof Dr J. Otte, Dr N. Jorch, Krankenanstalten Gilead Kinderklinik, Bielefeld; Dr H. J. Spaar, Dr T. Lieber, Zentralkrankenhaus St-Jürgen-Strasse, Prof Hess Kinderklinik, Bremen; Prof Dr U. Göbel, Dr G. Janßen, Universitätsklinikum Klinik für Kinder-Onkologie, -Hämatologie, und -Immunologie, Düsseldorf; Prof Dr J. F. Beck, Dr S. Weigel, Universitätsklinikum Zentrum für Kinder- und Jugendmedizin Abteilung Pädiatrische Onkologie und Hämatologie, Greifswald; Dr Streitberger, Kreiskrankenhaus Kinderabteilung, Heide; Dr W. Nürnberger, Klinik für KMT und Hämatologie/Onkologie, Pädiatrische Hämatologie/Onkologie, Idar-Oberstein; Dr C. von Klinggräff, Dr K. Westerbeck, Städtisches Krankenhaus Klinik für Kinder- und Jugendmedizin, Kiel; Dr P. Thomas, Dr S. Völpel, Städtisches Krankenhaus Zentrum für Kinder- und Jugendmedizin, Krefeld; Prof Dr D. Körholz, Dr U. Bierbach, Universitätsklinik für Kinder und Jugendliche Abteilung für Hämatologie/Onkologie, Leipzig; Dr P. Gutjahr, Universitätsklinikum Kinderklinik, Mainz; Dr W. Müller, Dr I. Althaus, Krankenhaus Neuwerk Klinik für Kinder- und Jugendmedizin, Mönchengladbach; Prof Dr R. Roos, Dr P. Klose, Städtisches Krankenhaus Harlaching Abteilung für Kinder- und Jugendmedizin, Munich; Dr U. Graubner, Dr I. Schmidt, Klinikum der Universität Dr von Haunersches Kinderspital, Munich; Dr H. Müller, Dr R. Kolb, Klinikum Oldenburg Zentrum für Kinder- und Jugendmedizin, Oldenburg; Dr J. Wolff, Dr O. Peters, Klinik St Hedwig Klinik für Kinder- und Jugendmedizin, Regensburg; Dr J. Weber, Dr-Horst-Schmidt-Kliniken Klinik für Kinder- und Jugendmedizin, Wiesbaden; and Dr Dohrn, Helios Klinikum Zentrum für Kinder- und Jugendmedizin Kinderonkologie, Wuppertal, Germany.
Prepublished online as Blood First Edition Paper, May 18, 2006; DOI 10.1182/blood-2006-04-015990.
A complete list of the members of the COALL study group appears in the “Appendix.”
Supported in part by grants from the National Institutes of Health (C.-H.P., W.E.E.), the American Cancer Society (C.-H.P.), the American Lebanese Syrian Associated Charities (C.-H.P., W.E.E.), USA; The Elterninitiative Kinderkrebsklinik, Düsseldorf, Germany; the Pediatric Oncology Foundation Rotterdam (A.H., M.L.d.B., R.P.), the Nijbakker-Morra Foundation (M.L.d.B.) and the René Vogels stipendium 2002 (A.H.), The Netherlands.
M.L.d.B., J.R.D., G.E.J.-S., U.B.G., U.G., C.-H.P., W.E.E., and R.P. participated in designing and performing the study; K.M.K. participated in performing the study; A.H., M.H.C., and D.P. controlled and analyzed data; A.H. and M.H.C. wrote the paper; and all authors checked the final version of the manuscript.
A.H. and M.H.C. contributed equally to this article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Pui CH, Campana D, Evans WE. Childhood acute lymphoblastic leukaemia: current status and future perspectives. Lancet Oncol. 2001;2: 597-607. [DOI] [PubMed] [Google Scholar]
- 2.Schrappe M, Reiter A, Riehm H. Cytoreduction and prognosis in childhood acute lymphoblastic leukemia. J Clin Oncol. 1996;14: 2403-2406. [DOI] [PubMed] [Google Scholar]
- 3.Den Boer ML, Harms DO, Pieters R, et al. Patient stratification based on prednisolone-vincristine-asparaginase resistance profiles in children with acute lymphoblastic leukemia. J Clin Oncol. 2003;21: 3262-3268. [DOI] [PubMed] [Google Scholar]
- 4.Campana D. Determination of minimal residual disease in leukaemia patients. Br J Haematol. 2003;121: 823-838. [DOI] [PubMed] [Google Scholar]
- 5.Szczepanski T, Orfao A, van der Velden VH, San Miguel JF, van Dongen JJ. Minimal residual disease in leukaemia patients. Lancet Oncol. 2001;2: 409-417. [DOI] [PubMed] [Google Scholar]
- 6.Ferrando AA, Armstrong SA, Neuberg DS, et al. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102: 262-268. [DOI] [PubMed] [Google Scholar]
- 7.Ross ME, Zhou X, Song G, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102: 2951-2959. [DOI] [PubMed] [Google Scholar]
- 8.Yeoh EJ, Ross ME, Shurtleff SA, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1: 133-143. [DOI] [PubMed] [Google Scholar]
- 9.Cheok MH, Yang W, Pui CH, et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003;34: 85-90. [Erratum, Nat Genet. 2003;2034:2231.] [DOI] [PubMed] [Google Scholar]
- 10.Holleman A, Cheok MH, den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351: 533-542. [DOI] [PubMed] [Google Scholar]
- 11.Carroll WL, Bhojwani D, Min DJ, et al. Pediatric acute lymphoblastic leukemia. Hematology (Am Soc Hematol Educ Program). 2003: 102-131. [DOI] [PubMed]
- 12.Mosquera-Caro M, Helman P, Veroff R, et al. Identification, validation and cloning of a novel gene (OPAL1) and associated genes highly predictive of outcome in pediatric acute lymphoblastic leukemia using gene expression profiling [abstract]. Blood. 2003;102: 4a. [Google Scholar]
- 13.Willman CL. Discovery of novel molecular classification schemes and genes predictive of outcome in leukemia. Hematol J. 2004;5: S138-S143. [DOI] [PubMed] [Google Scholar]
- 14.Hammond MP, Birney E. Genome information resources: developments at Ensembl. Trends Genet. 2004;20: 268-272. [DOI] [PubMed] [Google Scholar]
- 15.Harms DO, Janka-Schaub GE. Co-operative study group for childhood acute lymphoblastic leukemia (COALL): long-term follow-up of trials 82, 85, 89 and 92. Leukemia. 2000;14: 2234-2239. [DOI] [PubMed] [Google Scholar]
- 16.München, Tumorzentrum. Besonderheiten der akuten Leukämien im Kindesalter. http://www.krebsinfo.de/ki/empfehlung/leu/802_5_Besonder.pdf. Accessed February 15, 2006.
- 17.Pui CH, Sandlund JT, Pei D, et al. Improved outcome for children with acute lymphoblastic leukemia: results of Total Therapy Study XIIIB at St Jude Children's Research Hospital. Blood. 2004;104: 2690-2696. [DOI] [PubMed] [Google Scholar]
- 18.Relling MV, Boyett JM, Blanco JG, et al. Granulocyte colony-stimulating factor and the risk of secondary myeloid malignancy after etoposide treatment. Blood. 2003;101: 3862-3867. [DOI] [PubMed] [Google Scholar]
- 19.Kaspers GJL, Veerman AJP, Pieters R, et al. Mononuclear cells contaminating acute lymphoblastic leukaemic samples tested for cellular drug resistance using the methyl-thiazol-tetrazolium assay. Br J Cancer. 1994;70: 1047-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pieters R, Huismans DR, Loonen AH, et al. Relation of cellular drug resistance to long-term clinical outcome in childhood acute lymphoblastic leukaemia. Lancet. 1991;338: 399-403. [DOI] [PubMed] [Google Scholar]
- 21.Meijerink JPP, Mandigers C, van de Locht L, et al. A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J Mol Diagn. 2001;3: 55-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stam RW, den Boer ML, Meijerink JPP, et al. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood. 2003;101: 1270-1276. [DOI] [PubMed] [Google Scholar]
- 23.Stams WAG, Den Boer ML, Beverloo HB, et al. Sensitivity to L-asparaginase is not associated with expression levels of asparagine synthetase in t(12;21)+ pediatric ALL. Blood. 2003;101: 2743-2747. [DOI] [PubMed] [Google Scholar]
- 24.Holleman A, den Boer ML, de Menezes RX, et al. The expression of 70 apoptosis genes in relation to lineage, genetic subtype, cellular drug resistance, and outcome in childhood acute lymphoblastic leukemia. Blood. 2006;107: 769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. New York, NY: Wiley; 1980: 163-188.
- 26.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16: 1141-1154. [Google Scholar]
- 27.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94: 496-509. [Google Scholar]
- 28.Satagopan JM, Ben-Porat L, Berwick M, et al. A note on competing risks in survival data analysis. Br J Cancer. 2004;91: 1229-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hongo T, Yamada M, Yajima S, et al. Biological characteristics and prognostic value of in vitro three-drug resistance to prednisolone, L-asparaginase and vincristine in childhood acute lymphoblastic leukemia. Int J Hematol. 1999;70: 268-277. [PubMed] [Google Scholar]
- 30.Kaspers GJL, Pieters R, Van Zantwijk CH, et al. Prednisolone resistance in childhood acute lymphoblastic leukemia: vitro-vivo correlations and cross-resistance to other drugs. Blood. 1998;92: 259-266. [PubMed] [Google Scholar]