Abstract
Developmental expression at the β-globin locus is regulated in part by the locus control region, a region upstream of the genes containing at least five major DNase I hypersensitive sites (HSs) in mammalian erythrocytes. Sequences farther 5′ of these HSs are conserved in mouse and human, and both loci are embedded within a cluster of functional odorant receptor genes. In humans, distant upstream sequences have been implicated in regulation of the β-globin genes. In this study, the role of the 5′-most HSs and their adjacent sequence was investigated by deletion of an 11-kb region from the mouse locus, including 5′HS 4.2, 5′HS 5, 5′HS 6, and the 5′β1 odorant receptor gene. Mice that were homozygous for this deletion were fully viable, and no significant effect on adult β-globin gene expression was seen. 5′HSs 1–4, which are located downstream of the deletion, were still present in the mutant mice. In addition, two new upstream HSs, HS −60.7 and HS −62.5, were found in erythroid tissue of both wild-type and mutant mice. Therefore, although the possibility of a minor role still exists, neither the HSs nor the other regions deleted in this study are essential for β-globin gene expression, and it is unlikely that chromatin structure is affected either upstream or downstream of the deletion. This is the largest deletion at the mouse locus control region to show no apparent phenotype, and focuses attention on the possible contribution of sequences even farther upstream.
The genes encoding the β subunits found in hemoglobin are present in a cluster in the genome and are organized in the order they are expressed during development in mammals (1–3). Developmental expression at the β-globin locus is regulated in part by the locus control region (LCR) in erythroid cells, a region 5′ of the genes containing at least four major erythroid-specific DNase I hypersensitive sites (HSs) in mammals (5′HSs 1–4), and three sites in chicken (5′HSs 1–3) (3–8). At least one additional HS is present at the 5′ end of the β-globin domain: 5′HS 4 in chicken (7, 8), 5′HSs 5–7 in human (9), and 5′HS 4.2, 5′ HS 5, and 5′HS 6 in mouse (10). In chicken, 5′HS 4 defines the 5′ boundary of the locus in erythrocytes, separating the generally DNase I-sensitive and hyperacetylated chromatin of the β-globin domain from the generally DNase I-insensitive and underacetylated chromatin farther upstream (11). In addition, 5′HS 4 in chicken has been shown to function as an insulator that can block enhancer activity when placed between an enhancer and a reporter gene in transfected erythroid cells (12, 13). This insulator can also protect against position effects in cell culture (14), transgenic fruit flies (12), transgenic mice (15, 16), and transgenic rabbits (17). It is known that in vertebrates, a binding site for the zinc finger protein CTCF is responsible for the enhancer-blocking activity (18). Enhancer-blocking activity has also been observed with 5′HS 5 of human (12, 19). However, whereas this enhancer-blocking activity has been observed in transfected cells, there is evidence both for (20) and against (21) this when tested in the context of the LCR in transgenic mice. In addition, there is no evidence of an abrupt change in chromatin structure in the region immediately upstream of human 5′HS 5, nor at the equivalent region of the mouse locus, because additional HSs are present farther upstream (see above; ref. 9). In the case of the human locus, it is also known that a transcribed retroviral element exists upstream of 5′HS 5 (22), and a region 34-kb upstream of 5′HS 5 has acetylation levels similar to those in other nontranscribed regions of the β-globin domain (23). On the other hand, we have found that conserved CTCF sequences exist at 5′HS 5 of both human and mouse, giving rise to the possibility that the enhancer-blocking function may still be conserved in these species (C.F. and G.F., unpublished observations). If indeed boundary elements do exist at the human and mouse loci, it raises the possibility that a 5′ boundary could serve to protect the domain from upstream encroaching chromatin effects in erythrocytes and/or in other cell types. Alternatively, a boundary could serve to protect neighboring upstream domains from the strong enhancing properties of the LCR in erythroid cells.
Analyses of the sequences farther upstream of the mouse HSs have revealed the presence of at least five odorant receptor (OR) genes (9), and at least two of these (MOR5′β1 and MOR5′β2) are expressed in odorant tissue. A minimum of five related OR genes are also present 3′ of the β-globin genes in mouse. Three OR genes (again of the same subfamily) have also been found upstream of the human β-globin LCR, and two are known to exist 3′ of the human locus from analysis of the breakpoints of the naturally occurring deletions HPFH-1 and HPFH-6 (9, 24). Thus, both the human and mouse loci are embedded within a cluster of functional olfactory receptor genes. In addition to the presence of the OR genes, a region of general sequence homology exists between mouse and human and extends for at least 20 kb farther upstream of the HSs (9). At least two OR genes are also present at the 3′ side of the chicken β-globin locus (9, 25). However, unlike the mouse and human locus, the chicken locus has a preerythroid-specific folate receptor gene as its 5′ neighbor (26), and this is separated from the β-globin domain by a 16-kb condensed chromatin region. Thus, the chicken 5′ insulator may have a function different from that of any analogous region of the mouse and human loci.
To examine the role of the upstream hypersensitive sites of the mouse β-globin locus, an 11-kb deletion was carried out by gene targeting in this study. The deletion included 5′HS 4.2, 5′HS 5, 5′HS 6, and the 5′β1 OR gene. Mice that were heterozygous and homozygous for the mutation were generated, with homozygous mice being fully viable and having no apparent abnormalities. Both heterozygous and homozygous mice had at or near wild-type (wt) levels of transcription of the adult β-globin genes, but mice with a marker gene replacing the deleted region had reduced levels of expression. In addition, hypersensitive site formation at the β-globin locus was normal in the mutant mice, and the mutation did not affect the appearance of two new farther upstream HSs, which were found at −60.7 and −62.5 kb.
Materials and Methods
Constructs and Clones.
All clones used in this study were derived from either a P1 clone from a 129Sv mouse library (Genome Systems, St. Louis) or from two overlapping lambda clones encompassing the upstream region of the mouse β-globin locus (diffuse haplotype). The sequences to −59.6 kb relative to the ɛy gene transcription start site have been reported previously (GenBank accession no. AF071080). Various smaller inserts from these sequences were subsequently subcloned into the vector pBluescript SK II (Stratagene). The targeting construct, pMnΔIns-2, was based on the vector pPNT (27), except that the PGK-neo gene cassette was flanked by loxP sites (28). The 5′ flank, which contains an EcoRV to SacI fragment from positions −37,968 to −34,890 relative to the ɛy gene start site, was inserted upstream of the loxP-flanked marker gene. The 3′ flank, which contains a PvuII to SacI fragment from positions −23,934 to −21,882, was inserted downstream of the marker gene.
Gene Targeting and Generation of Mutant Mice.
Gene targeting in 129 R1 embryonic stem (ES) cells (29) was carried out according to standard procedures (30); 192 colonies were picked after positive/negative selection, and DNA was isolated (31) and screened by Southern blotting. Chimeric mice were subsequently generated with a positive clone (30), and subsequent matings were carried out as described in Results and Discussion to generate mice that were heterozygous and homozygous for the mutation. DNA was isolated from a tail sample of each mouse (30), and Southern blotting was performed to determine the genotype.
Reverse Transcription–PCR (RT-PCR) Analysis.
RNA was isolated from the circulating blood of adult mice by using TRIzol LS Reagent (Life Technologies, Grand Island, NY). The primers used for amplification of the α-globin products are those described by Weiss et al. (32). The primers βm-4 and βm-2R were used for amplification of the β-globin genes (33), and the RT-PCR reactions were carried out and quantified as described by Fiering et al. (33).
DNase I Hypersensitivity Mapping.
In the case of DNase I hypersensitivity mapping of mouse spleen and liver, mice were treated with N-acetylphenylhydrazine (4 mg/ml in water; Sigma A4626) to induce erythropoiesis of the spleen; 0.04 mg/g body weight was injected intraperitoneally on days 1, 2, and 3, followed by sacrifice on day 5. Spleens increased in weight from ≈90 ± 20 mg pretreatment to 300 ± 50 mg posttreatment. In the case of mutant mice that retained the marker gene, spleen weight increased to 525 ± 25 mg. Nuclei were isolated from dissected livers and spleens (34), and DNase I hypersensitivity mapping was carried out as described by Reitman et al. (34).
In the case of DNase I hypersensitivity mapping of cell lines, nuclei were isolated as follows. Between 5 × 107 to 1 × 108 cells were harvested and washed once in ice-cold PBS, followed by one wash in ice-cold 10 mM Tris⋅HCl (pH 7.5)/10 mM NaCl/3 mM MgCl2 plus 0.1 mM PMSF. Cells were then resuspended in 5 ml of 10 mM Tris⋅HCl (pH 7.5)/10 mM NaCl/3 mM MgCl2 plus PMSF, and 10% NP-40 solution was added dropwise while slowly vortexing to give a final concentration of 0.5% NP-40. Cells were incubated on ice for 10 min, gently vortexed again, then nuclei were spun for 10 min at 600 × g. Nuclei were washed once more with 10 mM Tris⋅HCl (pH 7.5)/10 mM NaCl/3 mM MgCl2 plus PMSF, then resuspended in 5 ml of DNase I buffer [15 mM Tris⋅HCl (pH 7.5)/60 mM KCl/15 mM NaCl/3 mM CaCl2/5 mM MgCl2/250 mM sucrose]. The nuclei were then aliquoted in 10 tubes at 500 μl each, and the following amounts of DNase I (Worthington) were added in a 40-μl volume: 0, 0.2, 0.43, 0.85, 1.7, 3.4, 6.8, 13.6, 27.2, and 54.4 Units. Digestion was for 1 min at room temperature, and reactions were stopped by addition of 50 μl of 5% SDS/100 mM EDTA (pH 8.0); 12.5 μl of 20 mg/ml proteinase K was added and nuclei were digested at 37°C overnight. DNA was then purified by organic extractions and alcohol precipitation. Hypersensitive sites were subsequently mapped by indirect end labeling. The probes used for the hypersensitivity assays are described in the figure legends.
Results and Discussion
Deletion of an 11-kb upstream region of the mouse β-globin locus was carried out by gene targeting in ES cells (Fig. 1). The deleted region included 5′HS 4.2, 5′HS 5, 5′HS 6, and the coding region of the 5′β1 OR gene, whereas the remaining regions of the LCR were still present in the mutant allele (Fig. 1A). The neomycin marker gene of the targeting construct was flanked by loxP sites to allow for its subsequent excision in the presence of Cre recombinase, because it has previously been demonstrated that marker genes can affect the phenotype of a knockout at a regulatory region (10, 33, 35, 36). The strategy for screening ES cells that were transfected with the targeting construct is shown in Fig. 1B, and the Southern blot shows a single clone that was positive for the mutant allele.
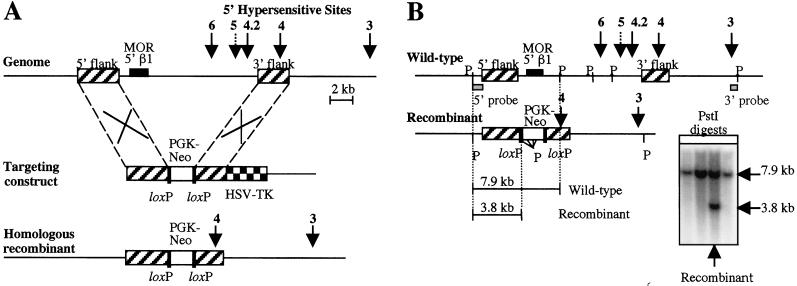
Figure 1.
Knockout strategy and screening of ES cells. (A) The upper part of the diagram shows the upstream region of the mouse β-globin locus. The positions of the 5′HSs are indicated by arrows. The dotted arrow indicates that 5′HS 5 is a relatively weak site (10). The boxes with diagonal stripes indicate the positions of the 5′ and 3′ flank sequences used in the targeting construct (center of diagram), where in vivo homologous recombination occurs with the corresponding regions of the genome. The targeting construct also contains a loxP-flanked PGK-neo marker gene for positive selection (white box), and an HSV-TK gene for negative selection (checkered box). The bottom of the diagram shows the expected recombinant allele after the correct targeting event, where the 10,956-bp region between the two flanks is deleted. (B) The diagram shows the screening strategy used to select for correctly targeted clones. The locations of PstI sites are indicated (P) for the wt and recombinant alleles, and the expected fragment sizes that are obtained after PstI digestion and 5′ probe screening are shown. The 5′ probe is a 670-bp PstI to EcoRV fragment indicated on the diagram (gray box). The Southern blot shows a single positive clone that was obtained. This clone was confirmed as positive by additional screening with a 3′ probe (a 605-bp SacI to PstI fragment shown in the diagram).
ES cells from the positive clone were subsequently injected into blastocysts to generate chimeric mice. These blastocysts were derived from the mouse strain C57BL/6, which contains the single haplotype of the β-globin locus, whereas the ES cells contain the diffuse haplotype of the β-globin locus (33). Thus, restriction enzyme polymorphisms between these two haplotypes are present throughout the β-globin locus and can be used to distinguish between the wt and mutant alleles (Fig. 2 and below). Chimeric males were subsequently bred with wt C57BL/6 females to generate mice that were heterozygous for the mutation (Fig. 2A), and heterozygous mice were then bred with each other to generate mice that were homozygous for the mutation (Fig. 2B). These mutant mice were fully viable, could breed normally, and had no obvious abnormalities, indicating a nondeleterious phenotype for the mutation. Because the mutant mice still contained the neomycin marker gene (Fig. 2A), these mice were further bred with a transgenic strain containing a Cre recombinase transgene that is expressed at the zygotic stage (28). Mutant mice with the marker gene excised were obtained (Fig. 2C) and then bred to homozygosity. Again, these mice showed no apparent defects.
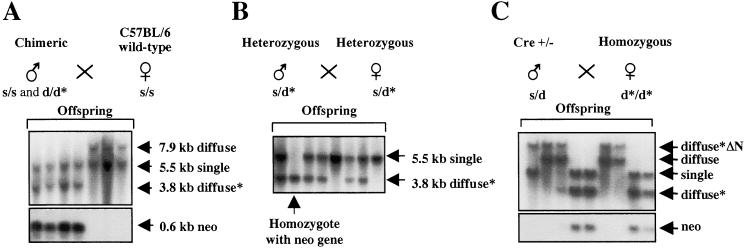
Figure 2.
Generation and screening of mice for the mutation. (A) Chimeric males, which were derived from C57BL/6 blastocysts and targeted ES cells (diffuse haplotype), were mated with wt C57BL/6 females (single haplotype indicated by s). The wt diffuse allele is indicated by d and the mutant allele by d*. Southern blotting of the resulting offspring is shown by using the same screening strategy as in Fig. 1B, and the fragment sizes of the different alleles are indicated. The wt single allele has a different fragment size from the wt diffuse allele because of a restriction site polymorphism. Heterozygous mice (s/d* genotype) also contain a 0.6-kb neomycin PstI fragment (lower blot). (B) Mating and screening of heterozygous mice. A mouse that is homozygous for the mutation (d*/d* genotype) is indicated. All offspring with the d* allele contain the neomycin marker gene. (C) Removal of the neomycin marker gene. Homozygous mice were bred with a mouse carrying a Cre recombinase transgene (28). Mice without the marker gene contain a 12-kb PstI fragment, indicated by diffuse*ΔN. These mice are negative for the neomycin fragment (lower blot).
To see whether the mutation affected gene expression at the β-globin locus, expression of the adult genes was quantified in wt and mutant mice (Fig. 3). RNA was extracted from the circulating blood of adult mice, and RT-PCR analysis was carried out by coamplification of the adult α- and β-globin genes. The α-globin mRNA levels (331-bp product) were used for normalization among the different mice samples. The β-globin primers, which amplified a 195-bp product, could amplify both the β-major and β-minor adult genes of the diffuse haplotype, as well as the homologous βS and βT adult genes of the single haplotype (33). Mice with and without the neomycin marker gene were analyzed. The average β-globin expression as a percentage of that found in wt mice after normalization to α-globin expression is shown in Fig. 3A. In the case of mutant mice that retained the marker gene, a significant decrease in β-globin gene expression was seen (up to 50%). This decrease was more severe in homozygotes. However, mutant mice with the marker gene removed showed little or no decrease in adult β-globin gene expression (Fig. 3A), indicating that the marker gene affects the phenotype of the deletion. Allele-specific expression was further examined in heterozygous mice by using a restriction enzyme polymorphism that is present in the RT-PCR product (Fig. 3B). In the case of heterozygotes that retained the marker gene, the mutant diffuse allele, which is digested by the BstXI enzyme, showed 2.5–3-fold lower levels of β-globin expression than the wt single allele. In the case of wt mice with a single/diffuse genotype or heterozygous mice with the marker gene removed, approximately equal levels were seen from both alleles. This confirms that the decrease in expression in mice that retain the marker gene is caused by down-regulation of their mutant allele. In addition, some erythrocytes of mutant mice that retained the marker gene had degenerative changes (hypochromasia, spherocytosis, and target cells) ranging from mild to moderate, especially in the case of homozygotes (data not shown). Examination of histologic sections of bone marrow indicated a mild to moderate increase in erythropoiesis in homozygotes with the marker gene. Mutant mice with the marker gene also showed increased erythropoiesis of the spleen in response to treatment with phenylhydrazine, compared with wt mice and mutant mice without the marker gene (Materials and Methods). Taken together, these results are consistent with previous reports of marker gene interference with other deletions at the β-globin locus, where severe phenotypes were also reported in the presence, but not the absence, of the marker gene (10, 33, 35, 36).
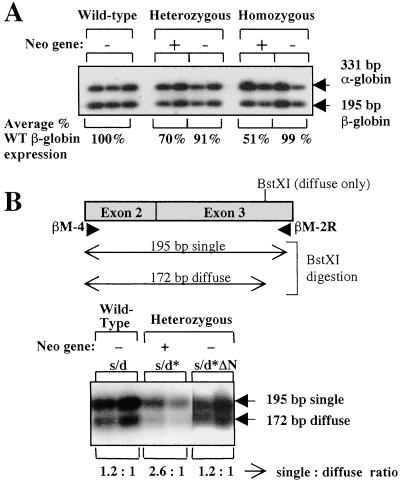
Figure 3.
Expression of adult globin genes. (A) RT-PCR analysis of adult globin genes in circulating blood of adult mice. The gel shows coamplification of adult β-globin mRNA (195-bp product) and α-globin mRNA (331-bp product). The genotypes of the mice and the presence or absence of the neomycin marker gene are indicated at the top of the gel. The average percentage of adult β-globin expression after normalization to α-globin expression is shown at the bottom of the gel. (B) Allelic expression of the adult β-globin genes in heterozygous mice. The diagram shows the expected product after amplification with the primers βM-4 and βM-2R. The position of the BstXI site in the diffuse allele is shown, and the expected fragment sizes after BstXI digestion are also indicated. The expression ratios from the single and diffuse alleles are shown underneath the gel.
The expression of the embryonic ɛy and βh1 genes was not examined in these mice. However, it is assumed that these genes are expressed at or near wt levels in the mutants, with perhaps some reduction in the case of mutant mice that retain the marker gene. It would be difficult to imagine that the mutant mice would survive beyond the embryonic/fetal stage if the expression levels of these genes were significantly reduced, unless there was a substantial compensatory up-regulation of the adult genes, which we do not expect here. We did not detect any abnormal embryonic lethality of the mutant mice, because the litter sizes of mutant and wt mice were similar. In addition, the newborn pups in mutant litters were not smaller than their wt counterparts, regardless of whether or not the marker gene was present.
To see whether the mutation affected formation of the HSs immediately downstream of the deletion, chromatin from the spleens and livers of mice that had been treated with phenylhydrazine was analyzed. The erythroid-specific 5′HS 4, which is within 1.3 kb of the 3′ breakpoint of the deletion, was found to be present in the erythroid spleen of wt and mutant mice (Fig. 4), but not in the nonerythroid liver (not shown). Similarly, 5′HSs 1–3 were detected in spleen chromatin of wt and mutant mice (Fig. 4). These HSs were detected in all mutant mice tested, regardless of whether or not the marker gene was present, indicating that the 11-kb deletion does not affect hypersensitive site formation.
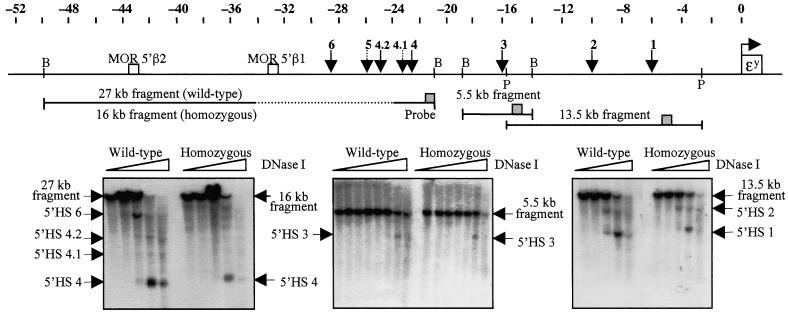
Figure 4.
DNase I hypersensitivity analysis downstream of the deleted region. The diagram shows a map of the upstream region of the mouse β-globin locus. The positions of the ɛy gene, the 5′ HSs, and two 5′ OR genes are indicated. The locations of the fragments used for hypersensitivity assays are shown, and the positions of BamHI (B) and PstI (P) sites are indicated. The dotted portion of the 27-kb BamHI fragment indicates the region that was deleted. The probe positions on the fragments are indicated by gray boxes. (Left) 5′HS 4 was detected by using a 27-kb BamHI parental fragment (16 kb for homozygotes with the deletion), with a 557-bp EcoRI to SacI fragment as a probe. (Center) 5′HS 3 was detected by using a 5.5-kb BamHI fragment, with a 359-bp SspI to BglII fragment as a probe. (Right) 5′HS 1 and 5′HS 2 were detected by using a 13.5-kb PstI fragment, with the previously described probe x PCR fragment as a probe (37). Mice used for the DNase I series shown were either wt (diffuse/diffuse genotype) or homozygous for the 11-kb mutation (diffuse*ΔN/diffuse*ΔN genotype).
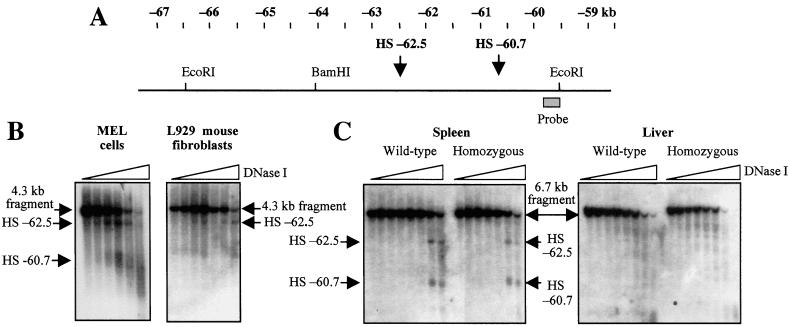
Because the deletion did not have a notable effect on the region to its 3′side, i.e., the β-globin domain, another possibility was that it could affect chromatin in a farther upstream region. We therefore analyzed the upstream chromatin of murine erythroleukemia cells to find the next hypersensitive site. Two new HSs were found at −60.7 and −62.5 kb, relative to the transcription start site of the ɛy gene (Fig. 5 A and B), with the strength of these sites being comparable to that of 5′HS 4 in murine erythroleukemia cells (not shown). To see whether these sites were present in other cell types, L929 mouse fibroblasts were also examined (Fig. 5B). HS −62.5 was present in these cells, but was considerably weaker than in murine erythroleukemia cells, and HS −60.7 was barely if at all detectable in L929 cells, indicating that this site is not ubiquitous. Spleen and liver chromatin from phenylhydrazine-treated mice were also examined for the presence of these upstream sites. Both of these sites were clearly visible in the erythroid spleen of wt and mutant mice (Fig. 5C), indicating that the mutation did not affect the formation of these sites. As was the case for 5′HSs 1–4, these sites were also present in mutant mice that retained the marker gene (not shown). The significance of these new HSs and their possible relationship to the β-globin genes is not currently known. It should be noted that whereas these sites were not present as discrete bands in the nonerythroid liver of these mice, some faint bands were detected in this region (Fig. 5C), indicating that in liver, the chromatin between −60 and −65 kb has some degree of DNase I sensitivity and is not condensed.
Figure 5.
DNase I hypersensitivity analysis upstream of the deleted region. (A) Map of the far upstream region of the mouse β-globin locus. The positions of the two new HSs are indicated. (B) Analysis of hypersensitive sites in murine erythroleukemia cells and L929 cells. The parental band is a 4.3-kb BamHI to EcoRI fragment. The position of the probe, a 285-bp AflIII to EcoRI fragment, is shown in A. (C) Analysis of upstream hypersensitive sites in spleen and liver chromatin of phenylhydrazine-treated mice. The parental band is a 6.7-kb EcoRI to EcoRI fragment, and the probe described in B was used. Mice used for the DNase I series shown were either wt (diffuse/diffuse genotype) or homozygous for the 11-kb mutation (diffuse*ΔN/diffuse*ΔN genotype).
The upstream region of the mouse β-globin locus also includes at least five OR genes, at least two of these functional in odorant tissue (9). The significance of the presence of these genes in relation to the β-globin locus and its LCR is not currently known, and there is thus far no evidence for the expression of these upstream genes in erythrocytes. It should be noted that the first OR gene upstream of the mouse β-globin locus, the 5′β1 gene (or at least its coding region which is located ≈5 kb upstream of 5′HS 6), has also been deleted in this study. However, because there are ≈1,000 OR genes in mouse located in several clusters throughout the genome (38–41), the loss of one such gene is not expected to show a detectable phenotype. This is pertinent given the fact that each OR gene is only expressed in ≈1–2% of olfactory neurons within a given zone, and each neuron expresses only a single OR gene (40, 41). The region between the β-globin HSs and the 5′β1 OR gene is also absent in these mice. If a putative boundary element had been deleted in these mice (either in this region or in one of the HSs), then it is possible that the strong enhancing properties of the LCR could activate the next upstream OR gene, the 5′β2 gene, in erythroid cells (see Fig. 4 for the location of this gene). However, RT-PCR analysis showed that this gene was not expressed in erythrocytes of either wt or mutant mice (data not shown).
In conclusion, none of the HSs or the regions deleted in this study are essential for β-globin gene expression. It is unlikely that this region is responsible for the open chromatin structure of the locus in erythrocytes, although the possibility of a minor role yet to be detected still exists. This result is in agreement with a recent series of knockouts by M. Groudine and colleagues, where deletion of any single HS did not show any major effect at the mouse β-globin locus (33, 35, 36, 42). A 3.5-kb region, including 5′HS 5 and 5′HS 6, was also deleted by this group (10), but again this deletion had only a minimal effect on expression of the β-globin genes. This deletion overlaps with the one described in this paper, but our deletion is larger and removes an additional 7.5 kb, including 5′HS 4.2 and ≈6 kb upstream of 5′HS 6. Both of these deletions together suggest that 5′HSs 5 and 6 and the region surrounding them do not play an important role at the β-globin locus. In addition, if any boundary elements are present in this region, such as the conserved CTCF site at 5′HS 5, they do not play a significant role in the maintenance and regulation of the domain. Although a larger deletion has been reported at the mouse LCR (43, 44), the deletion described in this paper is the largest one to date where no apparent phenotype was observed, and it suggests that the remaining regions of the LCR, 5′HSs 1–4, are sufficient for normal β-globin gene expression. This is consistent with the fact that human 5′HSs 1–4 are sufficient for full LCR activity and correct developmental regulation of β-globin genes in transgenic mice (45). In the larger deletion that was described in mouse, the entire region from 5′HS 1 to 5′HS 6 was removed, but this caused major decreases in β-globin gene expression (43, 44). However, the chromatin within the locus remained in an open conformation in erythrocytes, and a low level of gene expression with correct developmental switching was detected. These results suggest that other elements, either flanking or within the locus, are responsible for maintaining an open chromatin conformation. Possible upstream candidates include the two new HSs presented in Fig. 5, but no function has been attributed to these sites to date. A similar open chromatin conformation was also seen when 5′HS 2–5′HS 5 were deleted from the human β-globin locus, but in this case, no gene expression was detected (23, 46). This result is in contrast to the condensed chromatin conformation found in the naturally occurring Hispanic deletion on which the above deletion was modeled (23, 47), but the Hispanic deletion removes an additional 27 kb upstream of 5′HS 5. This suggests that this upstream region is important for regulation. Taken together, all of these deletions, including the one described in this paper, raise questions about the function and definition of the mammalian β-globin LCR and its boundaries, and its relationship, if any, to the olfactory receptor gene cluster. Future deletions should help shed more light on this.
Acknowledgments
We thank the staff of the Rodent and Rabbit Facility and the Laboratory Sciences Section, Veterinary Resources Program, National Institutes of Health, for animal care and technical assistance; Eric Lee for provision of the EIIa-cre transgenic mouse; and Cecelia Trainor and Michael Bulger for critical reading of the manuscript.
Abbreviations
- LCR
locus control region
- HS
DNase I hypersensitive site
- OR
odorant receptor
- ES
embryonic stem
- RT-PCR
reverse transcription–PCR
- wt
wild type
References
- 1.Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- 2.Martin D I, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 3.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 4.Tuan D, Solomon W, Li Q, London I M. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon A M, Ley T J. Proc Natl Acad Sci USA. 1990;87:7693–7697. doi: 10.1073/pnas.87.19.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiménez G, Gale K B, Enver T. Nucleic Acids Res. 1992;20:5797–5803. doi: 10.1093/nar/20.21.5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reitman M, Felsenfeld G. Mol Cell Biol. 1990;10:2774–2786. doi: 10.1128/mcb.10.6.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans T, Felsenfeld G, Reitman M. Annu Rev Cell Biol. 1990;6:95–124. doi: 10.1146/annurev.cb.06.110190.000523. [DOI] [PubMed] [Google Scholar]
- 9.Bulger M, von Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bender M A, Reik A, Close J, Telling A, Epner E, Fiering S, Hardison R, Groudine M. Blood. 1998;92:4394–4403. [PubMed] [Google Scholar]
- 11.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung J H, Whitely M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 13.Recillas-Targa F, Bell A C, Felsenfeld G. Proc Natl Acad Sci USA. 1999;96:14354–14359. doi: 10.1073/pnas.96.25.14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pikaart M J, Recillas-Targa F, Felsenfeld G. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, DeMayo F J, Tsai S Y, O'Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 16.Potts W, Tucker D, Wood H, Martin C. Biochem Biophys Res Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- 17.Taboit-Dameron F, Malassagne B, Viglietta C, Puissant C, Leroux-Coyau M, Chéreau C, Attal J, Weill B, Houdebine L-M. Transgenic Res. 1999;8:223–235. doi: 10.1023/a:1008919925303. [DOI] [PubMed] [Google Scholar]
- 18.Bell A C, West A G, Felsenfeld G. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Stamatoyannopoulos G. Blood. 1994;84:1399–1401. [PubMed] [Google Scholar]
- 20.Tanimoto K, Liu Q, Bungert J, Engel J D. Nature (London) 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- 21.Zafarana G, Raguz S, Pruzina S, Grosveld F, Meijer D. In: Molecular Biology of Hemoglobin Switching. Stammatoyannopoulos G, editor. Andover: Intercept; 1995. pp. 39–44. [Google Scholar]
- 22.Long Q, Bengra C, Li C, Kutlar F, Tuan D. Genomics. 1998;54:542–555. doi: 10.1006/geno.1998.5608. [DOI] [PubMed] [Google Scholar]
- 23.Schübeler D, Francastel C, Cimbora D M, Reik A, Martin D I K, Groudine M. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 24.Forget B G. Ann N Y Acad Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 25.Staines D M, Thomas J O. Gene. 1999;234:345–352. doi: 10.1016/s0378-1119(99)00186-9. [DOI] [PubMed] [Google Scholar]
- 26.Prioleau M-N, Nony P, Simpson M, Felsenfeld G. EMBO J. 1999;18:4035–4048. doi: 10.1093/emboj/18.14.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tybulewicz V L J, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 28.Lakso M, Pichel J G, Gorman J R, Sauer B, Okamoto Y, Lee E, Alt F W, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan B, Costantini F, Lacey E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1986. [Google Scholar]
- 31.Laird P W, Zijderveld A, Linders K, Rudnicki M A, Jaenisch R, Berns A. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 33.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I K, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 34.Reitman M, Lee E, Westphal H, Felsenfeld G. Mol Cell Biol. 1993;13:3990–3998. doi: 10.1128/mcb.13.7.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hug B, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ley T J, Hug B, Fiering S, Epner E, Bender M A, Groudine M. Ann N Y Acad Sci. 1998;850:45–53. doi: 10.1111/j.1749-6632.1998.tb10461.x. [DOI] [PubMed] [Google Scholar]
- 37.Jiménez G, Griffiths S D, Ford A M, Greaves M F, Enver T. Proc Natl Acad Sci USA. 1992;89:10618–10622. doi: 10.1073/pnas.89.22.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan S L, Adamson M C, Ressler K J, Kozak C A, Buck L B. Proc Natl Acad Sci USA. 1996;93:884–888. doi: 10.1073/pnas.93.2.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chess A. Adv Immunol. 1998;69:437–447. doi: 10.1016/s0065-2776(08)60613-6. [DOI] [PubMed] [Google Scholar]
- 41.Mombaerts P, Wang F, Dulac C, Vassar R, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R. Cold Spring Harbor Symp Quant Biol. 1996;61:135–145. [PubMed] [Google Scholar]
- 42.Bender M A, Mehaffey M G, Telling A, Hug B, Ley T J, Groudine M. Blood. 2000;95:3600–3604. [PubMed] [Google Scholar]
- 43.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I K, Kennedy M, Keller G, et al. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 44.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 45.Collis P, Antoniou M, Grosveld F. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]