Abstract
By sequencing regions flanking the β-globin gene complex in mouse (Hbbc) and human (HBBC), we have shown that the β-globin gene cluster is surrounded by a larger cluster of olfactory receptor genes (ORGs). To facilitate sequence comparisons and to investigate the regulation of ORG expression, we have mapped 5′ sequences of mRNA from olfactory epithelium encoding β-globin-proximal ORGs. We have found that several of these genes contain multiple noncoding exons that can be alternatively spliced. Surprisingly, the only common motifs found in the promoters of these genes are a “TATA” box and a purine-rich motif. Sequence comparisons between human and mouse reveal that most of the conserved regions are confined to the coding regions and transcription units of the genes themselves, but a few blocks of conserved sequence also are found outside of ORG transcription units. The possible influence of β-globin regulatory sequences on ORG expression in olfactory epithelium was tested in mice containing a deletion of the endogenous β-globin locus control region, but no change in expression of the neighboring ORGs was detected. We evaluate the implications of these results for possible mechanisms of regulation of ORG transcription.
Olfactory receptors are a family of seven-transmembrane G protein-coupled receptors expressed in sensory neurons of nasal epithelium, where they bind to odorant epitopes and transduce this primary signal into membrane potential (1–3). These molecules are encoded by a family of up to 1,000 genes in mouse and human, which are grouped into discrete clusters at different chromosomal locations (4–7). Production of a given olfactory receptor is restricted to one of four spatially defined domains within the olfactory epithelium (8, 9). Despite the large size of this gene family, each olfactory neuron appears to express only a single olfactory receptor gene (ORG) (10, 11) in an allele-specific manner (12). The mechanisms that underlie any of these regulatory decisions—expression in olfactory neurons, zonal specificity, one receptor per neuron, or allelic exclusion—are undefined. In addition, expression of some ORGs has been documented in nonolfactory tissues (13–17), but the significance of such expression is also unknown.
The characterization of DNA sequences required for proper gene expression, such as promoters and enhancers, represents a basic approach to such questions. Functional studies of putative ORG regulatory elements, however, are hindered by the lack of a viable cell culture model for olfactory neurons. Although analysis of transgenes in mice provides a suitable model system, the production of mouse lines is time-consuming and expensive. Thus, genomic approaches are particularly relevant to the identification of ORG regulatory sequences, in part to guide the selection of regions to be examined by functional studies in transgenic mice. Candidates for such regions can be recognized as noncoding sequences that are conserved between species (18).
Previously, we reported that ORGs surround the complex of β-like globin genes of humans (HBBC) and mice (Hbbc) (19). (We use the abbreviation HBBC to refer to the complex of genes containing HBE1, HBG2, HBG1, HBD, and HBB at 11p15.5 in humans, and Hbbc for the complex of genes containing Hbb-y, Hbb-bh1, Hbb-b1, and Hbb-b2 on chromosome 7 in mice.) The major enhancer of the β-globin locus, termed the locus control region (LCR), is a 20-kb segment of DNA containing several DNase-hypersensitive sites, located to the 5′ side of the genes (20–22). In contrast to the view that the LCR controls chromatin “opening” of the globin locus (23), deletion of the LCR from the endogenous Hbb locus leaves the globin genes in a DNase-sensitive domain in erythroid cells, albeit with much lower levels of expression (24–26). These results suggest that other DNA sequences may be involved in opening a chromatin domain and that these sequences might be found within the adjacent ORG clusters. The conserved juxtaposition of these two distinct gene clusters also poses the question of whether the β-globin LCR plays a functional role in ORG expression or whether sequence elements exist to segregate erythroid and neuronal-specific regulatory inputs.
We have sequenced additional DNA on both sides of the β-like globin genes in both mouse and human, analyzed the aligned sequences, and studied the expression of the ORGs in olfactory tissue. Our results show significant mouse/human sequence matches within and adjacent to the transcription units of orthologous pairs of mouse and human ORGs. Surprisingly, the ORG promoters that we have mapped exhibit no significant similarities other than the presence of a TATA box and a purine-rich motif. Many of the ORGs contain multiple noncoding exons, some of which are alternatively spliced in vivo. We also show that expression of the globin-proximal ORGs in olfactory epithelium appears normal in mice containing a deletion of the entire β-globin LCR. These results provide a more complete context for studies of ORG regulation in nasal epithelium, the extent of the open chromatin domain containing the globin genes in erythroid cells, and the distribution of sequences regulating these two gene clusters.
Methods
Subclones, Sequencing, and Annotations.
Sequences reported herein have been deposited in the GenBank sequence database. Mouse 5′ and 3′ sequences were obtained from a P1 clone (Genome Systems, St. Louis) and a bacterial artificial chromosome (BAC) clone (a gift from T. Ley, St. Louis). Human 5′ and 3′ sequences were obtained from several BAC clones (Research Genetics, Huntsville, AL), from published clones PAC148O22, BAC233K18, BAC141J14, and BAC44E16 (27), and from GenBank accession no. AC026083. Smaller inserts used for sequencing were obtained by restriction enzyme digestion and subcloning into pBluescript KSII (+) or SKII (+). Sequencing was performed by using Big Dye terminators and universal or custom-synthesized primers on an Applied Biosystems model 377XL automated sequencer; both strands of all regions were sequenced at least once.
Repetitive elements in the DNA sequences were identified with REPEATMASKER (http://ftp.genome.washington.edu/RM/RepeatMasker.html). ORGs and other genes were identified by a combination of BLAST2 searches against the databases of nonredundant gene sequences and expressed sequence tags (http://www.ncbi.nlm.nih.gov/blast/), gene prediction by GENSCAN (http://genes.mit.edu/GENSCAN.html) and analysis by the commercial program GENE CONSTRUCTION KIT 2.
Sequence Alignments.
Pairwise alignments of the human and mouse DNA sequences through the ORG and HBBC regions were computed and viewed by using the PIPMAKER package (http://bio.cse.psu.edu/pipmaker/). The multiple alignment of the ORG promoters was generated in several steps. First, the regions around the 5′ ends of the genes were examined in pairwise comparisons (HOR5′β7 vs. MOR5′β4, HOR5′β6 vs. MOR5′β3, HOR5′β1 vs. MOR5′β1, and HOR3′β2 vs. MOR3′β1; where HOR stands for human olfactory receptor and MOR stands for mouse olfactory receptor); the four pairs of orthologous sequences were truncated to begin 100 bp before the start site and aligned by CLUSTAL W (http://www.ebi.ac.uk/clustalw/). This alignment then was fixed on the A+T-rich region, rearranging the gaps to show the A+T-rich region and the purine-rich region.
5′ Rapid Amplification of cDNA Ends (RACE) and ORG Expression.
The cDNA for 5′ RACE, containing a SMARTII oligonucleotide (CLONTECH) at the 5′ end of the mRNA, was generated from mouse olfactory epithelium poly(A)-selected RNA by using the SMART RACE cDNA amplification kit from CLONTECH. The 5′ ends of the genes were determined by amplifying the 5′ RACE cDNA template, by using one primer from the coding region of each ORG plus the SMARTII oligonucleotide and the Advantage 2 polymerase mix from CLONTECH. These products were sequenced directly or sequenced after purification from agarose gels.
Gene-specific primer pairs were synthesized, with forward primers from the first exon and reverse primers from the coding exon; a nested PCR strategy was used for each gene except MOR3′β1. The primers used were as follows (5′ → 3′): GCCATTCTGGTCTACAGTACAAAC, GGCCAGCAAGGAAAGTAGATAG, ACAAACTGCTCTGACTTCATGGGT, GCAGACGAGGGCTGGTCTTAAT (MOR5′β4); ATCCTTCTCAAAGCTGAATATCTG, CCCTTGATGATGCTACTTGC, ATCTGAAGTTTCTAACAATGTCCC, GGTCCTGCTTTCCAATAACAAT (MOR5′β3); TGAGGGCATATGTAAAATCACA, AAGTAAGCAGTACTCTTCCTACCG, AGGGCATATGTAAAATCACAAAG, GTTCATGTTCTTATGCATCATTTC (MOR5′β1); GAATCTCCTTGCTTTTACTC, CTGCGTTCAGTCACTATCAG (MOR3′β1); ACCACAAAGATCCTATTCATGAGC, AGCCATTGAACTTGATCATGC, CTGTGTACATCTCACTAAATGGC, GCTGCTTCAACTTCTGTTCTATAC (MOR3′β3); TTCATCCTTTATAGAGGGAACAAC, GGAAGTAATACATGGGCTCG, TTCATCCTTTATAGAGGGAACAAC, CCTGTGAGGTAGAATGTAGAGGAC (MOR3′β4); and ATAGATGTGCAGATTATTAACAGG, CGCTCAACTTTGATGACAAC, TGCTGATTTTTCTCAGTCTAGAAG, CCTCAGCCTGTAGACATATGG (MOR3′β6). PCRs were carried out on cDNA made from total RNA from mouse olfactory tissue, generated by using random hexanucleotides for the first-strand synthesis. Products were amplified with the Advantage 2 polymerase mix as follows: For MOR3′β1, a single amplification was performed with a 60°C annealing temperature for 35 cycles. For the remaining genes, an initial round of amplification was performed with the outer primer pairs for 25 cycles at an annealing temperature of 55°C (50°C for MOR5′β4). After a 1:1,000 dilution of these reactions, a second round of amplification was performed with the inner primers at an annealing temperature of 60°C for 25 cycles, in the presence of 5% dimethyl sulfoxide. Trace amounts of [α-32P]dCTP were included in the PCRs. Products were separated on 5% polyacrylamide gels in 0.5× TBE (1× = 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3), and visualized by autoradiography.
In situ hybridization was performed as described (19).
Results
DNA Sequence of HBBC and Surrounding ORGs.
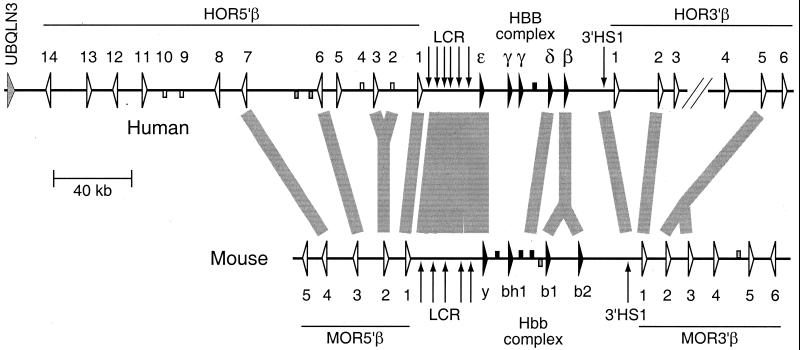
Our initial sequencing of regions flanking the HBBC locus in human and Hbbc in mouse demonstrated that the β-globin genes were embedded in a cluster of ORGs (19). We have extended our sequencing to include additional regions flanking the β-globin genes, which allowed us to generate a contiguous sequence of 224 kb from mouse and two contigs from human of 345 kb and 36 kb, separated by a gap of about 6 kb (Fig. 1). Analysis of the current sequence showed that at least 14 ORGs flank HBBC on the centromeric side (5′ to the β-globin genes) and at least six ORGs on the telomeric side. In mouse, Hbbc is flanked by at least five ORGs on the 5′ side and at least six on the 3′ side. The sequences we have obtained did not allow us to define the full extent of these ORG clusters, and it is likely that additional ORGs exist beyond the limits of our sequencing.
Figure 1.
β-Globin gene cluster and surrounding ORGs. Genes are shown as triangles pointing in the direction of transcription, and pseudogenes are boxes. Globin genes or pseudogenes are solid, ORGs are open, and genes that do not fall into either category are shaded. The left end of the human map is centromeric and the right end is telomeric (27). Erythroid DNase hypersensitive sites are indicated by vertical arrows. Broad shaded lines are drawn between orthologous regions; forks in these lines illustrate the occurrence of gene duplications after the divergence of the last common ancestor.
An exact match to human UBQLN3, encoding a gene expressed in testis (29), was found close to the centromeric end of the human sequence. Based on identity to the cDNA sequence and the fact that this gene has been mapped to 11p15.5, we conclude that this is the UBQLN3 gene, despite the absence of introns within the coding region (a feature shared with the coding regions of ORGs as well). The presence of this gene did not mark the end of the ORG cluster, however, because our analysis of sequence databases indicated the presence of at least two ORGs immediately 5′ of this gene (data not shown).
Definition of 5′ Ends and Noncoding Exons for Mouse ORGs.
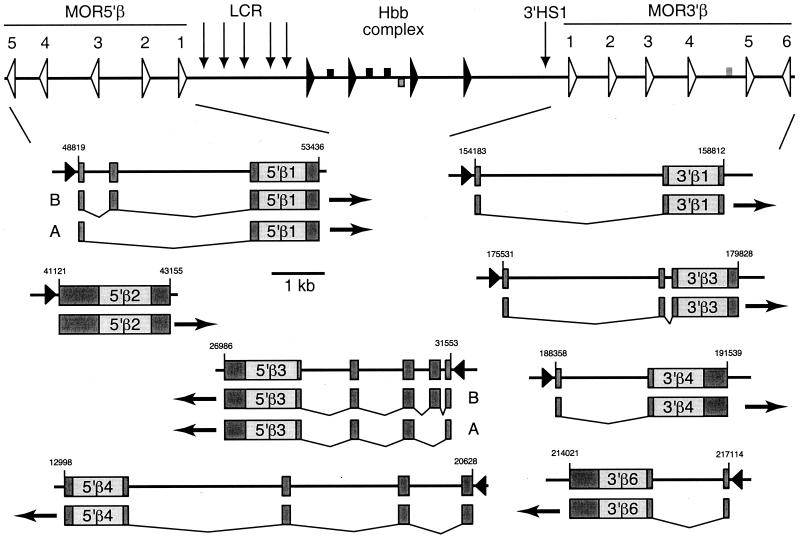
The coding regions of ORGs are not interrupted by introns, complicating PCR-based analysis of ORG expression—because of the small amount of RNA in olfactory epithelium that represents a given ORG, high cycle numbers must be used, which can reveal trace contamination of RNA samples by genomic DNA. To facilitate PCR-based expression studies, and also to aid in the interpretation of mouse/human sequence alignments, we mapped the 5′ ends of globin-proximal ORG mRNAs from mouse olfactory epithelium by using the 5′ RACE technique. The results are summarized in Fig. 2; detailed coordinates are in the GenBank annotations.
Figure 2.
Exon structures and alternative splicing of ORGs. The results of the 5′ RACE analysis are summarized. For each gene, the structure is shown on one line, with darker noncoding regions and lighter coding regions. The presumptive promoter at the 5′ ends is shown as a triangle. The 3′ untranslated region extends to the first AATAAA or ATTAAA on the nontemplate strand; the 3′ ends were not determined experimentally. Under the line with the gene structure, the pattern of spliced exons is shown, with exons connected by bent lines. Experimentally determined alternatively spliced products are labeled A and B; all of the alternatively spliced forms have not been identified.
Three of the mouse ORGs have a structure similar to that observed for other mammalian ORGs (15). A single small exon was found 5′ to the protein-coding exon for MOR3′β1, MOR3′β4, and MOR3′β6. Several of the ORGs located 5′ to the β-like globin genes have a more complex structure. MOR3′β3 had an additional exon separated from the protein-coding exon by a very short intron. The genes MOR5′β3 and MOR5′β4 had at least four and three additional exons, respectively, upstream of the protein-coding exon, and MOR5′β1 had two upstream noncoding exons; noncoding leader portions of the ORG mRNAs ranged as high as 900 bp. We searched the spliced upstream exons for in-frame ATGs or CTGs to identify alternative translation initiation codons but found none.
The 5′ RACE analysis also showed that ORG transcripts can be alternatively spliced. Isolation and sequencing of different RACE products for MOR5′β1 and MOR5′β3 showed differential splicing of the second exon. Several RACE products were obtained for MOR5′β4, but only one was sequenced. The major PCR product for this gene lacked the second exon of the 5′ RACE product (see below). All of the RACE products sequenced for MOR5′β2 were identical to the sequence of the genomic DNA 5′ to the coding region; no introns were detected for this gene.
Sequence Alignments Between Human and Mouse in the ORG-HBBC Region.
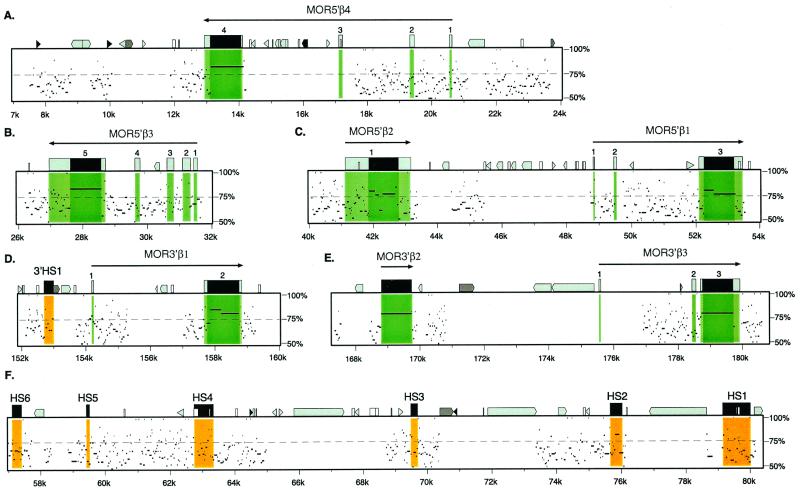
Comparison of the mouse and human ORG-HBBC regions revealed substantial blocks of conserved DNA sequences (data summarizing all of the matching sequences also can be found at the web site http://globin.cse.psu.edu). Aligning segments were found in both protein-coding and noncoding sequences, and these were easily visualized in the percent identity plots shown in Fig. 3, which display the positions and percent identity of aligning segments between human and mouse. The protein-coding regions within these segments aligned at 80–85% identity with no gaps (e.g., MOR5′β4 vs. HOR5′β7 and MOR5′β3 vs. HOR5′β6) or a single break in the alignment (MOR5′β1 vs. HOR5′β1 and MOR3′β1 vs. HOR3′β2). Other matches (broken by gaps to produce the many short, gap-free aligning segments displayed in the pips) extended through the transcription units, including introns and noncoding exons, and into the flanking regions.
Figure 3.
Percent identity plots of human vs. mouse comparisons for selected regions of HBBC and surrounding ORGs. The percent identity plot shows the positions in the mouse sequence and the percent identity of aligning segments from human. Features of the mouse sequence are indicated along the top line of each plot. Genes are shown as solid tall boxes for exons and shaded tall boxes for untranslated regions; exons are numbered and a long arrow shows the extent of the gene and the direction of transcription. Medium-size open boxes, simple repeats and low-complexity regions; solid triangles, MIRs; light shaded triangles, SINEs (such as Alu repeats); light shaded pointed boxes, LINE1 repeats; dark shaded pointed boxes, LINE2 repeats; and dark shaded triangles, other repeats. The percent identity plots are also shaded to emphasize coding and noncoding exons of each of the ORGs and regions that are known to contain DNase hypersensitive sites in erythroid cells. Segments aligning with the two mouse ORGs in E originate from the same human sequences (HOR3′β4), demonstrating that these ORGs in mouse represent a recent evolutionary duplication.
These large aligning fragments also coincided with the closest interspecies relationships between ORG coding regions. For example, the alignment of Fig. 3B demonstrates extensive sequence conservation between the regions containing MOR5′β3 and HOR5′β6. Similarly, the coding regions of these genes were more closely related to each other than they were to the coding regions of any other ORGs (data not shown). On this basis, we are able to assign orthologous relationships between large blocks of mouse and human sequence throughout the HBBC/ORG region (Fig. 1).
Some alignments between human and mouse sequences were found outside of ORG transcription units. These alignments included a region extending 3–6 kb from the 3′ end of the MOR5′β4 gene, another region located 1–3 kb from the transcription start site of MOR5′β4, a region 1.5–3 kb from the 3′ end of MOR5′β2, and an extended series of matches that corresponds to the β-globin LCR (Fig. 3 A, C, and F). In addition, a 1-kb block of sequence located 3′ of the globin genes also demonstrated significant homology with a similarly placed sequence in human (Fig. 3D), which corresponds to a nuclease hypersensitive site in erythroid cells (19, 30). Along with the sequence matches within the introns, noncoding exons, and promoters of the ORGs, all of these regions are candidates for regulatory sequences.
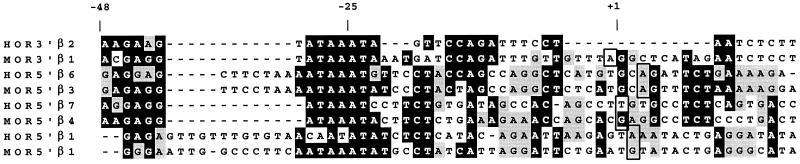
Sequence matches extended 5′ to the first exon for orthologous pairs of ORGs, whereas no matches outside the coding region were observed in other ORG comparisons, which argues against a highly conserved olfactory promoter sequence for the ORGs. However, short conserved motifs could be missed in this analysis, so we also examined an alignment of four orthologous pairs of ORGs. The start sites for transcription of the mouse genes were determined experimentally, and the homologous nucleotides were inferred as start sites for the human genes. Inspection of the alignment revealed an A+T-rich region 25–30 bp upstream from the 5′ end of the genes (Fig. 4), which could potentially function as a TATA box. A purine-rich motif was also apparent at the beginning of the alignment, but the distance of this motif from the TATA motif was not consistent among different ORGs. In addition, we have subjected the mouse ORG promoters we have mapped to analysis with transcription factor databases (results not shown) that predicted binding of TATA binding protein/TFIID to the A+T-rich regions at position −30 but failed to reveal any other motifs in common.
Figure 4.
Alignment of promoters for ORGs around HBBC. Single nucleotides that occur in at least 0.5 of the positions in each column are shaded black; purines or pyrimidines that occur in at least 0.5 of the positions in each column are shaded gray. The 5′ ends of the mouse genes, and the matching nucleotides in human, are boxes. These 5′ ends do not align precisely between paralogous genes, but vary only over a 4-nt region. The scale on the top line places +1 as the 5′ end of the MOR5′β4 gene, which is between the 5′ ends of the other genes. The positions in the alignment are numbered relative to this position as +1.
ORG Expression Is Unaffected by Deletion of the β-Globin LCR.
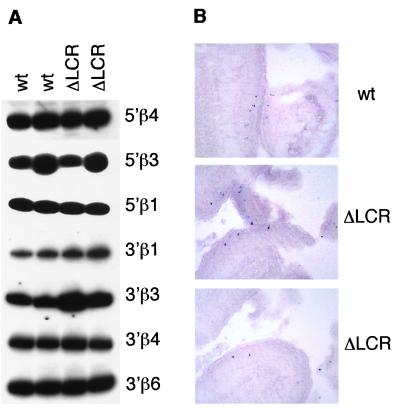
MOR5′β1 is as close to the β-globin LCR as is Hbb-y, the nearest globin gene target. The LCR is a major cis-regulatory sequence in erythroid cells, and so we examined the possibility that it also could play a role in regulation of linked genes in olfactory tissue by comparing ORG expression in olfactory epithelium of wild-type mice and mice homozygous for a deletion of the β-globin LCR (26). Using the exon assignments derived from the 5′ RACE products, we designed PCR primers to amplify cDNAs from spliced mRNA in mouse olfactory epithelium. The results in Fig. 5A show clear reverse transcription–PCR products for all of the ORGs tested, even in the absence of the β-globin LCR. Similarly, in situ hybridization to olfactory epithelium revealed no difference in the expression pattern of β-globin-proximal ORGs in comparisons of wild-type and LCR-deleted mice (Fig. 5B and data not shown). In both mouse strains, expression of these ORGs was observed in the most dorsal zone of olfactory epithelium and exhibited the punctate hybridization pattern that is characteristic of these genes.
Figure 5.
Expression of ORGs surrounding HBBC in olfactory epithelium. (A) Reverse transcription–PCR analysis of total RNA from mouse olfactory epithelium. An autoradiograph of the major PCR products for each of the ORGs tested is shown; sizes were derived by comparison with end-labeled φX174/HindIII standards. Total mRNA was derived from two wild-type mice and two mice homozygous for the deletion of the β-globin LCR (ΔLCR, ref. 26), as indicated. (B) In situ hybridization of olfactory epithelium in wild-type and LCR-deleted mice. Coronal sections through the mouse nasal cavity were hybridized with digoxigenin-labeled antisense RNA probes prepared from the mouse gene MOR5′β1.
The 5′RACE analysis indicated the presence of alternatively spliced forms of the ORGs in olfactory epithelium. Reverse transcription–PCR analysis suggested that mRNA for each of the ORGs tested occurred mostly in a single form; additional bands consistent with alternative splicing of the exons shown in Fig. 2 were observed but were significantly less abundant (data not shown).
Discussion
The genomic arrangement of ORGs surrounding HBBC raises questions of how these two clusters of tissue-specific genes are regulated, and how the signals that govern expression within each cluster are kept separate. In particular, little is known of how the distinctive pattern of ORG expression is accomplished. Interspecies sequence comparisons have been used to identify regulatory sequences within several loci, including the β-globin LCR (31) and the region containing the IL-4, IL-13, and IL-5 genes (32). The presence of extensive sequence conservation in the ORG clusters flanking HBBC in mouse and human, mostly between orthologous ORG transcription units, indicates the presence of regulatory elements that specify the pattern of expression of these genes.
Surprisingly, however, our analysis of the paralogous ORG promoters, noncoding exons, or introns that we have characterized revealed only an A+T-rich region 25–30 bp upstream from the apparent transcription start sites and a purine-rich motif a variable distance upstream from that. In contrast, comparisons of orthologous pairs of human and mouse ORGs reveal extensive conserved regions, including the proximal promoters. Thus, sequence comparisons detected conserved putative regulatory regions for these orthologous pairs, although conservation within this set of related ORGs was confined to a minimal basal promoter. These results suggest that a generalized regulatory motif for ORGs may not exist, despite the presence of sequence determinants of ORG expression that are conserved between orthologs. We suggest that these sequence differences may represent the basis for ORG selection in olfactory neurons.
We have found that transcripts of the ORGs near Hbbc have long 5′ untranslated regions, and some undergo alternative splicing. Alternative splicing of multiple noncoding exons (and lack of a generally conserved promoter sequence) also has been observed for members of a distinct cluster of ORGs located ∼1 megabase from HBBC in mouse (R. Lane, T. Cutforth, R. Axel, and B. Trask, personal communication), suggesting that this is a general feature of ORG expression. Possibilities for the role of these 5′ untranslated regions include control over translocation of the mRNA within the cell, translational control, or transcriptional control. Although no upstream, in-frame translation start sites were seen, it is possible that short out-of-frame translation products could be used to regulate protein synthesis, as has been observed for yeast GCN4 (33, 34). In addition, it has been observed that ORG mRNA is found both at the dendrite and at the axon (28). It is possible that such RNA localization could be regulated by alternative splicing; this could be tested by in situ hybridization with probes derived from specific noncoding exons.
The best candidates for elements involved in global regulation of the ORG and β-globin gene clusters are those conserved regions that occur outside of ORG transcription units. Several such regions exist (Fig. 3), the most striking of which is the β-globin LCR. Our results indicate that the β-globin LCR does not play a role in expression of ORGs in nasal epithelium. The remaining candidates are then several segments, located on either side of MOR5′β4/HOR5′β7, 3′ of MOR/HOR5′β2, and 3′ of the β-globin cluster. The last of these corresponds to a nuclease hypersensitive site (3′ hypersensitive site 1) that is present in erythroid cells but also reportedly in other cell types and that to date has no known function (30). We have found a strong erythroid-specific hypersensitive site at the conserved sequence located near the promoter of MOR5′β4/HOR5′β7 as well (unpublished results); furthermore, we have found that the “open” domain of nuclease-sensitive chromatin in erythroid cells extends through these regions and thus includes several of the ORGs. Given this observation and the presence of DNase-hypersensitive sites at both of these regions in erythroid cells, these sequences are candidates for erythroid regulatory elements. The results of the LCR deletions (24–26) suggest that formation of a generalized nuclease-sensitive chromatin structure in erythroid cells is accomplished by sequences other than the LCR, and one or both of these conserved hypersensitive sites may play a role in this process. We cannot at present eliminate the possibility, however, that either or both of these regions also may be involved in regulation of the proximal ORGs.
Acknowledgments
We thank T. Ley and G. Bepler for the gifts of BAC and PAC clones and L. Buck, T. Cutforth, and B. Trask for discussion and critical reading of the manuscript. This work was supported by National Institutes of Health Grants DK27635 (to R.H.) and DK44746 and DK54701 (to M.G.). M.B. is a fellow of the Helen Hay Whitney Foundation. M.A.B. is a J. S. McDonnell Foundation Scholar.
Abbreviations
- ORG
olfactory receptor gene
- LCR
locus control region
- RACE
rapid amplification of cDNA ends
- BAC
bacterial artificial chromosome
- HOR
human olfactory receptor
- MOR
mouse olfactory receptor
Footnotes
References
- 1.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Krautwurst D, Yau K W, Reed R R. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H, Ivic L, Otaki J M, Hashimoto M, Mikoshiba K, Firestein S. Science. 1998;279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 4.Rouquier S, Taviaux S, Trask B J, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Nat Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- 5.Sharon D, Glusman G, Pilpel Y, Horn-Saban S, Lancet D. Ann NY Acad Sci. 1998;855:182–193. doi: 10.1111/j.1749-6632.1998.tb10564.x. [DOI] [PubMed] [Google Scholar]
- 6.Trask B J, Friedman C, Martin-Gallardo A, Rowen L, Akinbami C, Blankenship J, Collins C, Giorgi D, Iadonato S, Johnson F, et al. Hum Mol Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- 7.Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. Genomics. 2000;63:227–245. doi: 10.1006/geno.1999.6030. [DOI] [PubMed] [Google Scholar]
- 8.Ressler K J, Sullivan S L, Buck L B. Cell. 1993;73:597–609. doi: 10.1016/0092-8674(93)90145-g. [DOI] [PubMed] [Google Scholar]
- 9.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 10.Malnic B, Hirono J, Sato T, Buck L B. Cell. 1999;96:713–723. doi: 10.1016/s0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 11.Mombaerts P, Wang F, Dulac C, Vassar R, Chao S K, Nemes A, Mendelsohn M, Edmondson J, Axel R. Cold Spring Harbor Symp Quant Biol. 1996;61:135–145. [PubMed] [Google Scholar]
- 12.Chess A, Simon I, Cedar H, Axel R. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 13.Feingold E A, Penny L A, Nienhuis A W, Forget B G. Genomics. 1999;61:15–23. doi: 10.1006/geno.1999.5935. [DOI] [PubMed] [Google Scholar]
- 14.Vanderhaeghen P, Schurmans S, Vassart G, Parmentier M. Genomics. 1997;39:239–246. doi: 10.1006/geno.1996.4490. [DOI] [PubMed] [Google Scholar]
- 15.Asai H, Kasai H, Matsuda Y, Yamazaki N, Nagawa F, Sakano H, Tsuboi A. Biochem Biophys Res Commun. 1996;221:240–247. doi: 10.1006/bbrc.1996.0580. [DOI] [PubMed] [Google Scholar]
- 16.Drutel G, Arrang J M, Diaz J, Wisnewsky C, Schwartz K, Schwartz J C. Recept Channels. 1996;3:33–40. [PubMed] [Google Scholar]
- 17.Nef S, Nef P. Proc Natl Acad Sci USA. 1997;94:4766–4771. doi: 10.1073/pnas.94.9.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardison R C. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- 19.Bulger M, von Doorninck J H, Saitoh N, Telling A, Farrell C, Bender M A, Felsenfeld G, Axel R, Groudine M. Proc Natl Acad Sci USA. 1999;96:5129–5134. doi: 10.1073/pnas.96.9.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardison R, Slightom J L, Gumucio D L, Goodman M, Stojanovic N, Miller W. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- 21.Bulger M, Groudine M. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Harju S, Peterson K R. Trends Genet. 1999;15:403–408. doi: 10.1016/s0168-9525(99)01780-1. [DOI] [PubMed] [Google Scholar]
- 23.Grosveld F. Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 24.Epner E, Reik A, Cimbora D, Telling A, Bender M, Fiering S, Enver T, Martin D, Kennedy M, Keller G, Groudine M. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 25.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bender M A, Bulger M, Close J, Groudine M. Mol Cell. 2000;5:387–393. doi: 10.1016/s1097-2765(00)80433-5. [DOI] [PubMed] [Google Scholar]
- 27.Bepler G, O'Briant K C, Kim Y-C, Schreiber G, Pitterle D M. Genomics. 1999;55:164–175. doi: 10.1006/geno.1998.5659. [DOI] [PubMed] [Google Scholar]
- 28.Vassar R, Chao S K, Sitcheran R, Nunez J M, Vosshall L B, Axel R. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 29.Conklin D, Holderman S, Whitmore T E, Maurer M, Feldhaus A L. Gene. 2000;249:91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 30.Fleenor D, Kaufman R E. Blood. 1993;81:2781–2790. [PubMed] [Google Scholar]
- 31.Jackson J D, Miller W, Hardison R C. Genomics. 1997;39:90–94. doi: 10.1006/geno.1996.4458. [DOI] [PubMed] [Google Scholar]
- 32.Loots G G, Locksley R M, Blankespoor C M, Wang Z E, Miller W, Rubin E M, Frazer K A. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 33.Hinnebusch A G, Jackson B M, Mueller P P. Proc Natl Acad Sci USA. 1988;85:7279–7283. doi: 10.1073/pnas.85.19.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cigan A M, Foiani M, Hannig E M, Hinnebusch A G. Mol Cell Biol. 1991;11:3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]