Abstract
Multiple myeloma (MM) is an incurable plasma cell malignancy. The 26S proteasome inhibitor, bortezomib, selectively induces apoptosis in MM cells; however, the nature of its selectivity remains unknown. Here we demonstrate that 5 different MM cell lines display similar patterns of sensitivity to 3 proteasome inhibitors (PIs) but respond differently to specific NF-κB inhibition. We further show that PIs initiate the unfolded protein response (UPR), a signaling pathway activated by the accumulation of misfolded proteins within the endoplasmic reticulum (ER). Consistent with reports that prosurvival/physiologic UPR components are required for B-cell differentiation into antibody-secreting cells, we found that MM cells inherently expressed the ER chaperones GRP78/Bip and GRP94/gp96. However, bortezomib rapidly induced components of the proapoptotic/terminal UPR, including PERK, the ER stress–specific eIF-2α kinase; ATF4, an ER stress–induced transcription factor; and its proapoptotic target, CHOP/GADD153. Consistent with our hypothesis that PIs induce the accumulation of misfolded ER-processed proteins, we found that the amount of immunoglobulin subunits retained within MM cells correlated with their sensitivity to PIs. These findings suggest that MM cells have a lower threshold for PI-induced UPR induction and ER stress–induced apoptosis because they constitutively express ER stress survival factors to function as secretory cells.
Introduction
Multiple myeloma (MM), the second most commonly diagnosed hematologic malignancy in the United States, is an essentially incurable malignancy of terminally differentiated B cells or plasma cells.1,2 Bortezomib (Velcade, PS-341) is a novel therapeutic agent that has been shown to selectively induce apoptosis in malignant cells.3,4 Bortezomib is particularly toxic to MM cells,5,6 but it has a favorable toxicity profile and was approved by the US Food and Drug Administration in 2003 for the treatment of relapsed refractory disease.7
Bortezomib is a potent and selective inhibitor of the 26S proteasome,8,9 a multisubunit protein complex present in the nucleus and the cytoplasm of all eukaryotic cells10 that is responsible for the degradation of ubiquitinated proteins.11 In addition to damaged or obsolete proteins, the proteasome is responsible for the degradation of proteins involved in cell-cycle regulation, oncogenesis, and apoptosis.12-20 Previous reports have demonstrated that proteasome inhibition by bortezomib abrogates degradation of IκB, leading to the cytoplasmic sequestration and inhibition of the transcription factor NF-κB.5,21-25 Although constitutive NF-κB activity in MM cells has been shown to increase MM cell survival and resistance to cytotoxic agents,26 bortezomib was shown to have more profound effects on MM cell proliferation than a specific IκB kinase inhibitor, PS-1145,22 suggesting that NF-κB inhibition cannot completely explain the nature of the selectivity of bortezomib for MM cells.
One of the defining features of plasma cells is an expansive and highly developed rough endoplasmic reticulum (ER) that is specialized for the production and secretion of thousands of antibody molecules per second.27 In fact the detection of large amounts of monoclonal immunoglobulin or light chain in the serum or urine is one of the diagnostic features of MM.28 Conditions that disrupt protein folding in the ER, such as a chemical insult or nutrient deprivation, activate a stress signaling pathway known as the unfolded protein response (UPR).29,30 UPR induction results in both an initial decrease in general protein synthesis, to reduce the influx of nascent proteins into the ER, and increased transcription of ER resident chaperones, folding enzymes, and components of the protein degradative machinery to prevent the aggregation of the accumulating misfolded proteins. These misfolded proteins are recognized by ER quality control systems and retained in the ER, preventing them from proceeding further through the protein maturation process.31-33 If these proteins cannot be properly refolded, they are targeted for ER-associated protein degradation (ERAD), which involves the retrograde translocation or dislocation of the misfolded proteins out of the ER and subsequent degradation by cytosolic 26S proteasomes.34,35 The UPR enables the cell to survive reversible environmental stresses. However, if the stress is severe or prolonged, UPR activation eventually leads to cell-cycle arrest36,37 and the induction of apoptosis.38-41
The retrograde translocation of misfolded proteins from the ER has been shown to be dependent on functioning cytosolic proteasomes.42-46 Thus, treatment of cells with proteasome inhibitors (PIs) results in the accumulation of misfolded proteins within the ER. We therefore hypothesized that treatment of MM cells with PIs initiates the UPR by inhibiting the retrograde translocation of misfolded proteins from the ER and that MM cells are highly sensitive to these agents because they produce large amounts of protein, namely immunoglobulin, that must be processed within the ER. Interestingly, we found that MM cells constitutively express high levels of UPR survival components, but that PI treatment leads to the rapid induction of proapoptotic UPR genes. We further demonstrate that the amount of immunoglobulin subunits retained in PI-treated MM cells correlates with their level of sensitivity to bortezomib. These data suggest that the secretory function of MM cells makes them more sensitive than other cell types to PI-induced UPR activation and ER stress-induced apoptosis.
Materials and methods
Multiple myeloma–derived cell lines
The 8226/S and U266 human MM cell lines were purchased from the American Type Culture Collection (Manassas, VA). The MM.1S cell line was obtained from Dr Steven Rosen (Northwestern University, Chicago, IL), and the KMS-11 and KMS-18 cell lines were provided by Dr P. Leif Bergsagel (Mayo Clinic, Scottsdale AZ). All cell lines were cultured in RMPI 1640 supplemented with 10% fetal bovine serum, 1% l-glutamine, and 1% penicillin/streptomycin (Mediatech, Herndon, VA).
Reagents
Bortezomib (PS-341, Velcade) was kindly provided by Millennium Pharmaceuticals (Cambridge, MA). Tunicamycin and melphalan were purchased from Sigma-Aldrich (St Louis, MO). Epoxomycin, PSI, brefeldin A, BAY 11-7082, and staurosporine were obtained from EMD Biosciences (La Jolla, CA).
Detection of cell death
For all cell death assays 2.5 × 105 cells/mL were treated with the indicated concentration of drug for up to 24 hours. Cells were washed twice in phosphate-buffered saline and then stained with Annexin V–FITC (Biovision, Mountainview, CA) and propidium iodide (Sigma-Aldrich) according to the manufacturer's instructions. Samples were acquired on a fluorescence-activated cell scanner (FACScan) flow cytometer (Becton Dickinson, Franklin Lakes, NJ) to assess viability.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assays for NF-κB–binding activity were performed as described previously.47 Briefly, cell lysates were prepared after 12 hours of the indicated treatment. Binding reactions containing 1.0 × 105 cpm γ32P-labeled double-stranded NF-κB consensus oligonucleotide probe (5′-GATCCAACGGCAGGGGAATTCCCCTCTCCTTA-3′) and 10 μg total protein were incubated at room temperature for 30 minutes. Samples were run on a 4% nondenaturing polyacrylamide gel, dried on Whatman paper, and visualized by autoradiography.
Immunoblotting
Whole-cell lysates (5.0 × 106 cells) were prepared in modified RIPA buffer containing protease inhibitors (170 μg/mL PMSF, 2 μg/mL aprotinin, and 1 μg/mL leupeptin; Sigma-Aldrich). Fifty micrograms of protein were resolved on 8% to 15% SDS–polyacrylamide gels and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, NH). The following primary antibodies were used: anti-GRP78/BiP (BD Biosciences, Pharmingen, San Diego, CA), anti-GRP94/gp96 (Stressgen Bioreagents, Victoria, BC), anti–total eIF-2α and anti–phosphorylated eIF-2α (Biosource International, Camarillo, CA), anti–phosphorylated PERK (Cell Signaling Technology, Danvers, MA), anti–total PERK, anti-ATF4, and anti-GADD153/CHOP (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-actin (Sigma-Aldrich). Secondary antibodies were obtained from Amersham Biosciences (Piscataway, NJ). Blots were developed by using enhanced chemiluminescence. Protein expression was quantified by using Bio-Rad Quantity One Software (Hercules, CA).
Enzyme-linked immunosorbent assay
The Human Lambda (bound and free) ELISA (enzyme-linked immunosorbent assay) Quantitation Kit (Bethyl Laboratories, Montgomery, TX) was used to detect secreted and intracellular λ light chain according to the manufacturer's instructions. For secreted λ, cells were cultured for 24 hours, and the supernatant from 1.0 × 106 pelleted cells was diluted 1:100 and 1:200 for use in the ELISA. For intracellular λ, whole-cell lysates were prepared as described under “Immunoblotting,” and 500 ng total protein was used in the assay.
Results
Different multiple myeloma cell lines have different levels of sensitivity to proteasome inhibitors
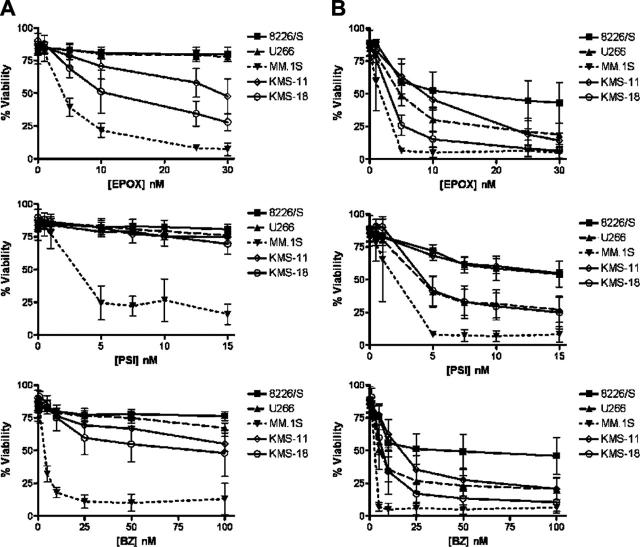
Previous studies have shown that bortezomib induces a caspase-dependent apoptotic cell death in different MM cell lines.5 We further expanded on these studies by examining apoptosis induction in MM cell lines treated with 3 different proteasome inhibitors. As nonsecretory MM occurs in less than 1% of patients, each of the MM cell lines examined secreted immunoglobulin components. The 8226/S, MM.1S, and KMS-18 myeloma lines are λ light chain–only secreting plasma cells; KMS-18 is a κ light chain–only secreting cell line; and U266 cells secrete a complete IgE antibody.48 Each cell line was treated for 12 and 24 hours with increasing concentrations of 1 of 3 inhibitors selective for the 26S proteasome: epoxomycin, PSI, or bortezomib (Velcade, PS-341).49,50 We found that the 5 MM cell lines had different levels of sensitivity to proteasome inhibition, but that they responded similarly to all 3 PIs. In response to treatment with each PI, the MM.1S cell line was the most sensitive, the 8226/S cell line was the most resistant, and the other 3 cell lines had intermediate levels of sensitivity (Figure 1). Profound decreases in MM.1S cell viability were detectable after as little as 6 to 12 hours of treatment (Figure 1A; data not shown). Thus, after 24 hours, 6.4% ± 1.1% of cells were viable in 5 nM epoxomycin, 8.1% ± 5.7% of cells were viable in 5 nM PSI, and 6.4 ± 4.2% of cells were viable in 10 nM bortezomib. In contrast, at higher concentrations of drug, approximately 50% of the 8226/S cells remained viable after 24 hours of treatment such that at 30 nM epoxomycin, 42.9% ± 15.5% of cells were viable, at 15 nM PSI 54.1% ± 9.9% of cells were viable, and 45.9% ± 14.1% of cells were viable at 100 nM bortezomib. However, the fact that very low (nM) concentrations of each agent could be used to induce 50% to 90% cell death after 24 hours of treatment indicates that MM cells are extremely sensitive to the inhibition of 26S proteasome activity.
Figure 1.
Different multiple myeloma cell lines have different levels of sensitivity to proteasome inhibitors. Five different human multiple myeloma cell lines were cultured for 12 (A) and 24 (B) hours in increasing concentrations of the proteasome inhibitors epoxomycin (EPOX), PSI, and Bortezomib (BZ). Cell viability was determined by flow cytometry following Annexin V–FITC and propidium iodide staining. The data are presented as the mean ± SD from at least 3 independent experiments.
Proteasome inhibition does not produce the same response as NF-κB inhibition in multiple myeloma cell lines
Constitutive NF-κB activity in MM cells has been reported to enhance MM cell survival and resistance to cytotoxic agents.26 Additionally, as proteasome inhibition sequesters NF-κB in the cytoplasm, it has been rationally hypothesized that the selectivity of PIs for malignant cells mainly involved the inhibition of NF-κB.5,21-25 However, growth inhibition studies have shown that the effects of nanomolar amounts of bortezomib were far more potent than up to 50 μM of the selective IκB kinase inhibitor PS-1145.22 Consistent with these studies, when cell viability was evaluated directly, we found that all 5 MM cell lines were nearly 100% viable after 24 hours of treatment with up to 100 μM PS-1145 (data not shown), although 10 μM of this drug has been reported to inhibit NF-κB activation in MM.1S cells.22
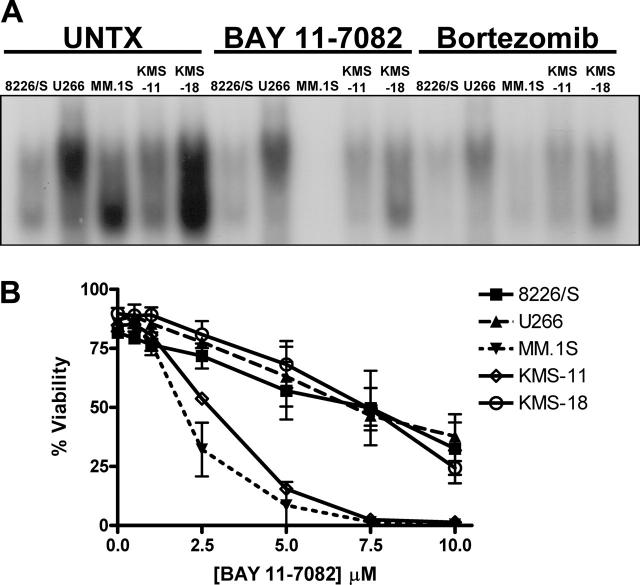
Similarly, the response of the 5 MM cell lines to the irreversible inhibitor of IκBα phosphorylation, BAY 11-7082, is not the same as their response to PIs. After 12 hours of treatment, NF-κB DNA binding is inhibited in all 5 MM cell lines treated with either 5 μM BAY 11-7082 or 10 to 100 nM bortezomib (Figure 2A). However, after 24 hours of treatment, significant reductions in cell viability are seen in bortezomib-treated cells (Figure 1), but at 5 μMofBAY 11-7082 only the MM.1S and KMS-11 cell lines are sensitive to the drug (Figure 2B). There is a slight decrease in the viability of the 8226/S, U266, and KMS-18 cell lines at this concentration, and these 3 cell lines progressively undergo apoptosis as the concentration of BAY 11-7082 is increased up to 10 μM. However, in contrast to PI treatment, their responses are indistinguishable. Taken together, these studies indicate that, although the inhibition of NF-κB has effects on MM cell viability, it cannot completely explain the selectivity of PIs for MM cells.
Figure 2.
Multiple myeloma cell lines respond differently to direct inhibition of NF-κB than they do to direct inhibition of the 26S proteasome. (A) Whole-cell lysates were prepared from untreated multiple myeloma cell lines, cells treated with 5 μM BAY 11-7082 or cells treated with 100 nM (8226/S and U266 cells), 25 nM (KMS-11, KMS-18), or 5 nM (MM.1S) bortezomib and incubated with a double-stranded NF-κB oligonucleotide probe. Samples were run on a native polyacrylamide gel, and NF-κB binding was visualized by autoradiography. (B) Five different human multiple myeloma cell lines were cultured for 24 hours in increasing concentrations of the NF-κB inhibitor BAY 11-7082. Cell viability, as assessed by Annexin V–FITC and propidium iodide staining, is presented as the mean ± SD from at least 3 independent experiments.
Proteasome inhibitors marginally increase the expression of physiologic UPR components in MM cell lines
MM cells are malignant plasma cells that, similar to their normal counterparts, produce and secrete large amounts of immunoglobulin.1,2,27,48 The ER is the site where integral membrane proteins and secretory proteins are folded into their proper tertiary structures, and multimeric proteins, such as immunoglobulins, are assembled.31-33 The inhibition of cytosolic proteasomes has been shown to cause the accumulation of misfolded proteins within the ER lumen.42-46 We therefore hypothesized that proteasome inhibition may lead to UPR induction in secretory MM cell lines, presumably by inhibiting the retrograde translocation of misfolded proteins from the ER.
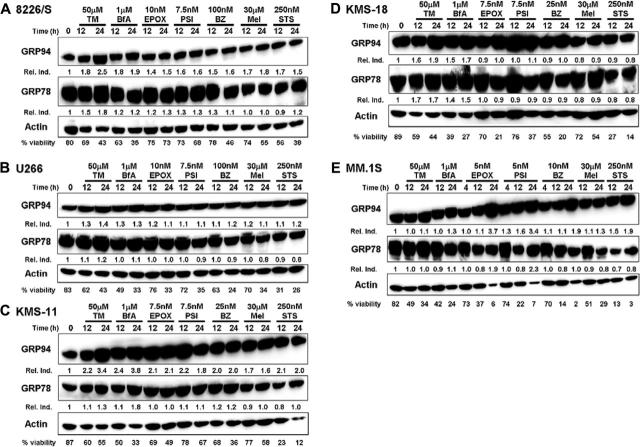
One of the ways the UPR enables any cell to respond to ER stress is by up-regulating the expression of ER molecular chaperones and folding enzymes51,52 to prevent the aggregation of misfolded proteins and to help them refold. ER chaperone induction was examined in each of the MM cell lines treated for 12 and 24 hours with the 3 different proteasome inhibitors. Because these cell lines have different levels of sensitivity to proteasome inhibition, the highest concentrations of the PIs were used to treat the 8226/S and U266 MM cell lines, intermediate concentrations of PIs were used to treat the KMS-11 and KMS-18 cell lines, and the lowest concentrations of PIs were used to treat the MM.1S cell line. Additionally, we compared the induction of the ER molecular chaperones GRP78/BiP and GRP94/gp96 in PI-treated cells with cells treated with known ER stress agents or other apoptosis-inducing agents. Tunicamycin, an inhibitor of N-linked glycosylation, and brefeldin A, an inhibitor of ER to Golgi transport, are 2 chemical agents known to induce ER stress and the UPR.29,30 The alkylating agent melphalan53,54 and the nonspecific kinase inhibitor staurosporine55 were used as negative controls. Interestingly, we found that all 5 MM cell lines constitutively expressed high levels of both ER molecular chaperones, and that the levels of these proteins remained unchanged or were marginally increased in response to treatment with PIs or known ER stress agents, even as these cells began to undergo apoptosis (Figure 3). Thus, GRP94 was induced 1.0- to 3.8-fold and GRP78 was induced 0.9- to 1.8-fold in MM cells treated with classical ER stress agents compared with a 0.9- to 3.7-fold GRP94 induction and a 0.8- to 2.3-fold GRP78 induction in PI-treated cells. Cytotoxic agents that do not induce ER stress also had little effect on ER chaperone expression, causing only a 0.7- to 2.1-fold induction in GRP94 and a 0.8- to 1.2-fold induction in GRP78.
Figure 3.
Multiple myeloma cell lines constitutively express high levels of physiologic UPR components. Five different multiple myeloma cell lines were treated for 12 and 24 hours with 50 μM tunicamycin (TM), 1 μM brefeldin A (BfA), 30 μM melphalan (Mel), 250 nM staurosporine (STS), or the indicated concentration of the proteasome inhibitors epoxomycin (EPOX), PSI, or bortezomib (BZ). Whole-cell lysates were isolated, subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes. The membranes were sequentially probed for GRP94, GRP78, and actin. The relative induction (Rel. Ind.) is the amount of protein present in treated samples relative to untreated cells after normalizing protein loading to actin using densitometry. Cell viability was assessed from an aliquot of the treated cells by Annexin V–FITC and propidium iodide staining. Representative blots from at least 3 independent experiments are shown.
Our finding that MM cells constitutively express ER chaperones is consistent with previous reports that certain components of the UPR are induced during plasma cell development and are required to be constitutively expressed for these cells to function properly.56-58 It has been demonstrated that the expression of GRP78 and GRP94 is induced and maintained in mature B cells as they differentiate into antibody secreting plasma cells, whereas UPR components associated with decreased protein synthesis and apoptosis were not induced under these conditions.56-59 The specific induction of UPR genes that enable cells to differentiate into professional secretory cells capable of tolerating the constitutive production of high amounts of ER-processed proteins has been defined as a “physiologic” UPR. This UPR is distinct from the “ER stress” or “terminal” UPR, which is induced by nutrient deprivation or chemical agents that cause severe or prolonged ER stress.56,57 Taken together, these data suggest that prosurvival/physiologic UPR components are already highly expressed in MM cells and that their expression cannot be significantly altered by agents that cause additional ER stress within the cell.
Upstream terminal UPR component and eIF-2α kinase, PERK, are rapidly activated in MM cells treated with proteasome inhibitors
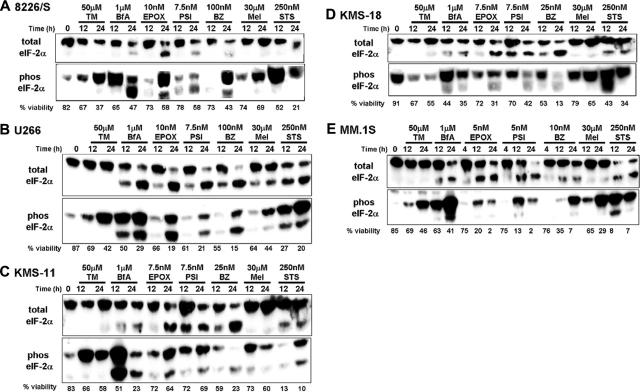
The UPR also enables cells to survive stressful conditions through a transient decrease in de novo protein synthesis to reduce the client protein load in the ER. This is accomplished through phosphorylation of eukaryotic translation initiation factor-2α (eIF-2α).60 To determine whether this aspect of the UPR was activated in MM cells treated with PIs, we examined the levels of phosphorylated eIF-2α in cells treated with PIs, classical ER stress-inducing agents, and other cytotoxic agents. Varying levels of phosphorylated eIF-2α were detected in untreated cells; however, all of the cytotoxic agents used were able to induce the phosphorylation of eIF-2α within 24 hours (Figure 4). Consistent with previous reports that eIF-2α is a caspase-3 target,61 both eIF-2α antibodies also detected a band that migrated slightly faster than full-length eIF-2α in treated MM cells that underwent apoptosis.
Figure 4.
eIF-2α is phosphorylated in multiple myeloma cells treated with a variety of cytotoxic agents. The indicated multiple myeloma cell lines were cultured in 50 μM tunicamycin (TM), 1 μM brefeldin A (BfA), 30 μM melphalan (Mel), 250 nM staurosporine (STS), or the indicated concentration of the proteasome inhibitors epoxomycin (EPOX), PSI, or bortezomib (BZ). After 12 and 24 hours of treatment, cell viability was determined by Annexin V–FITC and propidium iodide staining, and whole cell lysates were isolated to sequentially evaluate the amount of phosphorylated and total eIF-2α present by Western blot analysis. The data are representative of at least 3 different experiments.
In addition to ER stress, the phosphorylation of eIF-2α can be induced by other cellular stresses, such as amino acid starvation or viral infection.62,63 To determine whether the PI-induced phosphorylation of eIF-2α in MM cells was associated with ER stress, we examined the activation of PERK, the ER stress-associated eIF-2α kinase.40,64,65 PERK is rapidly and specifically activated by autophosphorylation in response to ER stress, leading to decreased protein translation as early as 30 minutes after ER stress agent treatment.66 Thus, we were able to detect PERK activation as early as 30 minutes after treatment of the KMS-11 and KMS-18 myeloma cell lines with the classical ER stress agent tunicamycin. Consistent with ER stress being the primary cause of PI-induced eIF-2α phosphorylation in MM cell lines, preliminary experiments suggest that PERK is also rapidly activated in MM cells treated with bortezomib (data not shown). These data suggest that, similar to classical ER stress agents, PIs induce the rapid activation of the ER lumenal sensor, PERK, the primary eIF-2α kinase that is activated during the terminal UPR.
Proteasome inhibitors induce the expression of transcription factors associated with ER stress–induced apoptosis
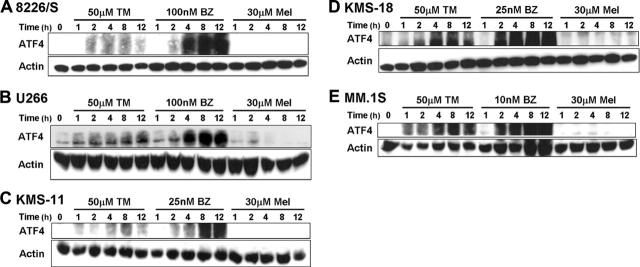
Although the phosphorylation of eIF-2α leads to the attenuation of global protein synthesis, it is also associated with stress-induced gene expression, and different cellular stresses can influence the types of genes that are induced.67,68 In response to ER or nutrient stress–induced phosphorylation of eIF-2α, there is a paradoxical increase in the translation of the transcription factor ATF4.67,69 Consistent with our hypothesis that PIs induce ER stress and the UPR in MM cells, ATF4 expression was rapidly increased after as little as 1 to 2 hours of treatment with bortezomib or tunicamycin in all 5 MM cell lines. However, ATF4 protein levels remained low to undetectable after up to 12 hours of melphalan treatment (Figure 5).
Figure 5.
Proteasome inhibitors specifically induce the expression of the transcription factor ATF4. The indicated multiple myeloma cell lines were treated for up to 12 hours with 50 μM tunicamycin (TM), 30 μM melphalan (Mel), or the indicated concentration of bortezomib (BZ). Whole-cell lysates were prepared, and Western blots for ATF4 were performed. The blots were then stripped and reprobed for actin to confirm equal protein loading. Representative blots from 3 independent experiments are shown.
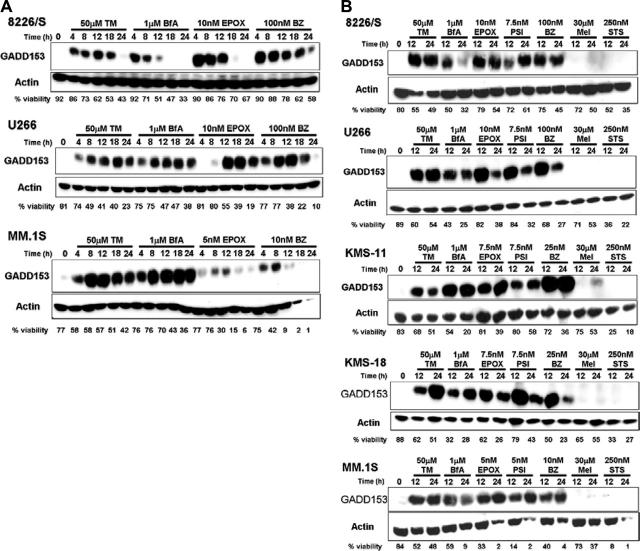
Although ATF4 is responsible for up-regulating the transcription of various prosurvival stress response genes involved in amino acid transport and the oxidative stress response, ATF4 also increases the transcription of the proapoptotic transcription factor GADD153/CHOP in response to ER stress and UPR induction.70 GADD153 induction has been shown to be specifically involved in ER stress–induced apoptosis,38,71 and mouse embryonic fibroblasts (MEFs) from GADD153-deficient mice have been shown to be resistant to PI- and ER stress–induced apoptosis.38,72 Consistent with these reports, GADD153 was rapidly induced after as little as 4 hours in MM cell lines treated with PIs or known ER stress agents (Figure 6A). Moreover, GADD153 induction is specific to ER stress agent and PI treatment because it is not induced after up to 24 hours of treatment with other cytotoxic agents (Figure 6B). Taken together, these results demonstrate that PIs cause the rapid and sequential induction of UPR components that are associated with severe or prolonged ER stress. These data further suggest that PIs, similar to classical ER stress agents, specifically induce proteins associated with a terminal UPR and ER stress–-induced apoptosis in MM cell lines.
Figure 6.
Proteasome inhibitors rapidly up-regulate a protein specifically involved in ER stress–induced apoptosis. (A) The 8226/S, U266, and MM.1S human multiple myeloma cell lines were cultured for up to 24 hours in 50 μM tunicamycin (TM), 1 μM brefeldin A (BfA), or the indicated concentration of epoxomycin (EPOX) or bortezomib (BZ). At the indicated time points, an aliquot of cells was taken to assess cell viability by Annexin V–FITC and propidium iodide staining, and the remainder of the cells was used to prepare whole-cell lysates. Induction of GADD153 and actin expression were determined by Western blot analysis. Blots representative of at least 3 independent experiments are shown. (B) Five different human multiple myeloma cell lines were treated for 12 and 24 hours with 50 μM tunicamycin (TM), 1 μM brefeldin A (BfA), 30 μM melphalan (Mel), 250 nM staurosporine (STS), or the indicated concentration of the proteasome inhibitors epoxomycin (EPOX), PSI, or bortezomib (BZ). Western blot and cell viability analyses were performed as described above. The data are representative of at least 3 independent experiments.
Accumulation of intracellular λ light chain correlates with sensitivity to proteasome inhibitors
Although PIs can induce the terminal UPR in each of the cell lines examined, the amount of the drug required for terminal UPR induction varies between different MM cell lines. Thus, a 10-fold higher concentration of bortezomib (100 nM) is required to induce apoptosis and the terminal UPR in the cell line that is most resistant to PIs (8226/S) compared with the cell line that is least resistant to PIs (MM.1S). Both of these cell lines are λ light chain–only secreting cells, which allowed us to investigate whether differences in immunoglobulin production correlate with PI sensitivity.
As we have hypothesized that MM cells are more sensitive to PIs than other malignant cells because they produce large amounts of ER-processed proteins, one explanation for the differences in sensitivity observed between MM cell lines is that cells that are more sensitive to PIs produce more immunoglobulin. However, when immunoglobulin secretion was quantitated by ELISA, we found that MM.1S cells secrete half as much light chain as 8226/S cells (Table 1). This assay measures the amount of immunoglobulin produced by an equal number of cells over a defined period of time without accounting for cell size, whereas visual inspection and forward scatter analysis have shown that 8226/S cells are much larger than MM.1S cells (data not shown).
Table 1.
Quantification of λ light chain secretion by multiple myeloma cell lines
| Cell line | Secreted λ, μg/1.0 × 106 cells |
|---|---|
| 8226/S | 29.96 ± 7.39 |
| MM.1S | 15.05 ± 4.84 |
MM cells were cultured at 5.0 × 105 cells/mL for 24 hours, after which the volume of media containing 1.0 × 106 cells was removed from the culture. Cells were pelleted, and the supernatant was removed for ELISA analysis. The data represent the mean ± the SD from 3 independent experiments.
In addition to cell size, ELISAs performed on cell supernatants also do not address the efficiency of light chain folding within MM cells. This is important because inhibition of the 26S proteasome leads to the accumulation of misfolded proteins in the ER lumen, whereas properly folded proteins can proceed along the secretory pathway. This is in contrast to the inhibition of ER-to-Golgi transport by brefeldin A, which inhibits all protein secretion. To distinguish between retained and presumably misfolded λ light chains and the total amount of light chain that is processed in the ER, whole-cell lysates were isolated from untreated, PI-treated, or brefeldin A–treated MM cells, and the amount of λ light chain present within the lysates was quantitated by ELISA. Additionally, the ratio of treated cells to untreated cells was used to normalize the data to make comparisons between cell lines. When compared with untreated controls, 10-fold less bortezomib caused the retention of 3- to 4-fold more λ light chain in MM.1S cells compared with 8226/S cells (Table 2), suggesting that higher amounts of misfolded light chains, which require the proteasome for degradation, are produced by the MM.1S cell line. The continued secretion of properly folded proteins combined with the activation of the PERK pathway (which decreases protein synthesis) is probably why the ratio is less than 1 in PI-treated cells. This is in contrast to brefeldin A–treated cells, in which the ratio of treated to untreated cells is greater than 1 because this drug inhibits protein secretion. Consistent with the secretion ELISA analysis, more protein accumulates within the brefeldin A–treated 8226/S cells compared with treated MM.1S cells. Taken together these data suggest that, although all MM cells are extremely sensitive to proteasome inhibition, the efficiency of protein folding within the ER can affect the level of sensitivity of individual MM cell lines to PIs.
Table 2.
Quantification of intracellular λ light chain retained in treated myeloma cell lines
|
Experiment 1
|
Experiment 2
|
Experiment 3
|
||||
|---|---|---|---|---|---|---|
| Ratio | 8226/S | MM.1S | 8226/S | MM.1S | 8226/S | MM.1S |
| BZ/UNTX | 0.257 | 0.854 | 0.481 | 1.387 | 0.304 | 1.316 |
| BfA/UNTX | 2.208 | 1.517 | 1.908 | 1.470 | 2.603 | 1.402 |
The 8226/S and MM.1S myeloma cell lines were left untreated, treated with 10 (MM.1S) to 100 nM (8226/S) bortezomib (BZ), or treated with 1 μM brefeldin A (BfA). Cell lysates were prepared after 7 hours of treatment, and 500 ng total protein was used to determine the amount of intracellular λ light chain present by ELISA. Three independent experiments are shown with the data presented as the ratio of the treated cells (BZ or BfA) to the untreated controls.
Discussion
NF-κB activity has been shown to enhance MM cell survival and resistance to chemotherapeutic agents,26 and functional proteasomes are required NF-κB activation.5,21-25 However, we found that despite that 5 human MM cell lines responded similarly to low concentrations of 3 different proteasome inhibitors (Figure 1), the effect of the irreversible inhibitor of IκBα phosphorylation, BAY 11-7082, on these MM cell lines differed from that of proteasome inhibition (Figure 2). These findings are similar to reports that bortezomib and a specific IκB kinase inhibitor, PS-1145, do not have the same effects on cell proliferation in MM cell lines22 and suggest that the inhibition of NF-κB cannot completely explain the selective anti-MM activity of proteasome inhibitors.
Because MM cells, similar to their normal counterparts, produce significant amounts of ER-processed proteins,27 we reasoned that these cells may be sensitive to perturbations in protein degradation, which would result in the activation of the UPR. One of the hallmarks of UPR induction is the increased transcription and translation of ER molecular chaperones.29,30,51,52 These genes are induced by the UPR transcription factors XBP1 and ATF6.52,73 Although XBP1 splicing and its resulting activation have been shown to be inhibited in PI-treated MM cells,74 our finding that the high constitutive expression of 2 XBP1 target genes products, GRP78 and GRP94, is not reduced by PI treatment (Figure 3) and the observation by Lee et al74 that the XBP1-dependent UPR target gene ERdj4 was normally induced by PIs suggest that the UPR remains functional in PI-treated MM cells. Because both XBP1 and ATF6 can bind to ER stress response elements in the promoters of UPR target genes,52,73 it is possible that ATF6 may compensate for decreased XBP1 activity in PI-treated MM cells.75 Consistent with this, it has been shown that the induction of GRP78 and GRP94 is only slightly impaired in XBP1–/– B cells59 and that the expression of GRP94 requires either, but not both, ATF6 or XBP1.75
The high constitutive expression of the ER resident chaperones GRP78 and GRP94 in MM cell lines is consistent with reports that physiologic UPR gene expression is required for professional secretory cell function.56,57,59,76 Elevated levels of ER chaperones are characteristic of plasma cells,77,78 and their expression is essential for proper antibody assembly and secretion. GRP78 has been shown to stably bind to immunoglobulin heavy chains that have not yet associated with immunoglobulin light chains and to assist in immunoglobulin assembly.79-81 Furthermore, both GRP78 and GRP94 are important for immunoglobulin light chain folding and targeting unassembled subunits for degradation.82-86 The fact that the expression of GRP78 and GRP94 is only slightly increased in MM cells treated with PIs and classical ER stress agents suggests they already express near-maximal levels of cytoprotective UPR proteins to function as secretory cells. Thus, these cells may have a lower threshold (compared with nonsecretory cells) for induction of the terminal UPR following any additional stress to the ER.
Although PI treatment of MM cells does not alter the expression levels of ER chaperones, it does lead to the sequential induction of terminal UPR components. Preliminary data suggest that both bortezomib and tunicamycin rapidly activated PERK, the ER stress-–specific eIF-2α kinase64,65 as early as 30 minutes after treatment (data not shown). Under conditions of ER stress or nutrient deprivation, eIF-2α phosphorylation leads to a general decrease in de novo protein synthesis40 and a paradoxical increase in the translation of ATF4.67,69 Consistent with terminal UPR induction by PIs, ATF4 protein levels rapidly increased in MM cells within 2 hours of treatment with bortezomib or tunicamycin (Figure 5). During severe ER stress, ATF4 induces the expression of CHOP/GADD153,67,87 a basic leucine zipper transcription factor associated with ER stress–induced apoptosis.38,39 In MM cells treated with PIs or classical ER stress agents, GADD153 induction was observed after as little as 4 hours (Figure 6A). The rapid activation and induction of terminal UPR proteins in MM cells was shown to be specific to ER stress–inducing agents, because melphalan and staurosporine did not induce any of these changes.
The fact that MM cell lines rapidly induce terminal UPR components without further increasing the expression of prosurvival UPR components when treated with PIs suggests that the secretory function of these cells makes them extremely sensitive to ER stress induced by this class of drug. This may explain the finding that murine myeloma cells can be sensitized to lower concentrations of PIs if they are also treated with tunicamycin,74 because the inhibition of N-linked glycosylation by tunicamycin would interfere with immunoglobulin/protein folding within the ER, creating an even larger amount of ERAD substrates. The selectivity of PIs for secretory cells is also supported by observations that PIs activate PERK much later in MEFs.72,88 Jiang and Wek72 were unable to detect PERK activation in MEFs after up to 6 hours of treatment with the proteasome inhibitor MG132, and Fribley et al88 only identified detectable levels of PERK activation in MEFs after 16 hours of bortezomib treatment.
Although the high levels of antibody synthesis in MM cells may indicate that myeloma cells, in general, have a greater need for the retrograde translocation of misfolded ERAD substrates, the differences in sensitivity observed between MM cell lines also appears to involve the efficiency of immunoglobulin folding. As each malignant myeloma clone arises from a mature B cell with a unique rearrangement of its immunoglobulin genes, one would expect that certain immunoglobulin protein subunits may fold or assemble more efficiently than others. Mutations that cause the inefficient or improper folding of ER-processed proteins have been associated with the pathology of diseases such as cystic fibrosis, and cytosolic proteasomes have been shown to be required for the removal and the degradation of these misfolded proteins from the ER.44,45 Consistent with the idea that immunoglobulin folding efficiency can affect the sensitivity of MM cells to PIs, higher amounts of λ light chain were found to accumulate within the more sensitive MM.1S cell line compared with the more resistant 8226/S cell line even when MM.1S cells were treated with 10-fold less drug (Table 2).
These studies further suggest that malignant cells can be sensitized to PIs by combining them with agents that affect protein folding within the ER or the cytoplasm. The accumulation of misfolded proteins in the ER lumen of PI-treated cells is thought to occur as a consequence of feedback inhibition, where the accumulation of ubiquitinated proteins that cannot be degraded in the cytosol prevents the retrograde translocation of additional unfolded proteins from the ER.89,90 The presence of misfolded proteins in the cytosol may partially explain why proteasome inhibitors also induce the expression of heat shock–factor proteins including HSP27, HSP70, and HSP90.6,91-95 HSP90 regulates the stability and function of many protein kinases, including IRE1α, the endoribonuclease that activates XBP1,96 and disrupting the ability of HSP90 to interact with its target proteins leads to their rapid destabilization and degradation by the proteasome.97,98 Thus, the ability of the HSP90 inhibitor NSC683664 to increase the sensitivity of MM.1S myeloma cells to bortezomib6 and the observation that treatment of MCF-7 breast cancer cells with either the HSP90 inhibitor, geldanamycin, or bortezomib alone was cytostatic, whereas combining these drugs is potently cytotoxic99 may both be explained by the fact that all cells require functional proteasomes for the degradation of misfolded ER proteins as well as destabilized cytosolic proteins.
Taken together, these data suggest that MM cells are inherently sensitive to PIs because of their large volume of immunoglobulin production, which requires the constitutive expression of physiologic UPR genes. This appears to lower their threshold for the induction of a terminal UPR in response to PI-induced ER stress. In addition to the amount, PI sensitivity also appears to involve the efficiency of immunoglobulin folding within MM cells. These parameters may lead to the more specific identification of MM tumors that will respond to bortezomib. Furthermore, more resistant myeloma clones as well as other nonsecretory malignancies may be sensitized to bortezomib by combining it with agents that interfere with the UPR or agents that destabilize cytosolic proteins.
Acknowledgments
We thank Bill Dalton, Leif Bergsagel, and Steve Rosen for kindly providing us with cell lines. We also thank Robert Levy, David McConkey, Linda Hendershot, and Alan Diehl for helpful advice.
Prepublished online as Blood First Edition Paper, February 28, 2006; DOI 10.1182/blood-2005-08-3531.
Supported in part by National Institutes of Health (grants R0-1 GM68513 and R0-1 CA97243; L.H.B. and K.P.L.), a Senior Research Award from the Multiple Myeloma Research Foundation (L.H.B.), and a Predoctoral Fellowship from the Howard Hughes Medical Institute (E.A.O.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Anderson KC. Bortezomib therapy for myeloma. Curr Hematol Rep. 2004;3: 65. [PubMed] [Google Scholar]
- 2.Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin. 2000;50: 7-33. [DOI] [PubMed] [Google Scholar]
- 3.Masdehors P, Omura S, Merle-Beral H, et al. Increased sensitivity of CLL-derived lymphocytes to apoptotic death activation by the proteasome-specific inhibitor lactacystin. Br J Haematol. 1999;105: 752-757. [DOI] [PubMed] [Google Scholar]
- 4.Adams J, Palombella VJ, Sausville EA, et al. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 1999;59: 2615-2622. [PubMed] [Google Scholar]
- 5.Hideshima T, Richardson P, Chauhan D, et al. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 2001;61: 3071-3076. [PubMed] [Google Scholar]
- 6.Mitsiades N, Mitsiades CS, Poulaki V, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci U S A. 2002;99: 14374-14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8: 508-513. [DOI] [PubMed] [Google Scholar]
- 8.Gardner RC, Assinder SJ, Christie G, et al. Characterization of peptidyl boronic acid inhibitors of mammalian 20 S and 26 S proteasomes and their inhibition of proteasomes in cultured cells. Biochem J. 2000;346(Pt 2): 447-454. [PMC free article] [PubMed] [Google Scholar]
- 9.Adams J, Behnke M, Chen S, et al. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg Med Chem Lett. 1998;8: 333-338. [DOI] [PubMed] [Google Scholar]
- 10.Brooks P, Fuertes G, Murray RZ, et al. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J. 2000;346(Pt 1): 155-161. [PMC free article] [PubMed] [Google Scholar]
- 11.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79: 13-21. [DOI] [PubMed] [Google Scholar]
- 12.Rock KL, Gramm C, Rothstein L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78: 761-771. [DOI] [PubMed] [Google Scholar]
- 13.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20: 2423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res. 1996;56: 2649-2654. [PubMed] [Google Scholar]
- 15.Blagosklonny MV, Wu GS, Omura S, el-Deiry WS. Proteasome-dependent regulation of p21WAF1/CIP1 expression. Biochem Biophys Res Commun. 1996;227: 564-569. [DOI] [PubMed] [Google Scholar]
- 16.An B, Goldfarb RH, Siman R, Dou QP. Novel dipeptidyl proteasome inhibitors overcome Bcl-2 protective function and selectively accumulate the cyclin-dependent kinase inhibitor p27 and induce apoptosis in transformed, but not normal, human fibroblasts. Cell Death Differ. 1998;5: 1062-1075. [DOI] [PubMed] [Google Scholar]
- 17.Pagano M, Tam SW, Theodoras AM, et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269: 682-685. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci U S A. 2000;97: 3850-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YC, Lee YS, Tejima T, et al. mdm2 and bax, downstream mediators of the p53 response, are degraded by the ubiquitin-proteasome pathway. Cell Growth Differ. 1998;9: 79-84. [PubMed] [Google Scholar]
- 20.Breitschopf K, Zeiher AM, Dimmeler S. Ubiquitin-mediated degradation of the proapoptotic active form of bid. A functional consequence on apoptosis induction. J Biol Chem. 2000;275: 21648-21652. [DOI] [PubMed] [Google Scholar]
- 21.Cusack JC Jr, Liu R, Houston M, et al. Enhanced chemosensitivity to CPT-11 with proteasome inhibitor PS-341: implications for systemic nuclear factor-kappaB inhibition. Cancer Res. 2001;61: 3535-3540. [PubMed] [Google Scholar]
- 22.Hideshima T, Chauhan D, Richardson P, et al. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem. 2002;277: 16639-16647. [DOI] [PubMed] [Google Scholar]
- 23.Ma MH, Yang HH, Parker K, et al. The proteasome inhibitor PS-341 markedly enhances sensitivity of multiple myeloma tumor cells to chemotherapeutic agents. Clin Cancer Res. 2003;9: 1136-1144. [PubMed] [Google Scholar]
- 24.Sunwoo JB, Chen Z, Dong G, et al. Novel proteasome inhibitor PS-341 inhibits activation of nuclear factor-kappa B, cell survival, tumor growth, and angiogenesis in squamous cell carcinoma. Clin Cancer Res. 2001;7: 1419-1428. [PubMed] [Google Scholar]
- 25.Russo SM, Tepper JE, Baldwin AS Jr, et al. Enhancement of radiosensitivity by proteasome inhibition: implications for a role of NF-kappaB. Int J Radiat Oncol Biol Phys. 2001;50: 183-193. [DOI] [PubMed] [Google Scholar]
- 26.Mitsiades N, Mitsiades CS, Poulaki V, et al. Biologic sequelae of nuclear factor-kappaB blockade in multiple myeloma: therapeutic applications. Blood. 2002;99: 4079-4086. [DOI] [PubMed] [Google Scholar]
- 27.Calame KL, Lin KI, Tunyaplin C. Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol. 2003;21: 205-230. [DOI] [PubMed] [Google Scholar]
- 28.Group IMW. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121: 749-757. [PubMed] [Google Scholar]
- 29.Patil C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol. 2001;13: 349-355. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13: 1211-1233. [DOI] [PubMed] [Google Scholar]
- 31.Ibba M, Soll D. Quality control mechanisms during translation. Science. 1999;286: 1893-1897. [DOI] [PubMed] [Google Scholar]
- 32.Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286: 1888-1893. [DOI] [PubMed] [Google Scholar]
- 33.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286: 1882-1888. [DOI] [PubMed] [Google Scholar]
- 34.Kostova Z, Wolf DH. For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J. 2003;22: 2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai B, Ye Y, Rapoport TA. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev Mol Cell Biol. 2002;3: 246-255. [DOI] [PubMed] [Google Scholar]
- 36.Brewer JW, Diehl JA. PERK mediates cell-cycle exit during the mammalian unfolded protein response. Proc Natl Acad Sci U S A. 2000;97: 12625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brewer JW, Hendershot LM, Sherr CJ, Diehl JA. Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc Natl Acad Sci U S A. 1999;96: 8505-8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zinszner H, Kuroda M, Wang X, et al. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12: 982-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21: 1249-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5: 897-904. [DOI] [PubMed] [Google Scholar]
- 41.Harding HP, Zeng H, Zhang Y, et al. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7: 1153-1163. [DOI] [PubMed] [Google Scholar]
- 42.O'Hare T, Wiens GD, Whitcomb EA, Enns CA, Rittenberg MB. Cutting edge: proteasome involvement in the degradation of unassembled Ig light chains. J Immunol. 1999;163: 11-14. [PubMed] [Google Scholar]
- 43.Mancini R, Fagioli C, Fra AM, Maggioni C, Sitia R. Degradation of unassembled soluble Ig subunits by cytosolic proteasomes: evidence that retrotranslocation and degradation are coupled events. FASEB J. 2000;14: 769-778. [DOI] [PubMed] [Google Scholar]
- 44.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83: 121-127. [DOI] [PubMed] [Google Scholar]
- 45.Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83: 129-135. [DOI] [PubMed] [Google Scholar]
- 46.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci U S A. 1996;93: 13797-13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St Louis DC, Woodcock JB, Franzoso G, et al. Evidence for distinct intracellular signaling pathways in CD34+ progenitor to dendritic cell differentiation from a human cell line model. J Immunol. 1999;162: 3237-3248. [PubMed] [Google Scholar]
- 48.Drexler HG, Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of multiple myeloma and plasma cell leukemia. Leuk Res. 2000;24: 681-703. [DOI] [PubMed] [Google Scholar]
- 49.Adams J. Proteasome inhibitors as new anticancer drugs. Curr Opin Oncol. 2002;14: 628-634. [DOI] [PubMed] [Google Scholar]
- 50.Almond JB, Cohen GM. The proteasome: a novel target for cancer chemotherapy. Leukemia. 2002;16: 433-443. [DOI] [PubMed] [Google Scholar]
- 51.Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332: 462-464. [DOI] [PubMed] [Google Scholar]
- 52.Lee K, Tirasophon W, Shen X, et al. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16: 452-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McElwain TJ, Powles RL. High-dose intravenous melphalan for plasma-cell leukaemia and myeloma. Lancet. 1983;2: 822-824. [DOI] [PubMed] [Google Scholar]
- 54.Alexanian R, Bergsagel DE, Migliore PJ, Vaughn WK, Howe CD. Melphalan therapy for plasma cell myeloma. Blood. 1968;31: 1-10. [PubMed] [Google Scholar]
- 55.Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid/Ca++ dependent protein kinase. Biochem Biophys Res Commun. 1986;135: 397-402. [DOI] [PubMed] [Google Scholar]
- 56.Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277: 49047-49054. [DOI] [PubMed] [Google Scholar]
- 57.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21: 81-93. [DOI] [PubMed] [Google Scholar]
- 58.van Anken E, Romijn EP, Maggioni C, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18: 243-253. [DOI] [PubMed] [Google Scholar]
- 59.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4: 321-329. [DOI] [PubMed] [Google Scholar]
- 60.Rowlands AG, Panniers R, Henshaw EC. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988;263: 5526-5533. [PubMed] [Google Scholar]
- 61.Satoh S, Hijikata M, Handa H, Shimotohno K. Caspase-mediated cleavage of eukaryotic translation initiation factor subunit 2alpha. Biochem J. 1999;342(Pt 1): 65-70. [PMC free article] [PubMed] [Google Scholar]
- 62.Kostura M, Mathews MB. Purification and activation of the double-stranded RNA-dependent eIF-2 kinase DAI. Mol Cell Biol. 1989;9: 1576-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68: 585-596. [DOI] [PubMed] [Google Scholar]
- 64.Shi Y, Vattem KM, Sood R, et al. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18: 7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397: 271-274. [DOI] [PubMed] [Google Scholar]
- 66.Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 2003;22: 1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6: 1099-1108. [DOI] [PubMed] [Google Scholar]
- 68.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366: 585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167: 27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11: 619-633. [DOI] [PubMed] [Google Scholar]
- 71.Wang XZ, Lawson B, Brewer JW, et al. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol Cell Biol. 1996;16: 4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem. 2005;280: 14189-14202. [DOI] [PubMed] [Google Scholar]
- 73.Yoshida H, Matsui T, Hosokawa N, Kaufman RJ, Nagata K, Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4: 265-271. [DOI] [PubMed] [Google Scholar]
- 74.Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A. 2003;100: 9946-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23: 7448-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reimold AM, Iwakoshi NN, Manis J, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412: 300-307. [DOI] [PubMed] [Google Scholar]
- 77.Lewis MJ, Mazzarella RA, Green M. Structure and assembly of the endoplasmic reticulum. The synthesis of three major endoplasmic reticulum proteins during lipopolysaccharide-induced differentiation of murine lymphocytes. J Biol Chem. 1985;260: 3050-3057. [PubMed] [Google Scholar]
- 78.Wiest DL, Burkhardt JK, Hester S, Hortsch M, Meyer DI, Argon Y. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J Cell Biol. 1990;110: 1501-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bole DG, Hendershot LM, Kearney JF. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J Cell Biol. 1986;102: 1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee YK, Brewer JW, Hellman R, Hendershot LM. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol Biol Cell. 1999;10: 2209-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature. 1983;306: 387-389. [DOI] [PubMed] [Google Scholar]
- 82.Chillaron J, Haas IG. Dissociation from BiP and retrotranslocation of unassembled immunoglobulin light chains are tightly coupled to proteasome activity. Mol Biol Cell. 2000;11: 217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knittler MR, Haas IG. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992;11: 1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Knittler MR, Dirks S, Haas IG. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995;92: 1764-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Melnick J, Aviel S, Argon Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J Biol Chem. 1992;267: 21303-21306. [PubMed] [Google Scholar]
- 86.Melnick J, Dul JL, Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994;370: 373-375. [DOI] [PubMed] [Google Scholar]
- 87.Ma Y, Brewer JW, Diehl JA, Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318: 1351-1365. [DOI] [PubMed] [Google Scholar]
- 88.Fribley A, Zeng Q, Wang CY. Proteasome inhibitor PS-341 induces apoptosis through induction of endoplasmic reticulum stress-reactive oxygen species in head and neck squamous cell carcinoma cells. Mol Cell Biol. 2004;24: 9695-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fiebiger E, Story C, Ploegh HL, Tortorella D. Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J. 2002;21: 1041-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.VanSlyke JK, Musil LS. Dislocation and degradation from the ER are regulated by cytosolic stress. J Cell Biol. 2002;157: 381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272: 9086-9092. [DOI] [PubMed] [Google Scholar]
- 92.Kawazoe Y, Nakai A, Tanabe M, Nagata K. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur J Biochem. 1998;255: 356-362. [DOI] [PubMed] [Google Scholar]
- 93.Kim D, Kim SH, Li GC. Proteasome inhibitors MG132 and lactacystin hyperphosphorylate HSF1 and induce hsp70 and hsp27 expression. Biochem Biophys Res Commun. 1999;254: 264-268. [DOI] [PubMed] [Google Scholar]
- 94.Mathew A, Mathur SK, Morimoto RI. Heat shock response and protein degradation: regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol. 1998;18: 5091-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pirkkala L, Alastalo TP, Zuo X, Benjamin IJ, Sistonen L. Disruption of heat shock factor 1 reveals an essential role in the ubiquitin proteolytic pathway. Mol Cell Biol. 2000;20: 2670-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marcu MG, Doyle M, Bertolotti A, Ron D, Hendershot L, Neckers L. Heat shock protein 90 modulates the unfolded protein response by stabilizing IRE1alpha. Mol Cell Biol. 2002;22: 8506-8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mimnaugh EG, Chavany C, Neckers L. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J Biol Chem. 1996;271: 22796-22801. [DOI] [PubMed] [Google Scholar]
- 98.Neckers L, Mimnaugh E, Schulte TW. Hsp90 as an anti-cancer target. Drug Resist Updat. 1999;2: 165-172. [DOI] [PubMed] [Google Scholar]
- 99.Mimnaugh EG, Xu W, Vos M, et al. Simultaneous inhibition of hsp 90 and the proteasome promotes protein ubiquitination, causes endoplasmic reticulum-derived cytosolic vacuolization, and enhances antitumor activity. Mol Cancer Ther. 2004;3: 551-566. [PubMed] [Google Scholar]