Abstract
The locus control region (LCR) was thought to be necessary and sufficient for establishing and maintaining an open β-globin locus chromatin domain in the repressive environment of the developing erythrocyte. However, deletion of the LCR from the endogenous locus had no significant effect on chromatin structure and did not silence transcription. Thus, the cis-regulatory elements that confer the open domain remain unidentified. The conserved DNaseI hypersensitivity sites (HSs) HS-62.5 and 3′HS1 that flank the locus, and the region upstream of the LCR have been implicated in globin gene regulation. The flanking HSs bind CCCTC binding factor (CTCF) and are thought to interact with the LCR to form a “chromatin hub” involved in β-globin gene activation. Hispanic thalassemia, a deletion of the LCR and 27 kb upstream, leads to heterochromatinization and silencing of the locus. Thus, the region upstream of the LCR deleted in Hispanic thalassemia (upstream Hispanic region [UHR]) may be required for expression. To determine the importance of the UHR and flanking HSs for β-globin expression, we generated and analyzed mice with targeted deletions of these elements. We demonstrate deletion of these regions alone, and in combination, do not affect transcription, bringing into question current models for the regulation of the β-globin locus.
Introduction
The β-globin locus remains a paradigm for vertebrate gene expression, as it contains multiple genes that are highly expressed in a tissue and developmentally specific manner, is highly conserved through evolution, and is a target for mutations affecting hundreds of thousands of patients worldwide.1 Despite intensive study, little is known of how the β-globin locus becomes and remains activated in the repressive environment of a differentiating erythroid nucleus. The transcriptional silencing of the human β-globin locus by naturally occurring deletions far upstream of the β-like globin gene cluster (γδβ thalassemias) suggested that this region contained regulatory elements capable of activating gene expression and/or protecting genes from the dramatic heterochromatinization that occurs during erythroid maturation.2-5 In particular, a deletion of 39.5 kb upstream of the β-globin genes, called the Hispanic deletion, results in profound alterations, including acquiring a DNaseI-resistant chromatin structure, changes in replication origin use and timing, and silencing of transcription.3,4,6
Analysis of β-globin transgenes in mice led to the identification of a regulatory element lying within this large deletion, the locus control region (LCR), defined by its ability to drive transgene expression at high and relatively consistent levels at nearly all genomic integration sites.7,8 This activity was long taken to suggest that the LCR was capable of counteracting the repressive effects of chromatin structure at different regions of the genome.1,9,10 Thus, it was thought that in addition to driving high-level gene expression, the LCR was necessary for the establishment of an “open” chromatin domain, as defined by DNaseI sensitivity and epigenetic modifications, such as histone acetylation, that are typically associated with active gene loci. However, targeted deletion of the endogenous mouse or human β-globin LCR resulted in a large decrease in β-globin gene expression, but failed to elicit any significant alterations in chromatin structure.11-13 Thus, in its native location, the sole essential function of the LCR is that of an enhancer of transcription, raising the question of what cis-acting sequences in the locus are essential for the initiation and maintenance of an open, active chromatin domain. As the Hispanic thalassemia deletion exhibits significant changes in chromatin structure, and since it removes an additional 27.5 kb of DNA upstream of the LCR, this upstream Hispanic region (UHR) represents a logical candidate for such a cis-acting sequence.
Two additional candidates for cis-acting elements regulating the chromatin structure and organization of the locus are the erythroid-specific DNaseI hypersensitive sites (HSs) located both upstream of the LCR (HS-62.5) and downstream of the β-globin genes (3′HS1).14-16 Studies using the chromatin conformation capture (3C) technique have led to a model in which these 2 regions and the 5′ end of the LCR interact to create an erythroid-specific, developmentally stable nuclear compartment designated the “chromatin hub” (CH) important for activation of β-like genes.17-20 Notably, all 3 regions are bound by CCCTC binding factor (CTCF), and 5′HS5 and 3′HS1 have been shown to function as CTCF-dependent enhancer-blocking elements when placed between enhancers and promoters.15,16 While the full spectrum of CTCF activities has not been elucidated, CTCF-dependent enhancer-blocking activity is a hallmark of some “boundary” elements that are believed to organize gene loci in eukaryotic genomes.21,22 Studies in yeast and Drosophila have led to models in which boundary elements anchor an intervening domain to a nuclear scaffold, isolating the domain from the effects of neighboring loci.23,24 Thus, cis-acting sequences (such as the LCR) within the flanked region would lead to epigenetic modifications permissive for full gene activation, while the flanking boundary elements define the extent of the modified domain and prevent interaction with surrounding regions. The paradigm for vertebrate systems is the chicken β-globin locus, flanked by c5′HS4 and c3′HS, which both bind CTCF in erythroid cells and demarcate sharp boundaries between the “open” DNaseI-sensitive locus, characterized by hyperacetylated core histones, and the flanking regions of DNaseI-resistant heterochromatin.25-28 While both HSs demonstrate CTCF-dependent enhancer-blocking activity at ectopic sites, cHS4 can protect randomly integrated reporter genes from silencing, and this activity is dependent on the binding of upstream stimulatory factor (USF) proteins.21,22,28-32 While the role of the flanking HSs at the endogenous locus in chickens has not been determined, it is hypothesized that these flanking HSs protect the locus from the spread of heterochromatin and transcriptional silencing, which may be particularly critical during terminal differentiation. Notably, the structural organization of the mouse and human β-globin loci is similar to that of chickens in that the LCR and genes are flanked by HSs that bind CTCF (HS-62.5 and 5′HS5 upstream, and 3′HS1 downstream).15,16,33
To determine the importance of the UHR and flanking HSs on β-globin gene expression at the endogenous locus, and to apply this to models of β-globin locus activation and organization, we have generated and analyzed several strains of mice in which these elements have been eliminated from the mouse β-globin locus by targeted deletion. To facilitate this analysis we have derived and validated the use of 2 embryonic stem (ES) cell lines heterozygotic for the β-globin locus that allow internally controlled experiments after in vitro differentiation. Using these cell lines, and mice generated from them, we show these regions alone, and in combination, do not affect transcription of genes in the locus, bringing into question current models for the regulation of the β-globin locus.
Materials and methods
Generation and screening of mutant mice
Targeted deletions of the HS-62.5 and 3′HS 1 regions and generation of mice were performed as described34,35 using LoxP-flanked PGK–neomycin phosphotransferase (NPT) or hygromycin cassettes and either D/S-2B or ΔLCR-D/S-1 ES cells (described in “Results”). These deletions remove nucleotides –63238 through –59669 and 66666 through 67467 relative to the Ey cap site, respectively. In each case the deletion removes the CTCF binding HS as well as other associated HSs. Cre-mediated excision of selectable markers and of the UHR was done by transient expression of the Cre recombinase followed by screening for clones with the desired structure.
RT-PCR assays
RNA isolation, reverse transcriptase–polymerase chain reaction (RT-PCR), and quantitation were done as described previously,35,36 with the following exception. For in vivo analysis flow cytometry sorted 100 cell pools of adult reticulocytes, or embryonic red blood cells (RBCs) were analyzed as described for our single-cell assay. For reticulocytes, a 1-step RT-PCR with 8 cycles was followed by a 20-fold dilution and radioactive PCR for an additional 16 cycles. In all cases we determined that PCR amplification was in the linear range, and no heteroduplex formation was detectable. For stress erythropoiesis studies spleen and blood bulk RNA was isolated while animals were recovering from a phenylhydrazine-induced anemia.27,34 Pools of sorted cells were not analyzed because phenylhydrazine-induced autofluorescence interferes with flow cytometry.
DNaseI hypersensitivity site mapping
Nuclei were isolated from spleens of mice during recovery from a phenylhydrazine-induced hemolytic anemia, at which time at least 75% of cells are erythroid.27,34 Phenylhydrazine treatment and DNaseI digestion were done as described.27,34 DNA was digested with EcoRV and hybridized with probes spanning from –54700 to –53803, 69993 to 70253, and –21222 to –20508 relative to the Ey cap for examining the HS-62.5, 3′HS1, and 5′HS4 regions, respectively.
Primary transcript FISH
Fetal liver cells from mice on day 13.5 after conception were simultaneously stained with TER-119,37 ERY-1,38 and anti-CD117 monoclonal antibodies.39 CD-117–, ERY-1–, TER-119+ cells consisting of nucleated RBCs with some orthochromatic normoblasts, and RBCs were isolated by flow cytometry and subjected to primary transcript RNA fluorescent in situ hybridization (FISH) as described.39
Results
Establishment of HbbD/HbbS (D/S) heterozygotic ES cell lines
To facilitate the analysis of targeted mutations of the endogenous murine β-globin locus after in vitro differentiation, we generated 2 ES cell lines heterozygotic for the HbbD (D) and HbbS (S) alleles. After targeting the D allele RT-PCR, 3C and chromatin immunoprecipitation (ChIP) assays exploiting polymorphisms can be used to quantitatively determine the effect of the mutation when compared to a wild-type (WT) S allele in the same cell. The S allele from C57 mice, and the X-linked mutant Hprt– gene from mice derived from BK4 ES cells40 were back-crossed onto a 129 background. Male 129 mice carrying a deletion of the endogenous LCR (ΔLCR-D/WT-D, Hprt+/0) were bred with 129 WT-S/WT-S Hprt–/– dams, and ES cell lines were generated from the resulting blastocysts using standard techniques.41 Genotype, karyotype, disomy for chromosome 8, and the potential to give good germ-line transmission were confirmed. Two lines were expanded for future use. D/S-2B cells are WT-D/WT-S, Hprt+/0 male cells. ΔLCR-D/S-1 cells are identical but contain the ΔLCR-D allele described previously instead of the WT-D allele.
Deletion of the UHR and HS-62.5 regions
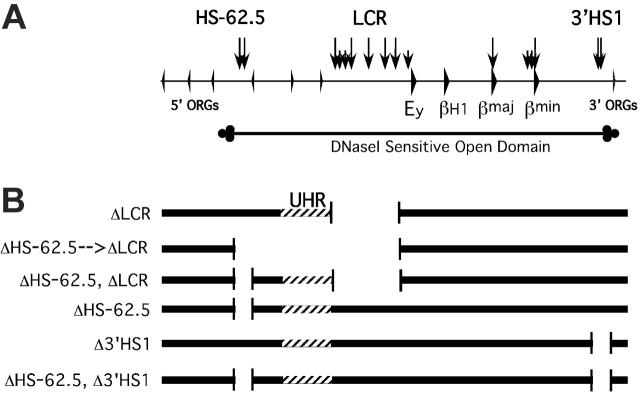
To determine whether the UHR and/or HS-62.5 element are required for the establishment or maintenance of an open, active chromatin domain and responsible for the residual transcriptional activity of the locus in the absence of the LCR, we generated 2 ES cell lines containing mutant alleles. ΔLCR-D/S-1 ES cells were targeted with a vector to replace the HS-62.5 region with a NPT gene flanked with lox sites. Restriction fragment–length polymorphism (RFLP) analysis revealed the D allele was correctly targeted; thus, this deletion is in cis to the LCR deletion (data not shown). Of note, a single lox site is present at the site of the LCR deletion. To avoid any effect of the selectable marker, and to excise the UHR, Cre recombinase was transiently expressed in ES cells and the structure of G418 sensitive clones was analyzed. Two types of mutant alleles were isolated and used for further analysis. Cre-mediated recombination between the far upstream and the LCR Lox P sites led to ΔHS-62.5 → LCR D/S cells that carry a deletion from –63238 to –2069 relative to the Ey cap (Figure 1). This allele lacks the LCR, HS-62.5, and all intervening sequences, which includes the UHR. If this extension of the LCR deletion led to silencing of the locus, it could be secondary to the loss of the UHR or the HS-62.5 region. To distinguish between these possibilities, we identified ΔHS-62.5, ΔLCR D/S clones that lack the LCR and HS-62.5 region but have an intact UHR on the D allele.
Figure 1.
Map of mutant D alleles generated by homologous recombination. (A) Map of the mouse β-globin locus. Arrowheads represent genes. Vertical arrows represent HSs. ORG indicates olfactory receptor gene. The extent of the DNaseI-sensitive domain is diagrammed below the locus map. (B) Diagrammatic representations of the targeted deletions generated. Striped region designates the UHR, the region upstream of the LCR that has homology to the region deleted in the Hispanic deletion.
The UHR is not essential for transcription in vitro
To determine the effects of the UHR and upstream HSs on transcription, ΔHS-62.5 → LCR/S and ΔHS-62.5, ΔLCR/S ES cells were induced to differentiate to the erythroid lineage in a semisynchronous manner on OP9 feeder cells.42 Bulk RNA was analyzed by quantitative RT-PCR assays that exploit RFLPs between the D and S alleles in order to determine allele-specific expression. For both cell lines the mutant D allele was expressed at 1% to 8% of the WT S allele (data not shown). This was true for the embryonic Ey and bH1 globin genes as well as the adult genes, and is similar to what we observed for the deletion of the LCR alone in vitro and in vivo. Thus, in vitro, additional deletion of the UHR and/or HS-62.5 region does not significantly alter transcription when compared to the deletion of the LCR alone.
Deletion of the LCR and UHR does not recapitulate the Hispanic thalassemia phenotype in vivo
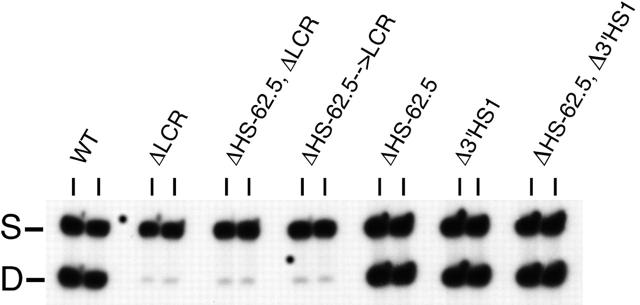
Differentiation in vitro may not faithfully recapitulate all epigenetic modifications that occur with passage through the germ line and normal development. To determine if the UHR and upstream element are necessary to prevent silencing of the β-globin locus after passage through the germ line, ΔHS-62.5 → LCR/S and ΔHS-62.5, ΔLCR/S ES cells were used to generate mice heterozygous for the mutant D allele and a WT S allele. For both strains the ratio of expression from the mutant adult β-globin genes to that from the WT allele was 4%, almost identical to that from the LCR mutation alone36 (Figure 2; Table 1). In theory, deletion of a boundary element or a domain-activating element could lead to variegated expression, with a subpopulation of mutant alleles being silenced. Heterochromatinization is known to exhibit stochastic behavior, resulting in the variegated expression of genes. Therefore, we examined β-globin gene expression by RT-PCR analysis of individual reticulocytes from each mouse line. Variegation was not evident in samples derived from any of the mutant mouse lines, as each cell expressed β-globin at the same low levels (Table 1) and no silenced or high-expressing alleles were found. Thus, in the context of a deleted LCR, we find no contribution of the UHR or upstream HS and CTCF site on the level or probability of expression. We demonstrate that deletion of the LCR, UHR, and upstream HS-62.5 element does not recapitulate the Hispanic thalassemia phenotype. In addition, these elements are not necessary for basal levels of expression, and thus are not required for the establishment or maintenance of an active chromatin domain.
Figure 2.
Expression of the adult β-globin genes in mice heterozygous for targeted deletions. The HbbD allele (D) carries the targeted mutation, while the HbbS allele (S) is wild-type. Pools of 100 reticulocytes were isolated from peripheral blood by flow cytometry and analyzed by RT-PCR with primers that amplify both adult genes from both alleles. Analysis of 2 representative independent pools is shown for each mouse strain. WT indicates wild-type D/S animals. Other strain designations are described in “Results.” S and D mark the RT-PCR product from the S and D alleles, respectively.
Table 1.
Adult β-globin expression measured in the peripheral blood of heterozygotic animals
| WT | ΔLCR | ΔHS-62.5, ΔLCR | ΔHS-62.5 →ΔLCR | Δ-62.5 | Δ3′HS1 | Δ-62.5, Δ3′HS1 | |
|---|---|---|---|---|---|---|---|
| 100-cell pools (n) | 0.97 ± 0.02 (4) | 0.05 ± 0.01 (3) | 0.04 ± 0.01 (3) | 0.04 ± 0.0 (3) | 1.06 ± 0.02 (8) | 1.04 ± 0.04 (8) | 1.02 ± 0.02 (8) |
| Single cells (n, no. cells analyzed) | 1.00 ± 0.12 (1.8) | 0.03 ± 0.01* (4, 26) | 0.05 ± 0.03 (1, 10) | 0.04 ± 0.02 (4, 16) | 1.15 ± 0.09 (3, 12) | 1.10 ± 0.08 (3, 11) | 1.02 ± 0.11 (1, 16) |
Values are expressed as the ratio of expression from a mutant D allele to that of a wild-type S allele measured in either 100-cell pools of reticulocytes or in individual reticulocytes ± SD.
Historical data.36
The HS-62.5 and 3′HS1 regions have no effect on transcription
The CTCF-binding elements HS-62.5 and 3′HS1 that flank the β-globin locus have been implicated in the regulation of an active chromatin domain. Support for this is derived from 3C studies that suggest these regions interact prior to activation of the locus, as well as orthology to the chicken locus.15,17,18,20,33 To determine the effects of the flanking HSs on transcription and to test the model that the ends of the locus must colocalize in a functionally significant way to define the ends of an active domain and mediate gene activation, we generated ES cells with targeted deletions of the HS-62.5 region and 3′HS1 individually and in combination (Figure 1). All mutations were made on the D allele of D/S-2B cells.
Despite loss of both flanking HSs, no alteration in transcription of the adult genes was noted after erythroid differentiation of mutant ES cells (data not shown). To confirm this in vivo, ES cells were used to generate heterozygotic mice, and the ratio of expression from the mutant D allele to the WT S allele in pools of 100 reticulocytes was determined (Figure 2; Table 1). Deletion of each region alone, or in combination, did not result in significant alteration of adult gene expression. Analysis of allele-specific expression in individual reticulocytes showed homogeneous levels of expression that were identical to WT, and no nonexpressing alleles were noted (Table 1). In addition, hematologic analysis of adult mice homozygous for the deletion of both flanking HSs (ΔHS-62.5 and Δ3′HS1) reveals a normal hematocrit, reticulocyte count, and cell size, demonstrating that overall hemoglobin production is not affected (Table 2). Thus, the flanking HSs of the β-globin locus are not needed for full expression of the β-like genes in the context of an intact chromosome in vitro or in vivo.
Table 2.
Red-cell indices of WT and homozygous ΔHS-62.5, Δ3′HS1 mice
| Genotype* | RBC, × 1012/L | Hgb, g/L | HCT, % | MCV, fL | MCH, μg | MCHC, % | Reticulocyte, % |
|---|---|---|---|---|---|---|---|
| WT | 10.11 ± 0.26 | 171.5 ± 3.1 | 50.35 ± 1.32 | 50.00 ± 0.00 | 17.0 ± 0.26 | 34.13 ± 0.63 | 2.54 ± 0.6 |
| ΔHS-62.5, Δ3′HS1 | 9.75 ± 0.72 | 173.3 ± 6.9 | 49.30 ± 3.59 | 50.75 ± 0.96 | 17.83 ± 0.85 | 35.25 ± 1.38 | 2.89 ± 0.44 |
Hgb indicates hemoglobin level; HCT, hematocrit; MCV, mean cell volume; MCH, mean cell hemoglobin; and MCHC, mean cell hemoglobin concentration. Values are expressed ± SD. Automated complete blood counts were obtained from WT and homozygous ΔHS-62.5, Δ3′HS1 mice; red-cell indices are displayed. Reticulocyte counts were determined by flow cytometry after staining with RetiCount (Becton Dickinson, San Jose, CA).
For each genotype, N = 4.
As adult β-globin gene expression is not altered from the mutant ΔHS-62.5, Δ3′HS1 allele in heterozygotes, we tested several hypotheses for the role of these conserved HSs. If, as hypothesized for the chicken locus, the flanking HSs function to prevent the spread of a heterochromatic state into the locus, resulting in gene silencing, we would expect to see the largest decrease in active transcription from ΔHS-62.5, Δ3′HS1 allele, compared with WT, in the most mature nucleated erythroid cells. To directly test this, nucleated RBCs and orthochromatic normoblasts were isolated from fetal liver by flow cytometry and subjected to primary transcript FISH using oligonucleotide probes specific to the adult β-major gene introns.39 This population represents approximately 5% of total fetal liver cells. No significant difference in the number of actively transcribing alleles was observed between the nucleated RBCs and normoblasts of WT and homozygous ΔHS-62.5, Δ3′HS1 cells (Table 3). Thus, the flanking HSs are not necessary to prevent gene silencing associated with terminal erythroid maturation. In addition, while the lack of transcriptional phenotype in heterozygous cells could be explained by pairing with, or complementation by, the WT allele (ie, transvection), the normal transcription pattern observed in these homozygous mutant cells rules out this potential explanation. In contrast to definitive erythropoiesis, primitive erythropoiesis is marked by a high proportion of long-lived circulating nucleated RBCs, many of which show active transcription of the Ey gene.43 To determine if the flanking HSs act to ensure transcription during primitive erythropoiesis, 100 cell pools of circulating cells were isolated from embryos 10.5 days after conception, and expression of the embryonic Ey gene was analyzed by RT-PCR. The ratio of expression from the mutant D allele to the WT S allele was 1.1 ± 0.05, similar to that observed in control animals. Finally, the flanking elements may protect the locus during stress erythropoiesis, when high rates of transcription in the context of altered patterns of cell division may affect the content of histone modifications and gene expression. To address this, heterozygous mutant ΔHS-62.5, Δ3′HS1-D/S, and control WT-D/S mice were analyzed during recovery from a phenylhydrazine-induced hemolytic anemia. Adult β-globin gene expression in peripheral blood and spleen (the major site of erythropoietic response to acute anemic stress) was analyzed by allele-specific quantitative RT-PCR. No significant difference was detected in the ratio of expression of the mutant D allele to the control S allele when compared with that obtained in WT heterozygotic controls (Table 4). Thus, these flanking HSs are not necessary for protecting the locus from silencing during acute erythropoietic stress.
Table 3.
Active β-major transcription in orthochromatic normoblasts and nucleated RBCs as determined by primary transcript RNA FISH
| Alleles analyzed, no. | Percentage expressing alleles | Standard error | |
|---|---|---|---|
| WT | 152 | 86 | 2.8 |
| ΔHS-62.5, Δ3′HS1 | 150 | 87 | 2.7 |
Fetal liver cells of WT and homozygous ΔHS-62.5, Δ3′HS1 mice were isolated, stained, fractionated, and subjected to primary transcript FISH as described.39 Cells staining with TER-119, but not with antibodies to CD117 or ERY-1, representing the most mature nucleated erythroid cells, were analyzed.
Table 4.
Adult β-globin expression in erythroid tissues during stress erythropoiesis
| Genotype* | Spleen | Peripheral blood |
|---|---|---|
| WT | 1.12 ± 0.04 | 1.07 ± 0.04 |
| ΔHS-62.5, Δ3′HS1 | 1.09 ± 0.03 | 0.99 ± 0.03 |
Tissues were isolated during recovery from a phenylhydrazine-induced hemolytic anemia. Values are expressed as the ratio of expression from a mutant D allele to that of a wild-type S allele followed by the standard deviation.
For each genotype, N = 4.
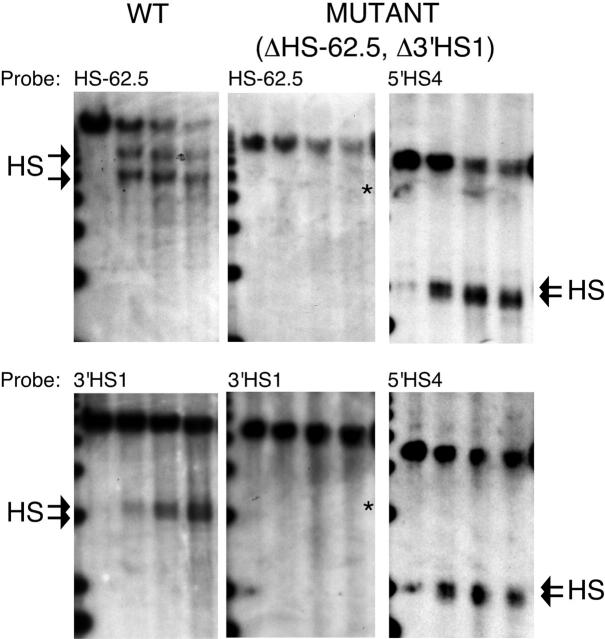
Although we removed the regions in which the flanking HSs form, it is formally possible that sequences outside of our deletions dictate HS formation, or that redundant sequence elements lead to formation of new HSs. To determine if HSs form in the region of our targeted deletions, DNaseI HSs were mapped in erythroid tissue of WT and homozygous ΔHS-62.5, Δ3′HS1 mice. Figure 3 shows that while characteristic HSs form in the WT mice, no HSs form within several kilobases of the HS-62.5 or 3′HS1 deletions in mutant mice. Rehybridization of the ΔHS-62.5, Δ3′HS1 blots demonstrates normal formation of 5′HS4 of the LCR; thus, this lack of flanking HS formation is not a result of inadequate DNaseI digestion conditions.
Figure 3.
Mapping of DNaseI HSs in the HS-62.5 and 3′HS1 regions of WT and ΔHS-62.5, Δ3′HS1 mice. DNaseI series from WT and homozygous mutant mice were digested with EcoRV and hybridized with probes to the 3′ end of each restriction fragment. After hybridization to HS-62.5 or 3′HS1 region probes, blots from mutant mice were stripped and rehybridized to a probe to the 5′HS4 region to demonstrate the quality of the DNase series. The edge of a 1-kb marker lane is observed on the far left. DNaseI digestion increases from left to right. The characteristic doublet of HSs for the HS-62.5 and 3′HS1 regions in WT samples are marked by arrows (left blots). The characteristic doublet of 5′HS4 is marked by arrows (right blots). An asterisk marks the size of expected fragments if a HS formed at the site of the targeted deletions in mutant samples (middle blots). Left blots are from WT mice. Middle and right blots are from homozygous ΔHS-62.5, Δ3′HS1 mice. Probes used are indicated above each panel.
Discussion
Heterozygous D/S-2B and ΔLCR-D/S-1 ES cells facilitate the in vitro analysis of targeted β-globin locus mutations
Previously, in vitro assessment of the transcriptional phenotype of β-globin mutations was dependent on the generation of homozygous mutations and relied on comparison to external reference genes. In addition, the effects of passage through the germ line were not known. The use of ES cells heterozygous for the HbbS (S) and HbbD (D) alleles allows quantitative comparison of transcriptional output from the mutant D allele compared with the wild-type control S allele on a per-cell basis throughout differentiation. Here we show that after in vitro differentiation the transcriptional phenotype of several mutant β-globin alleles parallels those seen in vivo. This is true for mutations that lead to no effect, or reduce expression to 3% to 5% of normal. In addition, using D/S-2B cells, we recently demonstrated that large, or core, deletions of 5′HS2 result in a 20% to 30% reduction in expression whether analyzed in vitro or in vivo.44 Thus, several mutations, displaying a wide range of severity, each have the same transcriptional phenotype in vitro as in vivo. While we cannot rule out subtle differences in the level of transcription or in globin gene switching, we have validated in vitro differentiation as a screening technique. The generation of ES-cell–derived erythroid progenitor lines (ES-EPs) with tremendous proliferative potential45 from these cell lines will further extend the potential for this approach by facilitating the attainment of sufficient cells to allow internally controlled chromatin immunoprecipitations and 3C analysis.
The Hispanic thalassemia phenotype may reflect chromosomal history
The deletion of the LCR from the endogenous mouse or human locus results in decreased expression, but not the profound heterochromatization and transcriptional silencing observed for the Hispanic thalassemia β-globin locus. To determine whether additional upstream sequences deleted in the Hispanic chromosome (UHR) were responsible for this discrepancy, we extended the deletion of the mouse LCR to remove all sequences homologous to those deleted in the Hispanic deletion in the human locus, as well as the conserved HS-62.5 region. Compared with the deletion of the LCR, deletion of this additional 35 kb did not further affect transcription when examined in bulk or in single cells. Thus, the UHR does not contribute to the LCR-independent basal transcription observed previously. Although we cannot formally rule out the possibility that the UHR may possess transcriptional regulatory activity, our results suggest that any such activity is dependent on the LCR or elements therein.
There are several possible explanations as to why excising all homology with the Hispanic deletion does not recapitulate the Hispanic phenotype. Inherent differences between the mouse and human chromosomes, and our not faithfully replicating the breakpoints of the Hispanic deletion are unlikely, as regenerating the Hispanic deletion in a human chromosome 11 transfer system silences transcription, but the nondeleted HSs (eg, 3′HS1) form normally (A. Reik and M.G., unpublished observation, December 1999). While cross-species effects cannot be ruled out, we believe it is more likely that the discrepancy in observed phenotype is a function of the cellular history of the Hispanic chromosome. The studies demonstrating that the Hispanic deletion resulted in heterochromatinization of the β-globin locus were performed after transfer of the patient's chromosomes from Epstein-Barr virus (EBV)–transformed lymphoblasts into MEL cells.4 After transfer from lymphoid to MEL cells the β-globin locus on the patient's wild-type chromosome 11 is in a DNaseI-sensitive conformation, is expressed in an inducible manner, and replicates early in S-phase using the normal replication origin. In marked contrast, the β-globin locus on the Hispanic allele, transferred in parallel, is DNaseI resistant, is transcriptionally silent, and replicates late in S-phase using an alternate origin. Prolonged passage of the Hispanic chromosome in an erythroid environment, and treatment with demethylating agents and histone deacetylase inhibitors have not altered this phenotype (Chul G. Kim, unpublished observations, February 1991). Thus, the Hispanic deletion may leave the β-globin locus uniquely susceptible to epigenetic modifications in a lymphoid environment that leads to irreversible gene silencing. The LCR alone or in combination with upstream sequences may protect the locus from such stable modifications in nonerythroid environments. Notably, β-globin LCR HS formation has been described in lymphoid cell lines,46 which could reflect a state whereby the locus is protected from irreversible silencing. This may have important implications for chromosome and nuclear transfer experiments where epigenetic modifications occurring in one cellular background may profoundly effect gene regulation after transfer, particularly on chromosomes with alterations in elements implicated in regulating gene expression.
Deletion of the flanking HSs has no affect on transcription
This is the first deletional, or loss-of-function, analysis of HSs flanking an endogenous β-globin locus and LCR. A seminal question in globin gene regulation is what prevents silencing of the locus during erythroid differentiation. It has been hypothesized that barrier elements flank the locus, protecting it from the encroachment of heterochromatin.22 Suggestions that HS-62.5 and 3′HS1 regions might be involved in preventing silencing of the locus comes from (1) sequence conservation between mouse and human; (2) conserved organization of these regions between species as disparate as mouse and chicken, in which these elements flank the LCR and genes of the locus and reside near the transition from DNase-sensitive to -resistant chromatin; and (3) studies of the well-characterized chicken β-globin locus. In the chicken, the flanking HSs mark an abrupt change in chromatin structure and presumably protect the locus from the spread of the surrounding heterochromatin while preventing the inadvertent activation of neighboring genes.25,26,29 Prior studies defining the activities of the c5′HS4 and 3′HS elements were performed using ectopic integration sites and demonstrated that while both elements have CTCF-dependent enhancer-blocking activity, only cHS4 has barrier activity, and this is dependent on binding of USF proteins.21,32,47 Notably, the HSs flanking the murine β-globin locus have multiple features similar to those flanking the chicken locus. Both HS-62.5 and 3′HS1 in the mouse bind CTCF, and 3′HS1 has enhancer-blocking activity.15 In addition, consistent with the suggestion that 3′HS1 may function as a boundary element, mouse 3′HS1 contains a USF consensus sequence (CACGTG) similar to c5′HS4 (data not shown). Although the phenotype of deleting the chicken elements in the context of the endogenous locus has not been determined, we observe no effect on the accumulation of stable β-globin transcripts upon deleting either, or both flanking HS regions from the endogenous mouse locus. We have addressed the hypothesis that these flanking HSs are important in preventing silencing during the terminal stages of erythroid maturation, and see no effect on nascent transcripts along the β-major gene in erythroid cells just prior to enucleation. Thus, we find no evidence that the flanking HSs protect the locus from heterochromatinization as hypothesized for the chicken locus. Similarly, we see no effect on transcription in response to acute erythropoietic stress upon deleting these HSs. There are several possibilities for this lack of transcriptional phenotype.
Functional redundancy. We cannot exclude the possibility that additional CTCF- and/or USF-binding sites, or additional elements that could substitute for the deleted regions, exist elsewhere in the locus. Of note, upon deletion of the HS-62.5 and 3′HS1 regions, no new HSs form within several kilobases of the deletion site. In addition, a gross rearrangement of the locus, deletion of the flanking HSs coupled with inversion of the entire region between them (LCR and genes), does not significantly affect transcription when assayed after ES differentiation in vitro (M.A.B., unpublished observation, January 2001). Taken together, these observations argue that if these elements have a critical function in erythroid cells, there is little stringency for their placement or context.
Divergent evolution. The flanking HSs may have a minimal or no role in the regulation of the mammalian β-globin locus. In mammals adult nucleated RBCs have a short half-life, and there is only a brief window of development when primitive, nucleated RBCs are essential. In contrast, in adult avian erythropoiesis red cells remain nucleated. Thus, there may be selective pressure in the chicken to protect the β-globin locus from becoming incorporated into heterochromatin, while in mammals, with enucleated RBCs, this selective pressure has been lost. Consistent with this, analysis using pipmaker48 reveals that although there is homology between mouse and human 3′HS1, it is less than that observed for the LCR HSs and β-like genes.49
The primary role of these elements is in nonerythroid cells. Although we cannot rule this out, homozygous ΔHS-62.5, Δ3′HS1 mice have no apparent phenotype and grow and breed normally. The β-globin locus resides within an array of olfactory receptor genes (ORGs), raising the question of whether these elements are required to insulate the surrounding ORGs from the effects of the LCR.49 This is unlikely, as normally regulated ORGs lie between the LCR and HS-62.5.
The role of the flanking HSs is limited to physiologic states yet to be identified. Regardless of the explanation for the lack of observed phenotype after deleting HS-62.5 and 3′HS1 from the endogenous β-globin locus in vivo, this finding has implications for current models for globin gene regulation.
Implications for models of chromatin structure
The lack of a detectable transcriptional phenotype and the presence of a normal erythroid cell size demonstrate that any potential change in chromatin structure or spatial conformation of the locus associated with deletion of the flanking HSs has little functional relevance to globin gene expression and hemoglobin accumulation. This has implications for both barrier element and chromatin hub (CH) models regarding the function of the flanking HSs. The CH model suggests 3 CTCF-containing elements (the 2 flanking HSs and 5′ end of the LCR) interact to create an erythroid-specific, developmentally stable nuclear compartment designated the core CH.18 It has been hypothesized that this core CH acts as an enzyme, facilitating a proximity-dependent activation of gene expression by the LCR in a structure designated the active CH (ACH).17-20 Here we demonstrate that at the endogenous locus, the absence of 2 of the 3 defined components of the core CH has no effect on transcription. As suggested for human transgenes containing the β-globin locus, the dispensability of the flanking HSs may be due to redundancy,19 but as noted in “Functional redundancy,” this would suggest that the requirements for sequence and positioning are not stringent. Alternatively, the colocalizations detected by 3C may have little functional importance in mammalian erythropoiesis but may rather reflect low frequency or nonspecific juxtaposition. A barrier element model would predict that deletion of the flanking HSs would lead to heterochromatinization and eventual silencing of the locus, yet we find no transcriptional phenotype associated with excision of both flanking HSs. This includes no effect on transcription in the terminal phases of maturation just prior to enucleation, when most genes are silenced, presumably due to incorporation into heterochromatin. While this could be related to functional redundancy, alternatively, the lack of transcriptional phenotype could be explained by these HSs not having a role in chromatin regulation in mammals. Predictions from barrier element models in the chicken may not be relevant to mammals because the loci may not be truly orthologous.15 Differences in the inherited gene duplication products of an ancestral β-globin gene, the organization of the locus with respect to flanking genes with divergent expression patterns, and differences in histone modifications of the surrounding regions suggest chicken and mammalian loci may have evolved in different contexts; thus, there may be a different role, if any, for the flanking HSs in the different phylogenetic classes.
In summary, although the 5 strains of mice described here harboring deletions of the UHR, HS-62.5 region, 3′HS1, and combinations thereof do not define functional roles for these regions, these results do bring into question several prominent models of β-globin locus domain regulation and suggest models where the primary determinants of an open, active mammalian β-globin locus are the genes themselves, and the flanking elements function primarily in a limited, yet undefined context.
Acknowledgments
We thank Oliver Smithies for the gift of BK-4 ES cells, Arthur Sytkowski for the antibody ERY-1, and the Fred Hutchison Cancer Research Center (FHCRC) flow cytometry, image analysis, and transgenic core facilities for assistance. We thank Tomo Sawado and Tony Krumm for review of the manuscript.
Prepublished online as Blood First Edition Paper, April 27, 2006; DOI 10.1182/blood-2006-04-014431.
Supported by National Institutes of Health grant DK44746 (M.G.), American Society of Hematology Scholar Award, and Cooley's Anemia Foundation Award (M.A.B.). M.B. is supported by a Career Award from the Burroughs Wellcome Fund.
M.A.B. designed research, performed research, analyzed data, and wrote the paper; R.B. performed research and analyzed data; T.R. performed research and analyzed data; A.T. performed research and analyzed data; M.B. contributed vital reagents, designed research, and wrote the paper; and M.G. designed research and wrote the paper.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
- 1.Stamatoyannopoulos G, Nienhus AW. Hemoglobin switching. In: Stamatoyannopoulos G, Nienhus AW, Majerus PW, Varmus H, eds. The Molecular Basis of Blood Diseases (2nd ed). Philadelphia, PA: Saunders; 1994: 107-155.
- 2.Curtin P, Pirastu M, Kan YW, Gobert-Jones JA, Stephens AD, Lehmann H. A distant gene deletion affects beta-globin gene function in an atypical gamma delta beta-thalassemia. J Clin Invest. 1985;76: 1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Driscoll MC, Dobkin CS, Alter BP. Gamma delta beta-thalassemia due to a de novo mutation deleting the 5′ beta-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci U S A. 1989;86: 7470-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forrester WC, Epner E, Driscoll MC, et al. A deletion of the human beta-globin locus activation region causes a major alteration in chromatin structure and replication across the entire beta-globin locus. Genes Dev. 1990;4: 1637-1649. [DOI] [PubMed] [Google Scholar]
- 5.Taramelli R, Kioussis D, Vanin E, et al. Gamma delta beta-thalassaemias 1 and 2 are the result of a 100 kbp deletion in the human beta-globin cluster. Nucleic Acids Res. 1986;14: 7017-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aladjem MI, Groudine M, Brody LL, et al. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270: 815-819. [DOI] [PubMed] [Google Scholar]
- 7.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51: 975-985. [DOI] [PubMed] [Google Scholar]
- 8.Ryan TM, Behringer RR, Martin NC, Townes TM, Palmiter RD, Brinster RL. A single erythroid-specific DNase I super-hypersensitive site activates high levels of human beta-globin gene expression in transgenic mice. Genes Dev. 1989;3: 314-323. [DOI] [PubMed] [Google Scholar]
- 9.Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13: 2465-2477. [DOI] [PubMed] [Google Scholar]
- 10.Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205: 73-94. [DOI] [PubMed] [Google Scholar]
- 11.Bender MA, Bulger M, Close J, Groudine M. Beta-globin gene switching and DNase I sensitivity of the endogenous beta-globin locus in mice do not require the locus control region. Mol Cell. 2000;5: 387-393. [DOI] [PubMed] [Google Scholar]
- 12.Epner E, Reik A, Cimbora D, et al. The beta-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse beta-globin locus. Mol Cell. 1998;2: 447-455. [DOI] [PubMed] [Google Scholar]
- 13.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human beta-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18: 5992-6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulger M, Bender MA, van Doorninck JH, et al. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse beta-globin gene clusters. Proc Natl Acad Sci U S A. 2000;97: 14560-14565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulger M, Schubeler D, Bender MA, et al. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol Cell Biol. 2003;23: 5234-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol Cell Biol. 2002;22: 3820-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Laat W, Grosveld F. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 2003;11: 447-459. [DOI] [PubMed] [Google Scholar]
- 18.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35: 190-194. [DOI] [PubMed] [Google Scholar]
- 19.Patrinos GP, de Krom M, de Boer E, et al. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18: 1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10: 1453-1465. [DOI] [PubMed] [Google Scholar]
- 21.Bell AC, Felsenfeld G. Stopped at the border: boundaries and insulators. Curr Opin Genet Dev. 1999;9: 191-198. [DOI] [PubMed] [Google Scholar]
- 22.Burgess-Beusse B, Farrell C, Gaszner M, et al. The insulation of genes from external enhancers and silencing chromatin. Proc Natl Acad Sci U S A. 2002;99: 16433-16437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111: 151-154. [DOI] [PubMed] [Google Scholar]
- 24.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16: 271-288. [DOI] [PubMed] [Google Scholar]
- 25.Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken beta-globin chromosomal domain. EMBO J. 1994;13: 1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20: 2224-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reitman M, Felsenfeld G. Developmental regulation of topoisomerase II sites and DNase I-hypersensitive sites in the chicken beta-globin locus. Mol Cell Biol. 1990;10: 2774-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitoh N, Bell AC, Recillas-Targa F, et al. Structural and functional conservation at the boundaries of the chicken beta-globin domain. EMBO J. 2000;19: 2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993; 74: 505-514. [DOI] [PubMed] [Google Scholar]
- 30.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12: 2852-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99: 6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16: 453-463. [DOI] [PubMed] [Google Scholar]
- 33.Bulger M, Sawado T, Schubeler D, Groudine M. ChIPs of the beta-globin locus: unraveling gene regulation within an active domain. Curr Opin Genet Dev. 2002;12: 170-177. [DOI] [PubMed] [Google Scholar]
- 34.Bender MA, Reik A, Close J, et al. Description and targeted deletion of 5′ hypersensitive site 5 and 6 of the mouse beta-globin locus control region. Blood. 1998;92: 4394-4403. [PubMed] [Google Scholar]
- 35.Fiering S, Epner E, Robinson K, et al. Targeted deletion of 5′HS2 of the murine beta-globin LCR reveals that it is not essential for proper regulation of the beta-globin locus. Genes Dev. 1995;9: 2203-2213. [DOI] [PubMed] [Google Scholar]
- 36.Schubeler D, Groudine M, Bender MA. The murine beta-globin locus control region regulates the rate of transcription but not the hyperacetylation of histones at the active genes. Proc Natl Acad Sci U S A. 2001;98: 11432-11437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kina T, Ikuta K, Takayama E, et al. The monoclonal antibody TER-119 recognizes a molecule associated with glycophorin A and specifically marks the late stages of murine erythroid lineage. Br J Haematol. 2000;109: 280-287. [DOI] [PubMed] [Google Scholar]
- 38.Bacon ER, Sytkowski AJ. Identification and characterization of a differentiation-specific antigen on normal and malignant murine erythroid cells. Blood. 1987;69: 103-108. [PubMed] [Google Scholar]
- 39.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. The murine beta-globin LCR positions the globin locus at engaged transcription factories during erythroid maturation. Genes Dev. 2006;20: 1447-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326: 292-295. [DOI] [PubMed] [Google Scholar]
- 41.Abbondanzo SJ, Gadi I, Stewart CL. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225: 803-823. [DOI] [PubMed] [Google Scholar]
- 42.Nakano T, Era T, Takahashi T, Kodama H, Honjo T. Development of erythroid cells from mouse embryonic stem cells in culture: potential use for erythroid transcription factor study. Leukemia. 1997;11: 496-500. [PubMed] [Google Scholar]
- 43.Trimborn T, Gribnau J, Grosveld F, Fraser P. Mechanisms of developmental control of transcription in the murine alpha- and beta-globin loci. Genes Dev. 1999;13: 112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu X, Bulger M, Bender MA, Fields J, Groudine M, Fiering S. Deletion of the core region of 5′ HS2 of the mouse beta-globin locus control region reveals a distinct effect in comparison with human beta-globin transgenes. Blood. 2006;107: 821-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carotta S, Pilat S, Mairhofer A, et al. Directed differentiation and mass cultivation of pure erythroid progenitors from mouse embryonic stem cells. Blood. 2004;104: 1873-1880. [DOI] [PubMed] [Google Scholar]
- 46.Dhar V, Nandi A, Schildkraut CL, Skoultchi AI. Erythroid-specific nuclease-hypersensitive sites flanking the human beta-globin domain. Mol Cell Biol. 1990;10: 4324-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell AC, West AG, Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98: 387-396. [DOI] [PubMed] [Google Scholar]
- 48.Schwartz S, Zhang Z, Frazer KA, et al. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res. 2000;10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bulger M, van Doorninck JH, Saitoh N, et al. Conservation of sequence and structure flanking the mouse and human beta-globin loci: the beta-globin genes are embedded within an array of odorant receptor genes [published erratum appears in Proc Natl Acad Sci U S A. 1999;96:8307]. Proc Natl Acad Sci U S A. 1999;96: 5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]