Abstract
The biophysical properties of oligodeoxyribonucleotides (ODNs) selectively modified with conformationally ‘locked’ bicyclo[3.1.0]hexane pseudosugars (Maier,M.A., Choi,Y., Gaus,H., Barchi,J.J. Jr, Marquez,V.E., Manoharan,M. (2004) Synthesis and characterization of oligonucleotides containing conformationally constrained bicyclo[3.1.0]hexane pseudosugar analogs Nucleic Acids Res., 32, 3642–3650) have been studied by various techniques. Six separate synthetic ODNs based on the Dickerson Drew dodecamer sequence (CGCGAAT*T*CGCG) were examined where each one (or both) of the thymidines (T*) were substituted with a bicyclic pseudosugar locked in either a North (2′-exo) or South (3′-exo) ring pucker. Circular dichroism spectroscopy, differential scanning calorimetry and 1H NMR spectroscopy were used to examine the duplex stability and conformational properties of the ODNs. Replacement of one or both thymidines with North-locked sugars (RNA-like) into the dodecamer did not greatly affect duplex formation or melt temperatures but distinct differences in thermodynamic parameters were observed. In contrast, incorporation of South-locked sugar derivatives that were predicted to stabilize this standard B-DNA, had the unexpected effect of causing a conformational equilibrium between different duplex forms at specific strand and salt concentrations. Our data and those of others suggest that although DNA can tolerate modifications with RNA-like (North) nucleotides, a more complicated spectrum of changes emerges with modifications restricted to South (DNA-like) puckers.
INTRODUCTION
The design and construction of novel DNA oligomers with tunable structural and hybridization properties have recently been the topics of intense research interest. Nucleotide building blocks with synthetic modification in either the furanose ring (1) or the nucleobase (2) have been used for the design of novel oligonucleotides (ONs) and oligodeoxyribonucleotides (ODNs). Many of these constructs have been found to be useful biochemical and pharmacological tools in antisense and antigene strategies. Basic research in this area has produced an increasing number of synthetically accessible nucleoside analogs and these building blocks will continue to lead to new oligomers with exciting biophysical and therapeutic features.
There are several structural features of ODNs that combine to form the characteristic global folds of DNA and RNA, including (a) base pair hydrogen bonding and stacking, (b) backbone torsion angle preferences and (c) the conformation of the ribo-or deoxyribofuranose ring. Regarding point (c), most nucleoside monomers exist in a dynamic equilibrium between two furanose puckering forms: The characteristic 3′-endo/2′-exo or ‘North’ (N) and 2′-endo/3′-exo or ‘South’ (S) ring conformations. When subjected to the constraints of an ODN duplex, the sugar rings adopt either of the two forms, B-DNA oligomers favoring S puckers while A-DNA and RNA favor N puckers. Furthermore, the crystal structures of enzymes involved in cellular nucleoside metabolism pathways in complex with their substrates reveal a distinct preference for a particular furanose pucker that is protein dependent. (3,4) Based on these findings, our laboratory (5–7) and others (8–11) have focused on the synthesis of novel nucleoside analogs that are biased toward one specific ring pucker as potential therapeutic agents. The concept of ‘locking’(12) the furanose ring pucker by various synthetic manipulations began about 13 years ago and gave rise to a range of different analogs (7,10,13,14). Notable examples in the field derive from the bridged nucleic acids (BNAs) concept (8), where analogs contain an added ring that bridges two atoms of the furanose. One of the most widely studied of these BNAs is one that contains a methylene bridge between the 2′ oxygen and the 4′ carbon that confers conformational restriction. These derivatives and ONs containing these monomers have been coined ‘locked nucleic acids’ (LNAs) (15) since the conformation of the bicyclic furanose system is ‘locked’ in the N quadrant of the pseudorotational cycle (16) (Figure 1). Extensive work has been carried out proving the added thermodynamic stability of LNA–RNA duplexes (17,18) as well as the triplex-forming ability of these constructs (19).
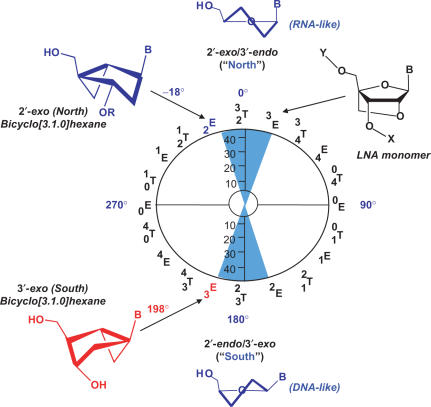
Figure 1.
Pseudorotational cycle for nucleoside furanose ring puckers. Ring puckers for standard nucleosides are typically either in the ‘Northern’ or ‘Southern’ hemispheres (illustrated by blue cones). Depictions of the N and S conventions along with the structure of an LNA monomer are shown. The pseudorotational positions for the N-MCT and S-MCT analogs are shown on the left half of the wheel.
In 1993, our laboratory published the first synthesis of a ‘locked’ (dideoxy) nucleoside analog based on the carbocyclic bicyclo[3.1.0]hexane (methanocarba (MC) nucleoside) systems (7,20) shown in Figure 1. An advantage of this system is that both N- and S-locked (21) platforms can be prepared by shifting the position of the fused cyclopropane ring (Figure 1). A range of nucleoside monomers based on this template have been synthesized and used to query the preferred substrate ring pucker of specific enzymes (3,22,23). In addition, a single-stranded DNA containing several N-locked MC thymidine (N-MCT) units has also been shown to hybridize efficiently with a complementary RNA strand (20).
Based on the hybridization efficiency of many N-locked ONs with RNA, there is great therapeutic potential for synthetic ONs in antisense and antigene technologies where regulation of specific gene translation may slow or halt the progression of disease. Recent work has shown that ONs containing N-locked LNA units can substitute for small interfering RNAs (siRNAs) in the process of post-transcriptional gene silencing (24,25) adding to the usefulness of these analogs. In addition, binding of ODNs to proteins such as transcription factors often leads to structural adjustments in the helical parameters that may induce bends or kinks in the duplex. During bending of B-DNA, there is an obligatory adjustment of the sugar pucker from S toward N to relieve torsional and angular strain (26–28). The ability to pre-organize ONs/ODNs to specific global folds that mimic these adjustments may lead to surrogates that could perturb protein production at the level of transcription. Recent data from our lab suggests that the addition of bicyclo[3.1.0]hexane nucleoside analogs to a standard B-DNA duplex can cause bending toward the minor groove (29).
In order to gain a better understanding of how structural variations in the sugar moiety can contribute to both the local and global structural fold of ODNs, both S- and N-MCT analogs were inserted into a structurally well-characterized self-complementary B-DNA sequence (Dickerson Drew dodecamer, DD) known to contain only S-disposed sugar rings. The conformational and thermodynamic changes resulting from inserting either a single modified thymidine or two modified thymidine residues in positions 7 and/or 8 of the CGCGAAT*T*CGCG sequence is the subject of this work.
MATERIALS AND METHODS
Synthesis of modified DNA
Thymidine monomers containing the bicyclo[3.1.0]hexane system were synthesized as described earlier (30,31) and incorporated into the DD by procedures that required modification of the coupling for the addition of the N-monomers due to some unexpected side reactions during the oligomer synthesis (32). Samples were purified by HPLC as described in (32) and further purified by EtOH precipitation. The precipitant was resuspended in water and lyophilized three times, then taken up in 10 mM phosphate buffer containing 0.1 mM EDTA and varying concentrations of Na+ (100, 500, or 1000 mM) at pH 7.0. DNA concentrations were determined by measuring the UV absorbance at 260 nm and fitting to Beer's Law using ε = 110 700 l/mol · cm (Schepartz Lab Biopolymer Calculator at http://paris.chem.yale.edu/extinct.html). The wild-type DD sequence was synthesized by the Laboratory of Molecular Technology Oligonucleotide Synthesis core facility at the NCI Frederick (Dr Leo Lee, Head, Frederick, Maryland).
Differential scanning calorimetry
Differential Scanning Calorimetry (DSC) data were acquired on a VP-DSC differential scanning calorimeter (Microcal, Inc., Northampton, MA). In a typical experiment, a 25–40 μM DNA (0.51 ml cell volume) solution was scanned from 10 to 90°C at a rate of 1°C/min. A 16-s filter period was used for all data acquisitions. A total of eight alternating scans (four up- and four down-scans) were recorded per experiment. A buffer versus buffer scan was obtained with 10 mM phosphate buffer containing 0.1 mM EDTA and NaCl (0.1, 0.5 or 1.0 M) placed in both of the calorimetric cells. This reference scan was subtracted from each corresponding experimental scan. The resulting thermogram was plotted as heat capacity (Cp) versus temperature. From the pre- and post-transition baselines, a cubic baseline was extrapolated under each melting transition. Values for Tm and ΔHcal were obtained after baseline subtraction, and by fitting the thermograms to a two-state model, according to the manufacturer's protocols (OriginLab, Northampton, MA). ΔCp was obtained as the difference between the pre- and post-transition baselines, prior to baseline subtraction, using a step at peak with no baseline subtraction. The Gibbs free energy of binding at 25°C (ΔGb25) was derived from ΔCp, ΔHcal, and Tm values using the following equations (33)
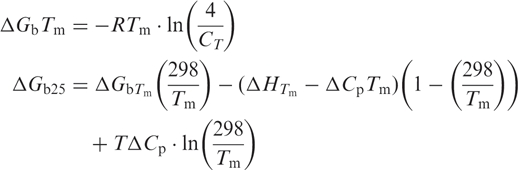 |
where CT is total strand concentration (29–40 μM) determined as described above and R is 1.987 cal/mol · K. Tm and T were in absolute temperature values (K). ΔScal was determined by taking the difference between ΔGb Tm and ΔHcal at the Tm.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a JASCO J-715 spectrometer using a quartz cell with a 1-mm optical path length. Data were recorded in the range of 190–350 nm at 1.0 nm bandwidth, 100 mdeg of sensitivity, 0.1 nm resolution, 1 s response time, a speed of 50 nm/min and an average of two scans. A Haake D1 constant temperature bath interfaced to a Jasco controller was used to adjust the sample temperature between 5 and 65°C. Sample concentrations were 90–135 μM DNA in a total volume of 250 μl of phosphate buffer (pH 7.0), with varying concentrations of Na+.
NMR spectroscopy
1H NMR spectra were acquired on a Varian Inova spectrometer operating at 500 MHz using an inverse triple resonance Nalorac (IDTG) variable temperature probe with z-gradients. DNA concentrations for NMR studies were 2 mM, 0.2 mM or 0.04 mM for the modified DNA species and 0.9 mM for the wild-type dodecamer DNA in pH 7.0 phosphate buffer containing 100 mM Na+ and 10% D2O. One-dimensional spectra were acquired with 128 transients using the ‘water’ macro from the Varian RNAPack suite of pulse sequences. Suppression of the H2O signal was performed with WATERGATE using a sinc shape for the 90° selective pulse on water. Spectra were acquired with a 12 000 Hz sweep width, an acquisition time of 2.7 s and a preacquisition delay of 1 s. Other parameters (power levels, offsets and pulse widths) were optimized for each individual sample. Spectra were acquired at intervals of 5°C from 5 to 65°C and were referenced to external 2,2-Dimethyl-2-silapentane-5-sulfonate sodium salt (DSS). Assignments of the imino protons were obtained by collecting standard COSY, TOCSY and NOESY spectra for each oligomer and using established ON sequential assignment techniques. The assignments were facilitated by comparison with spectra obtained for the wild-type DD. Raw NMR data were processed on a Sun Ultra10 440 MHz offline workstation using NMRPipe (34).
RESULTS
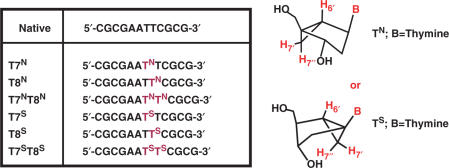
Six modified DD ODNs (Figure 2) were synthesized and studied by a variety of biophysical techniques. The self-complementary single-stranded ODNs contained either an N- or S-MCT nucleotide in either a single substitution at position T7 or T8, or a double substitution at both positions T7 and T8. N and S superscripts indicate the puckering of each thymidine substitution. After annealing, the resulting double-stranded helix would have an N- or S-MCT at positions T7/T19 (designated T7N or T7S, Figure 2), T8/T20 (designated T8N or T8S) and T7,8/T19,20 (designated T7NT8N or T7ST8S) paired with normal 2′-deoxyadenosines. Our assumptions were the following: The presence of one or two residues restricted in the ‘South’ conformation in an all ‘South’ DNA should, in effect, stabilize the duplex whereas perturbations to the local and/or global duplex structure may result from inclusion of nucleotides locked in the ‘North’ conformation. Various experiments were conducted to address the following questions: (1) Do the S-modifications help stabilize the duplex? (2) Do the N-modifications prevent formation of a stable duplex? (3) If a duplex is formed, do these perturbations remain localized to the site of modification? The following data supplied some intriguing answers to these questions.
Figure 2.
Structures used in this study. The bicyclo [3.1.0] hexane nucleosides that were incorporated into the DNA are shown along with an illustration and nomenclature of the extra protons of these systems (in red).
Circular dichroism
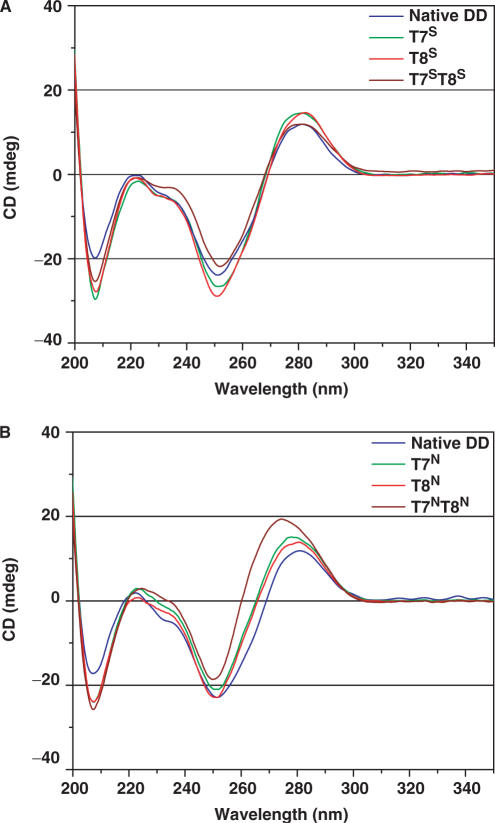
CD was used to qualitatively assess the conformations populated by the ODNs in Figure 2. To fully explore the duplex behavior of the analogs, we performed CD measurements at several temperatures and salt concentrations. The CD spectra of all ODNs exhibited characteristics typical of B-type duplex DNA with some minor differences (Figure 3). Typical CD signatures for B-DNA include a positive long wave (275–280 nm) band and negative shorter wave (∼250 nm) band of almost equal magnitude often followed by a second zero cross-over point and a small negative band at ∼210 nm (35). In CD spectra of A-DNA and RNA, the long-wave positive band shifts to lower wavelength and has significantly higher magnitude than the short-wave negative band; a concomitant increase in the negative band at 210 nm is also observed. The native DD, T7S, T8S and T7ST8S all have typical B-DNA signatures at 25°C (Figure 3A) with the S-modified ODNs displaying higher magnitude negative bands at 210 nm. The significance of this band with respect to the CD signatures of RNA and RNA-like ODNs (A-DNA) has been under-appreciated according to some (36) but in our case it is difficult to evaluate since all modified ODNs showed an increase in this band. Overall, except for the negative band at 210 nm, the CD features of all the S-modified ODNs showed little change from the native DD. On the other hand, the spectra of each of the N-modified duplexes tend slightly toward a more A-like signature as evidenced by the increased magnitude and hypsochromic shift of the first positive band as well as an increase in magnitude of the negative band at 210 nm. These features were shown to increase in the order T7N ∼ T8N < T7NT8N, with the double mutant showing the most significant change in both positive and negative band magnitudes as well as a more enhanced hypsochromic shift of the positive long-wavelength band (Figure 3B). The overall shape, cross-over points, wavelength at maximum ellipticity change and band maxima are similar to those shown previously for the DD (37). Temperature-dependent CD spectra also revealed that at temperatures above the Tm, the spectra acquired features that were reminiscent of other single-stranded ODNs (data not shown).
Figure 3.
CD spectra of S-MCT- (A) and N-MCT- (B) modified DNAs at 25°C, at 100 mM Na+, pH 7.0 (phosphate buffer).
Differential scanning calorimetry
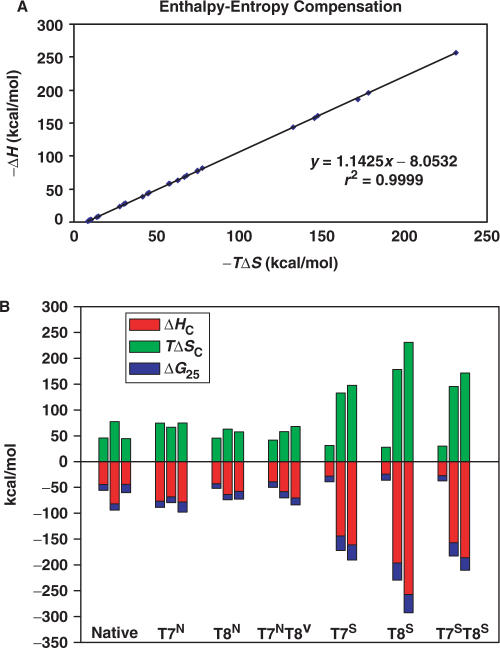
DSC was used to understand the thermodynamic consequences resulting from incorporation of the bicyclo[3.1.0]hexane-modified bases into the DD. DSC provides an accurate measurement of the thermodynamic quantities describing the melting or dissociation of DNA duplexes such as the change in heat capacity (ΔCp) that accompanies the transition, the melting transition temperature (Tm), and the calorimetric enthalpy of dissociation (ΔHcal). From these values, the Gibbs free energy of binding (ΔGb25) and the entropy of dissociation (ΔScal) can be derived. Previous studies of the native DD have shown that increased concentrations of Na+ ions lead to the stabilization of a duplex structure, while at ≤10 mM Na+ and DNA concentrations ≤50 μM, a biphasic transition in the melt curves was obtained with the tentative assignment of the first intermediate as a hairpin loop structure (37). Therefore, the effect of Na+ concentration was also examined. A summary of the thermodynamic parameters calculated from the calorimetric data is listed in Table 1. An enthalpy–entropy compensation plot (−ΔH versus –TΔS) showed a linear correlation (r2 = 0.99) and an observed slope of 1.14 (Figure 4A). This compensation demonstrated that the enthalpy and entropy terms were approximately equal in magnitude for all the ODN species, but that the enthalpies were favorable enough to offset the unfavorable dissociation entropies, resulting in moderately strong free energies of duplex formation for all ODN species (Table 1, Figure 4B).
Table 1.
Thermodynamic data for the ODNs in Figure 2.
| [Na+] (mM) | Tm (°C) | ΔCp (kcal/mol/°C) | −ΔHc (kcal/mol) | −ΔSc (cal/mol/K) | −ΔGb25 (kcal/mol) | |
|---|---|---|---|---|---|---|
| Native | 100 | 65.30 | 0.203 | 44.1 | 153.23 | 11.56 |
| 500 | 71.37 | 1.780 | 81.8 | 260.41 | 12.01 | |
| 1000 | 73.65 | –0.810 | 44.0 | 149.81 | 15.90 | |
| T7N | 100 | 62.86 | 1.430 | 76.3 | 250.34 | 12.33 |
| 500 | 68.14 | 1.540 | 68.3 | 223.48 | 11.18 | |
| 1000 | 69.80 | –0.820 | 78.1 | 251.10 | 19.65 | |
| T8N | 100 | 60.72 | 1.410 | 42.9 | 151.77 | 8.71 |
| 500 | 66.96 | 1.780 | 63.6 | 210.59 | 10.04 | |
| 1000 | 67.86 | –0.180 | 57.8 | 193.08 | 14.78 | |
| T7NT8N | 100 | 60.69 | 0.150 | 38.9 | 139.90 | 10.82 |
| 500 | 67.10 | 0.880 | 58.1 | 194.34 | 11.80 | |
| 1000 | 68.15 | 0.803 | 70.2 | 229.28 | 13.59 | |
| T7S | 100 | 35.65 | 0.187 | 8.1 | 49.04 | 7.06 |
| 71.56 | –0.064 | 28.3 | 104.99 | 10.85 | ||
| 500 | 34.07 | 0.115 | 3.9 | 35.59 | 6.91 | |
| 67.59 | –1.055 | 144.0 | 445.66 | 27.75 | ||
| 1000 | 32.38 | 0.280 | 2.6 | 31.28 | 6.85 | |
| 67.80 | –0.867 | 161.0 | 495.28 | 29.47 | ||
| T8S | 100 | 34.91 | 0.702 | 2.1 | 29.81 | 6.77 |
| 67.08 | –0.712 | 24.1 | 93.73 | 11.73 | ||
| 500 | 34.43 | –0.385 | 3.3 | 33.53 | 6.97 | |
| 67.61 | –0.807 | 196.0 | 598.30 | 33.58 | ||
| 1000 | 32.81 | –0.459 | 2.4 | 30.84 | 6.92 | |
| 68.45 | 1.440 | 257.0 | 775.53 | 35.35 | ||
| T7ST8S | 100 | 30.88 | 0.923 | 7.2 | 46.49 | 6.84 |
| 68.85 | –0.027 | 26.9 | 101.35 | 10.28 | ||
| 500 | 29.94 | –0.313 | 1.8 | 28.84 | 6.85 | |
| 64.25 | –0.202 | 157.0 | 488.39 | 25.57 | ||
| 1000 | 28.71 | –0.856 | 2.3 | 30.38 | 6.86 | |
| 63.46 | 1.649 | 186.0 | 575.68 | 24.30 |
Figure 4.
(A) Enthalpy–entropy compensation plot for the ODNs in Figure 2. (B) Bar plot of ΔHC, TΔSc and ΔG25. Each bar shows the comparison of the three thermodynamic parameters. The three bars in each cluster represent (from left to right) the data for the labeled ODN in 0.1, 0.5 and 1.0 M Na+
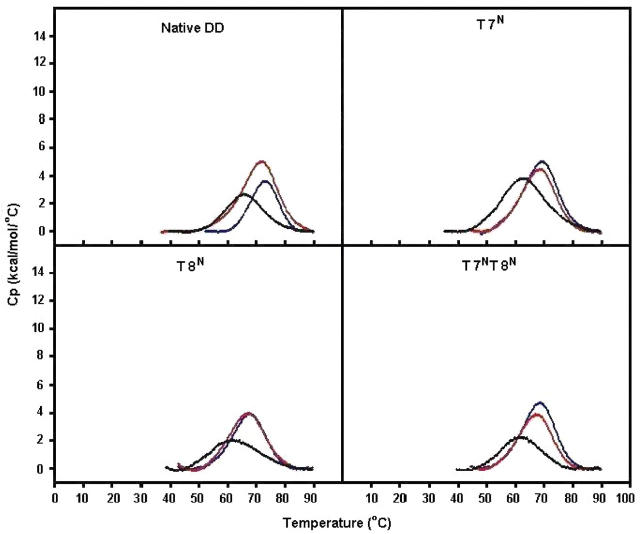
DSC of N-modified DDs
The thermodynamic properties of the native DD duplex in 100 mM Na+ were consistent with those obtained earlier (37). Incorporation of either one or two N-MCT units slightly destabilized the duplex with Tms of 60.7 to 62.9°C, while the Tm of the native DNA measured 65.3°C (Table 1, Figure 5). The T7N DNA was the most enthalpically stable species in that series with progressively lower negative values of ΔHcal for the T8N and T7NT8N molecule, respectively. In the case of T7N, although its enthalpy of duplex formation was almost 2-fold greater than that of the native DD species, the accompanying entropy change was less favorable, thereby yielding a comparable free energy change (ΔGb25(native) = −11.56 versus ΔGb25(T7N) = −12.33). Overall, the thermodynamic data were relatively similar for the native and N-modified species. The data indicated that stable duplexes were formed upon incorporation of the N-modified bases and that only minor disruptions in conformation were produced by substituting the central residues with RNA-like furanose surrogates.
Figure 5.
DSC data for the native DD, T7N, T8N and T7NT8N ODNs at 0.1 M Na+ (black), 0.5 M Na+ (red), 1.0 M Na+ (blue), pH 7.0 (phosphate buffer). Exact conditions are given in the experimental section.
As expected, at 0.5 M Na+ an increase in the Tm value of all N-modified DNA species was observed. There was also an associated increase in the calorimetric enthalpy and the change in heat capacity, leading to a more stabilized DNA species in 0.5 M Na+ compared to 0.1 M Na+. At 0.5 M Na+, the native DNA was slightly more stable (ΔGb25 of –12.01 kcal) than the modified DNA species (ΔGb25 = –10.04 to –11.80). Increasing the salt concentration to 1.0 M resulted in a concomitant increase in the Tm values for the DNA species (Figure 5A–D). However, the high salt concentration led to a paradoxical decrease in the calorimetric enthalpy for the native DD back near the value obtained at 0.1 M Na+ with a compensating increase in entropic stabilization. In addition, the change in heat capacity also decreased for all species, particularly for the native, T7N and T8N species (Figure 5A–C). Interestingly, the T7N species seemed to be the most stable ODN overall and at all salt concentrations. All DSC data were fit to a single transition (two-state model) and reasonably low errors to the fit were obtained. Data obtained earlier (38) had postulated the presence of a hairpin structure at low salt and DNA concentrations at a Tm higher than that of the duplex. Although we did not examine the ODNs under these conditions, both the calorimetric and NMR data (see below) obtained here did not indicate the presence of any hairpin structures.
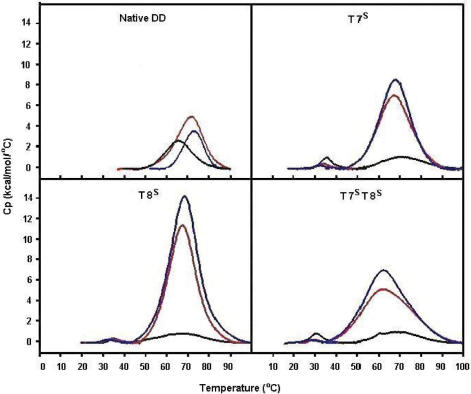
DSC of S-modified ODNs
As discussed above, there was nothing extraordinary in the CD data of the S-modified DDs and it was thought these analogs would stabilize the canonical, B-DNA duplex described previously for the DD. The most striking difference was in the melting behavior (Tm) of each duplex. Biphasic-type transitions were observed for all S-modified ODNs in standard duplex milieus (0.1 M Na+) where low melting species at temperatures between 30 and 36°C were observed followed by higher temperature transitions (Figure 6). In addition, these lower melting transitions were small with a decreased enthalpy of duplex dissociation on the order of 2–8 kcal/mol, almost 6-fold lower than that of the native DD. Interestingly, whereas biophysical measurements at varying strand concentrations of the N-modified ODN species did not alter the population of structures present (data not shown), the presence and stability of the low-melting species in the S-modified ODN species was concentration dependent. At concentrations that are typical for DSC (30–50 μM) or UV (1–10 μM) measurements, multiple species were observed. At higher concentrations that are standard in NMR spectroscopy of ODN species (1–2 mM), only the higher melting duplex form was observed (vide infra).
Figure 6.
DSC data for the native DD, T7S, T8S and T7ST8S ODNs at 0.1 M Na+ (black), 0.5 M Na+ (red), 1.0 M Na+ (blue), pH 7.0 (phosphate buffer). Exact conditions are given in the experimental section.
For all S-modified ODNs, the second melting transitions all had higher Tm values than that of the native DD by at least 1.7°C (Table 1, Figure 6). These data suggested that inclusion of the S-modified base pairs into the DD stabilized the ‘major’ duplex conformation formed, although additional species were present at equilibrium with incorporation of the S-MCT nucleotides into the strand. At higher salt concentrations (0.5 and 1.0 M NaCl) the higher melting transitions were enhanced dramatically, evidently at the expense of the lower temperature transitions which virtually disappeared. Hence, the calorimetric enthalpy increased 4–10-fold by increasing the salt concentration from 0.1 to 1.0 M Na+ for each S-modified species and as much as 4-fold over the ΔHcal of the native DD. In direct contrast to the cooperative unfolding events of the N-modified ODN species, the melting transitions were broad for the S-modified ODN species at the 0.5 and 1.0 M Na+. This suggested that multiple unfolding intermediates were present. Given such broad transitions at the 0.5 and 1.0 M Na+, the derived melting temperatures most probably reflected an average of multiple similar species melting at or near the same Tm value. Therefore, the Tm values for the S-modified ODNs appeared to remain fairly constant with increasing salt, with the exception of the T7ST8S species which showed a 4–5°C drop in Tm from 0.1 to 1.0 M Na+ concentrations.
As with the N-modified species, the duplex formation/dissociation of the South counterparts revealed an enthalpy–entropy compensation (Figure 4B). However, in the case of the T7ST8S species (in 0.5 and 1.0 M Na+; Table 1 and Figure 4B) greater entropy compensation resulted in a less favored ΔGb25 of duplex formation as compared to the T7S or the T8S species. This suggested that the incorporation of the modified carbocyclic sugars in the T7ST8S may have caused perturbation of structure and prevented the proper annealing of the duplex. This finding was supported by NMR results where the T7ST8S imino protons showed larger chemical shift changes as compared to the other modified ODN species.
Nuclear magnetic resonance (NMR)
1H NMR: global melting of DNA
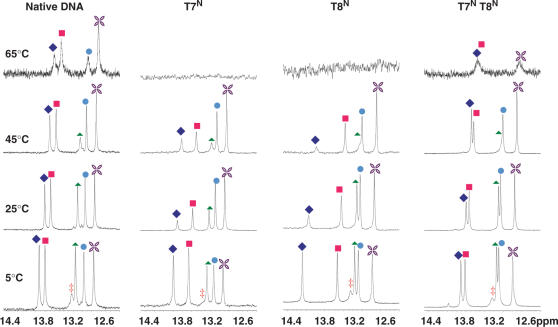
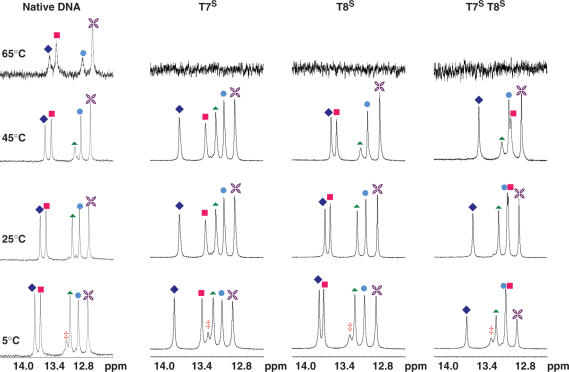
Proton NMR spectroscopy at variable temperatures was used to assess structural features and duplex stability of the modified ODN species. Assignments of imino protons and the entire proton network of all ODNs were determined based on standard 2D NMR data ((39) and Barchi and Maderia, unpublished results). At 5°C, each ODN from Figure 2 revealed all six imino protons (from base pairs G2, G4, T7, T8, G10 and G12) of those anticipated for a duplex formed from this self-complementary sequence (38,40), while warming to 25°C caused the imino proton assigned to G12 to fade, perhaps due to fraying at the end of the helix. All imino protons resonated between 12 and 14 ppm, chemical shifts that are indicative of duplex formation. Imino protons resonating in the 10–11 ppm range, which would serve as evidence for hairpin structures (40), were not observed at any temperature. Figures 7 and 8 show the temperature profiles of the imino resonances for the N- and S-modified ODNs, respectively. In general, trends for the disappearance of each proton followed those observed in previous studies of the DD (33). For example, the protons of base pairs closer to the ends of the duplex were only observed at low temperatures whereas those toward the center of the duplex persisted until close to the melting temperatures.
Figure 7.
NMR spectra of the imino 1H region of the N-MCT modified oligos at various temperatures. The assignments are as follows: filled diamond T8, filled square T7, filled triangle G2, circle G4, star G10 and double dagger, G12.
Figure 8.
NMR spectra of the imino 1H region of the S-MCT modified oligos at various temperatures. The assignments are as follows: filled diamond T8, filled square T7, filled triangle G2, filled circle G4, star G10 and double dagger, G12.
1H chemical shifts: effect of temperature
Differences in 1H NMR chemical shifts are exquisite indicators of structural/conformational changes. By monitoring the changes in the chemical shifts (Δδ) of the protons as a function of temperature, information can be obtained about changes in the chemical environment upon dissociation of the duplex. With increasing temperature, the imino protons primarily shifted upfield and broadened (Figures 7 and 8). In general, the T7 and T8 imino protons showed a larger change in chemical shift from 5 to 65°C (Δδ ≤ 0.4 ppm for N-ODNs; ≤0.8 for S-ODNs) compared to the G2, G4 and G10 imino protons (≤0.15 ppm, for N-ODNs and ≤0.4 ppm for S-ODNs) which was consistent with changes observed in the native DD. Non-exchangeable protons, such as the H6 aromatic proton of thymidine and the T7 and T8 methyl protons, which are near the center of the duplex at the site of modification exhibited less than 0.08 ppm changes in chemical shifts as a function of temperature from 5 to 65°C in both native and all six T-modified DNA species. This data suggests that the global unfolding of the DNA was not greatly affected by modification of the sugars of the thymidine residues.
1H chemical shifts: comparisons to native DD
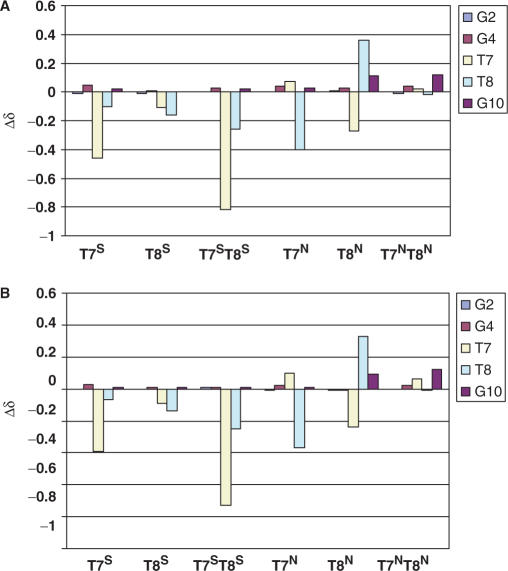
At 5°C, the chemical shift of each imino proton of the native DNA was compared to those of the imino protons of each T-modified DNA species (Figure 9). Upon incorporation of an N-modified thymidine at position 7 (T7N), there was a difference of −0.40 ppm in the chemical shift of the T8 imino proton, but little differences (Δδ < 0.1 ppm) were observed for other imino protons (Figure 9A). For the T8N DNA, there was a Δδ of −0.23 ppm in the chemical shift of T7 compared to the native imino proton as well as a +0.34 ppm difference for the δ of the T8 imino proton. There were only slight changes in the chemical shifts of the guanosine imino protons (less than 0.1 ppm) in the T8N DNA. However, the double TN modification (T7NT8N ODN) showed changes in chemical shifts for the T7 and T8 imino protons that are similar to the native DD, with the largest difference in chemical shift assigned to the G10 imino proton at Δδ = 0.17 ppm. These same trends were also observed at 25°C (Figure 9B).
Figure 9.
Changes in the chemical shift of the imino protons between the N-MCT- and S-MCT- modified ODN species and the native DD at 5°C (A) and at 25°C (B).
A contrasting picture emerged from the S-modified oligos. For the T7S structure, at 5°C, the T7 imino proton experienced a −0.42 ppm shift while for the T8 imino proton the shift was only 0.08 ppm (Figure 9A). For T8S, very little change was observed for most imino protons in contrast to the T8N ODN which showed both upfield and downfield changes for the T7 and T8 imino protons, respectively. The most startling change was seen for the T7ST8S compared to its N counterpart. Very little change was observed in the Δδ for T7NT8N ODN while the T7 imino proton of T7ST8S shifted by greater than −0.8 ppm and T8 shifted by −0.23 ppm relative to the native DD. The changes were consistent from 5 to 25°C (Figure 9A and B). Based on the DSC data described above, this result is perhaps not surprising. There seem to be extensive local structural rearrangements taking place due to the presence of the modified nucleotides. These may be due to accommodations made for the cyclopropane ring of the modified carbocyclic sugar in the unusual 1′-O4′ position (see discussion).
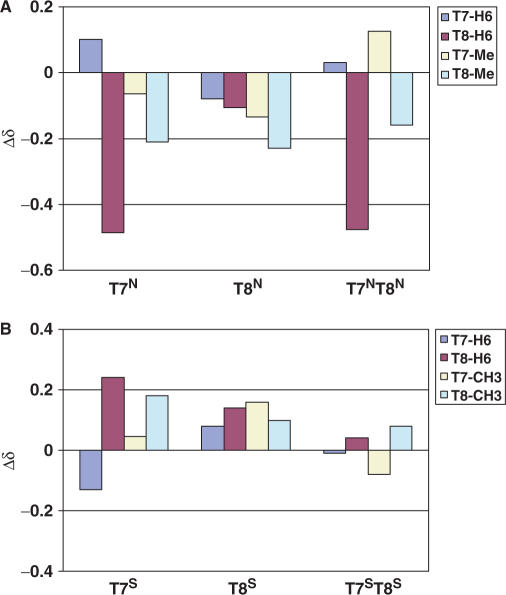
Changes in the chemical shift of the non-exchangeable protons between the native DNA and those from the modified DNAs are shown in Figure 10. The local environment around the aromatic H6 proton and methyl group of the T7 residue in each N-modified ODN was slightly modified with the largest Δδ observed for the methyl group of the T8 residue in the T7N and T8N ODNs (−0.23 and −0.21 ppm, respectively, Figure 10A). Significant shifts were seen for the H6 protons of the T8 residue in both the T7N (−0.49 ppm) and T7NT8N (0.48 ppm) ODNs. For the S-MCT-modified structures, the T7S ODN displayed the largest change in chemical shift, and this occurred in the T8 residue: H6 and the CH3 group shifted by 0.22 and 0.18 ppm, respectively (Figure 10B). Interestingly, the double-modified duplex, T7ST8S, showed very little change in the chemical shifts of the aromatic protons, which suggested that the magnetic environment of this duplex was similar to that of the native DD. These changes were consistent with temperature (5–25°C, data not shown).
Figure 10.
Changes in the chemical shift of select non-exchangeable protons between the N-MCT- (A) and S-MCT- (B) modified ODN species and the native DD at 25°C.
Concentration-dependent NMR of S-modified DNA
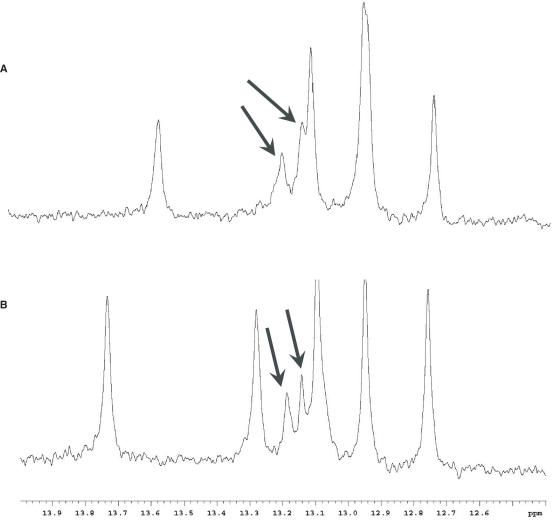
In an attempt to obtain more information regarding the various transitions obtained from the DSC data of the S-modified ODN species, NMR spectra were collected at the concentrations used to acquire the calorimetric data, i.e. 40 and 200 μM. Interestingly, new peaks in the imino proton region of the spectra were observed at these concentrations. Figure 11 shows the imino proton regions of the spectra for the T7S (Figure 11A) and T7ST8S (Figure 11B) ODNs with the additional peaks indicated by arrows. As before, there were no new resonances in any other region of the spectra that may indicate hairpin structures. No attempt was made to assign these new peaks to a particular structure, but this information corroborated the observed DSC behavior of the S-modified DNA species at low single-strand concentrations. As described above, when the DSC data was run at higher salt concentrations which is known to stabilize duplexes, clean transitions were observed at temperatures that were similar to the Tm's of the native DD. To confirm the presence of only one stable species, NMR spectra of the actual lyophilized DSC samples of the S-modified DNAs at these salt concentrations were collected at various temperatures. Resonances observed in the imino region of the spectra (12–14) were similar to those observed for the major species at other DNA and salt concentrations (not shown).
Figure 11.
1D NMR spectra of (A) T7S and (B) T7ST8S imino regions at 200 μM strand concentration. Additional peaks that were not observed in 1–2 mM solutions are indicated with arrows.
DISCUSSION
The bicyclo[3.1.0]hexane system has emerged as an extremely useful template for the study of nucleosides and nucleic acids with a rigidified five-membered ring employed as a surrogate for a standard (deoxy)ribose system. In this article, we presented several biophysical measurements on a series of Dickerson dodecamer DNA sequences where one or two thymidines have been substituted with nucleotides constructed on this template. The template effectively locks the ring pucker of the fused cyclopentane ring into either the Northern (N) or Southern (S) hemisphere of the pseudorotational cycle. The concept of conformational ‘locking’ furanose rings in this manner has greatly clarified many aspects of nucleoside and ON/ODN biochemistry for a variety of systems (21,41–44). These data confirm the utility of this template as a unique biochemical tool for probing structure–activity relationships in ON biology.
Since the ribose rings of the native DD are puckered primarily in the standard B-type (S) conformation, it was anticipated that these substitutions may either stabilize or disrupt the local/global structure accordingly. Our thought experiment was to use the S-locked nucleotides as a ‘control’ unit to stabilize an otherwise B-duplex by pre-organization of the proper sugar ring conformation prior to duplex formation. The N-type analogs were thought to disrupt the otherwise B-type helix and introduce a possible nucleation point for transmission of N-like conformational information along the sequence (45). Results from CD, NMR and DSC measurements all combined to paint a descriptive picture of the effect of either modified nucleoside on duplex formation and stability. As is shown here and in previous reports (46,47), the incorporation of N-locked analogs only minimally perturbed the DD system with regard to duplex formation and thermodynamics. However, we can now conclude that replacing the middle thymidine nucleotides with S-locked analogs leads to a more complex equilibrium of species rather than the idealized (and overly simplistic) assumption of an all B-DNA ‘stabilization’ in a manner that was predicted a priori. The data has made the interpretation of the effects of incorporating the S-locked analogs significantly more difficult, especially in light of the concentration dependence of the aforementioned equilibrium. However, the major conformation formed was a duplex with a slightly higher thermodynamic stability compared to the native DD.
An encouraging conclusion to the data described here was that all of the modified ODN species can form stable duplexes under either standard conditions or with slightly elevated salt concentrations. Thus, incorporation of N-locked nucleotides into an otherwise B-DNA (S-puckered sugars) does not greatly affect duplex formation, at least for the constructs described here. In addition, although the sequence of the DD is self-complimentary and under certain conditions has a tendency to form hairpin structures (37), there was no indication of these folds for any of the ODN species studied. However, we have shown earlier that N-modified ODNs have a propensity to bend at the central residues more than the native DD (29). A recent detailed molecular modeling study of the three N-locked DNAs (48) was in solid agreement with the experimental results in (29). Hence, it may be surmised that along with their ability to form stable helices, these analogs serve to adjust the local structural environment (bend) in response to conformational restriction in the 2′-exo puckering regime.
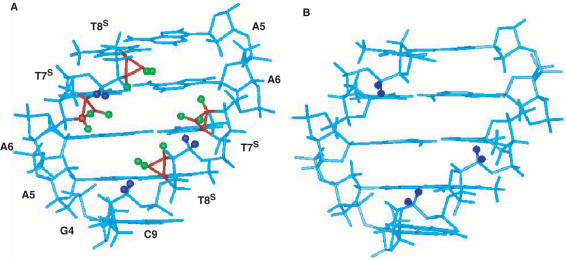
The data described here suggests that incorporation of the S-modified nucleotides into the DD complicate the conformational equilibrium of the ODN species. These were designed to ‘pre-organize’ a canonical, all B-DNA to a standard duplex fold. However, multiple transitions were observed at particular low salt and strand concentrations. We have not fully characterized the lower temperature transition from these ODN species but as suggested above, it is not a hairpin structure (49). NMR analysis showed that multiple species are present but they are limited to solutions that are of low micromolar strand concentrations at salt concentrations of 100 μM. At higher salt concentrations, these lower temperature melting species seem to be replaced solely by duplex structures that melt as broad transitions at a higher temperature and with a large change in ΔHcal. The specific disposition of this duplex may be important to its characteristic change in heat capacity. It may be surmised that the S-modified nucleotides actually do stabilize a B-like helix and that the more hydrated environment associated with this type of helix requires a larger enthalpic change upon melting (see Table 1). Concomitantly, the melting of the putative well-ordered hydration sphere of this type of helix would be accompanied by an unfavorable entropic contribution (Table 1, Figure 4). The validity of these proposals may be supported by recalling that the modified nucleotides are carbocyclic in nature, lacking the 4′-oxygen atom that may be important for DNA hydration; disruption by intervening carbon atoms could lead to destabilization. Equally important is the presence of additional atoms in the modified templates which may differentially influence the steric environment around the fused cyclopropane. Figure 12 shows a comparison of the central base pairs of the DD highlighting the cyclopropane rings of the T7S and T8S and the potential steric clashes they may engender with neighboring H5′, H5″ protons. These interactions could potentially cause adjustments in the backbone torsion angles that may serve to destabilize the initial folding of a stable helical conformation. In addition, both solid state and solution structural information on S-locked monomers (51,52) and DNA where an S-locked abasic site was incorporated (53) suggested that the rotamer distribution about the anomeric C-N bond (angle χ) prefers a syn orientation rather than anti. Thus, it is perhaps not surprising that ‘forcing’ a south conformation into the DD is not as well accommodated as a north-locked template. We may conclude that the S-modified ODNs resemble the native DD at low strand and Na+ concentrations; higher concentrations and additional salt is necessary to overcome thermodynamically unfavorable barriers and allow formation of highly stabilized duplexes.
Figure 12.
Side-by-side view of (A) T7ST8S (with residues labeled) and (B) the native dodecamer to illustrate the position and potential contacts of the additional atoms in the bicyclo[3.1.0]hexane systems compared to standard furanose rings. Only residues G4-T8 (3′-5′) and C9-A5 (5′-3′) are displayed for clarity. Backbone atoms are cyan; H5′, H5′′ of selected residues are dark blue; carbon and hydrogen atoms of the cyclopropane ring are shown in red and green, respectively.
The results obtained here for the N-locked DNAs complement the already highly studied systems of ‘locked’ nucleic acid oligomers constructed with the 2′–4′ bridged bicyclic nucleosides developed earlier (8,54). These constructs have been employed as hybridizing partners with RNA to potentially elicit a biological response akin to that of antigene therapy (8,55) or RNAi technology (24). Similar hybridization with RNA is achievable with the MCT ONs and it remains to be seen if this technology could be used for gene silencing in a similar manner or via other mechanisms. From our previous results that showed that the T7NT8N system can induce bending in the DD (29), we feel it is conceptually possible to design ODNs with specific bending properties that would mimic those induced by transcription factor binding to particular DNA sequences. Hence, a judiciously prepared N-modified oligo may inhibit this binding through highly specific substitution of the transcribing sequence. However, this will require the high-resolution structural characterization of each ODN separately to define the atomic coordinates of specific modified sequences. Work in this area is currently in progress.
ACKNOWLEDGEMENTS
We thank Professor Alex MacKerrell, University of Maryland, Baltimore, for model structures of the DD and /S/-modified DNA used for Figure 12. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charge was also provided under contract N01-CO-12400.
Conflict of interest statement. None declared.
REFERENCES
- 1.Herdewijn P. Conformationally restricted carbohydrate-modified nucleic acids and antisense technology. Biochim. Biophys. Acta. 1999;1489:167–179. doi: 10.1016/s0167-4781(99)00152-9. [DOI] [PubMed] [Google Scholar]
- 2.Kool ET. Replacing the nucleobases in DNA with designer molecules. Acc. Chem. Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 3.Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Feldman RJ, Mitsuya H, George C, Barchi JJ. Hiv-1 reverse transcriptase can discriminate between two conformationally locked carbocyclic AZT triphosphate analogs. J. Am. Chem. Soc. 1998;120:2780–2789. [Google Scholar]
- 4.Marquez VE, Ben Kasus T, Barchi JJ, Green KM, Nicklaus MC, Agbaria R. Experimental and structural evidence that herpes 1 kinase and cellular DNA polymerase(S) discriminate on the basis of sugar pucker. J. Am. Chem. Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 5.Siddiqui MA, Ford H, George C, Marquez VE. Synthesis, conformational analysis, and biological activity of a rigid carbocyclic analog Of 2′-deoxyaristeromycin built on a bicyclo[3.1.0]hexane template. Nucleosides Nucleotides. 1996;15:235–250. [Google Scholar]
- 6.Rodriguez JB, Marquez VE, Nicklaus MC, Mitsuya H, Barchi JJ. Conformationally locked nucleoside analogs – synthesis of dideoxycarbocyclic nucleoside analogs structurally related to neplanocin-C. J. Med. Chem. 1994;37:3389–3399. doi: 10.1021/jm00046a024. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez JB, Marquez VE, Nicklaus MC, Barchi JJ. Synthesis of cyclopropane-fused dideoxycarbocyclic nucleosides structurally related to neplanocin-C. Tetrahedron Lett. 1993;34:6233–6236. [Google Scholar]
- 8.Obika S. Development of bridged nucleic acid analogs for antigene technology. Chem. Pharma. Bull. 2004;52:1399–1404. doi: 10.1248/cpb.52.1399. [DOI] [PubMed] [Google Scholar]
- 9.Koshkin AA, Rajwanshi VK, Wengel J. Novel convenient syntheses of lna [2.2.1]bicyclo nucleosides. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 10.Opalinska JB, Kalota A, Gifford LK, Lu PZ, Jen KY, Pradeepkumar PI, Barman J, Kim TK, Swider CR, et al. Oxetane modified, conformationally constrained, antisense oligodeoxyribonucleotides function efficiently as gene silencing molecules. Nucleic Acids Res. 2004;32:5791–5799. doi: 10.1093/nar/gkh893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renneberg D, Bouliong E, Reber U, Schumperli D, Leumann CJ. Antisense properties of tricyclo-DNA. Nucleic Acids Res. 2002;30:2751–2757. doi: 10.1093/nar/gkf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jepsen JS, Sorensen MD, Wengel J. Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides. 2004;14:130–146. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 13.Bolli M, Lubini P, Leumann C. Nucleic-acid analogs with restricted conformational flexibility in the sugar-phosphate backbone (‘bicyclo-DNA’). 5. synthesis, characterization, and pairing properties of oligo-alpha-d-(bicyclodeoxynucleotides) of the bases adenine and thymine (Alpha-Bicyclo-DNA) Helv. Chim. Acta. 1995;78:2077–2096. [Google Scholar]
- 14.Rajwanshi VK, Kumar R, Kofodhansen M, Wengel J. Synthesis and restricted furanose conformations of three novel bicyclic thymine nucleosides: A xylo-lna nucleoside, a 3′-o,5′- c-methylene-linked nucleoside, and A 2′-O,5′-C- methylene- linked nucleoside. J. Chem. Soc. Perkin Trans. I. 1999:1407–1414. [Google Scholar]
- 15.Singh SK, Nielsen P, Koshkin AA, Wengel J. LNA (Locked Nucleic Acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998:455–456. [Google Scholar]
- 16.Altona C, Sundaral M. Conformational-analysis of sugar ring in nucleosides and nucleotides – improved method for interpretation of proton magnetic-resonance coupling-constants. J. Am. Chem. Soc. 1973;95:2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- 17.Singh SK, Wengel J. Universality of lna-mediated high-affinity nucleic acid recognition. Chem. Commun. 1998:1247–1248. [Google Scholar]
- 18.Petersen M, Bondensgaard K, Wengel J, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA:NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 19.Obika S, Hari Y, Morio K, Imanishi T. Triplex formation by an oligonucleotide containing conformationally locked C-nucleoside, 5-(2-O,4-C-methylene-beta-d-ribofuranosyl)oxazole. Tetrahedron Lett. 2000;41:221–224. [Google Scholar]
- 20.Marquez VE, Siddiqui MA, Ezzitouni A, Russ P, Wang JY, Wagner RW, Matteucci MD. Nucleosides with A twist. can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J. Med. Chem. 1996;39:3739–3747. doi: 10.1021/jm960306+. [DOI] [PubMed] [Google Scholar]
- 21.Ezzitouni A, Marquez VE. Conformationally locked carbocyclic nucleosides built on a bicyclo[3.1.0]hexane template with a fixed southern conformation. synthesis and antiviral activity. J. Chem. Soc. Perkin Trans. I. 1997:1073–1078. [Google Scholar]
- 22.Kim HS, Ohno M, Xu B, Kim HO, Choi YS, Ji XD, Maddileti S, Marquez VE, Harden TK, et al. 2-Substitution of adenine nucleotide analogs containing a bicyclo[3.1.0]hexane ring system locked in a northern conformation: enhanced potency as P2y(1) receptor antagonists. J. Med. Chem. 2003;46:4974–4987. doi: 10.1021/jm030127+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi Y, Moon HR, Yoshimura V, Marquez VE. Recent advances in the synthesis of conformationally locked nucleosides and their success in probing the critical question of conformational preferences by their biological targets. Nucleosides Nucleotides Nucleic Acids. 2003;22:547–557. doi: 10.1081/NCN-120021954. [DOI] [PubMed] [Google Scholar]
- 24.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu YH, Wahren B, Liang ZC, Urum H, et al. Locked nucleic acid (LNA) mediated improvements in sirna stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jepsen JS, Wengel J. LNA-antisense rivals sirna for gene silencing. Curr. Opin. Drug Discov. Devel. 2004;7:188–194. [PubMed] [Google Scholar]
- 26.Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 27.Olson WK, Zhurkin VB. Modeling DNA deformations. Curr. Opin. Struc. Biol. 2000;10:286–297. doi: 10.1016/s0959-440x(00)00086-5. [DOI] [PubMed] [Google Scholar]
- 28.Rice PA, Yang SW, Mizuuchi K, Nash HA. Crystal structure of an ihf-DNA complex: a protein-induced DNA u-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 29.Wu ZR, Maderia M, Barchi JJ, Marquez VE, Bax A. Changes in DNA bending induced by restricting nucleotide ring pucker studied by weak alignment NMR spectroscopy. Proc. Nat. Acad. Sci. U.S.A. 2005;102:24–28. doi: 10.1073/pnas.0408498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ezzitouni A, Barchi JJ, Marquez VE. A simple approach to 1′,1′a-methano carbocyclic thymidine. J. Chem. Soc. Chem. Commun. 1995:1345–1346. [Google Scholar]
- 31.Yoshimura Y, Moon HR, Choi Y, Marquez VE. Enantioselective synthesis of bicyclo[3.1.0]hexane carbocyclic nucleosides via a lipase-catalyzed asymmetric acetylation. characterization of an unusual acetal byproduct. J. Org. Chem. 2002;67:5938–5945. doi: 10.1021/jo020249u. [DOI] [PubMed] [Google Scholar]
- 32.Maier MA, Choi Y, Gaus H, Barchi JJ, Marquez VE, Manoharan M. Synthesis and characterization of oligonucleotides containing conformationally constrained bicyclo[3.1.0]hexane pseudosugar analogs. Nucleic Acids Res. 2004;32:3642–3650. doi: 10.1093/nar/gkh667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakrabarti MC, Schwarz FP. Thermal stability of PNA/DNA and DNA/DNA duplexes by differential scanning calorimetry. Nucleic Acids Res. 1999;27:4801–4806. doi: 10.1093/nar/27.24.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe – a multidimensional spectral processing system based on unix pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov VI, Minchenk LE, Schyolki AK, Poletaye AI. Different conformations of double-stranded nucleic-acid in solution as revealed by circular-dichroism. Biopolymers. 1973;12:89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- 36.Tinoco I, Bustamante C, Maestre MF. The optical-activity of nucleic-acids and their aggregates. Annu. Rev. Biophy. Bioeng. 1980;9:107–141. doi: 10.1146/annurev.bb.09.060180.000543. [DOI] [PubMed] [Google Scholar]
- 37.Marky LA, Blumenfeld KS, Kozlowski S, Breslauer KJ. Salt-dependent conformational transitions in the self-complementary deoxydodecanucleotide D(Cgcaattcgcg) – evidence for hairpin formation. Biopolymers. 1983;22:1247–1257. doi: 10.1002/bip.360220416. [DOI] [PubMed] [Google Scholar]
- 38.Patel DJ, Kozlowski SA, Marky LA, Broka C, Rice JA, Itakura K, Breslauer KJ. Premelting and melting transitions in the D(Cgcgaattcgcg) self-complementary duplex in solution. Biochemistry. 1982;21:428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- 39.Wu ZR, Maderia M, Barchi JJ, Marquez VE, Bax A. Changes in DNA bending induced by restricting nucleotide ring pucker studied by weak alignment NMR spectroscopy. Proc. Nat. Acad. Sci. U.S.A. 2005;102:24–28. doi: 10.1073/pnas.0408498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blommers MJJ, Walters JALI, Haasnoot CAG, Aelen JMA, Vandermarel GA, Vanboom JH, Hilbers CW. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989;28:7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- 41.Jacobson KA, Ravi RG, Nandanan E, Kim HS, Moro S, Kim YC, Lee K, Barak D, Marquez VE, et al. Ribose modified nucleosides and nucleotides as ligands for purine receptors. Nucleosides Nucleotides Nucleic Acids. 2001;20:333–341. doi: 10.1081/NCN-100002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobson KA, Ji XD, Li AH, Melman N, Siddiqui MA, Shin KJ, Marquez VE, Ravi RG. Methanocarba analogs of purine nucleosides as potent and selective adenosine receptor agonists. J. Med. Chem. 2000;43:2196–2203. doi: 10.1021/jm9905965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marquez VE, Wang PY, Nicklaus MC, Maier M, Manoharan M, Christman JK, Banavali NK, Mackerell AD. Inhibition of (Cytosine C5)-methyltransferase by oligonucleotides containing flexible (Cyclopentane) and conformationally constrained (Bicyclo[3.1.0]Hexane) abasic sites. Nucleosides Nucleotides Nucleic Acids. 2001;20:451–459. doi: 10.1081/NCN-100002319. [DOI] [PubMed] [Google Scholar]
- 44.Wang PY, Brank AS, Banavali NK, Nicklaus MC, Marquez VE, Christman JK, Mackerell AD. Use of oligodeoxyribonucleotides with conformationally constrained abasic sugar targets to probe the mechanism of base flipping by hhai DNA (Cytosine C5)-methyltransferase. J. Am. Chem. Soc. 2000;122:12422–12434. [Google Scholar]
- 45.Pradeepkumar PI, Zamaratski E, Foldesi A, Chattopadhyaya J. Conformation-specific cleavage of antisense oligonucleotide-rna duplexes by rnase H. J. Chem. Soc. Perkin Trans. II. 2001:402–408. [Google Scholar]
- 46.Maderia M, Wu J, Bax A, Shenoy S, O'keefe B, Marquez VE, Barchi JJ. Engineering DNA topology with locked nucleosides: a structural study. Nucleosides Nucleotides Nucleic Acids. 2005;24:687–690. doi: 10.1081/ncn-200060256. [DOI] [PubMed] [Google Scholar]
- 47.Maderia M, Shenoy S, O'keefe B, Wu J, Bax A, Marquez V, Barchi JJ. Manipulating DNA structure with ‘locked’ nucleosides. Biophys. J. 2004;86:188a–189a. [Google Scholar]
- 48.Macias AT, Banavali NK, MacKerell AD. DNA bending induced by carbocyclic sugar analogues constrained to the north conformation. Biopolymers. 2007 doi: 10.1002/bip.20673. (Early View Online, 8 January 2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Braunlin WH, Giri I, Beadling L, Breslauer KJ. Conformational screening of oligonucleotides by variable-temperature high performance liquid chromatography: dissecting the duplex-hairpin-coil equilibria of D(Cgcgaattcgcg) Biopolymers. 2004;74:221–231. doi: 10.1002/bip.20079. [DOI] [PubMed] [Google Scholar]
- 50.Marquez VE, Ben Kasus T, Barchi JJ, Green KM, Nicklaus MC, Agbaria R. Experimental and structural evidence that herpes 1 kinase and cellular DNA polymerase(S) discriminate on the basis of sugar pucker. J. Am. Chem. Soc. 2004;126:543–549. doi: 10.1021/ja037929e. [DOI] [PubMed] [Google Scholar]
- 51.Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Feldman RJ, Mitsuya H, George C, Barchi JJ. Hiv-1 reverse transcriptase can discriminate between two conformationally locked carbocyclic azt triphosphate analogs. J. Am. Chem. Soc. 1998;120:2780–2789. [Google Scholar]
- 52.Horton JR, Ratner G, Banavali NK, Huang N, Choi Y, Maier MA, Marquez VE, Mackerell AD, Cheng XD. Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by hhal methyltransferase. Nucleic Acids Res. 2004;32:3877–3886. doi: 10.1093/nar/gkh701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wengel J, Koshkin A, Singh SK, Nielsen P, Meldgaard M, Rajwanshi VK, Kumar R, Skouv J, Nielsen CB, et al. LNA (locked nucleic acid) Nucleosides Nucleotides Nucleic Acids. 1999;18:1365–1370. [Google Scholar]
- 54.Buchini S, Leumann CJ. Recent improvements in antigene technology. Curr. Opin. Chem. Biol. 2003;7:717–726. doi: 10.1016/j.cbpa.2003.10.007. [DOI] [PubMed] [Google Scholar]